Abstract
Axonal protein synthesis is a complex process involving selective mRNA localization and translational regulation. In this study, using in situ hybridization and metabolic labeling, we show that the mRNAs encoding eukaryotic translation initiation factors eIF2B2 and eIF4G2 are present in the axons of rat sympathetic neurons and are locally translated. We also report that a noncoding microRNA, miR16, modulates the axonal expression of eIF2B2 and eIF4G2. Transfection of axons with precursor miR16 and anti-miR16 showed that local miR16 levels modulated axonal eIF2B2 and eIF4G2 mRNA and protein levels, as well as axon outgrowth. siRNA-mediated knock-down of axonal eIF2B2 and eIF4G2 mRNA also resulted in a significant decrease in axonal eIF2B2 and eIF4G2 protein. Moreover, results of metabolic labeling studies showed that downregulation of axonal eIF2B2 and eIF4G2 expression also inhibited local protein synthesis and axon growth. Together, these data provide evidence that miR16 mediates axonal growth, at least in part, by regulating the local protein synthesis of eukaryotic translation initiation factors eIF2B2 and eIF4G2 in the axon.
Introduction
Early studies, conducted in both invertebrate and vertebrate animal species, have identified numerous axonally localized mRNAs encoding a functionally diverse set of proteins, such as cytoskeletal elements, translation factors, ribosomal proteins, molecular chaperones, signaling molecules, transcription factors, and nuclear-encoded mitochondrial proteins (Perrone-Capano et al., 1987; Moccia et al., 2003; Willis et al., 2007; Vogelaar et al., 2009; Tcherkezian et al., 2010; Zivraj et al., 2010; Gumy et al., 2011). It has also been shown that local translation plays a key role in axonal functions, such as axon growth, regeneration, synaptic plasticity, signal transduction, and long-term viability (Martin et al., 1997; Campbell and Holt, 2001; Zhang and Poo, 2002; Hanz et al., 2003; Si et al., 2003; Verma et al., 2005; Wu et al., 2005; Leung et al., 2006; Hillefors et al., 2007; Cox et al., 2008; Dubacq et al., 2009; Yoon et al., 2012; Donnelly et al., 2013).
In addition to these mRNAs, axons contain a diverse population of small, noncoding RNAs, namely, microRNAs (miRs), which play key roles in the regulation of local protein synthesis (Schratt et al., 2006; Aschrafi et al., 2008; Muddashetty et al., 2011; Olde Loohuis et al., 2012). Recently, it has been reported that ∼130 different miRNAs are present in the axons of sympathetic neurons of the rat superior cervical ganglia (SCG) (Natera-Naranjo et al., 2010). One of the most abundant miRs identified in SCG axons was miR16. A bioinformatic search for genes encoding mRNAs that contain miR16-binding sites in the 3′ untranslated region (3′UTR) revealed eukaryotic translation initiation factors (eIFs), eIF2B2 and eIF4G2, as potential candidates. In this communication, we report that the mRNAs encoding eIF2B2 and eIF4G2, components involved in eukaryotic translation initiation pathway, are present in the axon and that axonal translation of these factors is regulated by miRNA16. In this communication, we also demonstrate that local translation of eIF2B2 and eIF4G2 plays an important role in the regulation of the axonal protein synthetic system and axon growth.
Materials and Methods
Neuronal cell cultures.
SCGs were obtained from 3-day-old Harlan Sprague Dawley rats of either sex. Neurons were dissociated using Miltenyi Biotec gentle MACS Dissociator and Neuronal Tissue Dissociation Kit according to the manufacturer's protocol. Dissociated primary neurons were plated into the center compartment of a three-compartmented Campenot culture chamber (Hillefors et al., 2007). Cells were grown in serum-free Neurobasal medium (Invitrogen) containing nerve growth factor (NGF; 50 ng/ml), 20 mm KCl, and 20 U/ml penicillin and 20 mg/ml streptomycin (Hyclone) for 2–7 d before use. The culture medium was replaced every 3–4 d. Two days after plating, 5-fluoro-2′-deoxyuridine (50 μm) was added to the culture medium to inhibit the growth of non-neuronal cells and remained in the medium for the duration of the experiments. Culture media also contained NGF at all times. The side compartments, which contained the distal axons used in the experiments, were devoid of neuronal soma and non-neuronal cells, as judged by phase-contrast microscopy and 5-bromodeoxyuridine staining.
Fluorescence in situ hybridization (FISH).
In situ hybridization for miR16, eIF2B2, and eIF4G2 was performed according to the protocol described previously (Natera-Naranjo et al., 2012). Briefly, 7-day-old SCG axons were fixed in 4% formalin for 15 min and then washed three times in TBS-Triton X-100 (1 × TBS, 0.1% Triton X-100) for 5 min. Axons were subsequently permeabilized in 0.5% Triton X-100 in TBS for 10 min and postfixed with 4% formalin. After washes with TBS-T for 5 min, acetylation was performed in 25% acetic anhydride, 0.1 m HEPES for 10 min followed by equilibration in 4× sodium–saline citrate (SSC), 50% formamide for 20 min. FITC-conjugated locked nucleic acid eIF2B2 antisense (5′ GAAACAAACAUAGCCUAGUCAC 3′), eIF4G2 antisense (5′ ACAUUUCUGGUUCGGUUUUCAA 3′), and negative control probes (5′ GUGUAACACGUCUAUACGCCCA 3′) were hybridized at 55°C overnight (25 nm probes in 10% dextran sulfate, 4 × SSC, 1 × Denhardt's solution, 40% formamide, 1 mm DTT, 0.1 mg/ml yeast tRNA, AND 0.1 mg/ml salmon sperm DNA). After incubation, axons were washed once in 40% formamide, 1 × SSC at 55°C for 20 min, and three times in 1 × SSC at 55°C, 5 min each. The signal intensity of the locked nucleic acid probes was amplified with AlexaFluor-488 signal amplification kit (Invitrogen).
Luciferase reporter assay.
The pEZX-Reporter containing the eIF2B2 and eIF4G2 3′UTR constructs and the eIF2B2 and eIF4G2 miR16 miR targeting sequence (MTS) deletion vectors were obtained from GeneCopoeia. SCG neurons were cotransfected with pEZX-reporter as control, or the dual luciferase reporter constructs containing either the full eIF2B2 or eIF4G2 3′UTR or eIF2B2 and eIF4G2 3′UTR with miR16 MTS deletion, together with either precursor miR16 or inhibitors for miR16 (anti-miR16) or a nontargeting control (NT), respectively. Twenty-four hours after transfection, cells were assayed for firefly luciferase and Renilla luciferase expression: Renilla luciferase levels were used to normalize differences in transfection efficiency. The Dual-Light luminescent reporter gene assay kit (Promega) was used for the detection of firefly and Renilla luciferase in the same sample, using the GloMax 96 Microplate Luminometer (Promega).
Metabolic labeling of newly synthesized proteins.
To label newly synthesized proteins, the Click-iT Protein Labeling Kit (Invitrogen) was used. SCG axons were incubated in methionine-free Neurobasal medium for 1 h. The medium was then replaced with methionine-free Neurobasal medium containing 0.2 μm of the methionine analog, l- azidohomoalanine (AHA). The axons were incubated in AHA-containing medium for 6 h, allowing incorporation of AHA into nascent proteins. Negative control cultures were grown in Neurobasal medium containing methionine instead of AHA. After incubation, cells were lysed in 50 mm Tris-HCl, pH 8.0, 1% SDS, and complete protease inhibitor mix (Sigma). The newly synthesized, AHA-incorporated protein was crosslinked to alkyne-derivatized biotin (Invitrogen) by a copper (I)–catalyzed cycloaddition (Click-iT) according to the manufacturer's recommendation using the Click-iT protein reaction buffer kit (Invitrogen). After crosslinking, the AHA-labeled, biotin-crosslinked proteins were isolated by affinity pulldown with streptavidin coated Dynabeads M-280 (Invitrogen). Matrix-bound proteins were eluted into LDS sample buffer (Invitrogen) by boiling for 5 min. Total protein inputs and affinity-purified fractions were separated by SDS-PAGE and eIF2B2, eIF4G2, β-actin, COXIV, and hypoxia-inducible factor 1α (HIF1α) proteins were detected by Western analysis. Antibodies against eIF2B2 and eIF4G2 were obtained from Sigma. HIF1α antibody was purchased from Novus Biological, and COXIV antibody was obtained from Cell Signaling Technology. To rule out leakage of AHA to the parental cell bodies in the center compartment during incubation of the axons located in the side compartment of the Campenot chambers, lysates from cell bodies of AHA-treated axons were also used for biotin crosslinking, streptavidin affinity purification, and Western analyses.
For fluorescence studies, AHA-labeled axons and corresponding cell body compartments were fixed in 4% formalin, permeabilized with Triton X-100, and incubated with alkyne-derivatized AlexaFluor-594 and Click-iT cell reaction buffer kit (Invitrogen) according to the manufacturer's instructions. For time course experiments, axons were labeled with AHA for 1, 4, and 6 h. After incubation, axons were fixed and permeabilized, and AHA incorporation was measured by labeling with Click-iT AlexaFluor-594. To assess the effects of the protein synthesis inhibitor, emetine, on axonal translation, 7-day-old SCG axons were treated with AHA and 50 or 100 μm emetine for 6 h. AHA incorporation was measured by monitoring the Click-iT AlexaFluor-594 signal. Fluorescence images were captured using a Nikon Eclipse TE 300 fluorescence microscope, and images were deconvoluted using the Deconvolution Lab Plugin (Vonesch and Unser, 2008).
All fluorescence quantification used original unprocessed image data, with no pixels at zero intensity or saturated. Fluorescence levels were quantified using NIH software ImageJ. For quantification of fluorescence, the thick axon bundles in the field were analyzed. The boundary of the axon bundle was selected by the drawing/selection tool, and the measurement function of the analysis tool was used to measure the area, integrated density, and the mean gray value for each bundle. The integrated density value was normalized for the area by dividing with the area of the selected bundle. To determine the background, several regions near the bundle were selected and the mean fluorescence of background reading was obtained. The relative fluorescence intensity for each bundle was determined as the difference between normalized integrated density value and the product of mean background fluorescence intensity and the area of the selected bundle. All axon bundles in each culture dish were measured, and statistical comparisons were made by measuring at least 35–40 axon bundles per treatment.
Transfection of neurons with miR precursor, anti-miR, or siRNAs.
The rat precursor miR16, nontargeting precursor control miR-NT (pre-NT), inhibitor anti-miR16, and nontargeting control anti-NT were obtained from Ambion. eIF2B2- and eIF4G2-specific and NT-siRNAs were purchased from Dharmacon. The small RNAs (miRs or siRNAs, each at final concentration of 25 nm), were transfected into distal axons located in the lateral compartments of Campenot cultures using siPORT NeoFX (Ambion). Axons were exposed to media containing the transfection reagents for 16 h. The two lateral compartments that harbored the distal axons were transfected independently, and the total RNA was processed separately. Two DIV cultures were used for axon growth experiments, and 6–8 DIV were used for all other experiments.
Western analysis.
Distal axons, were harvested and lysed in 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1% NP-40, and complete protease inhibitor mixture (Sigma). Equal amounts of each lysate were fractionated by 4–12% gradient SDS-PAGE (Invitrogen) and electroblotted onto Hybond-LFP PVDF membrane (GE Healthcare). Membranes were blocked with ECL Advance Blocking Agent (GE Healthcare) for 1 h, followed by overnight incubation at 4°C with antibodies against either eIF2B2, eIF4G2 (Sigma) or β-actin (Cell Signaling Technology). Membranes were washed three times with 1 × TBS-Tween 20 (1 × TBS and 0.1% Tween 20) and incubated with HRP-labeled secondary antibody for 1 h at room temperature. After washing, membranes were developed with ECL Advance Blotting Detection Kit reagents (GE Healthcare).
Measurement of axonal growth.
Images of axons were obtained using an inverted phase-contrast microscope (Nikon). Distal axon length was measured using the neurite tracing and quantification tool, Neuron J, from ImageJ software (NIH). All axons in each culture dish were measured, and statistical comparisons were made by measuring at least 35–40 axons per treatment.
Statistical analyses.
All statistical analyses were performed in Excel. Results are expressed as mean ± SEM. p values were calculated by Student's two-tailed t test.
Results
As mentioned in the Introduction, we previously reported the presence of a heterogeneous population of miRs in SCG axons. One of the most abundant miRNAs found in these axons was miR16 (Natera-Naranjo et al., 2010). A bioinformatic search for potential targets of miR16, using various miRNA target prediction software, revealed eIF2B2 and eIF4G2 mRNAs as candidates (Fig 1A).
Figure 1.
miR16 targets rat eIF2B2 and eIF4G2 mRNAs. A, Putative MTS in the eIF2B2 and eIF4G2 3′UTRs. B, Schematic of the vector backbone of miRNA 3′UTR target clone (GeneCopoeia) carrying dual luciferase reporters. Constructs contained the firefly luciferase open reading frame (hluc ORF) upstream of either the complete eIF2B2 or eIF4G2 3′UTR, or the eIF2B2 and eIF4G2 3′UTR in which the MTSs were deleted (δ16). The dual-reporter construct also contained a Renilla luciferase ORF (hRluc ORF). The hluc luciferase expression was driven by the Simian virus 40 (SV40) promoter, whereas hRluc expression was driven by the cytomegalovirus (CMV) promoter. C, D, Luciferase activity of the chimeric eIF2B2 and eIF4G2 3′UTR reporter genes, as well as the Renilla tracking gene, was measured after transfection of pre-miR16 or anti-miR16. pre-NT or anti-NT was used as control. Luciferase reporter constructs were cotransfected with the pre-miR16 or anti-miR16 or nontargeting controls, and luciferase activity was assessed 24 h after transfection. The ratio of reporter gene to tracking gene was normalized for each reporter and expressed as relative luciferase intensity. Error bars indicate the SEM (n = 9). The experiment was repeated three times with similar results. *p < 0.01 (Student's t test).
miR16 targets rat eIF2B2 and eIF4G2
To assess whether miR16 specifically targets the 3′UTRs of eIF2B2 and eIF4G2, SCG neurons were transfected at 3 DIV with a dual luciferase reporter plasmid carrying both the eIF2B2/eIF4G2 3′UTR downstream of the firefly luciferase coding sequence and the Renilla luciferase open reading frame alone, which served as a tracking gene (Fig. 1B). Firefly luciferase activity was quantified 24 h after cotransfection with the dual reporter plasmid and either pre-miR16 or anti-miR16. The ratio of firefly reporter gene to Renilla tracking gene expression was normalized for each reporter and was displayed as relative luciferase intensity. Overexpression of miR16 by treatment with pre-miR16 reduced luciferase activity by ∼40–50% in the context of both eIF2B2 and eIF4G2 3′UTRs compared with the nontargeting control, pre-NT (Fig. 1C). Conversely, treatment with the miR16 inhibitor, anti-miR16, resulted in an approximately twofold increase in relative luciferase intensity compared with the nontargeting control, anti-NT (Fig. 1C). Deletion of the putative miR16 MTS from the eIF2B2 and eIF4G2 3′UTR in the dual luciferase reporter constructs eliminated the response of the reporter gene to increased miR16 levels (Fig 1D). Together, the results of these experiments confirmed that the putative MTS in the eIF2B2 and eIF4G2 3′UTRs are functional and targeted by miR16.
eIF2B2 and eIF4G2 are expressed in rat SCG axons
To visualize miR16 and the mRNAs encoding eIF2B2 and eIF4G2 in rat SCG axons, we used FISH. Consistent with our earlier results (Natera-Naranjo et al., 2010), the miR16-specific probe showed punctate hybridization signals in the axons, compared with a scrambled probe used as a negative control (Fig. 2A). In subsequent experiments, eIF2B2- and eIF4G2-specific probes showed punctate signals along the length of the axon with regions of high density labeling. No fluorescent signals were observed with the nonspecific hybridization probe (Fig. 2B). Consistent with the compartmentalized culture results, FISH performed on single primary dissociated sympathetic neurons with probes specific to miR16 and mRNAs encoding eIF2B2 and eIF4G2 showed punctate fluorescent labeling (Fig. 2C and Fig. 2D, respectively). These results established the axonal localization of miR16 and eIF2B2 and eIF4G2 mRNAs in SCG neurons.
Figure 2.
miR16 and mRNAs encoding eIF2B2 and eIF4G2 are present in distal axons of sympathetic neurons. FISH was performed in primary SCG neurons (7 DIV) using FITC-conjugated locked nucleic acid probes for miR16 (A), eIF2B2, eIF4G2 (B), and scrambled probes as negative controls. Insets, Enlargement of regions of the axons in A and B. C, D, Phase-contrast images of the axon fibers are provided in the corresponding panels. miR16 and mRNAs encoding eIF2B2 and eIF4G2 were also visualized in primary dissociated sympathetic neurons by FISH. Bottom, Magnified images of distal regions of the axon showing specific punctate labeling. Arrows indicate the locations of punctate FISH signals in axons.
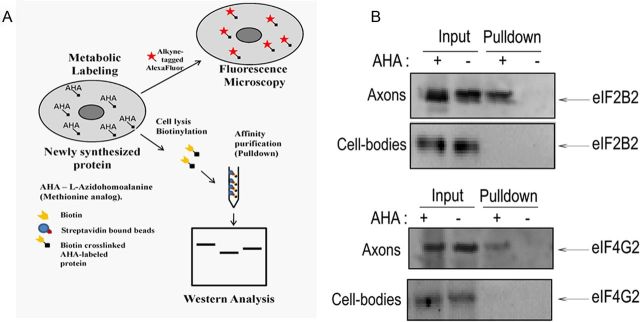
Based upon the above findings, we evaluated the hypothesis that eIF2B2 and eIF4G2 proteins are locally synthesized in the axon. To test this postulate, we used bio-orthogonal labeling with AHA coupled with Click-iT technology. In this experiment, SCG axons in the lateral compartments were exposed to AHA in Neurobasal medium without methionine for 6 h. After incubation, axons were lysed, and the newly synthesized, AHA-labeled proteins were crosslinked to the alkyne-derivatized biotin using the Click-iT reaction (Fig. 3A; see Materials and Methods). The AHA-labeled, biotin-crosslinked proteins were then affinity purified with streptavidin beads, and Western analysis was performed to detect the presence of eIF2B2 and eIF4G2 proteins. The results of Western analyses showed that both eIF2B2 and eIF4G2 were present in the streptavidin-purified fraction of the AHA-labeled axons, whereas no bands were observed in the nonlabeled (−AHA) axons (Fig. 3B). Additionally, to exclude the possibility of leakage of AHA into the cell body compartment, soma lysates from central compartments of AHA-treated axons were also subjected to biotin-crosslinking and streptavidin affinity purification followed by Western analyses. No specific AHA-labeled eIF2B2 or eIF4G2 bands were present in the pulldown fraction obtained from the cell bodies, indicating that eIF2B2 and eIF4G2 bands in the AHA-labeled axonal lysates were indeed derived from de novo protein synthesis in the axon (Fig. 3B).
Figure 3.
mRNAs encoding eIF2B2 and eIF4G2 are translated in the distal axons of SCG neurons. A, A schematic diagram showing the metabolic labeling strategy used to detect newly synthesized axonal proteins. AHA, an analog of l-methionine, was added to axonal compartments, biosynthetically incorporated into newly synthesized proteins, and detected by a copper (I)–catalyzed cycloaddition (Click-iT) reaction to either an alkyne-derivatized fluorophore or biotin-alkyne followed by streptavidin affinity absorption and Western analysis. Local synthesis of eIF2B2 and eIF4G2 proteins was assessed by metabolic labeling of axonally synthesized proteins with AHA for 6 h, followed by biotinylation and affinity purification by streptavidin beads. As control, soma lysates from center compartments whose axonal compartments were labeled with AHA were also subjected to biotinylation and affinity purification by streptavidin beads. B, A Western analysis was performed on the affinity-purified fraction to detect the presence of eIF2B2 and eIF4G2 using monoclonal antibodies. There is a lack of AHA-labeled (pulldown fraction) eIF2B2 and eIF4G2 in the lysates from the cell bodies. Ten percent of the total input and 50% of the pulldown were loaded in each lane of the SDS gel for the Western analysis.
miR16 regulates axonal eIF2B2 and eIF4G2 expression
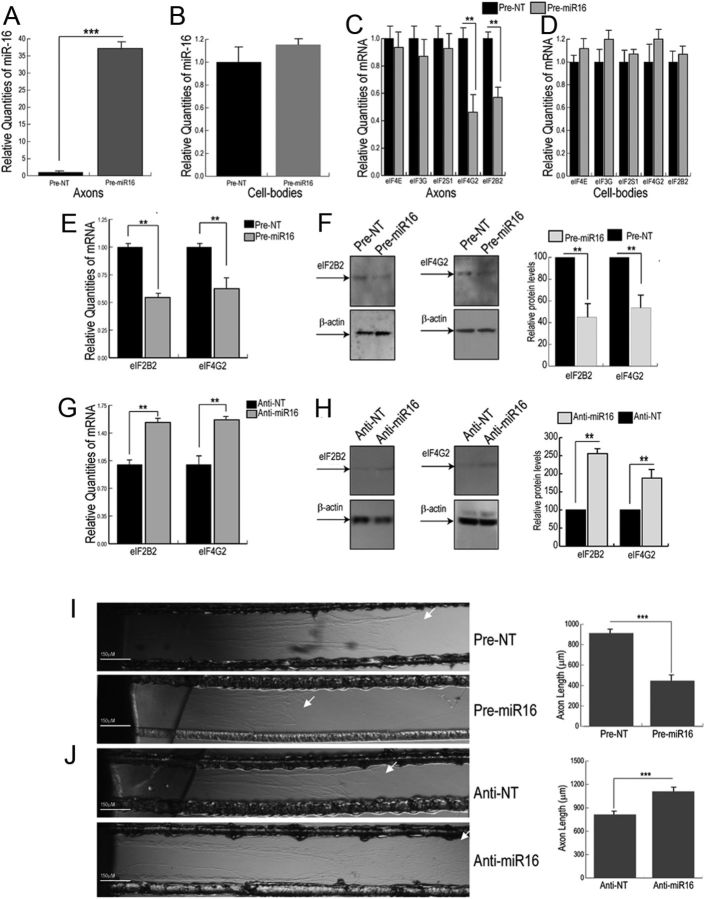
To evaluate the role of miR16 in the regulation of axonal eIF2B2 and eIF4G2 expression, we assessed the effects of pre-miR16 transfection on axonal eIF2B2 and eIF4G2 mRNA and protein levels. Axonal compartments were transfected with either pre-miR16 or anti-miR16, and mRNA and protein levels were measured by quantitative PCR and Western analysis, respectively. Transfection of the lateral compartment with pre-miR16 for 16 h lead to an ∼40-fold increase in mature miR16 levels in the axons (Fig. 4A), whereas no significant change in miR16 levels was observed in the corresponding cell bodies (Fig. 4B). At 16 h after transfection, eIF2B2 and eIF4G2 mRNA levels in the axon decreased ∼40–50% (Fig. 4C,E), whereas no effect on parental somal levels of eIF2B2 and eIF4G2 transcripts were observed (Fig. 4D). To evaluate the specificity of these effects, we compared the effects of miR16 levels on the relative abundance of eIF4E, eIF3G, and eIF2S1, three eukaryotic translation initiation factor mRNAs that do not contain a miR16 binding site in their 3′UTRs. As shown in Figure 4C, D, the introduction of pre-miR16 into the distal axons had no effect on the levels of these mRNAs either in the distal axon or parental cell bodies. Consistent with the decrease in mRNA levels, introduction of pre-miR16 into axons lead to a 50–60% decrease in eIF2B2 and eIF4G2 protein levels (Fig. 4F). Conversely, transfection of anti-miR16 directly into the distal axons resulted in a 50% increase in eIF2B2 and eIF4G2 mRNA and a 60% increase in eIF2B2 and eIF4G2 protein (Fig. 4G,H) compared with the nontargeting anti-NT control. These results demonstrate that miR16 can regulate the local expression of eIF2B2 and eIF4G2.
Figure 4.
miR16 regulates axonal mRNA and protein levels of eIF2B2 and eIF4G2 and influences axon outgrowth. Quantification of mature miR16 levels or transcripts encoding eukaryotic translation initiation factors eIF4E, eIF3S, eIF2S1, eIF4G2, and eIF2B2 in axons (A and C, respectively) and cell bodies (B and D, respectively) 16 h after transfection of distal axons with pre-miR16. Quantification of eIF2B2 and eIF4G2 mRNA (E) and protein (F) levels in the distal axons, after 16 h transfection with pre-miR16, as determined by real-time quantitative PCR and Western analysis, respectively. The mRNA (G) and protein (H) levels for eIF2B2 and eIF4G2 in distal axons treated with anti-miR16 for 16 h. mRNA levels were plotted relative to β-actin mRNA, and β-actin protein levels were used as a loading control. Error bars represent the SEM (n = 6). To assess the effect of miR16 on axonal growth, SCG neurons were cultured for 2 d, and axons were subsequently transfected with pre-miR16 (I), anti-miR16 (J), or nontargeting (NT) control. The direction of axon elongation is from left (central compartment) to right (lateral compartment). Arrows indicate the location of axon terminals. After 16 h treatment, axon length was measured in the lateral sides of the compartmentalized cultures. Data are mean ± SEM from 50 to 60 axons. The experiment was performed three times with similar results. **p < 0.001, ***p < 0.0001 (Student's t test).
Modulation of axonal miR16 levels alters axon growth rate in SCG neurons
Next, we evaluated the functional consequences of the modulation of miR16 levels by assessing its effects on axon elongation. In this experiment, we transfected distal axons of 2-day-old SCG cultures with pre-miR16 or anti-miR16. Sixteen hours after transfection, axon length in the lateral compartments was measured by phase-contrast microscopy. The results of these experiments showed that transfection of the axons with pre-miR16 decreased axon length by ∼40% compared with nontargeting controls (Fig. 4I). Interestingly, the inhibition of endogenous miR16 activity by anti-miR16 transfection resulted in an increase in axon length (Fig. 4J). Together, these results indicate that modulation of the local levels of miR16 can regulate the growth of the axon.
siRNA-mediated downregulation of axonal eIF2B2 and eIF4G2 leads to an inhibition of local protein synthesis
To further evaluate the postulate that the effects of miR16 on axonal growth are mediated, at least in part, by the local synthesis of eIF2B2 and eIF4G2, we reasoned that inhibition of eIF2B2 and eIF4G2 expression would affect local, axonal translation. The levels of local protein synthesis in the axons were directly measured using Click-IT assay (Invitrogen). To determine the length of AHA labeling required for the study, we first performed a time course experiment by labeling newly synthesized, axonal proteins with AHA for 1, 4, or 6 h time intervals. The newly synthesized, AHA-labeled proteins were visualized by covalent crosslinking of the incorporated AHA to an AlexaFluor-594 tag (Fig. 5A). The results of the time course experiment showed that incorporation of AHA into SCG axons was time-dependent, with higher AHA incorporation observed in 4–6 h time periods compared with 1 h AHA exposure. AHA labeling for longer than 6 h did not show a significant increase in AHA incorporation in the axon (data not shown). Importantly, covalent crosslinking of the AlexaFluor-594 tag in the parental cell bodies after AHA labeling of the axonal compartment showed no AHA signal in the cell body compartments during the labeling periods used in this study (Fig. 5A,B). These results suggest that, within the experimental paradigm used in this study, no significant leakage of AHA occurred into the central compartment of Campenot cultures.
Figure 5.

Nascent protein synthesis can be visualized by fluorescent AHA labeling in rat sympathetic axons. A, Time course for the detection of newly synthesized proteins in axons. Axonal compartments of SCG cultures were incubated with culture medium containing the methionine analog AHA or methionine, for the time intervals indicated. The AHA-containing, newly synthesized proteins were visualized by crosslinking to alkyne-derivatized AlexaFluor-594 using the Click-iT Cell reaction kit (Invitrogen). Arrow indicates fluorescently labeled, newly synthesized protein in the treated axon bundles. There is absence of AHA fluorescence in the parental cell bodies of AHA-treated axons. Bottom, Quantitation of the fluorescence signals in the axon bundles at specified time points. B, SCG distal axons were incubated with growth medium containing either AHA or methionine (control) in the presence or absence of either 50 or 100 μm emetine for 6 h. Arrows indicate fluorescently labeled, newly synthesized protein in the treated axon bundles. Fluorescence intensity was measured for each treatment. Bottom, Quantitative data. Data are mean ± SEM from 30 axon bundles. **p < 0.001 (Student's t test).
In the subsequent studies, we evaluated whether AHA incorporation into axonal protein was sensitive to perturbations in the protein synthetic machinery. In these experiments, we measured AHA incorporation in SCG axons labeled (6 h) in the presence of emetine dosages of either 50 or 100 μm. The reagent vehicle containing DMSO served as a negative control. The data indicated that increasing concentrations of emetine reduced AHA labeling to near background levels (Fig. 5B). Together, the results of these experiments showed that fluorogenic labeling of incorporated AHA can be used to document levels of local translation in SCG axons by fluorescence microscopy.
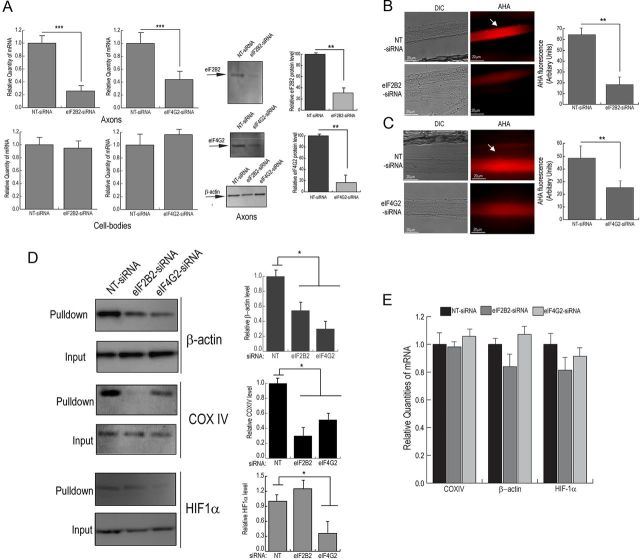
To test the hypothesis that inhibition of the local expression of eIF2B2 or eIF4G2 alters total axonal protein synthesis, eIF2B2- or eIF4G2-specific siRNA was introduced into distal axons (5–7 DIV). Sixteen hours after transfection, eIF2B2 and eIF4G2 mRNA and protein levels were measured by quantitative PCR and Western analyses, respectively. The results of these experiments showed that eIF2B2 mRNA and protein levels decreased ∼80%, whereas eIF4G2 mRNA and protein levels decreased ∼50–60% (Fig. 6A). No change in eIF2B2 or eIF4G2 mRNA levels was observed in parental soma after the axonal compartment was transfected with either eIF2B2- or eIF4G2-specific siRNA (Fig. 6A). Next, levels of local translation in SCG axons were assessed by measuring fluorescent labeling of incorporated AHA after 16 h treatment with eIF2B2- or eIF4G2-specific siRNA. We observed a marked decrease in fluorescence intensity of eIF2B2- and eIF4G2-siRNA-treated axons compared with NT-siRNA-treated axons (Fig. 6B,C). Quantification of the fluorescent signal in eIF2B2 or eIF4G2 siRNA-treated axons revealed that levels of local translation decreased by 70% or 50%, respectively (Fig. 6B,C). Together, these results demonstrate that local levels of eIF2B2 and eIF4G2 affect the activity of the axonal protein synthetic system.
Figure 6.
siRNA-mediated knock-down of eIF2B2 and eIF4G2 expression inhibits nascent protein synthesis in rat sympathetic axons. siRNA oligonucleotides targeted against either eIF2B2 or eIF4G2 or nontargeting NT-siRNA were transfected into distal axons of SCG neurons. A, The eIF2B2 and eIF4G2 mRNA levels from axons and cell bodies were measured by quantitative PCR and axonal protein levels quantified by Western analysis. The mRNA levels are expressed relative to β-actin mRNA, and β-actin protein levels served a loading control in the Western analyses. The axonal compartments of 1-week-old SCG cultures were treated with siRNA targeted against either eIF2B2 (B) or eIF4G2 (C) or NT control for 16 h and were subsequently labeled with AHA for 6 h. After incubation, cells were either fixed, and the newly synthesized AHA-labeled proteins were visualized by fluorescence microscopy (B, C), or alternatively cells were lysed, and the newly synthesized AHA-labeled proteins were isolated by affinity purification with streptavidin beads and specific proteins detected by Western analyses (D). B, C, Arrows indicate fluorescently labeled, newly synthesized protein in the treated axon bundles. Mean fluorescence intensities of eIF2B2 or eIF4G2 siRNA-treated distal axon bundle segments relative to nontargeted control axons are shown. Data are mean ± SEM of 30 axon bundles per treatment. D, Western analyses of affinity purified pulldown fractions show the levels of newly synthesized β-actin, COXIV, and HIF1α proteins. Quantification of immunoblots with ImageJ showed changes in β-actin, COXIV, and HIF1α signal in pulldown from AHA-labeled axons treated with eIF2B2 or eIF4G2 siRNA compared with NT-siRNA treatment. E, Axonal β-actin, COXIV, and HIF1α mRNA levels were quantified after transfection of eIF2B2, eIF4G2, and NT-siRNA. Values represent mean ± SEM from three independent experiments. *p < 0.05, **p < 0.001, ***p < 0.0001 (Student's t test).
Downregulation of eIF2B2 and eIF4G2 expression inhibits the translation of specific axonally localized mRNAs β-actin, COXIV, and HIF1α
Because siRNA-mediated downregulation of eIF2B2 or eIF4G2 led to a reduction in total axonal protein synthesis, we explored whether the knock-down of eIF2B2 or eIF4G2 inhibits the translation of specific mRNAs. Earlier studies have established that β-actin mRNA is localized and translated in axons cultured in various systems, such as squid giant axon, superior cervical ganglia, dorsal root ganglia, cortical neurons, and retinal ganglion cells, and this local translation plays an important role in axon growth and development (Kaplan et al., 1992; Olink-Coux and Hollenbeck, 1996; Bassell et al., 1998; Zhang et al., 1999; Leung et al., 2006; Yao et al., 2006; Donnelly et al., 2013). Additionally, we previously reported the presence and functionality of COXIV mRNA in the axons of SCG neurons (Hillefors et al., 2007; Aschrafi et al., 2008). Another subset of mRNAs that have been shown to be present in axons is that encoding transcription factors (Willis et al., 2007; Gumy et al., 2011). Locally synthesized transcription factors have been reported to be involved in retrograde signaling in response to stimuli, such as exposure to NGF or peripheral nerve injury (Cox et al., 2008; Michaelevski et al., 2010; Ben-Yaakov et al., 2012). In light of these findings, we assessed the effect of siRNA-mediated downregulation of eIF2B2 or eIF4G2 on axonal translation of β-actin, COXIV mRNA, and HIF1α, a transcription factor whose mRNA has been shown to be present in the axons (Ben-Yaakov et al., 2012). SCG axons (5–7 DIV) were treated with siRNA targeted against eIF2B2 or eIF4G2 for 16 h and were subsequently labeled with AHA for 6 h. After labeling, AHA-incorporated proteins were crosslinked to biotin and purified with streptavidin beads. Western analyses were performed to assess the levels of locally synthesized β-actin or COXIV or HIF1α. As shown in Figure 6D, eIF2B2 and eIF4G2 siRNA-treated axons exhibited a 50–60% decrease in axonal β-actin translation compared with NT-siRNA-treated controls. It is important to note that the decrease in β-actin protein levels observed after eIF2B2 or eIF4G2 knock-down only corresponded to the locally synthesized pool, and the total levels of β-actin protein in the axon did not change significantly. Interestingly, we observed an ∼70% decrease in COXIV translation in eIF2B2-siRNA treated axons, whereas eIF4G2-siRNA treatment showed a 40–50% decrease in axonal COXIV synthesis. The knock-down of eIF4G2 led to a 60–70% decrease in axonal translation of HIF1α, whereas eIF2B2 knock-down did not show a statistically significant effect on HIF1α levels (Fig. 6D). Additionally, no significant effect was observed on axonal mRNA levels of β-actin, COXIV, and HIF1α after treatment of axons with eIF2B2 or eIF4G2-specific siRNA (Fig. 6E). These findings establish that, in addition to inhibiting global axonal protein synthesis, siRNA-mediated downregulation of eIF2B2 or eIF4G2 reduces the translation of specific axonally localized mRNAs.
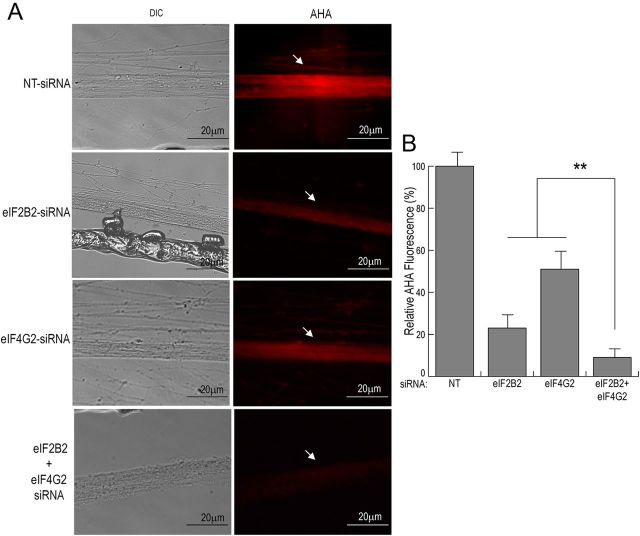
Downregulation of both eIF2B2 and eIF4G2 has an additive inhibitory effect on axonal protein synthesis
To investigate whether the knock-down of axonal eIF2B2 and eIF4G2 has an additive inhibitory effect on local protein synthesis, we transfected SCG axons (5–7 DIV) with siRNAs targeted against eIF2B2, eIF4G2, or both eIF2B2 and eIF4G2. After transfection, axons were treated with AHA for 6 h, and AHA incorporation was assessed by fluorescence microscopy. AHA labeling in axons showed that knock-down of local levels of both eIF2B2 and eIF4G2 together resulted in a significant decrease in axonal translation, compared with axons treated with either eIF2B2- or eIF4G2-specific siRNA alone (Fig. 7A,B). Interestingly, simultaneous knock-down of both eIF2B2 and eIF4G2 levels in SCG axons also resulted in a marked change in axon morphology (Fig. 7A).
Figure 7.
Downregulation of both eIF2B2 and eIF4G2 in SCG axons shows additive inhibitory effects on local protein synthesis. Axonal compartments of 1-week-old SCG cultures were transfected with siRNA targeted against eIF2B2, eIF4G2, a combination of eIF2B2 and eIF4G2, or nontargeting NT control for 16 h. After transfection, axons were labeled with AHA for 6 h. A, Axons were fixed, and newly synthesized, AHA-containing proteins were labeled with AlexaFluor-594. Arrows indicate fluorescently labeled, newly synthesized protein in the treated axon bundles. B, Mean fluorescence intensities of treated distal axon bundle segments relative to nontargeted control neurons are shown. Data are mean ± SEM from 30 axons. **p < 0.001 (Student's t test).
Knock-down of local eIF2B2 and eIF4G2 expression reduces axonal growth
To evaluate the postulate that inhibition of local protein synthesis could have deleterious effects on the development and/or viability of the axon, distal axons located in the lateral compartments of 2-day-old SCG neuron cultures were transfected with either eIF2B2- or eIF4G2-specific siRNA or a combination of both siRNAs. Axons in the contralateral compartment of the culture dish were treated with NT-siRNA and served as a negative control. Axon length in both side compartments of the Campenot chambers was measured 16 h transfection by phase-contrast microscopy. Consistent with the findings obtained from pre-miR16 treatment, siRNA-mediated downregulation of the axonal expression of eIF2B2 or eIF4G2 resulted in a significant decrease in axonal elongation compared with the distal axons transfected with nontargeting siRNA (Fig. 8A,B). These data provide evidence that the local translation of eIF2B2 and eIF4G2 mRNAs plays a key role in regulating axonal growth.
Figure 8.
Knock-down of local eIF2B2 and eIF4G2 expression attenuates axon elongation in sympathetic neurons. A, SCGs were cultured for 2 d, and axons located in the lateral compartments were transfected with siRNA targeted against eIF2B2, eIF4G2, both eIF2B2 and eIF4G2-siRNA together, or nontargeted NT control siRNA. Axon elongation is from left to right; arrows indicate the location of axonal terminals. B, Axon length was measured 16 h after transfection. Data represent the mean ± SEM of 50–60 axons. The experiments were repeated three times with similar results. ***p < 0.0001 (Student's t test).
Because knock-down of both eIF2B2 and eIF4G2 together showed an additive inhibitory effect on local protein synthesis, we also evaluated the effect of the eIF2B2 and eIF4G2 double knock-down on axon growth. Consistent with the findings of the metabolic labeling experiments, the results of the axon growth experiment showed that simultaneous knock-down of both eIF2B2 and eIF4G2 led to a greater decrease in axon length, compared with axons treated with eIF2B2 or eIF4G2-specific siRNAs individually (Fig. 8A,B). Together, these findings suggest that these two translation initiation factors may regulate local protein synthesis and axonal growth. In addition, these findings suggest that the effects of miR16 on axonal growth are mediated, at least in part, by the local translation of eIF2B2 and eIF4G2.
Discussion
Studies using both invertebrate and vertebrate model systems have reported the presence of a number of axonally localized mRNAs encoding ribosomal proteins and factors involved in the regulation of protein synthesis (Perrone-Capano et al., 1987; Moccia et al., 2003; Willis et al., 2007; Taylor et al., 2009; Vogelaar et al., 2009; Zivraj et al., 2010). In this study, we investigated the role of locally synthesized translation factors in the regulation of axonal protein synthesis. To monitor axonal translation activity, we used metabolic labeling studies with AHA, an amino acid analog incorporated into proteins in place of methionine. The incorporation of AHA into newly synthesized proteins can be visualized after cell fixation by addition of a tagged fluorescent alkyne or can be affinity purified by tagging AHA-labeled proteins with biotin after cell lysis. Consistent with earlier experimental approaches, we observed a time-dependent accumulation of the AHA label in the axons (Dieterich et al., 2006, 2010; Tcherkezian et al., 2010). This labeling of axons with AHA was sensitive to treatment with the translation inhibitor, emetine. Also, within our experimental paradigm, in which only the axonal compartments were labeled with AHA, no axonally applied AHA label was observed in the cell bodies within the short-labeling periods (6 h) used in these studies. This finding suggests that metabolic labeling with AHA in the axon is a valid technique to assess local translation and that the newly synthesized, AHA-labeled proteins we observed in the distal axons were not derived from the cell body during the labeling time periods used in this study.
Interestingly, the AHA labeling experiments showed that downregulation of axonal eIF2B2 and eIF4G2 expression led to a marked decrease in local mRNA translation. Consistent with the decrease in global axonal protein synthesis after eIF2B2 or eIF4G2 knock-down, local translation of the axonally localized mRNAs, β-actin, COXIV, and HIF1α was also inhibited. The local translation of β-actin mRNA, in response to chemotactic cues, has been shown to be important for axon guidance (Bassell et al., 1998; Leung et al., 2006; Yao et al., 2006) and elongation (Zhang et al., 1999). It bears mention here that studies from the Letourneau and Ervasti laboratories have argued against the role of β-actin in axon pathfinding and motor neuron development respectively (Roche et al., 2009; Cheever et al., 2011). In addition, studies have also shown that intra-axonal translation of nuclear-encoded mitochondrial mRNAs, such as COXIV and ATP synthase subunit 9 (ATP5G1), affects axon outgrowth (Aschrafi et al., 2008; Natera-Naranjo et al., 2012). These findings suggest that the decrease in axon growth observed in our studies after downregulation of eIF2B2 or eIF4G2 may be a consequence, at least in part, of the decrease in local β-actin, as well as COXIV and ATP synthase protein levels. Additionally, our results also suggest that local synthesis of transcription factors, such as HIF1α, can be modulated by axonal levels of eukaryotic translation initiation factors. Together, our results raise the possibility that regulation of local levels of translation initiation factors can have broad secondary effects on axonal function by affecting the expression of multiple axonally translated mRNAs.
In this communication, we also show that a noncoding RNA, miR16, can play a role in regulating the local expression of two eukaryotic translation initiation factors, eIF2B2 and eIF4G2. Using luciferase gene reporter constructs containing the eIF2B2 or eIF4G2 3′UTR, we showed that, in the presence of the 3′UTR of either eIF2B2 or eIF4G2, cotransfection of the pre-miR16 led to a repression in luciferase activity, whereas transfection of the specific inhibitor anti-miR16 increased the activity of the reporter chimeric gene construct in SCG neurons. Deletion of the cognate miR16 targeting sequence from the eIF2B2 and eIF4G2 3′UTR blocked the mir16-mediated modulation of luciferase activity. In addition, we found that modulation of miR16 levels in the axon, by transfection of pre-miR16 or anti-miR16, significantly affected local eIF2B2 and eIF4G2 mRNA and protein levels. This miR16-mediated regulation of eIF2B2 and eIF4G2 levels was specific to these transcripts, as we observed no significant effect of miR16 modulation on the axonal levels of other translation factors, such as eIF4E or eIF3G. Interestingly, we also observed that the modulation of local levels of miR16 influenced the rate of axon outgrowth. Together with the observation that specific knockdown of eIF2B2 and eIF4G2 using siRNA methodology also affected axon growth, our results show that the effects of miR16 on axon growth are mediated, at least in part, by the axonal translation of eIF2B2 and eIF4G2. These findings demonstrate that noncoding RNAs present in the axon can influence eukaryotic translation initiation by modulating levels of translation factors, which are important contributors to the maintenance and function of the axon. It bears mention that earlier studies have suggested that axonal protein synthesis is not required for axonal growth (Eng et al., 1999; Campbell and Holt, 2001). However, the miR16-mediated axon growth modulation observed in this study is consistent with previous work from our laboratory, which showed that blockage of local protein synthesis affected both axonal growth and maintenance on SCG axons grown in compartmentalized cultures (Hillefors et al., 2007; Manns et al., 2012). As previously discussed, differences between the results reported in these studies could be attributed to differences in the neurons used as well as differences in cell culture conditions, such as NGF concentrations, culture substratum, and the use of serum-free culture medium.
In conclusion, the findings reported here indicate that local synthesis of proteins involved in the translation machinery play an important role in the regulation of local protein synthesis in the axon and axonal function. Finally, we show that a noncoding mRNA regulates axonal protein synthesis by modulating the local levels of proteins involved in the control of mRNA translation.
Footnotes
This work was supported by the Division of Intramural Research Programs of the National Institute of Mental Health. We thank Ms. Sanah Vohra for invaluable technical assistance and critical reading of the manuscript.
The authors declare no competing financial interests.
References
- Aschrafi A, Schwechter AD, Mameza MG, Natera-Naranjo O, Gioio AE, Kaplan BB. MicroRNA-338 regulates local cytochrome c oxidase IV mRNA levels and oxidative phosphorylation in the axons of sympathetic neurons. J Neurosci. 2008;28:12581–12590. doi: 10.1523/JNEUROSCI.3338-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassell GJ, Zhang H, Byrd AL, Femino AM, Singer RH, Taneja KL, Lifshitz LM, Herman IM, Kosik KS. Sorting of β-actin mRNA and protein to neurites and growth cones in culture. J Neurosci. 1998;18:251–265. doi: 10.1523/JNEUROSCI.18-01-00251.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yaakov K, Dagan SY, Segal-Ruder Y, Shalem O, Vuppalanchi D, Willis DE, Yudin D, Rishal I, Rother F, Bader M, Blesch A, Pilpel Y, Twiss JL, Fainzilber M. Axonal transcription factors signal retrogradely in lesioned peripheral nerve. EMBO J. 2012;31:1350–1363. doi: 10.1038/emboj.2011.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell DS, Holt CE. Chemotropic responses of retinal growth cones mediated by rapid local protein synthesis and degradation. Neuron. 2001;32:1013–1026. doi: 10.1016/S0896-6273(01)00551-7. [DOI] [PubMed] [Google Scholar]
- Cheever TR, Olson EA, Ervasti JM. Axonal regeneration and neuronal function are preserved in motor neurons lacking β-actin in vivo. PLoS One. 2011;6:e17768. doi: 10.1371/journal.pone.0017768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox LJ, Hengst U, Gurskaya NG, Lukyanov KA, Jaffrey SR. Intra-axonal translation and retrograde trafficking of CREB promotes neuronal survival. Nat Cell Biol. 2008;10:149–159. doi: 10.1038/ncb1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich DC, Link AJ, Graumann J, Tirrell DA, Schuman EM. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT) Proc Natl Acad Sci U S A. 2006;103:9482–9487. doi: 10.1073/pnas.0601637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich DC, Hodas JJ, Gouzer G, Shadrin IY, Ngo JT, Triller A, Tirrell DA, Schuman EM. In situ visualization and dynamics of newly synthesized proteins in rat hippocampal neurons. Nat Neurosci. 2010;13:897–905. doi: 10.1038/nn.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly CJ, Park M, Spillane M, Yoo S, Pacheco A, Gomes C, Vuppalanchi D, McDonald M, Kim HK, Merianda TT, Gallo G, Twiss JL. Axonally synthesized beta-actin and GAP-43 proteins support distinct modes of axonal growth. J Neurosci. 2013;33:3311–3322. doi: 10.1523/JNEUROSCI.1722-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubacq C, Jamet S, Trembleau A. Evidence for developmentally regulated local translation of odorant receptor mRNAs in the axons of olfactory sensory neurons. J Neurosci. 2009;29:10184–10190. doi: 10.1523/JNEUROSCI.2443-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng H, Lund K, Campenot RB. Synthesis of β-tubulin, actin, and other proteins in axons of sympathetic neurons in compartmented cultures. J Neurosci. 1999;19:1–9. doi: 10.1523/JNEUROSCI.19-01-00001.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumy LF, Yeo GS, Tung YC, Zivraj KH, Willis D, Coppola G, Lam BY, Twiss JL, Holt CE, Fawcett JW. Transcriptome analysis of embryonic and adult sensory axons reveals changes in mRNA repertoire localization. RNA. 2011;17:85–98. doi: 10.1261/rna.2386111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanz S, Perlson E, Willis D, Zheng JQ, Massarwa R, Huerta JJ, Koltzenburg M, Kohler M, van-Minnen J, Twiss JL, Fainzilber M. Axoplasmic importins enable retrograde injury signaling in lesioned nerve. Neuron. 2003;40:1095–1104. doi: 10.1016/S0896-6273(03)00770-0. [DOI] [PubMed] [Google Scholar]
- Hillefors M, Gioio AE, Mameza MG, Kaplan BB. Axon viability and mitochondrial function are dependent on local protein synthesis in sympathetic neurons. Cell Mol Neurobiol. 2007;27:701–716. doi: 10.1007/s10571-007-9148-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan BB, Gioio AE, Capano CP, Crispino M, Giuditta A. β-Actin and β-tubulin are components of a heterogeneous mRNA population present in the squid giant axon. Mol Cell Neurosci. 1992;3:133–144. doi: 10.1016/1044-7431(92)90017-V. [DOI] [PubMed] [Google Scholar]
- Leung KM, van Horck FP, Lin AC, Allison R, Standart N, Holt CE. Asymmetrical β-actin mRNA translation in growth cones mediates attractive turning to netrin-1. Nat Neurosci. 2006;9:1247–1256. doi: 10.1038/nn1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manns RP, Cook GM, Holt CE, Keynes RJ. Differing semaphorin 3A concentrations trigger distinct signaling mechanisms in growth cone collapse. J Neurosci. 2012;32:8554–8559. doi: 10.1523/JNEUROSCI.5964-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin KC, Casadio A, Zhu H, Yaping E, Rose JC, Chen M, Bailey CH, Kandel ER. Synapse-specific, long-term facilitation of aplysia sensory to motor synapses: a function for local protein synthesis in memory storage. Cell. 1997;91:927–938. doi: 10.1016/S0092-8674(00)80484-5. [DOI] [PubMed] [Google Scholar]
- Michaelevski I, Segal-Ruder Y, Rozenbaum M, Medzihradszky KF, Shalem O, Coppola G, Horn-Saban S, Ben-Yaakov K, Dagan SY, Rishal I, Geschwind DH, Pilpel Y, Burlingame AL, Fainzilber M. Signaling to transcription networks in the neuronal retrograde injury response. Sci Signal. 2010;3:ra53. doi: 10.1126/scisignal.2000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moccia R, Chen D, Lyles V, Kapuya E, Kalachikov S, Spahn CM, Frank J, Kandel ER, Barad M, Martin KC. An unbiased cDNA library prepared from isolated Aplysia sensory neuron processes is enriched for cytoskeletal and translational mRNAs. J Neurosci. 2003;23:9409–9417. doi: 10.1523/JNEUROSCI.23-28-09409.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muddashetty RS, Nalavadi VC, Gross C, Yao X, Xing L, Laur O, Warren ST, Bassell GJ. Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Mol Cell. 2011;42:673–688. doi: 10.1016/j.molcel.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natera-Naranjo O, Aschrafi A, Gioio AE, Kaplan BB. Identification and quantitative analyses of microRNAs located in the distal axons of sympathetic neurons. RNA. 2010;16:1516–1529. doi: 10.1261/rna.1833310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natera-Naranjo O, Kar AN, Aschrafi A, Gervasi NM, Macgibeny MA, Gioio AE, Kaplan BB. Local translation of ATP synthase subunit 9 mRNA alters ATP levels and the production of ROS in the axon. Mol Cell Neurosci. 2012;49:263–270. doi: 10.1016/j.mcn.2011.12.006. [DOI] [PubMed] [Google Scholar]
- Olde Loohuis NF, Kos A, Martens GJ, Van Bokhoven H, Nadif Kasri N, Aschrafi A. MicroRNA networks direct neuronal development and plasticity. Cell Mol Life Sci. 2012;69:89–102. doi: 10.1007/s00018-011-0788-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olink-Coux M, Hollenbeck PJ. Localization and active transport of mRNA in axons of sympathetic neurons in culture. J Neurosci. 1996;16:1346–1358. doi: 10.1523/JNEUROSCI.16-04-01346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone-Capano CP, Giuditta A, Castigli E, Kaplan BB. Occurrence and sequence complexity of polyadenylated RNA in squid axoplasm. J Neurochem. 1987;49:698–704. doi: 10.1111/j.1471-4159.1987.tb00950.x. [DOI] [PubMed] [Google Scholar]
- Roche FK, Marsick BM, Letourneau PC. Protein synthesis in distal axons is not required for growth cone responses to guidance cues. J Neurosci. 2009;29:638–652. doi: 10.1523/JNEUROSCI.3845-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Si K, Giustetto M, Etkin A, Hsu R, Janisiewicz AM, Miniaci MC, Kim JH, Zhu H, Kandel ER. A neuronal isoform of CPEB regulates local protein synthesis and stabilizes synapse-specific long-term facilitation in aplysia. Cell. 2003;115:893–904. doi: 10.1016/S0092-8674(03)01021-3. [DOI] [PubMed] [Google Scholar]
- Taylor AM, Berchtold NC, Perreau VM, Tu CH, Li Jeon N, Cotman CW. Axonal mRNA in uninjured and regenerating cortical mammalian axons. J Neurosci. 2009;29:4697–4707. doi: 10.1523/JNEUROSCI.6130-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcherkezian J, Brittis PA, Thomas F, Roux PP, Flanagan JG. Transmembrane receptor DCC associates with protein synthesis machinery and regulates translation. Cell. 2010;141:632–644. doi: 10.1016/j.cell.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma P, Chierzi S, Codd AM, Campbell DS, Meyer RL, Holt CE, Fawcett JW. Axonal protein synthesis and degradation are necessary for efficient growth cone regeneration. J Neurosci. 2005;25:331–342. doi: 10.1523/JNEUROSCI.3073-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelaar CF, Gervasi NM, Gumy LF, Story DJ, Raha-Chowdhury R, Leung KM, Holt CE, Fawcett JW. Axonal mRNAs, characterization and role in the growth and regeneration of dorsal root ganglion axons and growth cones. Mol Cell Neurosci. 2009;42:102–115. doi: 10.1016/j.mcn.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonesch C, Unser M. A fast thresholded landweber algorithm for wavelet-regularized multidimensional deconvolution. IEEE Trans Image Process. 2008;17:539–549. doi: 10.1109/TIP.2008.917103. [DOI] [PubMed] [Google Scholar]
- Willis DE, van Nierkerk EA, Sasaki Y, Mesngon M, Merianda TT, Williams GG, Kendall M, Smith DS, Bassell GJ, Twiss JL. Extracellular stimuli specifically regulate localized levels of individual neuronal mRNAs. J Cell Biol. 2007;178:965–980. doi: 10.1083/jcb.200703209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu KY, Hengst U, Cox LJ, Macosko EZ, Jeromin A, Urquhart ER, Jaffrey SR. Local translation of RhoA regulates growth cone collapse. Nature. 2005;436:1020–1024. doi: 10.1038/nature03885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Sasaki Y, Wen Z, Bassell GJ, Zheng JQ. An essential role for β-actin mRNA localization and translation in Ca2+-dependent growth cone guidance. Nat Neurosci. 2006;9:1265–1273. doi: 10.1038/nn1773. [DOI] [PubMed] [Google Scholar]
- Yoon BC, Jung H, Dwivedy A, O'Hare CM, Zivraj KH, Holt CE. Local translation of extranuclear lamin B promotes axon maintenance. Cell. 2012;148:752–764. doi: 10.1016/j.cell.2011.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HL, Singer RH, Bassell GJ. Neurotrophin regulation of β-actin mRNA and protein localization within growth cones. J Cell Biol. 1999;147:59–70. doi: 10.1083/jcb.147.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Xh, Poo MM. Localized synaptic potentiation by BDNF requires local protein synthesis in the developing axon. Neuron. 2002;36:675–688. doi: 10.1016/S0896-6273(02)01023-1. [DOI] [PubMed] [Google Scholar]
- Zivraj KH, Tung YC, Piper M, Gumy L, Fawcett JW, Yeo GS, Holt CE. Subcellular profiling reveals distinct and developmentally regulated repertoire of growth cone mRNAs. J Neurosci. 2010;30:15464–15478. doi: 10.1523/JNEUROSCI.1800-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]