Abstract
The p75 neurotrophin receptor (p75NTR) is a member of the tumor necrosis factor receptor superfamily with a widespread pattern of expression in tissues such as the brain, liver, lung, and muscle. The mechanisms that regulate p75NTR transcription in the nervous system and its expression in other tissues remain largely unknown. Here we show that p75NTR is an oscillating gene regulated by the helix-loop-helix transcription factors CLOCK and BMAL1. The p75NTR promoter contains evolutionarily conserved noncanonical E-box enhancers. Deletion mutagenesis of the p75NTR-luciferase reporter identified the −1039 conserved E-box necessary for the regulation of p75NTR by CLOCK and BMAL1. Accordingly, gel-shift assays confirmed the binding of CLOCK and BMAL1 to the p75NTR−1039 E-box. Studies in mice revealed that p75NTR transcription oscillates during dark and light cycles not only in the suprachiasmatic nucleus (SCN), but also in peripheral tissues including the liver. Oscillation of p75NTR is disrupted in Clock-deficient and mutant mice, is E-box dependent, and is in phase with clock genes, such as Per1 and Per2. Intriguingly, p75NTR is required for circadian clock oscillation, since loss of p75NTR alters the circadian oscillation of clock genes in the SCN, liver, and fibroblasts. Consistent with this, Per2::Luc/p75NTR−/− liver explants showed reduced circadian oscillation amplitude compared with those of Per2::Luc/p75NTR+/+. Moreover, deletion of p75NTR also alters the circadian oscillation of glucose and lipid homeostasis genes. Overall, our findings reveal that the transcriptional activation of p75NTR is under circadian regulation in the nervous system and peripheral tissues, and plays an important role in the maintenance of clock and metabolic gene oscillation.
Introduction
The p75 neurotrophin receptor (p75NTR), a member of the tumor necrosis factor receptor (TNFR) superfamily, is expressed in the nervous system and many peripheral tissues (Lomen-Hoerth and Shooter, 1995) such as white adipose tissue (Peeraully et al., 2004), skeletal muscle (Deponti et al., 2009), and liver (Passino et al., 2007). p75NTR participates in multiple intracellular signaling pathways to regulate a wide range of biologic functions, including sensory neuron development (Lee et al., 1992), liver and muscle regeneration (Passino et al., 2007; Deponti et al., 2009), extracellular matrix remodeling (Sachs et al., 2007), hypoxia and angiogenesis (Le Moan et al., 2011), and glucose metabolism (Baeza-Raja et al., 2012). Although p75NTR has a wide range of expression profiles and biological functions, the physiologic mechanisms of its transcriptional regulation in the brain and peripheral tissues are still unknown. Previous studies of the p75NTR promoter revealed the presence of multiple GC elements (Sehgal et al., 1988) that can be activated by Specificity protein 1 (Sp1) transcription factors (Poukka et al., 1996). Sp1 binding induces p75NTR expression after osmotic swelling (Peterson and Bogenmann, 2003; Ramos et al., 2007). In addition to Sp1, early growth response factors 1 and 3 have been identified as direct modulators of p75NTR (Gao et al., 2007). However, regulation by these transcription factors alone is not sufficient to explain the pleiotropic expression pattern of p75NTR in central and peripheral tissues.
We recently demonstrated that p75NTR is a unique regulator of metabolic functions, including glucose homeostasis and insulin sensitivity (Baeza-Raja et al., 2012). A common denominator of genes regulating metabolic functions is their regulation by circadian clocks, and conversely metabolic dysfunction is strongly associated with the disruption of circadian clocks (Dibner et al., 2010; Asher and Schibler, 2011; Solt et al., 2012). Similar to p75NTR−/− mice, mutant mice of the main circadian regulators, Clock and Bmal1, display increased insulin sensitivity and altered gluconeogenesis (Rudic et al., 2004), while mice lacking Bmal1, specifically in liver, show hypoglycemia and increased glucose tolerance (Lamia et al., 2008). Moreover, knock-out mice for Clock and Bmal1 target genes Cry1 and Cry2 also show impaired liver regeneration (Matsuo et al., 2003). Because p75NTR is widely expressed, and regulates glucose homeostasis and insulin sensitivity—metabolic processes highly influenced by circadian clocks—we hypothesized that the transcriptional regulation of p75NTR might be controlled by circadian oscillators.
In this study, we show that p75NTR is an oscillating gene regulated by the circadian regulators CLOCK and BMAL1. The 5′ regulatory region of p75NTR contains evolutionarily conserved noncanonical E-boxes to which the CLOCK/BMAL1 complex binds and induces p75NTR transcriptional activity. We find that p75NTR RNA expression oscillates, in an E-box-dependent manner, not only in fibroblasts after serum shock, but also in vivo in the suprachiasmatic nucleus (SCN) and liver. In accordance, we show that p75NTR oscillation is disrupted in the SCN and liver from Clock−/− (DeBruyne et al., 2006) and ClockΔ19 (Vitaterna et al., 1994) mutant mice. Although genetic loss of p75NTR does not affect locomotor activity rhythms, it disrupts the expression of circadian clock genes in fibroblasts, SCN, and liver, and also affects the oscillation of genes involved in glucose and lipid homeostasis. Overall, our study identifies p75NTR as a first-order clock-controlled gene that oscillates in central and peripheral tissues, and its expression is necessary to maintain circadian and metabolic gene oscillation.
Materials and Methods
Animals.
Wild-type (WT) C57BL/6J, p75NTR−/− (Lee et al., 1992), and Per2::Luc (Yoo et al., 2004) mice were obtained from The Jackson Laboratory. p75NTR−/− and Per2::Luc mice were in C57BL/6 background. Clock−/− (DeBruyne et al., 2006) and ClockΔ19 (Vitaterna et al., 1994) mice in C57BL/6 and BALB/cJ background, respectively, were also used. Crossings between Per2::Luc and p75NTR−/− mice were performed to generate Per2::Luc/p75NTR−/− and their littermate controls Per2::Luc/p75NTR+/+. In vivo studies were performed using male mice between 8 and 12 weeks of age. Mice were housed under a 12 h light/dark cycle, were fed standard chow, and had access to food and water ad libitum. All animal experiments were performed under the guidelines set by the University of California San Francisco and Irvine Institutional Animal Care and Use Committees and are in accordance with those set by the National Institutes of Health.
Cell culture.
HEK293 cells and primary mouse embryonic fibroblasts (MEFs) derived from WT and p75NTR−/− embryos were cultured in DMEM supplemented with 10% fetal bovine serum (FBS; Invitrogen) and 1% penicillin/streptomycin (Invitrogen). Isolation of MEFs was performed as described previously (Russell et al., 2002). Briefly, embryos were isolated at E12–E14, the heads and internal organs were removed, and the remaining embryo was minced with scissors and kept on ice in PBS. Minced embryos were centrifuged for 5 min at 500 × g and trypsinized for 20 min at 37°C with occasional shaking with 1× Trypsin-EDTA. Trypsin was deactivated by the addition of DMEM with 10% FBS. MEFs were then pelleted by centrifugation at 500 × g for 5 min. Pellets were resuspended in DMEM with 10% FBS and 1% penicillin/streptomycin and then poured through a 70 μm filter onto a 10 cm culture plate.
DNA transfection and luciferase assays.
HEK293 cells were transiently transfected with LipofectAMINE (Invitrogen) according to the manufacturer's protocol. After transfection, either with luciferase reporters with or without expression plasmids or with expression plasmids alone, the transfection medium was replaced by growth medium for 24 h. All transfections included constant amounts of β-galactosidase reporter plasmid as a control for transfection efficiency. The total amount of DNA for each transfection was kept constant using an empty expression vector. Cells were then harvested to measure luciferase activity (Promega). Luciferase activities were normalized by dividing luciferase activity by β-galactosidase activity. For each experimental group, a minimum of three independent transfections (in triplicate) was performed.
DNA constructs.
The human p75NTR promoter was subcloned into pGL4.17 (Promega) luciferase reporter vector to generate pGL4-p75Luc after PCR using oligonucleotides containing enzymatic restriction sites for XhoI and HindIII. Deletion constructs (pGL4-p75LucΔ948, pGL4-p75LucΔ775, pGL4-p75LucΔ745, pGL4-p75LucΔ697, pGL4-p75LucΔ651, and pGL4-p75LucΔ553) were also generated by PCR using oligonucleotides containing XhoI and HindIII restriction sites. The luciferase reporter vector containing the human Per2 promoter and the expression vectors for the human CLOCK and BMAL1 were used.
Serum shock treatment.
Circadian induction of MEFs from WT and p75NTR−/− mice was performed on confluent cells as previously described (Balsalobre et al., 1998). Briefly, 50% of horse serum was added to the cells for 2 h and after the cells were incubated in DMEM without serum and collected at the indicated time points. Cells were then used for total RNA and nuclear extract isolation.
RNA isolation and RT-qPCR.
Total RNA was isolated from MEFs, SCN, and liver using the standard RNeasy Mini Kit (Qiagen) according to the manufacturer's instructions. RNA was reverse transcribed to cDNA using the GeneAmp RNA PCR Core Kit (Applied Biosystems) with random hexamers as primers according to the manufacturer's instructions. Real-time PCR was performed on a StepOnePlus real-time PCR system (Applied Biosystems) using an SYBR green PCR master mix (Applied Biosystems). Real-time PCR experiments were performed with the following primers: Per1, 5′-TGACACTGATGCAAACAGCA-3′ (forward) and 5′-AGCCACTGGTAGATGGGTTG-3′ (reverse); Per2, 5′-GGTGGACAGCCTTTCGATTA-3′ (forward) and 5′-AGGGCCTACCCTGACTTTGT-3′ (reverse); Clock, 5′-CACAGGGCACCACCAATAAT-3′ (forward) and 5′-CATATCCACTGCTGGCCTTT-3′ (reverse); Rorα, 5′-ACGCCCACCTACAACATCTC-3′ (forward) and 5′-TCACATATGGGTTCGGGTTT-3′ (reverse); Bmal1, 5′-CGAAGACAATGAGCCAGACA-3′ (forward) and 5′-AAATAGCTGTCGCCCTCTGA-3′ (reverse); reverse erythroblastosis virus-α (Rev-Erbα), 5′-CAGCTTCCAGTCCCTGACTC-3′ (forward) and 5′-GGAGGAGGAGGATGAAGAGG-3′ (reverse); Gapdh, 5′-CAAGGCCGAGAATGGGAAG-3′ (forward) and 5′-GGCCTCACCCCATTTGATGT-3′ (reverse); Pepck, 5′-CTTCTCTGCCAAGGTCATCC-3′ (forward) and 5′-AGTGAGAGCCAGCCAACAGT-3′ (reverse); Fas, 5′-TTGCTGGCACTACAGAATGC-3′ (forward) and 5′-AACAGCCTCAGAGCGACAAT-3′ (reverse); Glut4, 5′-GTGACTGGAACACTGGTCCTA-3′ (forward) and 5′-CCAGCCACGTTGCATTGTAG-3′ (reverse); D-site albumin promoter-binding protein (Dbp), 5′-ACCGTGGAGGTGCTAATGAC-3′ (forward) and 5′-TGGCTGCTTCATTGTTCTTG-3′ (reverse); nerve growth factor (NGF), 5′-CATGGGGGAGTTCTCAGTGT-3′ (forward) and 5′-GCACCCACTCTCAACAGGAT-3′ (reverse); brain-derived neurotrophic factor (BDNF), 5′-GCGGCAGATAAAAAGACTGC-3′ (forward) and 5′-CTTATGAATCGCCAGCCAAT-3′ (reverse); tropomyosin-related kinase A (TrkA), 5′-AGGTCTTTCTCGCTGAGTGC-3′ (forward) and 5′-GGTGCAGACTCCAAAGAAGC-3′ (reverse); TrkB, 5′-GACCTGATCCTGACGGGTAA-3′ (forward) and 5′-TTCCTCCACGGTGAGGTTAG-3′ (reverse); TrkC, 5′-CTTCGGGAATTGAGACTGGA-3′ (forward) and 5′-TGGCTCACACTGATCTCTGG-3′ (reverse), FAS-TNFα, 5′-TGTGAACATGGAACCCTTGA-3′ (forward) and 5′-TTCAGGGTCATCCTGTCTCC-3′ (reverse), and p75NTR was determined using oligonucleotides obtained from SABiosciences. PCR efficiencies of the primers were calculated by serial dilution of the template, and no significant differences in efficiency were found between the target genes and the housekeeping genes. Results were analyzed with the Opticon 2 Software using the comparative CT method, as described previously (Livak and Schmittgen, 2001). Data were expressed as 2−ΔΔCT for the experimental gene of interest normalized against the housekeeping gene and presented as the fold change versus the relevant control. Each real-time PCR was performed in triplicate and repeated at least three times.
Electrophoretic mobility shift assays.
Nuclear extracts were obtained from HEK293 cell lines transfected with various plasmids and MEFS derived from WT mice after serum treatment. Nuclear protein extraction was performed as described previously (De Cesare et al., 1995). Briefly, cells were washed twice in cold PBS and scraped, and the cellular pellet was resuspended in 10 mm HEPES, pH 7.9, 10 mm KCl, 1.5 mm MgCl2, 0.1 mm EGTA, and 0.5 mm dithiothreitol (DTT) on ice. Cells were passed five times through a 26-gauge needle and centrifuged to collect nuclei, which were subsequently resuspended in an equal volume of 10 mm HEPES, pH 7.9, 0.4 m NaCl, 1.5 mm MgCl2, 0.1 mm EGTA, 0.5 mm DTT, and 5% glycerol, to allow elution of nuclear proteins by gentle shaking at 4°C for 30 min. Nuclei were pelleted at 14,000 rpm for 5 min at 4°C, and the supernatant was aliquoted, snap frozen in liquid nitrogen, and stored at −80°C until use. All solutions contained protease and phosphatase inhibitors (Calbiochem). A protein assay (Bio-Rad) was used to determine protein concentration. For electrophoretic mobility shift assays, 10 μg of nuclear extracts were incubated in 50 mm Tris-HCl, pH 7.9, 12.5 mm MgCl2, 1 mm EDTA, 1 mm DTT, 20% glycerol, 0.5 mm phenylmethylsulfonyl fluoride, and 2 μg of polydI-dC for 10 min at room temperature to titrate out nonspecific binding before the addition of a 15,000–20,000 cpm labeled oligonucleotide. The reaction was then further incubated for 20 min at room temperature. When unlabeled competing oligonucleotides were added, nuclear extracts were preincubated for 30 min or 1 h at room temperature before addition of the labeled probe. Samples were loaded onto a prerun polyacrilamide gel (29:1 in 0.25× Tris borate-EDTA) and electrophoresed at 200 V. Gels were dried and autoradiographed at −80°C. For control electrophoretic mobility shift assay (EMSA) probes, we used the huPer1 E-box probe 5′-CAC CCA CCG GTC ACA CGT GGA CCC TTA ACT GT-3′ and the unrelated probe NF-κB probe: 5′-AGT TGA GGG GGA CTT TCC CAG GC-3′. For the hup75NTR E-box EMSA probes, we designed probes for E3, 5′-CTA ATA ATA GTA ACA ATT GCT GTT TGT AAT TG-3′; E2, 5′-ATT GTT GAA GAT GCA TCT GTT TGT TTG TTG AT-3′; E1, 5′-AAT GTC CGG ATC ACA TAT GTG CCC GTG TGC AT-3′; and ENC, 5′-TGC ACT GTG TGC ACA CCT GTG ACC CCT TCA AA-3′.
Per2::Luc bioluminescence experiments and data analysis.
SCN and liver explants from Per2::Luc/p75NTR+/+ and Per2::Luc/p75NTR−/− mice were cultured as described previously (Yamazaki and Takahashi, 2005). Briefly, mice were killed 1 h into the light phase, and the livers were isolated and dissected into small pieces. Luciferase activity from sealed cultures maintained at 37°C was monitored for several days with a 32-channel Luminometer (Actimetrics). Period lengths and amplitude were determined in plates with good oscillation using Lumicycle Analysis software. All explants from each animal were monitored simultaneously, and all experiments included explants from Per2::Luc/p75NTR+/+ and Per2::Luc/p75NTR−/− mice to allow for direct comparisons between genotypes.
Mouse behavioral analysis.
Wheel-running activity monitoring was performed as described previously (Xu et al., 2005). Briefly, 3- to 4-month-old mice were housed individually in cages equipped with running wheels, and exposed to a 12 h light/dark cycle for >14 d before being released into constant darkness. Using ClockLab (Actimetrics) software, the period and amplitude of activity rhythms was calculated from χ2 periodogram.
Statistics.
Statistical significance was calculated using JMP2 Software by unpaired t test for isolated pairs or by one-way ANOVA for multiple comparisons followed by Bonferroni's correction for comparisons of means. Data are shown as the mean ± SEM.
Results
The 5′ regulatory region of p75NTR contains evolutionarily conserved noncanonical E-boxes that are activated by CLOCK and BMAL1
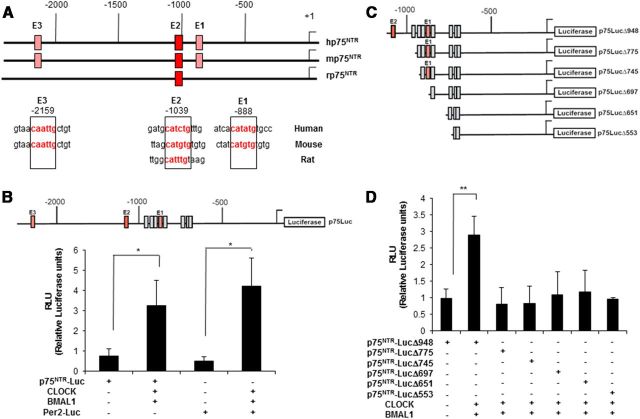
To examine the physiologic mechanisms that regulate p75NTR, we analyzed the p75NTR promoter using MatInspector 7.7.3, a program for the analysis and prediction of transcription factor binding sites. Computational analysis revealed the presence of several putative E-boxes (5′-CANNTG-3′) in the human promoter sequence of p75NTR. Using the multiple sequence alignment program Clustal 2.0.3, alignment of the p75NTR promoter sequences from human, mouse, and rat identified three noncanonical E-boxes that were conserved at base pair positions −888 (E1), −1039 (E2), and −2159 (E3) in the human p75NTR promoter region (Fig. 1A). E-boxes E1 and E3 were conserved between human and mouse, while E-box E2 at position −1039 was the only one conserved in human, mouse, and rat (Fig. 1A).
Figure 1.
The 5′ regulatory region of human, mouse, and rat p75NTR contains evolutionarily conserved E-boxes that are activated by CLOCK and BMAL1. A, Top, Schematic representation of the 2.5 kb upstream region of the human, mouse, and rat p75NTR promoter. Putative evolutionarily conserved E-box positions are shown with red boxes. Bottom, Aligned nucleotide sequences of the evolutionarily conserved human, mouse, and rat E-boxes. Alignment of the p75NTR promoter was performed using the Clustal 2.0.3 multiple sequence alignment program. B, Top, Schematic representation of the 2.2 kb p75NTR promoter cloned in the luciferase reporter vector pGL4.17. Red indicates the evolutionarily conserved E-boxes, while the gray boxes identify other nonconserved E-boxes. Bottom, Histogram of luciferase activity in HEK293 cells transfected with pGL4-p75NTR-Luc or Per2-luc in the presence or absence of CLOCK and BMAL1 expression vectors. Luciferase activities are expressed relative to the activity of the reporter constructs alone, which were given a value of 1 (*p < 0.05, t test). C, Schematic representation of the deletion constructs generated from the 2.2 kb hp75NTR promoter and cloned into the luciferase reporter vector pGL4.17. D, Histogram of luciferase activity in HEK293 cells transfected with the different constructs of the hp75NTR promoter in the presence of CLOCK and BMAL1 expression vectors. Luciferase activities are expressed relative to the activity of the reporter constructs alone, which were given a value of 1. Results of at least three independent experiments with duplicate measurements are shown. Error bars represent the SEM of the mean value. Statistical significance denoted by asterisks is relative to respective control (*p < 0.05; **p < 0.01, t test).
E-boxes are 5′-CANNTG-3′ DNA core sequences that bind transcription factors of the basic helix-loop-helix (bHLH) family. Binding of the bHLH transcription factors CLOCK and BMAL1 to E-box elements in the promoters of clock genes drives their rhythmic transcription (Gekakis et al., 1998; Travnickova-Bendova et al., 2002; Yoo et al., 2005). Since the p75NTR promoter contains three putative, evolutionarily conserved noncanonical E-boxes, we hypothesized that p75NTR could be under circadian regulation. To investigate this further, we generated a p75NTR promoter–luciferase reporter construct, which contains both conserved and nonconserved noncanonical E-boxes (p75-Luc). To examine whether CLOCK/BMAL1 activates the p75NTR promoter, cells were transfected with p75NTR-Luc and cotransfected with CLOCK and BMAL1 expression vectors (Fig. 1B). A Per2 promoter-luciferase reporter (Per2-Luc) was used as a positive control. As expected, cotransfection with CLOCK and BMAL1 activated the circadian oscillating gene Per2 (Fig. 1B). Strikingly, cotransfection of CLOCK and BMAL1 increased transcription of the p75NTR reporter (Fig. 1B). To identify which E-boxes contribute to the regulation of p75NTR transcription by CLOCK/BMAL1, we generated luciferase–reporter deletion constructs of the p75NTR promoter (Fig. 1C). CLOCK- and BMAL1-induced transcription of the full-length p75NTR-luciferase reporter that contains the −1039 E2-box, but not the deletion constructs that contain either the −888 E1-box or the nonconserved E-boxes (Fig. 1D). These results suggest that the evolutionarily conserved noncanonical E2-box is necessary for the activation of p75NTR by the transcription factors CLOCK and BMAL1.
The CLOCK/BMAL1 heterodimer binds to the E2-box of the p75NTR promoter
To determine whether CLOCK and BMAL1 bind to the p75NTR E2-box, we generated oligonucleotide probes corresponding to the E2 region and performed EMSAs using nuclear extracts of cells transfected with CLOCK and BMAL1. CLOCK/BMAL1 heterodimers showed strong binding to the p75NTR E2-box probe (Fig. 2A). Binding was reduced by the addition of a molar excess of unlabeled p75NTR E2-box probe but not by an unrelated probe, thereby confirming specificity (Fig. 2A). Also, CLOCK/BMAL1 heterodimers showed binding to the Per1 E-box probe (Fig. 2B), and the binding was reduced by the addition of a molar excess of unlabeled Per1 E-box probe, but not by exposure to an unrelated probe (Fig. 2B). In contrast, CLOCK/BMAL1 heterodimers bound weakly to p75NTR E1-box and E3-box probes (Fig. 2C). Moreover, unlabeled p75NTR E2-box probe was sufficient to compete for the binding of CLOCK/BMAL1 to a Per1 E-box probe (Fig. 2D).
Figure 2.
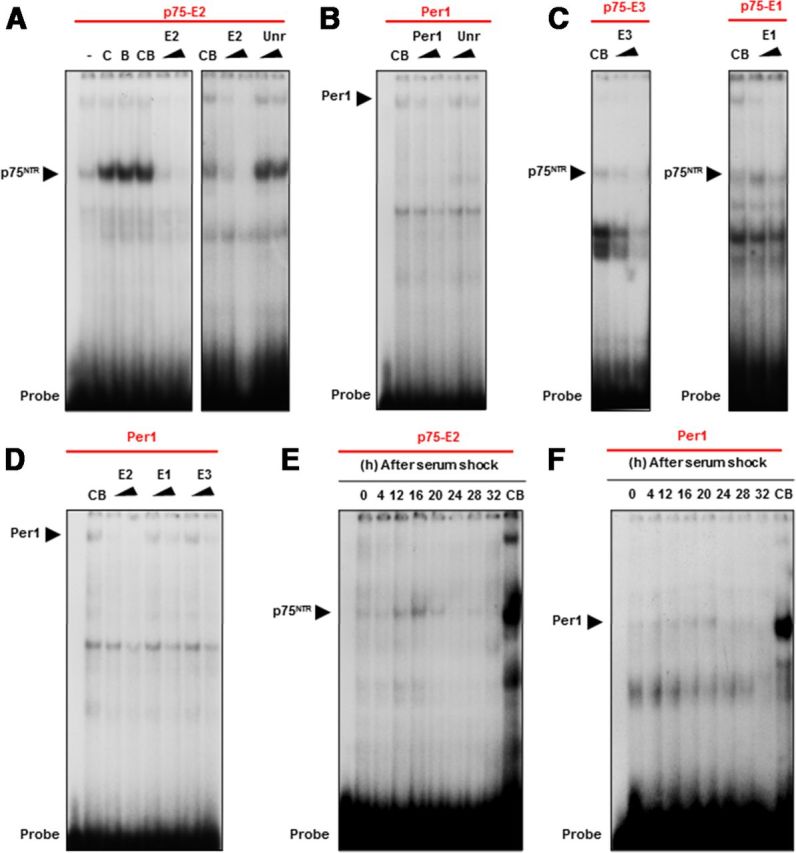
CLOCK/BMAL1 heterodimers bind to the E-boxes of the p75NTR promoter in vitro. A–F, EMSAs were performed using nuclear extracts of HEK293 cells cotransfected with CLOCK and BMAL1 (A–D) or MEFs derived from WT mice (E, F) on the different E-boxes (−2159, −1039 and −888 bp) from the human p75NTR and Per1 promoter as probes. CB means CLOCK and BMAL1 (CB) coexpression, a triangle indicates increasing amount of competitor (twofold or 10-fold) of the relevant E-box as indicated by the triangles, and a specific complex is indicated by black arrow. A, EMSAs performed on the E-box located at −1039 bp (E2) in the human p75NTR promoter as probe. Nuclear extracts from HEK293 cells transfected with an empty vector (-), CLOCK (C), BMAL1 (B) and CLOCK/BMAL1 (CB) expression vectors were used. B, EMSA performed on the E-box from the human Per1 promoter as probe. C, EMSAs performed on the E-boxes −2159 (E3) and −888 bp (E1) from the human p75NTR promoter as probe. D, EMSA performed on the E-box from the human Per1 promoter as probe. E, EMSA performed on the E-box located at −1039 bp (E2) in the human p75NTR promoter as probe. Nuclear extracts from WT MEFs were collected at the indicated times (h) after serum treatment. F, EMSA performed on the E-box from the human Per1 promoter as probe. Nuclear extracts from WT MEFs derived from WT mice were collected at the indicated times (h) after serum treatment.
MEFs are commonly used for studies of mammalian circadian gene regulation, as serum shock treatment induces oscillatory expression of clock genes in these cells (Balsalobre et al., 1998). The p75NTR E2-box probe bound strongly to nuclear extracts of primary MEFs after serum shock (Fig. 2E). Strikingly, the p75NTR E2-box showed an oscillating binding pattern with the strongest signal observed 16 h after serum shock (Fig. 2E). The oscillating binding pattern of the p75NTR E2-box probe showed similar temporal regulation as the Per1 E-box probe in MEFs (Fig. 2F). Altogether, these results demonstrate that CLOCK and BMAL1 bind to the evolutionarily conserved p75NTR E2-box in an oscillating manner.
p75NTR oscillates in the SCN and peripheral tissues
CLOCK/BMAL1 heterodimer regulates the oscillatory transcription of clock genes, such as Per1, Per2, and Rev-erbα (Darlington et al., 1998; Gekakis et al., 1998). Since the p75NTR E2-box probe showed an oscillating binding pattern in MEFs, we examined whether p75NTR expression oscillates. We induced clock gene oscillation in MEFs by serum shock and performed real-time PCR at different time points. Indeed, p75NTR showed an oscillating transcription pattern in phase with the clock genes Per1 and Rev-erbα (Fig. 3A). As expected, the expression of Rorα showed no circadian oscillation after serum shock (Fig. 3A).
Figure 3.
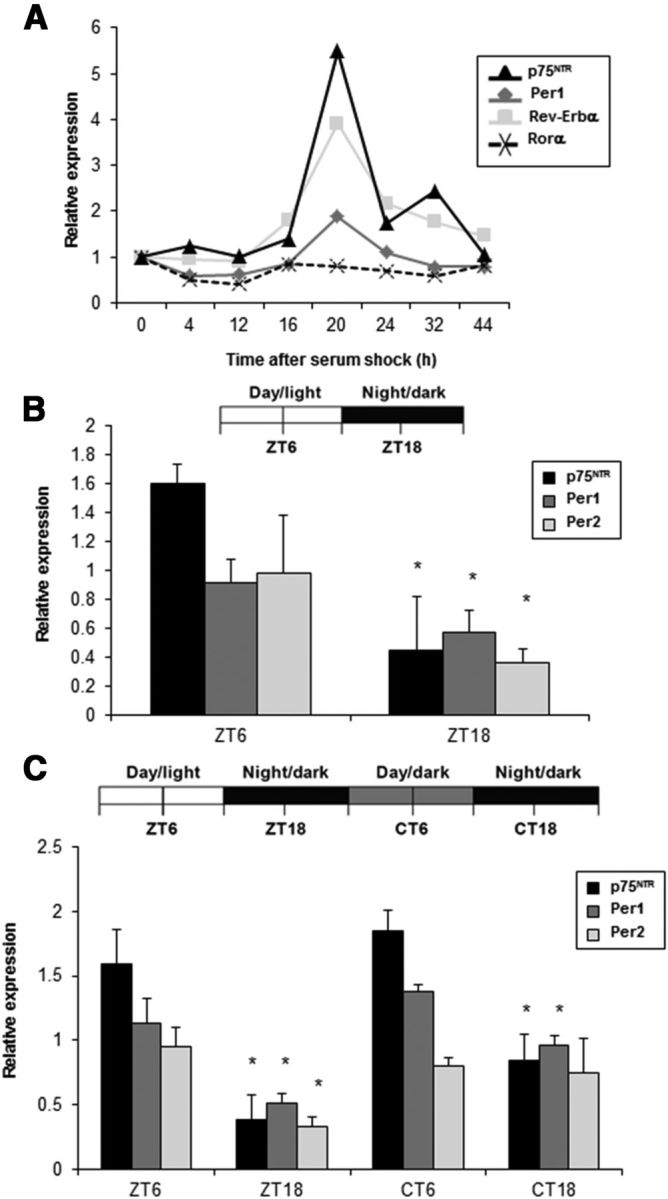
p75NTR RNA oscillates in vitro and in vivo. A, Serum shock treatment was performed on MEFs derived from WT mice, and cells were collected at the indicated times (h). p75NTR, Per1, Rev-Erbα, and RORα mRNA levels were analyzed by real-time PCR. Data are presented as the relative expression level with respect to time 0, which was given a value of 1. B, Top, Schematic representation of the light schedule. Mice were entrained in a 12 h light/dark cycle and killed at ZT6 and ZT18 (n = 5 per group). Bottom, At the indicated times, the SCN was dissected; RNA was extracted; and p75NTR, Per1, and Per2 levels analyzed by real-time PCR. C, Top, Schematic representation of the light schedule. Mice entrained in a 12 h light/dark cycle were transferred to DD. Mice were killed at ZT6, ZT18, CT6, and CT18 (n = 5 per group). Bottom, At the indicated times, the SCN was dissected, mRNA extracted and p75NTR, Per1, and Per2 levels analyzed by real-time PCR. Results of at least three independent experiments with duplicate measurements are shown. Error bars represent the SEM of the mean value. Statistical significance denoted by asterisks is relative to the expression obtained during the day/subjective day (*p < 0.05, t test). ZT, Zeitgeber time; CT, circadian time.
In mammals, the circadian timing system has a hierarchical architecture composed of a central clock in the SCN and subsidiary oscillators in virtually all cells of the body (Dibner et al., 2010). External cues like light signals stimulate and synchronize clock gene expression in the SCN (Quintero et al., 2003). Since p75NTR is expressed in the SCN (Moga, 1998a,b), we examined whether p75NTR oscillates in a light/dark cycle. Expression of p75NTR was higher during the light period and lower during the dark period, oscillating in phase with the clock genes Per1 and Per2 (Fig. 3B). To test whether the observed oscillation is driven by the endogenous clock system and not by exposure to light signals, p75NTR expression was analyzed in mice under constant darkness condition. p75NTR transcript levels displayed a robust circadian oscillation that was high during the subjective day and low during the subjective night (Fig. 3C), suggesting that p75NTR oscillation in the SCN is E-box dependent, and not light dependent in accordance with previous studies (Beaulé and Amir, 2001, 2002). As expected, similar results were obtained with E-box-dependent clock genes Per1 and Per2 (Fig. 3C).
Clock genes are not only expressed in the brain, but also in peripheral tissues (Schibler and Sassone-Corsi, 2002; Schibler, 2009). p75NTR is known to be expressed in several peripheral tissues such as the liver (Trim et al., 2000; Cassiman et al., 2001; Passino et al., 2007), lung (Sachs et al., 2007), and kidney (Wheeler and Bothwell, 1992). To examine whether p75NTR oscillates in the periphery, we compared the expression of p75NTR and clock genes in the liver and SCN every 4 h during the light/dark cycle. In the SCN, p75NTR oscillates in phase with Per1 (Fig. 4). Similar to the SCN, oscillation of p75NTR in liver was in phase with both Per1 and Per2 (Fig. 4). As expected, Clock in the SCN and Rorα in the liver did not oscillate (Fig. 4). These results indicate that p75NTR oscillates in a circadian time-specific manner in MEFs after serum shock as well as in the SCN and liver.
Figure 4.
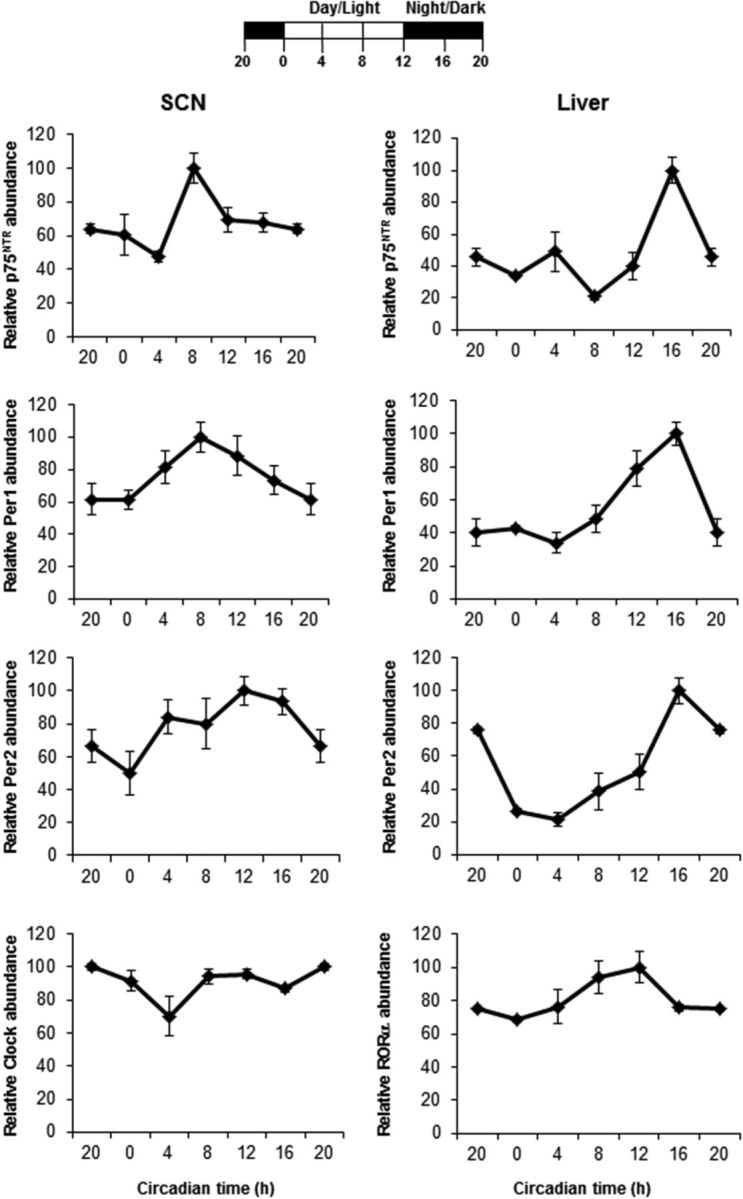
p75NTR RNA oscillates in a circadian manner in the SCN and liver. Top, Schematic representation of the light schedule. Mice were entrained in a 12 h light/dark cycle and were killed at 4 h intervals (n = 4 per group). Bottom, At the indicated times, the SCN (left) and liver (right) were dissected; RNA was extracted; and the levels of p75NTR, Per1, and Per2, together with nonoscillating control genes such as Clock for the SCN and Rorα for the liver, were analyzed by real-time PCR. The results of at least three independent experiments with duplicate measurements are shown. Data from CT20 are double plotted. Error bars represent the SEM of the mean, where the maximum RNA amount was set to 100. CT, Circadian time.
Oscillation of p75NTR is in phase with NGF and TrkA oscillations in the SCN
Since p75NTR binds to neurotrophin ligands (Roux and Barker, 2002; Chao, 2003; Reichardt, 2006; Underwood and Coulson, 2008), we examined the expression of NGF, BDNF, and their tropomyosin-related kinase (Trk) receptors A–C in the SCN and liver every 4 h during the light/dark cycle. Interestingly, in the SCN, NGF and TrkA oscillate in phase with p75NTR and Per1 (Figs. 4, 5A), while TrkB and TrkC did not oscillate (Fig. 5A). In accordance with previous studies (Liang et al., 1998), BDNF expression in the SCN showed an oscillatory pattern with higher expression during the subjective night and lower during the subjective day (Fig. 5A). On the other hand, NGF and TrkA expression in liver did not oscillate (Fig. 5A), BDNF was undetectable, and TrkB and TrkC expression showed an oscillatory pattern with similar phase to Per1 and Per2 clock genes, peaking at 12 h (Fig. 5A), suggesting diverse oscillatory patterns for neurotrophins and their Trk receptors in the SCN and peripheral tissues. Since p75NTR is a member of the TNFR superfamily, we also examined whether the expression of other TNF receptors such as TNFR member 6 or FAS, and TNFR1B oscillate in the SCN. FAS expression showed an oscillatory pattern peaking at 12 h, while TNFR1B was undetectable (Fig. 5B), suggesting that oscillation of p75NTR is not in phase with other members of the TNFR superfamily. Overall, these results show that while neurotrophin ligands and Trk receptors differentially oscillate in the SCN and liver, p75NTR is unique among neurotrophin receptors in oscillating in phase with clock genes in both central and peripheral tissues.
Figure 5.
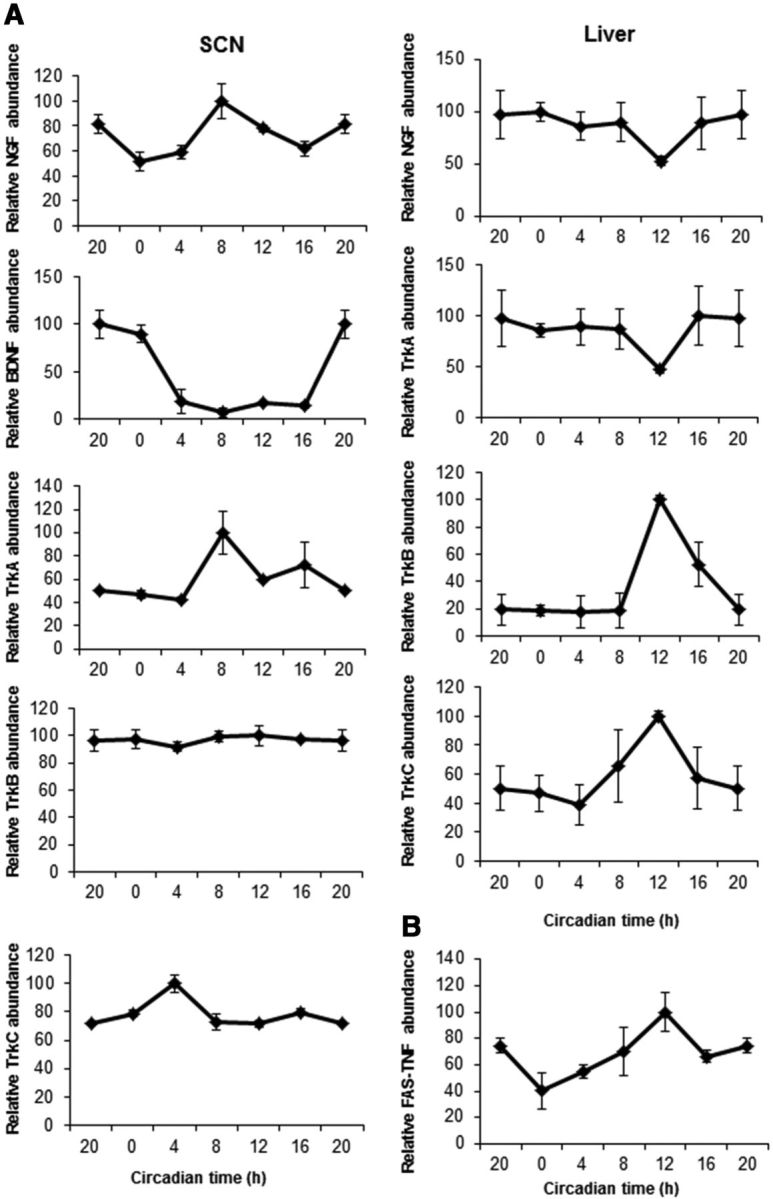
NGF and TrkA RNA oscillate in a circadian manner in the SCN and TrkB and TrkC in the liver. Mice were entrained in a 12 h light/dark cycle and were killed at 4 h intervals (n = 4 per group and genotype). A, At the indicated times, the SCN (left) and liver (right) were dissected; RNA was extracted; and the levels of NGF, BDNF, TrkA, TrkB, and TrkC were analyzed by real-time PCR. B, RNA from SCN was extracted and the level of FAS-TNFα was analyzed by real-time PCR at the indicated times. Results of at least three independent experiments with duplicate measurements are shown. Data from CT20 are double plotted. Error bars represent the SEM of the mean, where the maximum RNA amount was set to 100. CT, Circadian time.
Genetic deletion of Clock disrupts p75NTR oscillation in the SCN and liver
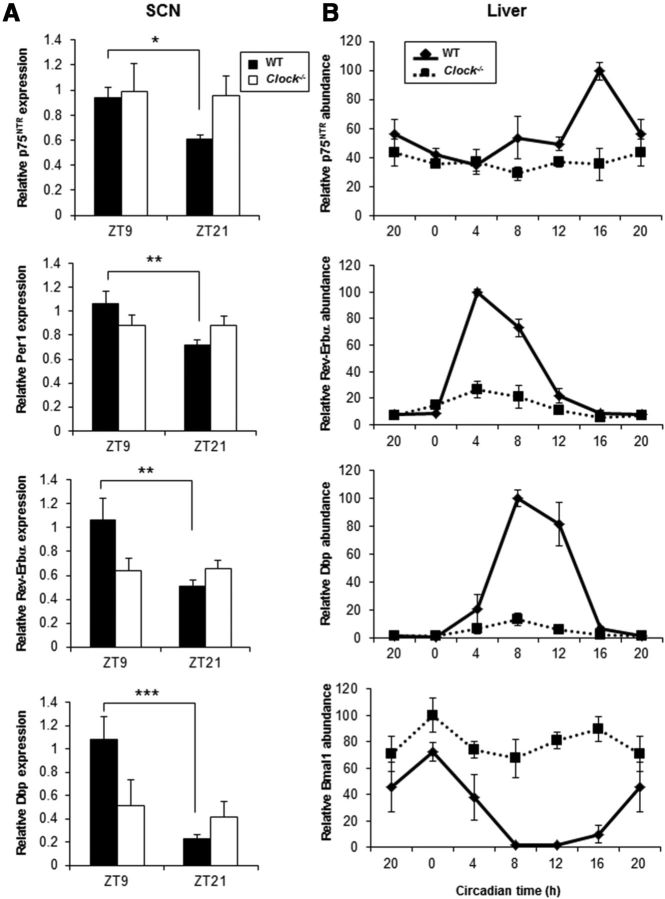
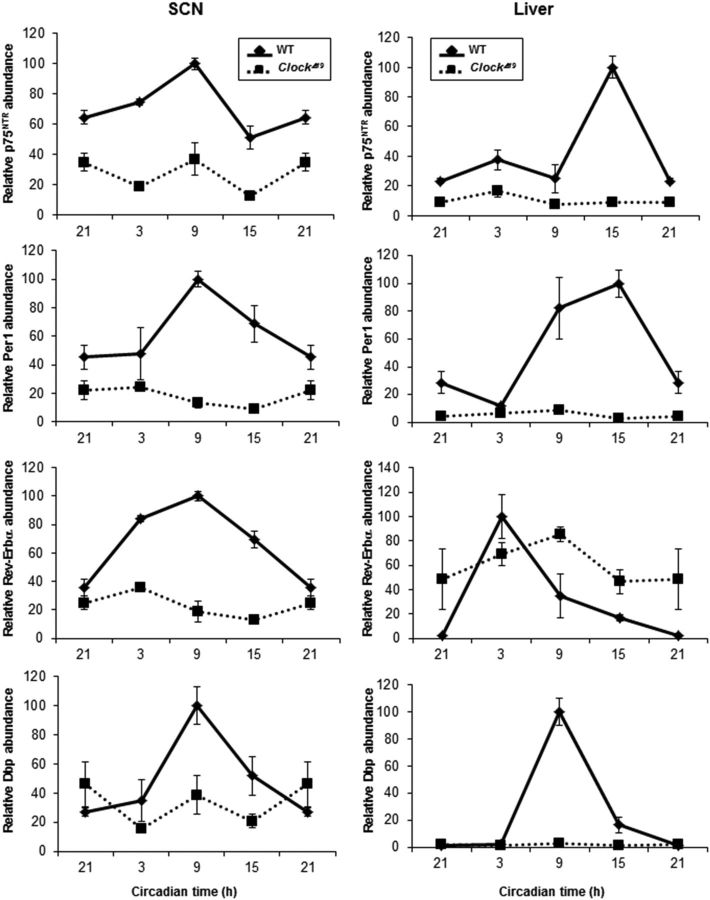
To study whether CLOCK/BMAL1 heterodimer is required for the circadian oscillation of p75NTR, we analyzed p75NTR expression in SCN and liver from Clock−/− (DeBruyne et al., 2006) and ClockΔ19 (Vitaterna et al., 1994) mice during the light/dark cycle. Consistent with binding of CLOCK/BMAL1 heterodimer to the p75NTR promoter (Fig. 2), p75NTR oscillation was disrupted in the SCN and liver from Clock−/− mice (Fig. 6A,B), suggesting that CLOCK is a critical regulator of p75NTR transcriptional regulation. We also tested known CLOCK-regulated genes as positive controls. In accordance with previous studies in Clock−/− mice (Preitner et al., 2002; DeBruyne et al., 2006), in the SCN Per1, Rev-Erbα, and Dbp showed reduced expression and disrupted oscillation (Fig. 6A), while in the liver Per1 and Rev-Erbα were reduced and Bmal1 was increased compared with WT mice (Fig. 6B). Although Clock−/− mice have defects in the amplitude of rhythmic gene expression of a subset of oscillatory genes (DeBruyne, 2008), ClockΔ19 mutant mice show global deficits in circadian gene oscillation (Jin et al., 1999; Kume et al., 1999; Ripperger et al., 2000) because it functions as a dominant negative due to impaired CLOCKΔ19/BMAL1 heterodimers (DeBruyne, 2008). Therefore, we also analyzed whether p75NTR expression and oscillation in SCN and liver from ClockΔ19 mice were disrupted. Similar to the Clock−/− mice, p75NTR expression and oscillation was disrupted in the SCN and liver from ClockΔ19 mice (Fig. 7). As expected, Per1, Rev-Erbα, and Dbp expression and oscillations were also disrupted (Fig. 7). Together, these results demonstrate that p75NTR is an oscillatory gene regulated by CLOCK/BMAL1 heterodimer in the SCN and liver.
Figure 6.
Genetic deletion of Clock disrupts p75NTR expression in the SCN and liver. A, Wild-type mice (Clock+/+) and Clock−/− mice were entrained in a 12 h light/dark cycle and killed at ZT9 and ZT21 (n = 4 per group and genotype). SCN was dissected; RNA was extracted; and p75NTR, Per1, Rev-Erbα, and Dbp levels were analyzed by real-time PCR. B, Wild-type mice (Clock+/+) and Clock−/− mice were entrained in a 12 h light/dark cycle and killed at 4 h intervals (n = 3 per group and genotype). At the indicated times, liver was dissected; RNA was extracted; and p75NTR, Rev-Erbα, Dbp, and Bmal1 levels were analyzed by real-time PCR. Data from CT20 are double plotted. Error bars represent the SEM, where the maximum RNA amount was set to 100. Statistical significance denoted by asterisks is relative to the expression obtained during the day/light phase ZT9 (*p < 0.05, **p < 0.01, ***p < 0.001, one-way ANOVA). ZT, Zeitgeber time; CT, circadian time.
Figure 7.
Genetic mutation of Clock disrupts p75NTR expression in the SCN and liver. Wild-type mice and ClockΔ19 mice were entrained in a 12 h light/dark cycle and killed at 6 h intervals (n = 3 per group and genotype). At the indicated times, SCN (left) and liver (right) were dissected; RNA was extracted; and p75NTR, Per1, Rev-Erbα, and Dbp levels were analyzed by real-time PCR. Data from CT21 are double plotted. Error bars represent the SEM of the mean, where the maximum RNA amount was set to 100. CT, Circadian time.
Loss of p75NTR disrupts clock gene oscillation in the SCN, liver, and MEFs after serum shock
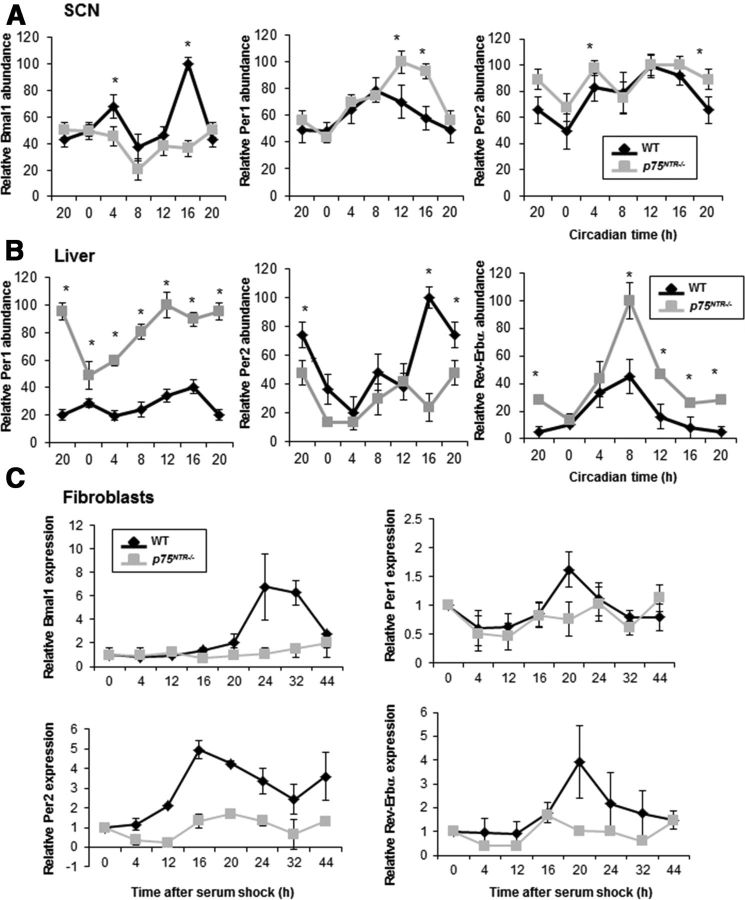
Although gene expression profiles obtained from microarray studies have revealed that 10–15% of all transcripts in different tissues display circadian oscillation (Akhtar et al., 2002; Duffield et al., 2002; Panda et al., 2002; Storch et al., 2002), only a subset of genes under circadian control are directly regulated by the binding of CLOCK/BMAL1 to their promoters. Therefore, the finding that p75NTR is an oscillatory gene regulated by CLOCK and BMAL1 suggests that p75NTR might play critical roles in circadian rhythms, either as a core clock protein or transducer of core oscillation. To examine the role of p75NTR in the regulation of circadian rhythms, we compared the expression of several clock genes in the SCN and liver of WT and p75NTR−/− mice every 4 h during the light/dark cycle, and also in MEFs derived from these mice after serum shock. Intriguingly, genetic deletion of p75NTR disrupted the transcriptional oscillations of clock genes such as Per1 and Per2 not only in the SCN (Fig. 8A), but also in liver (Fig. 8B). Consistent with these results, transcriptional clock gene oscillation of Bmal1, Per1, Per2, and Rev-Erbα in serum-shocked p75NTR−/− MEFs also caused a loss of the transcriptional oscillatory patterns (Fig. 8C), suggesting that p75NTR might be involved in the maintenance of the central and peripheral transcriptional oscillation of clock genes.
Figure 8.
Genetic deletion of p75NTR disrupts clock genes in SCN, liver, and fibroblasts. A, Top, Schematic representation of the circadian entrainment of the mice. Mice were entrained in a 12 h light/dark cycle and were killed at 4 h intervals (n = 4 per group and genotype). Bottom, At the indicated times, the SCN was dissected; RNA was extracted; and Bmal1, Per1, and Per2 levels were analyzed by real-time PCR. Error bars represent the SEM, where the maximum RNA amount was set to 100. B, Mice were entrained in a 12 h light/dark cycle and killed at 4 h intervals. At the indicated times, the liver was dissected; RNA was extracted; and Per1, Per2, and Rev-Erbα levels were analyzed by real-time PCR. Error bars represent the SEM, where the maximum RNA amount was set to 100. C, Serum shock treatment was performed on MEFs derived from WT and p75NTR−/− mice; cells were collected at the indicated times (h); and Bmal1, Per1, Per2, and Rev-Erbα mRNA levels were analyzed by real-time PCR. Data are presented as the relative expression level with respect to time 0, which was given a value of 1. Results of at least three independent experiments with duplicate measurements are shown. Data from CT20 are double plotted. Statistical significance denoted by asterisks is relative to the respective WT time point (*p < 0.05, one-way ANOVA). CT, Circadian time.
p75NTR−/− mice maintain robust locomotor activity rhythms
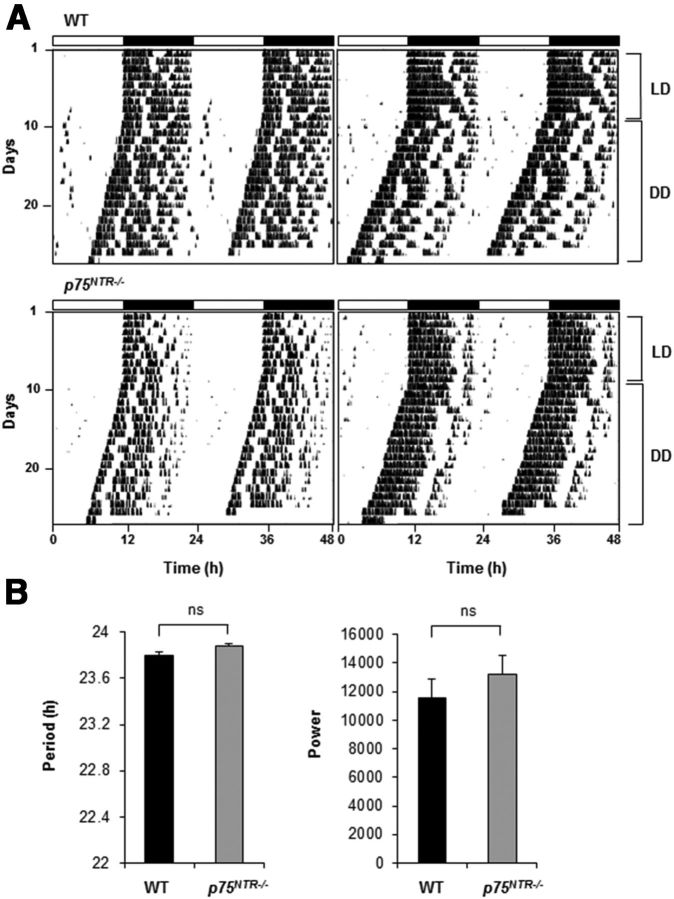
The SCN is responsible for regulating locomotor activity rhythms (Reppert and Weaver, 2002; Lowrey and Takahashi, 2004). Mice with disrupted clock gene expression, such as ClockΔ19 and Bmal1−/− show changes in period and amplitude in locomotor activity rhythms (Vitaterna et al., 1994; Bunger et al., 2000). On the other hand, several other mutant mice, such as Clock−/− and NPAS2−/− [neuronal Per-Arnt-Sim (PAS) domain containing protein 2], despite changes in gene expression, do not exhibit locomotor activity deficits (Dudley et al., 2003; DeBruyne et al., 2006). To determine the potential effects of p75NTR deficiency on behavioral rhythmicity, we monitored wheel-running activity under constant darkness (DD) for ∼2 weeks, following an initial 2 weeks in a 12 h light/dark cycle. p75NTR−/− mice displayed normal circadian patterns of behavior, with periodicities of 23.88 ± 0.03 (mean ± SEM), similar to those of WT mice (23.80 ± 0.02) (Fig. 9A,B). Despite the fact that p75NTR−/− mice showed increased activity levels, these differences were not statistically significant (Fig. 9B). Thus, similar to other clock mutants, such as Clock−/− and NPAS2−/−, p75NTR is not required for regulating locomotor activity rhythms.
Figure 9.
Circadian locomotor activity of p75NTR−/− mice. A, Representative activity records (actograms) of wild-type mice and p75NTR−/− mice are shown in double-plotted format. Each horizontal line represents 48 h. Vertical bars represents periods of voluntary wheel running activity. B, Periodogram estimates of period (h) for each genotype and power corresponding to circadian amplitude (power-significance from periodogram analyses). Each bar is mean × SEM (n = 14 per genotype). Statistical comparisons were made with Student's t test (ns, not significant).
Loss of p75NTR decreases Per2 amplitude in the liver, but not the SCN
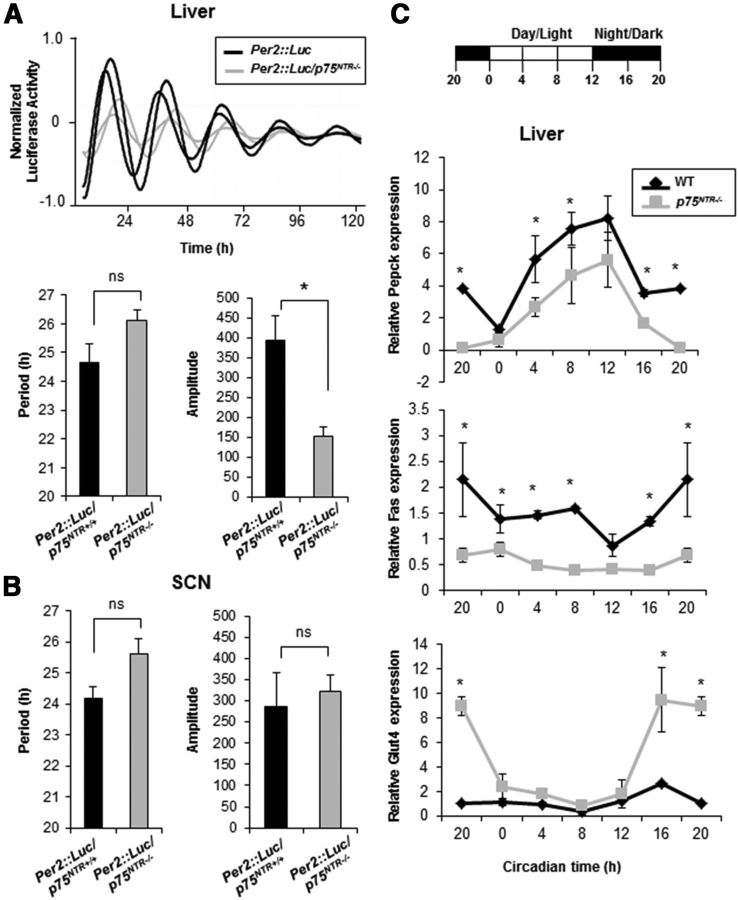
The circadian system is composed of several endogenous clocks, and the SCN is the main clock setting the phase of cell-autonomous and self-sustained cellular oscillators in peripheral tissues (Asher and Schibler, 2011). To examine the dynamic effects of the p75NTR deletion on circadian rhythms, p75NTR−/− mice were crossed with the Per2::Luc reporter mice (Yoo et al., 2004), which express Per2::Luciferase fusion protein and allow the recording of oscillatory pattern in real time. Interestingly, the amplitude of Per2 oscillation from Per2::Luc/p75NTR−/− liver explants was significantly decreased compared with that observed in Per2::Luc/p75NTR+/+ littermates (Fig. 10A). No differences in period were observed between Per2::Luc/p75NTR+/+ and Per2::Luc/p75NTR−/− liver explants (Fig. 10A). In accordance with the robust locomotor activity rhythms in the p75NTR−/− mice (Fig. 9), Per2::Luc/p75NTR−/− SCN explants exhibited robust circadian oscillations of bioluminescence, and no significant differences were observed in period or amplitude (Fig. 10B). Altogether, these results indicate that p75NTR is necessary to maintain Per2 oscillation in the liver, suggesting that p75NTR plays an important role in clock gene expression.
Figure 10.
Deletion of p75NTR reduces rhythm amplitudes and affects the expression of genes involved in glucose and lipid metabolism in liver. A, Top, Representative bioluminescence profiles of tissue explants from liver. The two representative profiles shown for each genotype (Per2::Luc p75NTR+/+ and Per2::Luc p75NTR−/−) are from separate experiments. Bottom, Period and amplitude of the bioluminescence profiles from liver explants. Each bar is the mean ± SEM of n (explants)/n (animals); Per2::Luc p75NTR+/+ = 21/7; Per2::Luc p75NTR−/− = 21/7 (*p < 0.05; t test). B, Period and amplitude of the bioluminescence profiles from SCN explants. Each bar is the mean ± SEM of n (explants)/n (animals); Per2::Luc p75NTR+/+ = 28/14; Per2::Luc p75NTR−/− = 28/14 (ns, not significant). C, Top, Schematic representation of the light schedule. Mice were entrained in a 12:12 h light/dark cycle and were killed at 4 h intervals (n = 4 per group). Bottom, At the indicated times, the liver was dissected, RNA extracted, and Pepck, Fas, and Glut4 levels analyzed by real-time PCR. Results of at least three independent experiments with duplicate measurements are shown. Data from CT20 are double plotted. Error bars represent the SEM of the mean value, where the maximum RNA amount was set to 100. Statistical significance denoted by asterisks is relative to the respective WT time point (*p < 0.05, one-way ANOVA).
Loss of p75NTR alters oscillation of glucose homeostasis genes in liver
Previous studies have reported that alterations in the period, phase, or amplitude of clock gene expression can trigger metabolic disorders (Bass and Takahashi, 2010). Since Per2::Luc/p75NTR−/− liver explants—and the livers of p75NTR−/− mice in vivo—showed reduced amplitude and altered transcriptional oscillation of clock genes, we analyzed whether the lack of p75NTR altered the oscillatory transcriptional expression of genes involved in glucose and lipid homeostasis. Gene expression analysis in the livers of p75NTR−/− mice showed reduced expression of the gluconeogenic gene phosphoenolpyruvate carboxykinase (Pepck) and the lipogenesis gene fatty-acid synthase (Fas), but increased expression of the glucose transporter 4 (Glut4) (Fig. 10C). These results show that deletion of p75NTR alters the circadian expression of genes involved in glucose and lipid metabolism in liver. Together, our results suggest that circadian regulation of p75NTR by CLOCK and BMAL1 might play an important role in the circadian expression of genes involved in glucose and lipid homeostasis in liver.
Discussion
In this study, we report a novel mechanism for the transcriptional regulation of p75NTR by investigating the molecular events that regulate its expression in the CNS and peripheral tissues. Our results demonstrated that p75NTR expression oscillates via the direct binding of CLOCK/BMAL1 to noncanonical E-box elements present in the p75NTR promoter. CLOCK and BMAL1 transcription factors are expressed not only in the SCN and other brain regions, but also in all peripheral tissues such as liver, heart, kidneys, lungs, and skeletal muscle (Schibler and Sassone-Corsi, 2002; Dibner et al., 2010). The widespread expression pattern of clock genes correlates with the variety of cell types and different organs that express p75NTR in the brain and peripheral tissues, suggesting that transcriptional regulation of p75NTR by CLOCK/BMAL1 could be a general mechanism to regulate p75NTR expression. Therefore, it is likely that CLOCK/BMAL1 is involved in the circadian regulation of p75NTR, while upon injury or other stressors, transcription factors such as Sp1 could drive the expression of p75NTR via binding to different elements of the promoter.
CLOCK and BMAL1 are known to regulate clock gene expression by interacting with a promoter element termed the E-box (CACGTG) (Hogenesch et al., 1998). However, noncanonical E-boxes or E-box-like sequences have also been reported for several clock-controlled genes. For instance, analysis of the gene Dbp demonstrated CLOCK and BMAL1 binding at multiple E-boxes, including one noncanonical E-box important for circadian expression (Ripperger and Schibler, 2006). In Per2, CLOCK and BMAL1 bind to an E-box-like sequence shown to be sufficient and indispensable for the circadian oscillation of Per2 expression (Yoo et al., 2005). Therefore, noncanonical E-boxes found in the p75NTR promoter are common targets for CLOCK and BMAL1 binding. In addition to Per and Cry genes (Darlington et al., 1998; Griffin et al., 1999; Kume et al., 1999; Sato et al., 2004), CLOCK/BMAL1 regulates genes that encode essential regulators of hormonal and metabolic control such as Vasopressin (Jin et al., 1999), Dbp (Ripperger and Schibler, 2006), and the transcription factors Rev-Erbα and Rev-Erbβ (Preitner et al., 2002). Therefore, the presence of functional noncanonical E-boxes in the p75NTR promoter directly regulated by CLOCK and BMAL1 suggests that p75NTR is a clock-controlled gene. Importantly, our results showed altered p75NTR rhythmic expression in the SCN and liver from Clock−/− and ClockΔ19 mice. Indeed, Clock−/− and ClockΔ19 mice show altered rhythmic expression of clock genes in the SCN and peripheral tissues (Vitaterna et al., 1994; DeBruyne, 2008). Therefore, the alteration of p75NTR rhythmic expression in the SCN and liver in Clock−/− and ClockΔ19 mice further supports the identification of p75NTR as a novel clock-controlled gene.
Previous studies showed that BDNF and its receptor TrkB oscillate in the SCN (Bova et al., 1998; Liang et al., 1998). Indeed, alteration in the expression levels of BDNF and TrkB into the SCN affects circadian pacemaker responses to light (Liang et al., 2000; Allen et al., 2005). Because p75NTR binds all neurotrophins (Chao, 2003), it is possible that neurotrophin and Trk receptor expression could be also regulated by CLOCK/BMAL1. Indeed, our results show that NGF and TrkA expression oscillate in a similar pattern to clock genes in the SCN, while TrkB and TrkC oscillate in the liver. Therefore, it is possible that neurotrophins and their receptors might play diverse roles in central and peripheral circadian oscillations. However, p75NTR is unique among neurotrophin receptors in oscillating in phase with circadian genes in the SCN and the periphery. This expression pattern is consistent with its pleiotropic functions in peripheral tissues, such as metabolism and liver regeneration (Passino et al., 2007; Baeza-Raja et al., 2012). Expression of Trks can determine the biological outcome of p75NTR signaling (Hempstead et al., 1991; Reichardt, 2006). Therefore, it is likely that the regulation of circadian gene oscillation by p75NTR could also be modulated by the oscillatory expression of its Trk coreceptors. Future studies will shed light on the contribution of neurotrophins in the regulation of circadian rhythms.
The SCN is important for rhythmic locomotor activity (Lehman et al., 1987). Although p75NTR−/− mice displayed a robust circadian locomotor activity similar to those of WT mice, similar results have been reported for several clock gene-deficient mice. For instance, Clock−/− (DeBruyne et al., 2006) and Per3-null mutant mice (Shearman et al., 2000) also display a robust circadian locomotor activity without substantial differences in period or amplitude. Moreover, single gene mutations in Per1, Per2, Cry1, and Cry2 have little effect on the locomotor activity period, while only disruption of Per1 and Per2 or Cry1 and Cry2 genes together causes behavioral and molecular arrhythmicity (Okamura et al., 1999; Vitaterna et al., 1999; Zheng et al., 1999, 2001; Bae et al., 2001; Cermakian et al., 2001). Therefore, it is possible that deletion of p75NTR alone is not enough to disrupt circadian locomotor activity in mice. It is also likely that p75NTR might be compensated by Trk receptor expression, similar to neuronal PAS domain protein 2 implicated to compensate for loss of Clock (Rutter et al., 2001; DeBruyne et al., 2006). Additionally, locomotor activity is a polygenic trait that varies widely among inbred strains of mice (Flint et al., 1995). Therefore, it is possible that p75NTR deletion in other mouse strains could display differences in the locomotor activity.
Clock-controlled genes such as the Per genes (Per1, Per2, and Per3) and cryptochrome genes (Cry1 and Cry2) form a heterodimeric repressor complex that translocates into the nucleus to inhibit CLOCK/BMAL1-mediated activation of clock-controlled genes (Darlington et al., 1998; Griffin et al., 1999; Kume et al., 1999; Sato et al., 2006). Previous studies showed that ubiquitination and proteasomal degradation of PER and CRY proteins play an essential role in the maintenance of clock genes oscillation (Eide et al., 2005; Busino et al., 2007; Godinho et al., 2007; Reischl et al., 2007; Siepka et al., 2007; Yoo et al., 2013). Since our results show altered rhythmic expression of clock genes not only in the SCN and liver from p75NTR−/− mice, but also in serum-shocked p75NTR−/− MEFs, it is possible that p75NTR might play a role in the maintenance of clock gene oscillation through the regulation of Per and Cry proteolytic degradation. Indeed, p75NTR directly interacts with E3 ubiquitin ligases (Le Moan et al., 2011), which govern circadian periodicity by degradation of Cry (Yoo et al., 2013). Therefore, it is possible that p75NTR could regulate proteolytic degradation of Per and Cry proteins by regulating E3 ubiquitin ligases. Future studies will determine whether interaction of p75NTR with E3 ubiquitin ligases plays a role in the regulation of clock gene oscillation.
Our results show altered rhythmic expression of clock genes such as Bmal1 and Per1, and to a lesser extent Per2, in SCN from p75NTR−/− mice, suggesting that p75NTR might play an important regulatory function for the maintenance of normal clock gene oscillatory patterns in the SCN. Accordingly, analysis of circadian oscillations of Per2 in SCN explants revealed no differences in period or amplitude. It is possible that the minor differences of Per2 expression observed in SCN from p75NTR−/− were not sufficient to induce changes in period or amplitude. Additionally, factors such as entraining signals or responsiveness to entraining signals, and communication between different areas of the brain or between different peripheral tissues could contribute to the differences in circadian properties of Per2 observed in SCN between explants and at the organismal level (Schibler et al., 2003). Similar to p75NTR−/− mice, Clock−/− mice show an altered expression of clock genes in the SCN (DeBruyne et al., 2006), although Per2::Luc/Clock−/− SCN explants show normal maintenance of Per2::Luc bioluminescence rhythmicity and amplitude (DeBruyne et al., 2007). Future studies will address the specific role of p75NTR in the regulation of clock gene oscillation in the SCN and/or other brain regions involved in the maintenance of clock gene expression.
Our study shows that loss of p75NTR alters the oscillation of clock and metabolic genes in the periphery, thus raising the question whether the peripheral effects of p75NTR are due to its peripheral expression or its effects in the SCN. On one hand, p75NTR is expressed in peripheral tissues such as liver (Trim et al., 2000; Cassiman et al., 2001; Passino et al., 2007), lung (Sachs et al., 2007), and kidney (Wheeler and Bothwell, 1992). Moreover, we demonstrated in cell-autonomous systems that p75NTR regulates glucose uptake in adipocytes and muscle cells (Baeza-Raja et al., 2012) and hepatic stellate cell differentiation (Passino et al., 2007). Therefore, it is possible that p75NTR alters the oscillation of clock and metabolic genes through its expression in the periphery. On the other hand, several studies have suggested an important role of the autonomic nervous system in the circadian regulation of metabolic functions such as glucose homeostasis (Kalsbeek et al., 2010a,b). Indeed, p75NTR regulates glucose uptake (Baeza-Raja et al., 2012) and is highly expressed by sensory and sympathetic neurons where it plays an essential role in the regulation of innervation during development (Reichardt, 2006). Interestingly, the SCN not only controls daily circadian rhythms, but also uses its projections to sympathetic preautonomic neurons in the paraventricular nucleus to control hepatic glucose production (Kalsbeek et al., 2004; Cailotto et al., 2005, 2008; Yi et al., 2010). Therefore, it is possible that p75NTR might be involved in the communication between the SCN and the periphery to control circadian and metabolic functions. Although p75NTR regulates clock gene oscillation in cell-autonomous systems of MEFs, our study does not exclude potential effects of p75NTR deletion on circadian regulation due to its major role in nervous system development (Reichardt, 2006). Although loss of p75NTR does not affect liver development, as the p75NTR−/− mice need to be challenged either by injury or fibrosis to exhibit defects in liver functions (Passino et al., 2007; Kendall et al., 2009), our study does not exclude a role for p75NTR in the control of peripheral clocks via peripheral innervation. Future studies using tissue-specific deletion of p75NTR will demonstrate the contribution of the central and peripheral expression of p75NTR in the regulation of circadian and metabolic functions.
Our study reveals p75NTR as an important link between the disruption of endogenous clocks in mice and metabolic dysfunction. Circadian clocks participate in the daily regulation of metabolic functions, and this is accomplished by the rhythmic expression of genes encoding regulators and enzymes of various metabolic pathways such as Fas and Pepck (Froy, 2007). Previous studies showed that CLOCK/BMAL1 directly regulates the expression of the orphan receptor Rev-Erbα and the peroxisome proliferator-activated receptors (Preitner et al., 2003; Oishi et al., 2005; Canaple et al., 2006), important regulators of triglyceride mobilization and lipid metabolism, respectively. Furthermore, Clock mutant mice present severe metabolic dysfunction including hyperglycemia, hypertriglyceridemia, and hepatic steatosis (Turek et al., 2005). Clock mutant and Bmal1−/− mice show increased insulin sensitivity and altered gluconeogenesis (Rudic et al., 2004). Mice with liver-specific deletion of Bmal1 show hypoglycemia and increased glucose tolerance (Lamia et al., 2008), and liver- and muscle-specific Clock mutant mice show impaired glucose homeostasis (Kennaway et al., 2007). In accordance, p75NTR−/− mice show exacerbated hepatic disease (Passino et al., 2007; Kendall et al., 2009) and increased insulin sensitivity (Baeza-Raja et al., 2012). Because p75NTR is an important regulator of glucose homeostasis (Baeza-Raja et al., 2012), it is therefore possible that p75NTR might regulate metabolic processes through the SCN, and by regulating clock genes in the periphery.
Overall, the regulation of p75NTR by CLOCK and BMAL1 identifies a novel first-order clock-controlled gene that oscillates in central and peripheral tissues and is important for the circadian oscillation of clock and metabolic genes. Therefore, these results may reveal p75NTR as a therapeutic target for diseases with altered circadian oscillation such as sleep disorders, neurologic and behavioral disorders, and metabolic diseases.
Footnotes
This work was supported by a Spanish Ministry of Education and Science postdoctoral fellowship (to B.B.-R.); and Pilot/Feasibility grants from the University of California San Francisco Liver Center (Grant P30 DK026743) and Diabetes and Endocrinology Center (Grant P30 DK63720); and National Institutes of Health Grant NS051470 (to K.A.). We thank Catherine Bedard and Saurabh Sahar for technical assistance.
References
- Akhtar RA, Reddy AB, Maywood ES, Clayton JD, King VM, Smith AG, Gant TW, Hastings MH, Kyriacou CP. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol. 2002;12:540–550. doi: 10.1016/S0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- Allen GC, Qu X, Earnest DJ. TrkB-deficient mice show diminished phase shifts of the circadian activity rhythm in response to light. Neurosci Lett. 2005;378:150–155. doi: 10.1016/j.neulet.2004.12.023. [DOI] [PubMed] [Google Scholar]
- Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 2011;13:125–137. doi: 10.1016/j.cmet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- Bae K, Jin X, Maywood ES, Hastings MH, Reppert SM, Weaver DR. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron. 2001;30:525–536. doi: 10.1016/S0896-6273(01)00302-6. [DOI] [PubMed] [Google Scholar]
- Baeza-Raja B, Li P, Le Moan N, Sachs BD, Schachtrup C, Davalos D, Vagena E, Bridges D, Kim C, Saltiel AR, Olefsky JM, Akassoglou K. p75 neurotrophin receptor regulates glucose homeostasis and insulin sensitivity. Proc Natl Acad Sci U S A. 2012;109:5838–5843. doi: 10.1073/pnas.1103638109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/S0092-8674(00)81199-X. [DOI] [PubMed] [Google Scholar]
- Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330:1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulé C, Amir S. Photic regulation of circadian rhythms and the expression of p75 neurotrophin receptor immunoreactivity in the suprachiasmatic nucleus in rats. Brain Res. 2001;894:301–306. doi: 10.1016/S0006-8993(01)02021-2. [DOI] [PubMed] [Google Scholar]
- Beaulé C, Amir S. Effect of 192 IgG-saporin on circadian activity rhythms, expression of P75 neurotrophin receptors, calbindin-D28K, and light-induced Fos in the suprachiasmatic nucleus in rats. Exp Neurol. 2002;176:377–389. doi: 10.1006/exnr.2002.7969. [DOI] [PubMed] [Google Scholar]
- Bova R, Micheli MR, Qualadrucci P, Zucconi GG. BDNF and trkB mRNAs oscillate in rat brain during the light-dark cycle. Brain Res Mol Brain Res. 1998;57:321–324. doi: 10.1016/S0169-328X(98)00092-8. [DOI] [PubMed] [Google Scholar]
- Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103:1009–1017. doi: 10.1016/S0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busino L, Bassermann F, Maiolica A, Lee C, Nolan PM, Godinho SI, Draetta GF, Pagano M. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science. 2007;316:900–904. doi: 10.1126/science.1141194. [DOI] [PubMed] [Google Scholar]
- Cailotto C, La Fleur SE, Van Heijningen C, Wortel J, Kalsbeek A, Feenstra M, Pévet P, Buijs RM. The suprachiasmatic nucleus controls the daily variation of plasma glucose via the autonomic output to the liver: are the clock genes involved? Eur J Neurosci. 2005;22:2531–2540. doi: 10.1111/j.1460-9568.2005.04439.x. [DOI] [PubMed] [Google Scholar]
- Cailotto C, van Heijningen C, van der Vliet J, van der Plasse G, Habold C, Kalsbeek A, Pévet P, Buijs RM. Daily rhythms in metabolic liver enzymes and plasma glucose require a balance in the autonomic output to the liver. Endocrinology. 2008;149:1914–1925. doi: 10.1210/en.2007-0816. [DOI] [PubMed] [Google Scholar]
- Canaple L, Rambaud J, Dkhissi-Benyahya O, Rayet B, Tan NS, Michalik L, Delaunay F, Wahli W, Laudet V. Reciprocal regulation of brain and muscle Arnt-like protein 1 and peroxisome proliferator-activated receptor alpha defines a novel positive feedback loop in the rodent liver circadian clock. Mol Endocrinol. 2006;20:1715–1727. doi: 10.1210/me.2006-0052. [DOI] [PubMed] [Google Scholar]
- Cassiman D, Denef C, Desmet VJ, Roskams T. Human and rat hepatic stellate cells express neurotrophins and neurotrophin receptors. Hepatology. 2001;33:148–158. doi: 10.1053/jhep.2001.20793. [DOI] [PubMed] [Google Scholar]
- Cermakian N, Monaco L, Pando MP, Dierich A, Sassone-Corsi P. Altered behavioral rhythms and clock gene expression in mice with a targeted mutation in the Period1 gene. EMBO J. 2001;20:3967–3974. doi: 10.1093/emboj/20.15.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao MV. Neurotrophins and their receptors: a convergence point for many signalling pathways. Nat Rev Neurosci. 2003;4:299–309. doi: 10.1038/nrn1078. [DOI] [PubMed] [Google Scholar]
- Darlington TK, Wager-Smith K, Ceriani MF, Staknis D, Gekakis N, Steeves TD, Weitz CJ, Takahashi JS, Kay SA. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science. 1998;280:1599–1603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- DeBruyne JP. Oscillating perceptions: the ups and downs of the CLOCK protein in the mouse circadian system. J Genet. 2008;87:437–446. doi: 10.1007/s12041-008-0066-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBruyne JP, Noton E, Lambert CM, Maywood ES, Weaver DR, Reppert SM. A clock shock: mouse CLOCK is not required for circadian oscillator function. Neuron. 2006;50:465–477. doi: 10.1016/j.neuron.2006.03.041. [DOI] [PubMed] [Google Scholar]
- DeBruyne JP, Weaver DR, Reppert SM. Peripheral circadian oscillators require CLOCK. Curr Biol. 2007;17:R538–R539. doi: 10.1016/j.cub.2007.05.067. [DOI] [PubMed] [Google Scholar]
- De Cesare D, Vallone D, Caracciolo A, Sassone-Corsi P, Nerlov C, Verde P. Heterodimerization of c-Jun with ATF-2 and c-Fos is required for positive and negative regulation of the human urokinase enhancer. Oncogene. 1995;11:365–376. [PubMed] [Google Scholar]
- Deponti D, Buono R, Catanzaro G, De Palma C, Longhi R, Meneveri R, Bresolin N, Bassi MT, Cossu G, Clementi E, Brunelli S. The low-affinity receptor for neurotrophins p75NTR plays a key role for satellite cell function in muscle repair acting via RhoA. Mol Biol Cell. 2009;20:3620–3627. doi: 10.1091/mbc.E09-01-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- Dudley CA, Erbel-Sieler C, Estill SJ, Reick M, Franken P, Pitts S, McKnight SL. Altered patterns of sleep and behavioral adaptability in NPAS2-deficient mice. Science. 2003;301:379–383. doi: 10.1126/science.1082795. [DOI] [PubMed] [Google Scholar]
- Duffield GE, Best JD, Meurers BH, Bittner A, Loros JJ, Dunlap JC. Circadian programs of transcriptional activation, signaling, and protein turnover revealed by microarray analysis of mammalian cells. Curr Biol. 2002;12:551–557. doi: 10.1016/S0960-9822(02)00765-0. [DOI] [PubMed] [Google Scholar]
- Eide EJ, Woolf MF, Kang H, Woolf P, Hurst W, Camacho F, Vielhaber EL, Giovanni A, Virshup DM. Control of mammalian circadian rhythm by CKIepsilon-regulated proteasome-mediated PER2 degradation. Mol Cell Biol. 2005;25:2795–2807. doi: 10.1128/MCB.25.7.2795-2807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J, Corley R, DeFries JC, Fulker DW, Gray JA, Miller S, Collins AC. A simple genetic basis for a complex psychological trait in laboratory mice. Science. 1995;269:1432–1435. doi: 10.1126/science.7660127. [DOI] [PubMed] [Google Scholar]
- Froy O. The relationship between nutrition and circadian rhythms in mammals. Front Neuroendocrinol. 2007;28:61–71. doi: 10.1016/j.yfrne.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Gao X, Daugherty RL, Tourtellotte WG. Regulation of low affinity neurotrophin receptor (p75(NTR)) by early growth response (Egr) transcriptional regulators. Mol Cell Neurosci. 2007;36:501–514. doi: 10.1016/j.mcn.2007.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- Godinho SI, Maywood ES, Shaw L, Tucci V, Barnard AR, Busino L, Pagano M, Kendall R, Quwailid MM, Romero MR, O'neill J, Chesham JE, Brooker D, Lalanne Z, Hastings MH, Nolan PM. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science. 2007;316:897–900. doi: 10.1126/science.1141138. [DOI] [PubMed] [Google Scholar]
- Griffin EA, Jr, Staknis D, Weitz CJ. Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science. 1999;286:768–771. doi: 10.1126/science.286.5440.768. [DOI] [PubMed] [Google Scholar]
- Hempstead BL, Martin-Zanca D, Kaplan DR, Parada LF, Chao MV. High-affinity NGF binding requires coexpression of the trk proto-oncogene and the low-affinity NGF receptor. Nature. 1991;350:678–683. doi: 10.1038/350678a0. [DOI] [PubMed] [Google Scholar]
- Hogenesch JB, Gu YZ, Jain S, Bradfield CA. The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc Natl Acad Sci U S A. 1998;95:5474–5479. doi: 10.1073/pnas.95.10.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Shearman LP, Weaver DR, Zylka MJ, de Vries GJ, Reppert SM. A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell. 1999;96:57–68. doi: 10.1016/S0092-8674(00)80959-9. [DOI] [PubMed] [Google Scholar]
- Kalsbeek A, La Fleur S, Van Heijningen C, Buijs RM. Suprachiasmatic GABAergic inputs to the paraventricular nucleus control plasma glucose concentrations in the rat via sympathetic innervation of the liver. J Neurosci. 2004;24:7604–7613. doi: 10.1523/JNEUROSCI.5328-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsbeek A, Yi CX, La Fleur SE, Fliers E. The hypothalamic clock and its control of glucose homeostasis. Trends Endocrinol Metab. 2010a;21:402–410. doi: 10.1016/j.tem.2010.02.005. [DOI] [PubMed] [Google Scholar]
- Kalsbeek A, Bruinstroop E, Yi CX, Klieverik LP, La Fleur SE, Fliers E. Hypothalamic control of energy metabolism via the autonomic nervous system. Ann N Y Acad Sci. 2010b;1212:114–129. doi: 10.1111/j.1749-6632.2010.05800.x. [DOI] [PubMed] [Google Scholar]
- Kendall TJ, Hennedige S, Aucott RL, Hartland SN, Vernon MA, Benyon RC, Iredale JP. p75 Neurotrophin receptor signaling regulates hepatic myofibroblast proliferation and apoptosis in recovery from rodent liver fibrosis. Hepatology. 2009;49:901–910. doi: 10.1002/hep.22701. [DOI] [PubMed] [Google Scholar]
- Kennaway DJ, Owens JA, Voultsios A, Boden MJ, Varcoe TJ. Metabolic homeostasis in mice with disrupted Clock gene expression in peripheral tissues. Am J Physiol Regul Integr Comp Physiol. 2007;293:R1528–R1537. doi: 10.1152/ajpregu.00018.2007. [DOI] [PubMed] [Google Scholar]
- Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/S0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci U S A. 2008;105:15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KF, Li E, Huber LJ, Landis SC, Sharpe AH, Chao MV, Jaenisch R. Targeted mutation of the gene encoding the low affinity NGF receptor p75 leads to deficits in the peripheral sensory nervous system. Cell. 1992;69:737–749. doi: 10.1016/0092-8674(92)90286-L. [DOI] [PubMed] [Google Scholar]
- Lehman MN, Silver R, Gladstone WR, Kahn RM, Gibson M, Bittman EL. Circadian rhythmicity restored by neural transplant. Immunocytochemical characterization of the graft and its integration with the host brain. J Neurosci. 1987;7:1626–1638. doi: 10.1523/JNEUROSCI.07-06-01626.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Moan N, Houslay DM, Christian F, Houslay MD, Akassoglou K. Oxygen-dependent cleavage of p75 neurotrophin receptor triggers stabilization of HIF-1 alpha. Mol Cell. 2011;44:476–490. doi: 10.1016/j.molcel.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang FQ, Walline R, Earnest DJ. Circadian rhythm of brain-derived neurotrophic factor in the rat suprachiasmatic nucleus. Neurosci Lett. 1998;242:89–92. doi: 10.1016/S0304-3940(98)00062-7. [DOI] [PubMed] [Google Scholar]
- Liang FQ, Allen G, Earnest D. Role of brain-derived neurotrophic factor in the circadian regulation of the suprachiasmatic pacemaker by light. J Neurosci. 2000;20:2978–2987. doi: 10.1523/JNEUROSCI.20-08-02978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lomen-Hoerth C, Shooter EM. Widespread neurotrophin receptor expression in the immune system and other nonneuronal rat tissues. J Neurochem. 1995;64:1780–1789. doi: 10.1046/j.1471-4159.1995.64041780.x. [DOI] [PubMed] [Google Scholar]
- Lowrey PL, Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo T, Yamaguchi S, Mitsui S, Emi A, Shimoda F, Okamura H. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302:255–259. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- Moga MM. Delayed loss of p75 neurotrophin receptor-immunoreactivity in the rat suprachiasmatic nucleus and intergeniculate leaflet after binocular enucleation. Neurosci Lett. 1998a;253:187–190. doi: 10.1016/S0304-3940(98)00658-2. [DOI] [PubMed] [Google Scholar]
- Moga MM. 192 IgG-saporin abolishes p75 neurotrophin receptor immunoreactivity in rat SCN. Neuroreport. 1998b;9:3197–3200. doi: 10.1097/00001756-199810050-00012. [DOI] [PubMed] [Google Scholar]
- Oishi K, Shirai H, Ishida N. CLOCK is involved in the circadian transactivation of peroxisome-proliferator-activated receptor alpha (PPARalpha) in mice. Biochem J. 2005;386:575–581. doi: 10.1042/BJ20041150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura H, Miyake S, Sumi Y, Yamaguchi S, Yasui A, Muijtjens M, Hoeijmakers JH, van der Horst GT. Photic induction of mPer1 and mPer2 in cry-deficient mice lacking a biological clock. Science. 1999;286:2531–2534. doi: 10.1126/science.286.5449.2531. [DOI] [PubMed] [Google Scholar]
- Panda S, Hogenesch JB, Kay SA. Circadian rhythms from flies to human. Nature. 2002;417:329–335. doi: 10.1038/417329a. [DOI] [PubMed] [Google Scholar]
- Passino MA, Adams RA, Sikorski SL, Akassoglou K. Regulation of hepatic stellate cell differentiation by the neurotrophin receptor p75NTR. Science. 2007;315:1853–1856. doi: 10.1126/science.1137603. [DOI] [PubMed] [Google Scholar]
- Peeraully MR, Jenkins JR, Trayhurn P. NGF gene expression and secretion in white adipose tissue: regulation in 3T3-L1 adipocytes by hormones and inflammatory cytokines. Am J Physiol Endocrinol Metab. 2004;287:E331–339. doi: 10.1152/ajpendo.00076.2004. [DOI] [PubMed] [Google Scholar]
- Peterson S, Bogenmann E. Osmotic swelling induces p75 neurotrophin receptor (p75NTR) expression via nitric oxide. J Biol Chem. 2003;278:33943–33950. doi: 10.1074/jbc.M302376200. [DOI] [PubMed] [Google Scholar]
- Poukka H, Kallio PJ, Jänne OA, Palvimo JJ. Regulation of the rat p75 neurotrophin receptor promoter by GC element binding proteins. Biochem Biophys Res Commun. 1996;229:565–570. doi: 10.1006/bbrc.1996.1844. [DOI] [PubMed] [Google Scholar]
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/S0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- Preitner N, Brown S, Ripperger J, Le-Minh N, Damiola F, Schibler U. Orphan nuclear receptors, molecular clockwork, and the entrainment of peripheral oscillators. Novartis Found Symp. 2003;253:89–99. doi: 10.1002/0470090839.ch7. [DOI] [PubMed] [Google Scholar]
- Quintero JE, Kuhlman SJ, McMahon DG. The biological clock nucleus: a multiphasic oscillator network regulated by light. J Neurosci. 2003;23:8070–8076. doi: 10.1523/JNEUROSCI.23-22-08070.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos A, Ho WC, Forte S, Dickson K, Boutilier J, Favell K, Barker PA. Hypo-osmolar stress induces p75NTR expression by activating Sp1-dependent transcription. J Neurosci. 2007;27:1498–1506. doi: 10.1523/JNEUROSCI.4806-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichardt LF. Neurotrophin-regulated signalling pathways. Philos Trans R Soc Lond B Biol Sci. 2006;361:1545–1564. doi: 10.1098/rstb.2006.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reischl S, Vanselow K, Westermark PO, Thierfelder N, Maier B, Herzel H, Kramer A. Beta-TrCP1-mediated degradation of PERIOD2 is essential for circadian dynamics. J Biol Rhythms. 2007;22:375–386. doi: 10.1177/0748730407303926. [DOI] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- Ripperger JA, Schibler U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet. 2006;38:369–374. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- Ripperger JA, Shearman LP, Reppert SM, Schibler U. CLOCK, an essential pacemaker component, controls expression of the circadian transcription factor DBP. Genes Dev. 2000;14:679–689. [PMC free article] [PubMed] [Google Scholar]
- Roux PP, Barker PA. Neurotrophin signaling through the p75 neurotrophin receptor. Prog Neurobiol. 2002;67:203–233. doi: 10.1016/S0301-0082(02)00016-3. [DOI] [PubMed] [Google Scholar]
- Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, Hogenesch JB, Fitzgerald GA. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2:e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell JL, Powers JT, Rounbehler RJ, Rogers PM, Conti CJ, Johnson DG. ARF differentially modulates apoptosis induced by E2F1 and Myc. Mol Cell Biol. 2002;22:1360–1368. doi: 10.1128/MCB.22.5.1360-1368.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter J, Reick M, Wu LC, McKnight SL. Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science. 2001;293:510–514. doi: 10.1126/science.1060698. [DOI] [PubMed] [Google Scholar]
- Sachs BD, Baillie GS, McCall JR, Passino MA, Schachtrup C, Wallace DA, Dunlop AJ, MacKenzie KF, Klussmann E, Lynch MJ, Sikorski SL, Nuriel T, Tsigelny I, Zhang J, Houslay MD, Chao MV, Akassoglou K. p75 neurotrophin receptor regulates tissue fibrosis through inhibition of plasminogen activation via a PDE4/cAMP/PKA pathway. J Cell Biol. 2007;177:1119–1132. doi: 10.1083/jcb.200701040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA, Hogenesch JB. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Sato TK, Yamada RG, Ukai H, Baggs JE, Miraglia LJ, Kobayashi TJ, Welsh DK, Kay SA, Ueda HR, Hogenesch JB. Feedback repression is required for mammalian circadian clock function. Nat Genet. 2006;38:312–319. doi: 10.1038/ng1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibler U. The 2008 Pittendrigh/Aschoff lecture: peripheral phase coordination in the mammalian circadian timing system. J Biol Rhythms. 2009;24:3–15. doi: 10.1177/0748730408329383. [DOI] [PubMed] [Google Scholar]
- Schibler U, Sassone-Corsi P. A web of circadian pacemakers. Cell. 2002;111:919–922. doi: 10.1016/S0092-8674(02)01225-4. [DOI] [PubMed] [Google Scholar]
- Schibler U, Ripperger J, Brown SA. Peripheral circadian oscillators in mammals: time and food. J Biol Rhythms. 2003;18:250–260. doi: 10.1177/0748730403018003007. [DOI] [PubMed] [Google Scholar]
- Sehgal A, Patil N, Chao M. A constitutive promoter directs expression of the nerve growth factor receptor gene. Mol Cell Biol. 1988;8:3160–3167. doi: 10.1128/mcb.8.8.3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearman LP, Jin X, Lee C, Reppert SM, Weaver DR. Targeted disruption of the mPer3 gene: subtle effects on circadian clock function. Mol Cell Biol. 2000;20:6269–6275. doi: 10.1128/MCB.20.17.6269-6275.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepka SM, Yoo SH, Park J, Song W, Kumar V, Hu Y, Lee C, Takahashi JS. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell. 2007;129:1011–1023. doi: 10.1016/j.cell.2007.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solt LA, Wang Y, Banerjee S, Hughes T, Kojetin DJ, Lundasen T, Shin Y, Liu J, Cameron MD, Noel R, Yoo SH, Takahashi JS, Butler AA, Kamenecka TM, Burris TP. Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature. 2012;485:62–68. doi: 10.1038/nature11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- Travnickova-Bendova Z, Cermakian N, Reppert SM, Sassone-Corsi P. Bimodal regulation of mPeriod promoters by CREB-dependent signaling and CLOCK/BMAL1 activity. Proc Natl Acad Sci U S A. 2002;99:7728–7733. doi: 10.1073/pnas.102075599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trim N, Morgan S, Evans M, Issa R, Fine D, Afford S, Wilkins B, Iredale J. Hepatic stellate cells express the low affinity nerve growth factor receptor p75 and undergo apoptosis in response to nerve growth factor stimulation. Am J Pathol. 2000;156:1235–1243. doi: 10.1016/S0002-9440(10)64994-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, Eckel RH, Takahashi JS, Bass J. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood CK, Coulson EJ. The p75 neurotrophin receptor. Int J Biochem Cell Biol. 2008;40:1664–1668. doi: 10.1016/j.biocel.2007.06.010. [DOI] [PubMed] [Google Scholar]
- Vitaterna MH, King DP, Chang AM, Kornhauser JM, Lowrey PL, McDonald JD, Dove WF, Pinto LH, Turek FW, Takahashi JS. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science. 1994;264:719–725. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitaterna MH, Selby CP, Todo T, Niwa H, Thompson C, Fruechte EM, Hitomi K, Thresher RJ, Ishikawa T, Miyazaki J, Takahashi JS, Sancar A. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci U S A. 1999;96:12114–12119. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler EF, Bothwell M. Spatiotemporal patterns of expression of NGF and the low-affinity NGF receptor in rat embryos suggest functional roles in tissue morphogenesis and myogenesis. J Neurosci. 1992;12:930–945. doi: 10.1523/JNEUROSCI.12-03-00930.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Padiath QS, Shapiro RE, Jones CR, Wu SC, Saigoh N, Saigoh K, Ptácek LJ, Fu YH. Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature. 2005;434:640–644. doi: 10.1038/nature03453. [DOI] [PubMed] [Google Scholar]
- Yamazaki S, Takahashi JS. Real-time luminescence reporting of circadian gene expression in mammals. Methods Enzymol. 2005;393:288–301. doi: 10.1016/S0076-6879(05)93012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi CX, la Fleur SE, Fliers E, Kalsbeek A. The role of the autonomic nervous liver innervation in the control of energy metabolism. Biochim Biophys Acta. 2010;1802:416–431. doi: 10.1016/j.bbadis.2010.01.006. [DOI] [PubMed] [Google Scholar]
- Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, Menaker M, Takahashi JS. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SH, Ko CH, Lowrey PL, Buhr ED, Song EJ, Chang S, Yoo OJ, Yamazaki S, Lee C, Takahashi JS. A noncanonical E-box enhancer drives mouse Period2 circadian oscillations in vivo. Proc Natl Acad Sci U S A. 2005;102:2608–2613. doi: 10.1073/pnas.0409763102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SH, Mohawk JA, Siepka SM, Shan Y, Huh SK, Hong HK, Kornblum I, Kumar V, Koike N, Xu M, Nussbaum J, Liu X, Chen Z, Chen ZJ, Green CB, Takahashi JS. Competing E3 ubiquitin ligases govern circadian periodicity by degradation of CRY in nucleus and cytoplasm. Cell. 2013;152:1091–1105. doi: 10.1016/j.cell.2013.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Larkin DW, Albrecht U, Sun ZS, Sage M, Eichele G, Lee CC, Bradley A. The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature. 1999;400:169–173. doi: 10.1038/22118. [DOI] [PubMed] [Google Scholar]
- Zheng B, Albrecht U, Kaasik K, Sage M, Lu W, Vaishnav S, Li Q, Sun ZS, Eichele G, Bradley A, Lee CC. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell. 2001;105:683–694. doi: 10.1016/S0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]