Abstract
The notochord is the distinctive characteristic of chordates; however, the knowledge of the complement of transcription factors governing the development of this structure is still incomplete. Here we present the expression patterns of seven transcription factor genes detected in the notochord of the ascidian Ciona intestinalis at various stages of embryonic development. Four of these transcription factors, Fos-a, NFAT5, AFF and Klf15, have not been directly associated with the notochord in previous studies, while the others, including Spalt-like-a, Lmx-like and STAT5/6-b, display evolutionarily conserved expression in this structure as well as in other domains. We examined the hierarchical relationships between these genes and the transcription factor Brachyury, which is necessary for notochord development in all chordates. We found that Ciona Brachyury regulates the expression of most, although not all, of these genes. These results shed light on the genetic regulatory program underlying notochord formation in Ciona and possibly other chordates.
Keywords: ascidian, Brachyury, Ciona intestinalis, gene network, notochord, transcription factor, tunicate
INTRODUCTION
The notochord is a midline mesodermal structure whose presence is one of the defining features of the chordate body plan. This organ is critical for the embryonic development of all chordates, where it serves as the main axial support for the growing embryos (Stemple, 2005; Jiang and Smith, 2007). In vertebrates, the notochord is necessary for patterning the neural tube, specification of the cardiac field and formation of the endoderm (Cleaver and Krieg, 2001; Wilson and Maden, 2005). In vertebrate embryos, as ossification of the vertebral column proceeds, the notochord gradually disappears and its remnants become incorporated into the nucleus pulposus, the central portion of the intervertebral discs located between the vertebrae of the spinal column; these notochord residues can form malignant chordomas (Risbud et al., 2010).
The ascidian embryo provides an ideal model for studies of notochord development and differentiation. The notochord of these translucent embryos consists of just 40 cells and forms similarly to that of vertebrates. For example, notochord cells intercalate during convergent extension (Munro and Odell, 2002) relying upon the planar cell polarity (PCP) pathway, and subsequently stretch to allow tail extension (Jiang et al., 2005). Additionally, in both Ciona and vertebrates, the notochord is surrounded by a basement membrane consisting of extracellular matrix proteins, including laminins (Scott and Stemple, 2005; Veeman et al., 2008), and a collagen-rich notochordal sheath (Miyamoto and Crowther, 1985; Stemple, 2005). As development of the notochord proceeds, intercellular pockets of extracellular matrix (lumens) form in ascidians (Jiang and Smith, 2007), and intracellular vacuoles form in vertebrates (Stemple, 2005); the mechanical pressure exerted by lumens and vacuoles against the stiff notochordal sheath provides the rigidity necessary for the embryo to elongate (Stemple, 2005).
Despite the importance of notochord formation in shaping the chordate body plan, the network of transcriptional regulators controlling its development remains incompletely characterized in any model system. Nevertheless, it is generally acknowledged that one transcriptional regulator necessary for notochord formation in all chordates is the T-box transcription factor Brachyury. The crucial role for Brachyury in notochord development is underscored by recent data that have shown this gene to be a biomarker of chordomas (Vujovic et al., 2006) and to be duplicated in familial chordoma (Yang et al., 2009). In Ciona, Brachyury (Ci-Bra) is expressed exclusively in the notochord and its precursors beginning at the 64-cell stage, concurrent with notochord fate restriction (Corbo et al., 1997). The upstream regulatory cascade leading to Ci-Bra expression is well characterized (Yagi et al., 2004; Imai et al., 2006; Matsumoto et al., 2007), and at least 50 Ciona genes have been found to be controlled by Ci-Bra (Di Gregorio and Levine, 1999; Takahashi et al., 1999; Hotta et al., 2000; Oda-Ishii and Di Gregorio, 2007; Hotta et al., 2008; Kugler et al., 2008). A surprisingly low number of transcription factors was included within this first set of transcriptional targets; however, this number has been sharply increased by recent whole-genome studies of the in vivo occupancy of chromatin by Ci-Bra in early Ciona embryos (Kubo et al., 2010).
Nevertheless, a fraction of these presumptive Ci-Bra-downstream regulatory genes have not been shown to be expressed in notochord cells by previous studies. Furthermore, while it has been suggested that Ci-Bra might be controlling expression of its late target genes via transcriptional intermediaries (e.g., Hotta et al., 1999), the knowledge of the precise temporal windows of notochord expression and the presumptive functions of these Ci-Bra-downstream factors are still fragmentary. This represents a point of interest in studies of notochord formation, since these intermediaries of Ci-Bra, alongside Ci-Bra-independent notochord transcription factors, are likely to collectively direct the morphogenetic processes that begin at later developmental stages. To begin filling this gap, we undertook an alternative approach, as we sought to first identify additional transcription factors expressed in the Ciona notochord and secondly to study their hierarchical relationship with Ci-Bra.
Here we report the previously uncharacterized notochord expression of seven transcription factors in Ciona intestinalis. While for some of these genes this analysis provides the first evidence of expression in this domain in any chordate, for others it underscores the evolutionary conservation of notochord expression across the chordate phylum. In either case, understanding the hierarchical relationships of these factors with other components of the notochord gene regulatory network should enhance our knowledge of molecular mechanisms fundamental to notochord development and to the evolution of the chordate body plan. Toward this aim, we began examining the relationship between these genes and Ci-Bra. We found that loss of Ci-Bra function affects the expression of some, but not all, of these genes. Together, our results suggest that the notochord gene regulatory network in the simple chordate Ciona is complex and multifaceted.
RESULTS AND DISCUSSION
Identification of novel notochord transcription factors in the Ciona embryo
A comprehensive and detailed list of the transcription factor genes found in the Ciona intestinalis genome and their expression patterns has been published by Imai et al. (2004) and is publicly available in a searchable format (http://hoya.zool.kyoto-u.ac.jp/TF_KH.html).
We performed a microarray screen on neurula and mid-tailbud FACS-sorted notochord cells aimed at identifying genes enriched in the notochord lineage, and we found that most of the genes with the highest scores were previously characterized notochord markers, including Ci-Bra (Corbo et al., 1997), Ci-leprecan (Dunn and Di Gregorio, 2009), several Ci-Noto genes (Hotta et al., 2000) and Ci-tune (Passamaneck et al., 2009), among others (manuscript in preparation). We noticed that in addition to Ci-Bra and its target genes, the provisional notochord transcriptome also included a number of transcription factor genes whose expression in notochord cells had not been reported previously. Prompted by these observations, we prioritized the analysis of these candidate notochord transcriptional regulators, as they would likely broaden the current knowledge of the Ciona notochord gene regulatory network by shedding light on some of its yet undiscovered branches.
We began the study of these candidate notochord factors by performing whole-mount in situ hybridization on embryos ranging from the 64-cell through the mid-tailbud II stages (Hotta et al., 2007). This analysis allowed us to validate that seven genes, representing various families of transcription factors, are expressed in the Ciona notochord (Table 1; Figs. 1,2,3 and S1). As expected, the majority of these genes are expressed after notochord fate determination is complete; this observation raises the possibility that they may govern some of the morphogenetic processes required for subsequent stages of notochord development and differentiation.
Table 1.
Gene Models and ESTs for the Genes Examined in the Present Study
| Gene Name | Alternative Name(s) | KH Gene Modela | JGI v1.0 Gene Model(s) | JGI v1.0 Location | JGI v2.0 Location | cDNA Clone Used | Source(s) of Previously Published Patterns |
|---|---|---|---|---|---|---|---|
| Ci-Sall-a |
Ci-Spalt-like1 ZF(C2H2)-18 |
KH.L4.17 | ci0100141112 | scaffold_177 | scaffold_67 | cieg50j03 | Imai et al., 2004 |
| Ci-Lmx-like | N/A | KH.C9.485 | ci0100149991 ci0100145187 |
scaffold_89 | chr_09q | cinc025n05 | Imai et al., 2004 |
| Ci-Lmx | Ci-lmx1.2 | KH.C9.616 | ci0100150069 | scaffold_89 | chr_09q | citb30f24 |
Imai et al., 2004, 2009 Ishibashi et al, 2005 Tassy et al., 2010 |
| Ci-Fos-a | Ci-Fos | KH.C11.314 | ci0100130316 | scaffold_63 | scaffold_1690 scaffold_2122 |
cima839226 | Imai et al., 2004 |
| Ci-NFAT5 | N/A | KH.C3.133 | ci0100140442 ci0100141121 |
scaffold_128 scaffold_2370 |
chr_03q | cieg069a11 | Imai et al., 2004 |
| Ci-AFF | N/A | KH.C2.327 | ci0100131909 | scaffold_875 scaffold_117 |
chr_02q | cien95799 | N/A |
| Ci-STAT5/6-b | Ci-STAT-b | KH.C1.275 | ci0100154492 | scaffold_276 | chr_01q | cien84927 |
Imai et al., 2004 Hotta et al., 2008 |
| Ci-Klf15 | Ci-ZF148 | KH.C5.430 | N/A | scaffold_96 | chr_05q | cien222151 | Miwata et al., 2006 |
Satou et al., 2008
ZF, zinc-finger; v1.0, version 1.0; v2.0, version 2.0; N/A, not applicable
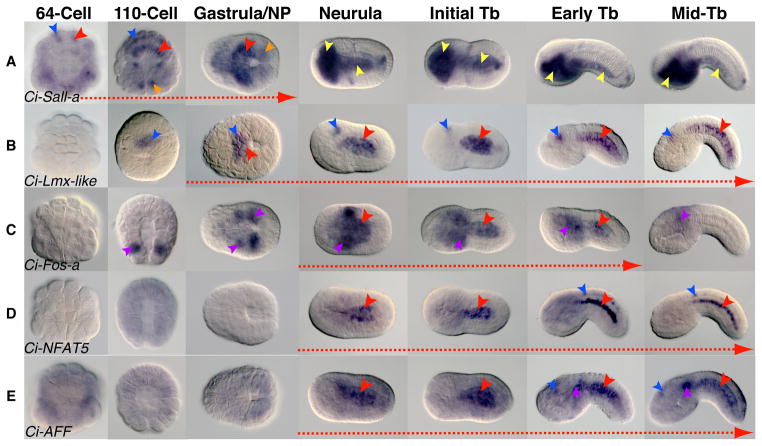
Figure 1. Expression patterns of novel Ciona notochord transcription factors during embryonic development.
(A–E) Whole-mount Ciona intestinalis embryos hybridized in situ with antisense RNA probes against the genes indicated in the lower left corner of each row. Developmental stages are indicated at the top of each column. Expression domains are highlighted with arrowheads colored as follows: red: notochord; orange: muscle; blue: CNS; yellow: endoderm; purple: mesenchyme. Red dashed arrows at the bottom of each row emphasize the stages when notochord expression is detected. In most panels, dorsal is up and anterior to the left with the exception of the 64- and 110-cell stage embryos, where anterior is up. NP: neural plate. Tb: tailbud.
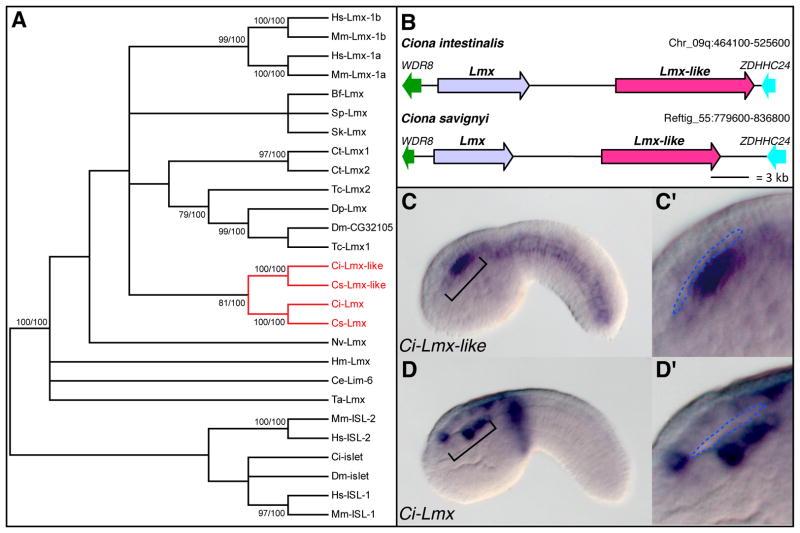
Figure 2. Ciona contains two Lmx paralogs with both overlapping and distinct expression domains.
(A) Phylogenetic reconstruction for the Lmx genes found in Ciona. The tree was obtained from neighbor-joining analyses and rooted using Islet (Isl) protein sequences as outgroups. Statistical support values ≥75% obtained with different methods are included over conserved nodes; the first number indicates the bootstrap support in neighbor-joining analysis (1000 bootstrap replicates) and the second number reports the posterior probabilities in Bayesian inference analysis. Abbreviations: Bf: Branchiostoma floridae; Ce: Caenorhabditis elegans; Ci: Ciona intestinalis; Cs: Ciona savignyi; Ct: Capitella teleta; Dm: Drosophila melanogaster; Dp: Daphnia pulex; Hm: Hydra magnipapillata; Hs: Homo sapiens; Mm: Mus musculus; Nv: Nematostella vectensis; Sk: Saccoglossus kowalevskii; Sp: Strongylocentrotus purpuratus; Ta: Trichoplax adhaerens; Tc: Tribolium castaneum. (B) Schematic depiction of the genomic regions encompassing the two Lmx genes found in the ascidians Ciona intestinalis and Ciona savignyi. The names of the genes flanking the Lmx genes are those indicated in each species’ genome browsers and were identified using the reciprocal best BLAST hit method (Bork et al., 1998). For simplicity, intron/exon structures of the genes are not depicted. (C–D′) Mid-tailbud stage embryos showing expression of Ci-Lmx-like (C,C′) and Ci-Lmx (D,D′) as detected by WMISH. The ventral portion of the sensory vesicle, indicated by a bracket, is shown in increased magnification in C′ and D′. The boundary between dorsal and ventral sensory vesicle is outlined by dashed blue lines in C′ and D′.
Figure 3. Ci-STAT5/6-b and Ci-Klf15 are both ubiquitously expressed during early embryogenesis and become localized to specific expression domains at later stages.
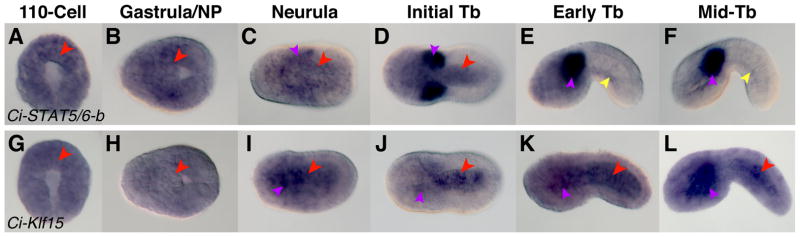
WMISH of Ciona embryos performed using antisense probes for Ci-STAT5/6-b (A–F) and Ci-Klf15 (G–L). Developmental stages are indicated at the top of each column. The notochord territory is denoted by red arrowheads. Purple arrowheads designate mesenchyme staining, while yellow arrowheads correspond to expression in the endoderm.
Ci-Sall-a
Spalt-like (Sall) proteins are zinc-finger transcription factors related to the product of Drosophila spalt major (salm) and spalt-related (salr) genes (de Celis and Barrio, 2009). While mammalian genomes contain four Sall family members, Ciona intestinalis has two. Through phylogenetic reconstructions, we found that the two Ciona Sall genes likely arose from a lineage-specific duplication event (Fig. S2A) and are both equally related to their vertebrate counterparts. Based on this observation, here we refer to these paralogs as Ci-Sall-a and Ci-Sall-b.
Ci-Sall-a is expressed in a number of spatial domains during Ciona embryonic development (Fig. 1A). Weak expression in notochord cells is detected starting at the 64-cell stage, however Ci-Sall-a transcripts become more evident in this territory by the 110-cell stage (Fig. 1A, red arrowheads). At these stages, faint expression is also detected in some CNS precursors (Fig. 1A, blue arrowheads). Notochord expression continues through gastrulation, but begins to be down-regulated during the neurula stage. Ci-Sall-a is also expressed in the trunk ventral cells (TVCs), the precursors of the Ciona heart (Fig. 1A, orange arrowheads), beginning at the 110-cell stage and additionally in endoderm beginning at the neurula stage (Fig. 1A, yellow arrowheads). Expression is retained in the trunk endoderm and endodermal strand through the mid-tailbud II stage (Fig. S1A). It has been shown that spalt genes, such as the chick csall3 gene, undergo alternative splicing (Sweetman and Munsterberg, 2006). Interestingly, Ci-Sall-a mRNAs also appear to be alternatively spliced (Satou and Satoh, 2005). Nevertheless, in our analysis, we found that probes synthesized from both Ci-Sall-a cDNAs exhibited identical expression patterns (Fig. 1A and data not shown).
Some members of the Sall1 group in various vertebrates are expressed in the notochord, as is the case for mouse Sall1 (Ott et al., 2001), zebrafish sall1a (Camp et al., 2003) and chick csal1 (Sweetman et al., 2005), while the other Spalt family members are absent from this tissue. Notably, each of the Sall1 genes mentioned above is also present in the developing heart and the neural tube (Ott et al., 2001; Camp et al., 2003; Sweetman et al., 2005). Interestingly, the composite expression pattern, which encompasses notochord, neural tube and heart, is nearly recapitulated by the combined expression patterns of the two Ciona spalt-like paralogs; Ci-Sall-a is detected in the notochord and heart precursors, and transiently in CNS precursors (Fig. 1A) while our previous work has shown that Ci-Sall-b is present in the posterior neural tube in tailbud embryos although it is absent from the notochord (Kugler et al., 2008). The roles of Spalt proteins in notochord development have not been examined; therefore, Ciona represents a simplified model system in which to assess Spalt function in the notochord.
Ci-Lmx-like
The LIM-homeodomain transcription factor gene Ciona LIM-homeobox like (Ci-Lmx-like) begins to be expressed at the 110-cell stage, where it is present in cells of the neural lineage (Fig. 1B, blue arrowheads). Beginning at gastrulation, transcripts are also detected in notochord cells, which gradually become the predominant expression domain (Fig. 1B, red arrowheads); Ci-Lmx-like expression persists in these two areas through the mid-tailbud stage (Fig. 1B) and begins to fade from the notochord around the mid-tailbud II stage (Fig. S1B).
Two members of the Lmx gene family are found in mammalian genomes, Lmx1a and Lmx1b (Hunter and Rhodes, 2005), and two Lmx genes are also found in Ciona (Wada et al., 2003; Imai et al., 2004). Since the molecular evolutionary history of these genes was incomplete (Wada et al., 2003; Srivastava et al., 2010), we performed a phylogenetic reconstruction of the Lmx family to ascertain the relationship between each Ciona Lmx and its vertebrate counterparts. After a thorough search, we concluded that most metazoan genomes contain only a single Lmx gene, with the notable exceptions of vertebrates, ascidians, the annelid Capitella teleta and the insect Tribolium castaneum, in which two Lmx genes were found. To analyze these duplications, we constructed a multiple alignment of the conserved domains of 21 Lmx-related proteins identified from 15 different metazoan species and used it to construct a phylogenetic tree whereby the closely related LIM-homeodomain Islet (Isl) proteins served as an outgroup (Fig. 2A). A similar topology was obtained when the full protein sequences were aligned (data not shown). Surprisingly, we found that the ascidian, vertebrate and annelid duplicates bundled into separate monophyletic groups with statistical supports over 80% using the neighbor-joining method and 100% when the Bayesian inference method was utilized, indicating that the Lmx duplicates in each of these species likely arose from independent lineage-specific duplication events. This, in turn, suggests that the last Deuterostome common ancestor had only one Lmx gene that was subsequently independently duplicated in the vertebrate and ascidian lineages (Fig. 2A). The clustered genomic localization and orientation of these two genes suggest that Ci-Lmx and Ci-Lmx-like are the result of a tandem duplication event (Fig. 2B).
In vertebrates, the Lmx family members have partially overlapping expression domains. For example, in mouse embryos both Lmx1a and Lmx1b are expressed in part of the mesencephalic dopaminergic neurons (Smidt et al., 2000; Failli et al., 2002), and play overlapping functions in the developing cerebellum (Mishima et al., 2009), while only Lmx1a is present in the notochord (Failli et al., 2002). In the course of this study, we noticed that the neural expression of Ci-Lmx-like was specific to a portion of the ventral posterior sensory vesicle (Fig. 2C,C′). Since Ci-Lmx was also previously reported to be expressed in the sensory vesicle (Imai et al., 2004), we carried out in situ hybridizations with a probe specific for this gene to determine whether the expression domains of the two Ciona Lmx genes overlap in this region of the ascidian CNS. The results showed that Ci-Lmx is present in various regions of the CNS, including the ventral posterior sensory vesicle in a region that overlaps with the expression territory of Ci-Lmx-like (Fig. 2D,D′). Therefore, it appears that both Ciona Lmx paralogs, like their vertebrate counterparts, exhibit complementary as well as overlapping expression patterns in the CNS, while only one family member is found in the notochord.
Interestingly, the ventral sensory vesicle of the ascidian embryo has been compared to the vertebrate hypothalamus (Moret et al., 2005; Hamada et al., 2011), and both Lmx1a and Lmx1b have been shown to be expressed in the developing mouse hypothalamus (Asbreuk et al., 2002). These results suggest that functional studies on Ci-Lmx and Ci-Lmx-like in a simplified model system such as Ciona could enhance our knowledge of the evolutionary origins of this structure.
In mammals, Lmx proteins have been found to serve important developmental functions outside of the CNS. The dreher mouse carries a mutation in Lmx1a which is responsible for numerous abnormalities, including skeletal defects such as a short tail (Bergstrom et al., 1999). Mutation of Lmx1b leads to aberrant collagen fibril formation in mouse corneas (Pressman et al., 2000) and also to a decrease in the levels of type IV collagens in glomerular basement membranes of the kidney (Morello et al., 2001). Given the involvement of Lmx1b in the expression and maturation of collagen in different tissues and the expression of various collagen genes in the Ciona notochord (Wada et al., 2006; Kugler et al., 2010), it is possible that Ci-Lmx-like could play an analogous role in the formation of the notochordal sheath, a possibility that has yet to be explored in any chordate.
Ci-Fos-a
Fos belongs to a highly conserved family of bZIP transcription factors with single-copy representatives in both Drosophila and C. elegans and four representatives in mammals. Ciona appears to have two Fos genes within its genome; the family member examined in the present study, which we refer to as Ci-Fos-a, is equally related to all four mammalian paralogs, while the other, Ci-Fos-b, is a highly divergent member of the Fos family (Amoutzias et al., 2007).
Expression of Ci-Fos-a begins at the 110-cell stage in precursors of the mesenchyme and persists in this tissue through the mid-tailbud stages (Figs. 1C and S1C, purple arrowheads). Transcripts appear in the notochord between the neurula and early tailbud stages (Fig. 1C, red arrowheads). However, as tailbud development progresses and intercalation is completed, expression in the notochord is down-regulated and only a residual patchy expression in the mesenchyme is observed (Figs. 1C, S1C). Ci-Fos-b, also known as orphan bZIP-4, is also expressed in the mesenchyme; however, it has not been reported to be expressed in the notochord (Imai et al., 2004).
In Drosophila, Fos is crucial for dorsal closure (Zeitlinger et al., 1997), while in C. elegans this gene is necessary for cell invasion through the basement membrane during vulvar development (Sherwood et al., 2005); therefore, Fos genes appear to be critical for cell motility. Given this role for Fos in other animals and the time of its expression during Ciona embryogenesis, it seems possible that Ci-Fos-a targets could be involved in the cell movements required for notochord intercalation (Munro and Odell, 2002; Jiang et al., 2005; Shi et al., 2009). Furthermore, c-Fos has been found to be expressed in both nucleus pulposus cells (Lee et al., 2007) and chordomas (Schwab et al., 2009), which suggests that vertebrate Fos family members might play as yet unexplored roles in notochord formation that could be elucidated in Ciona.
In vertebrates, Fos is known to heterodimerize with another bZIP transcription factor, Jun, to form the AP-1 complex (e.g., Woodgett, 1990). The AP-1 complex is involved in a variety of processes, ranging from the transcriptional regulation of skeletogenesis and bone remodeling (Karsenty, 2008) to various steps of tumorigenesis (Matthews et al., 2007). Interestingly, a Ciona Jun ortholog has been previously reported to be expressed exclusively in part of the B-line mesenchyme (Imai et al., 2004); this observation excludes the possibility that an AP-1-related complex might be formed in the Ciona notochord. On the other hand, Fos has been shown to heterodimerize with additional bZIP proteins, or even with members of other transcription factor families, including bHLH proteins (Chinenov and Kerppola, 2001). Therefore, it is possibile that Ci-Fos-a might interact with other notochord transcription factors, such as the bZIP factor XBPa (Kugler et al., 2008) or the Orphan bHLH-1 factor (Imai et al., 2004).
Ci-NFAT5
Nuclear Factor of Activated T-cells (NFAT) proteins are part of the Rel family of transcription factors that include NF-κB (Aramburu et al., 2006). While vertebrate genomes encode five NFAT genes (Hogan et al., 2003), Ciona has a single NFAT that appears to be the ortholog of NFAT5 (Yagi et al., 2003). We first detected Ci-NFAT5 transcripts at the time of neurulation (Fig. 1D). Ci-NFAT5 expression appears to be primarily confined to the notochord (Fig. 1D, red arrowheads), although a weak signal is also present in cells of the nerve cord (Fig. 1D, blue arrowheads). In contrast to what is seen in the case of the other genes that we analyzed, Ci-NFAT5 transcripts are localized to a narrow perinuclear area; this is particularly evident in the mid-tailbud stages (Figs. 1D and S1D).
In mammals, NFAT5 has been shown to be crucial for the regulation of osmotic stress, allowing the adaptation of cells to hypertonic environments (Aramburu et al., 2006). Because of this, perturbation of NFAT5 affects the function and development of tissues sensitive to osmotic fluctuations, including the kidney (Lopez-Rodriguez et al., 2004) and eye lens (Wang et al., 2005). Of note, intervertebral discs are surrounded by an extracellular matrix of high osmolarity (Kraemer et al., 1985) that is critical for counteracting pressure from the vertebrae (Risbud et al., 2010). Interestingly, NFAT5 is expressed in the nucleus pulposus of the intervertebral discs, where it has been shown to be important for the adaptation and survival of these cells to hyperosmotic environments (Tsai et al., 2006).
Supporting the hypothesis that the ancestral role of NFAT5 may be osmotic regulation, fly embryos deficient in the single-copy Drosophila NFAT are sensitive to high salt concentrations (Keyser et al., 2007). Because of this, we can speculate that Ci-NFAT5 may contribute to the regulation of later stages of notochord development, such as lumen formation, where maintenance of osmolarity is critical; this hypothesis represents an intriguing avenue to be explored in the future. It is also possible that Ci-NFAT5 plays multiple roles during notochord development. For example, it has been reported that overexpression of NFAT5 in breast and colon cancer cell cultures increased their mobility, thus implicating this protein in cellular migration (Jauliac et al., 2002). Since its expression in Ciona begins at the neurula stage, Ci-NFAT5 could contribute to generating the mobility required for notochord intercalation. Additionally, knock-down of NFAT5 in chondrocytes resulted in decreased expression of chondrocytic markers, including type II collagen (van der Windt et al., 2010). The notochord is thought to represent a primitive form of cartilage, as both structures share similar structural and morphological features, such as the expression of fibrillar collagens in vertebrates as well as in ascidians (Wada, 2010). This suggests that Ci-NFAT5 could also be regulating the expression of collagen genes in the notochord. As knowledge of the functions of NFAT5 during embryonic development is still fragmentary, the examination of the role(s) of Ci-NFAT5 in notochord formation can offer new insights into its uncharacterized functions.
Ci-AFF
AFF (AF4/FMR2) proteins belong to a family of transcriptional regulators, which in mammalian genomes consists of four genes. Recently, AF4 (acute lymphoblastic leukemia 1-fused gene from chromosome 4) has been shown to promote transcriptional elongation (Bitoun et al., 2007); however, members of the AFF family have been studied more extensively in relation to human disease. Most notably, mutations in FMR2 have been indicated as causative agents of mental retardation and all three other family members have been linked, although to different extents, to acute lymphoblastic leukemia (Gu and Nelson, 2003; Marschalek, 2010). Ciona, on the other hand, appears to have a single gene belonging to this family (Fig. S2B). Figure 1E shows the expression pattern of Ci-AFF. Hybridization signal is seen in the notochord precursors beginning at the neurula stage and persists through the mid-tailbud stage (Fig. 1E, red arrowheads). In addition to staining in the notochord, we often detected expression of Ci-AFF in cells of the mesenchyme and the sensory vesicle, predominantly in the dorsal anterior sensory vesicle (Fig. 1E, purple and blue arrowheads, respectively). This pattern was also observed in mid-tailbud II embryos (Fig. S1E).
Interestingly, in mice all AFF family members are expressed in the brain (Bitoun and Davies, 2005), which is suggestive that AFF genes may have an evolutionarily conserved function in the development of the CNS. Given that mammalian AFF genes are thought to have overlapping roles during brain development (Bitoun and Davies, 2005), studying the function of the single Ciona AFF in the CNS could help overcome the functional redundancy seen in higher chordates.
While the roles of AFF family members have been more readily studied in the CNS, their additional expression territories suggest that these genes might have a wider range of unexplored functions. For example, AFF3 is expressed in the cartilage of mice (Britanova et al., 2002) and this gene was also found to be enriched in intervertebral discs of E13.5 embryos (Sohn et al., 2010). Unfortunately, no knock-out mouse model exists for AFF3 that might help in assessing the importance of this expression domain, and the complete early embryonic expression patterns of the other AFFs remain largely unknown. Given the evolutionary relationship between notochord and cartilage (Stemple, 2005), elucidation of the function of Ci-AFF in notochord formation could inform future studies on both of these structures.
Ci-STAT5/6-b
Signal transducers and activators of transcription (STATs) are transcription factors activated in response to cytokine and growth factor receptor signaling, most notably through the action of Janus kinases (JAKs) (Hennighausen and Robinson, 2008). Mammalian genomes contain seven STAT family genes while Drosophila has a single STAT ortholog, dSTAT, which most closely resembles the mammalian STAT5a/b genes (Yan et al., 1996). Two STAT genes have been found in Ciona, Ci-STAT-a and Ci-STAT-b (Imai et al., 2004), both related to vertebrate STAT5a/b and STAT6 (Hino et al., 2003); Ci-STAT-b has also been reported as Ci-STAT5/6-b (Hotta et al., 2003; Hotta et al., 2008).
We found that Ci-STAT5/6-b is ubiquitously expressed at the 110-cell and gastrula stages (Fig. 3A,B), consistent with previous work (Imai et al., 2004; Hotta et al., 2008); however, we observed that the expression of this gene became progressively restricted at later stages (Fig. 3C–F). Staining in the mesenchyme and notochord was seen in neurula and initial tailbud embryos (Fig. 3C,D, purple and red arrowheads, respectively), but only mesenchyme expression persisted at the early and mid-tailbud stages (Fig. 3E,F, purple arrowheads). Embryos at the mid-tailbud stage also exhibited weak expression of Ci-STAT5/6-b in the endodermal strand (Fig. 3E,F, yellow arrowheads). In mid-tailbud II embryos, expression becomes fully restricted to the mesenchyme (Fig. S1F, purple arrowhead). This staining pattern appears to be specific, as no signal was observed when embryos were hybridized with a Ci-STAT5/6-b sense probe (Fig. S3A).
The presence of STAT proteins in the notochord is not unique to Ciona. STAT5 was found to be weakly expressed in the notochord of Xenopus embryos (Pascal et al., 2001). Additionally, zebrafish STAT3, which is thought to have arisen from a duplication of STAT5 (Lewis and Ward, 2004), has also been reported to be expressed in the notochord (Oates et al., 1999). The roles of these genes in notochord development remain unexplored. Of note, in fruit flies, dSTAT is required for several processes, including germ cell migration, establishment of planar polarity in the eye and convergent extension during hindgut elongation (Hou et al., 2002). Interestingly, zebrafish STAT3 morphants exhibit defects in convergent extension due to an impairment of the PCP pathway (Miyagi et al., 2004). Ci-STAT5/6-b is down-regulated in the notochord after intercalation is completed, suggesting that one of the ancestral roles of STAT signaling could be the establishment of PCP during a variety of cellular processes.
Ci-Klf15
Kruppel-like factors (Klf) are members of a subclass of zinc-finger transcription factors with varied roles in development, which in mammals includes 17 members (Pearson et al., 2008). In the Ciona genome we found six Klfs, including one gene of the Klf15 group (Fig. S4).
We determined that between the 110-cell and gastrula stages, Ci-Klf15 is expressed weakly and ubiquitously (Fig. 3G,H). Beginning at neurula, transcripts are refined to the notochord and the mesenchyme (Fig. 3I–L). Notochord expression is most prevalent at the initial-tailbud stage (Fig. 3J) and is down-regulated by the mid-tailbud II stage, when the hybridization signal becomes concentrated in the mesenchyme (Fig. S1G). No signal was observed when a Ci-Klf15 sense probe was used (Fig. S3B).
During mouse development, Klf15 is expressed in numerous embryonic structures, including the CNS (van der Zwaag et al., 2005) and the heart (Fisch et al., 2007), and Klf15 null mice are viable but show abnormal heart morphology (Fisch et al., 2007). In addition to acting as a negative regulator of cardiomyocyte hypertrophy, Klf15 has been shown to control adipogenesis in human cell lines (Yamamoto et al., 2010).
This study provides the first evidence for notochord expression of Klf15 thus far. Of note, another member of the Ciona Klf gene family, Ci-Klf6, is also found in the notochord (Imai et al., 2004). It is not unprecedented for members of this transcription factor family to work synergistically or antagonistically, and some Klf factors have been shown to regulate one another in other model organisms (Suzuki et al., 2005; Pearson et al., 2008); this suggests that Ci-Klf15 and Ci-Klf6 might influence each other during notochord formation.
Hierarchical relationships between newly identified notochord transcription factors and Ci-Bra
In mouse embryos, Brachyury and Foxa2 have been shown to act upstream of another notochord transcription factor, Not, which is necessary for the development of the caudal notochord (Abdelkhalek et al., 2004); similarly, in zebrafish, one of the two Brachyury orthologs, No tail, has been shown to control expression of floating head, a homeodomain transcription factor related to Not (Morley et al., 2009). In Xenopus, screens for Xbra transcriptional targets led to the identification of the paired-like homeobox genes Bix1 and Bix2-4 (Tada et al., 1998; Casey et al., 1999). Previously published work in Ciona has identified Ci-STAT5/6-b as a Ci-Bra target (Hotta et al., 2008), and with the present analysis we have uncovered the notochord expression of this gene.
In Ciona, we have the opportunity to study the relationship between Ci-Bra and the newly identified notochord transcription factors by analyzing their expression in embryos obtained from an ENU-induced ascidian line carrying a recessive mutation in the Ci-Bra locus (Chiba et al., 2009). This mutation inserts an early stop codon into the Ci-Bra coding sequence, which results in a predicted protein that lacks most of the DNA-binding domain and the C-terminal amino acid residues; for this reason, the mutation is considered a null (Chiba et al., 2009).
At the early tailbud stage, Ci-Bra−/− embryos are characterized by a shorter tail than wild-type or Ci-Bra+/− animals; this phenotype is due to visible alterations in the morphology of the notochord cells, which fail to intercalate and fail to form lumens later on. These characteristics render the Ci-Bra−/− embryos morphologically distinguishable from their wild-type and heterozygous siblings starting from approximately the early tailbud stage (Chiba et al., 2009). Hence, for our analysis, we chose to look at the expression of Ci-Lmx-like, Ci-NFAT5 and Ci-AFF in Ci-Bra−/− embryos, given their robust notochord expression at the early to mid-tailbud stages (Fig. 4). Embryos from a heterozygous mating of Ci-Bra mutants were hybridized in situ in parallel with wild-type control embryos. In all cases, approximately a quarter of the embryos deriving from the heterozygous cross showed a shorter tail and lack of a well-developed notochord, consistent with what could be expected in the case of Mendelian inheritance of the mutation (Fig. 4C,F,I). On the other hand, the remaining embryos (~75% of the total; a mixture of wild-type and heterozygotes) (Fig. 4B,E,H) were morphologically indistinguishable from the wild-type control embryos (Fig. 4A,D,G).
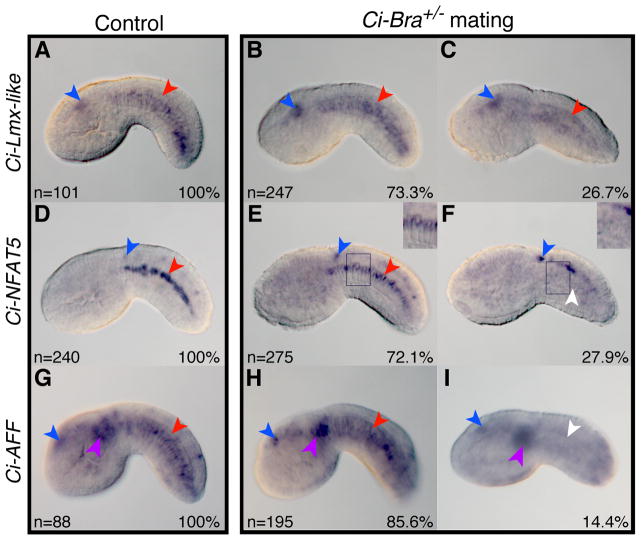
Figure 4. Expression of Ci-Lmx-like, Ci-NFAT5 and Ci-AFF in Ci-Bra mutant embryos.
Expression of Ci-Lmx-like (A–C), Ci-NFAT5 (D–F) and Ci-AFF (G–I) assessed by WMISH on wild-type control embryos (left) and on the offspring of animals heterozygous mutant for Ci-Bra (right). Numbers in the lower left corners report the number of embryos scored; the percentage of embryos exhibiting each phenotype is reported in the lower right corners. Expression in cells of the notochord lineage is indicated by a red arrowhead, while notochord cells lacking staining are denoted by a white arrowhead. Insets in the upper right hand corners of (E) and (F) show a closer view of the notochord cells boxed by the dark blue rectangles. Expression in other domains is denoted as follows: blue arrowheads: CNS expression; purple arrowheads: mesenchyme expression.
In the case of Ci-Lmx-like, Ci-Bra−/− embryos (Fig. 4C) showed a staining pattern similar to that of the remaining embryos and wild-type controls (Fig. 4A,B), with staining in cells of the notochord lineage as well as in the sensory vesicle. This result suggests that Ci-Lmx-like transcription in the notochord might be influenced by Ci-Bra only marginally, and that a notochord activator that belongs to a Ci-Bra-independent branch of the notochord gene regulatory network plays a main role in the regulation of the expression of this gene.
Conversely, the expression of Ci-NFAT5 in cells of the notochord lineage does appear to be affected by the loss of Ci-Bra (Fig. 4D–F). In fact, expression of this gene in wild-type control embryos and in the majority of the embryos resulting from the heterozygous mating is seen in the notochord and CNS (Fig. 4D,E), while in Ci-Bra−/− embryos, expression is specifically lost from the cells of the notochord lineage but is retained in the CNS (Fig. 4F). These results indicate that the loss of Ci-Bra function specifically affects Ci-NFAT5 expression in the notochord.
Similar to the case of Ci-NFAT5, expression of Ci-AFF (Fig. 4G–I) is also lost from the notochord in Ci-Bra−/− embryos and is detected only in cells of the sensory vesicle and mesenchyme (Fig. 4I). Based upon these data, therefore, we can begin to tentatively place Ci-NFAT5 and Ci-AFF downstream of Ci-Bra in the notochord gene regulatory network and suggest that they may function as transcriptional intermediaries for this transcription factor. Given the presence of Brachyury in the nucleus pulposus (Shapiro and Risbud, 2010), it is conceivable that control of NFAT5 and AFF genes by Brachyury might be an evolutionarily conserved feature.
Since morphological differences are not readily observed between Ci-Bra−/− and wild-type embryos before the mid-tailbud stage, we employed an alternative approach to unravel the hierarchical relationships between Ci-Bra and the transcription factors that are expressed prior to this developmental period. To this end, we examined the expression patterns of Ci-Fos-a and Ci-Klf15 in initial tailbud embryos electroporated at the one-cell stage with the Ci-Bra>Ci-Bra::enRD construct (abbreviated as Bra>Bra::enRD), which directs expression of a repressor form of Ci-Bra in the notochord and phenocopies the loss of function of Ci-Bra (Kugler et al., 2008); therefore, akin to the situation with Ci-Bra−/− animals, targets of Ci-Bra are expected to be down-regulated in the notochord of those embryos expressing the Bra>Bra::enRD transgene. This approach also offers the possibility of selecting transgenic embryos for further analyses through the co-electroporation of the Bra>Bra::enRD construct with an appropriate marker construct, such as Ci-Bra>eGFP (Corbo et al., 1997) (data not shown). Compared to stage-matched controls (Fig. 5A,C), the Bra>Bra::enRD transgenic embryos display a shorter tail and a malformed notochord (Fig. 5B,D), with fewer distinguishable notochord cells than the control (compare Fig. 5A′,B′ with 5B′,D′).
Figure 5. Ci-Fos-a and Ci-Klf15 are down-regulated in embryos expressing a repressor form of Ci-Bra.
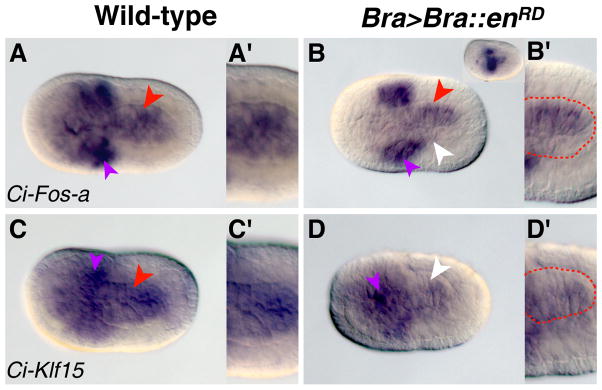
Wild-type (A,C) or Bra>Bra::enRD-carrying (B,D) initial tailbud stage embryos analyzed by WMISH for expression of Ci-Fos-a (A–B′) or Ci-Klf15 (C–D′). Purple arrowheads indicate mesenchyme expression; red arrowheads indicate expression in notochord cells, while white arrowheads denote a lack of notochord staining. Panels A′, B′, C′ and D′ show the notochord cells in greater detail, and dashed red lines outline the notochord territory in (B′) and (D′) for clarity. The inset in (B) shows a Bra>Bra::enRD transgenic embryo with a more severe notochord phenotype probed for Ci-Fos-a. (A–C) are dorsal views, while (D) is a dorsal-lateral view.
Figure 5B displays a Bra>Bra::enRD embryo probed for Ci-Fos-a expression. While notochord cells are generally organized as two rows along the midline at the initial tailbud stage, this embryo appears to have one normal and one aberrant row of cells (Fig. 5B), likely due to mosaic incorporation of the transgene. Interestingly, the unaffected cells are positive for Ci-Fos-a (Fig. 5B, red arrowhead) while the altered notochord cells are not (Fig. 5B, white arrowhead), indicating that this gene is down-regulated in cells expressing the repressor form of Ci-Bra. This point is supported by the observation that notochord staining is virtually absent in Bra>Bra::enRD transgenic embryos that exhibit a more severe notochord defect as a consequence of a higher incorporation of the transgene (Fig. 5B, inset).
Ci-Klf15 expression in the notochord also appears to be influenced by the levels of Ci-Bra, as expression of this gene is considerably down-regulated in Bra>Bra::enRD transgenic embryos (Fig. 5D) compared to wild-type controls (Fig. 5C), while mesenchyme expression remains unperturbed.
The expression of Ci-Sall-a was too transient to be assessed in either mutant background or by PCR methods (data not shown); however, this gene has been previously indicated as a putative Ci-Bra early target, based upon the occupancy of its genomic locus by Ci-Bra in early embryos (Kubo et al., 2010). The early occupancy of the Ci-Sall-a locus by Ci-Bra is consistent with our detection of Ci-Sall-a in notochord precursors starting from the 64-cell stage.
Conclusions
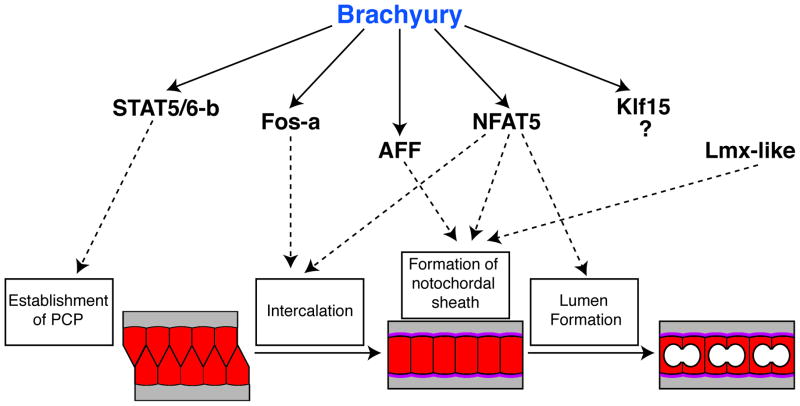
Our findings are summarized by the model shown in Figure 6. Ci-Bra might employ some of the notochord transcription factors identified in this study to control the various morphogenetic steps required for notochord development and differentiation. The function of each transcriptional intermediary of Ci-Bra has been tentatively inferred, whenever possible, from the functions previously assessed in other model organisms. The model suggests that the early-onset Ci-STAT5/6-b might be involved in PCP establishment; later transcription factors, such as Ci-Fos-a, might regulate intercalation, while Ci-AFF and the Ci-Bra-independent factor Ci-Lmx-like may contribute to the formation of the notochordal sheath. Lastly, the multifunctional transcription factor Ci-NFAT5 could control one or more of the previous steps, and/or the formation of extracellular lumens.
Figure 6. Newly identified notochord transcription factors, their relationship with Ci-Bra and their putative functions in notochord development.
Solid black arrows lead from Brachyury to the transcription factors that it regulates. Dashed black arrows point to the possible roles of each factor in notochord formation, as described in the text. Notochord cells are depicted in red while the notochordal sheath is shown in purple; white circles represent intercellular lumens.
Remarkably, the notochord expression of Ci-Fos-a, Ci-STAT5/6-b, and Ci-Klf15 is down-regulated in notochord cells at the tailbud stages, when the Ci-Bra protein is still present in the nuclei of notochord cells (our unpublished results). This suggests that the activation of these genes by Ci-Bra might be counterbalanced by repressive events.
In conclusion, this study has contributed to increasing the knowledge of the notochord gene complement in Ciona with information that is likely applicable to other chordates. These results add new depth to the Ci-Bra-downstream gene regulatory network through the efficient read-out of notochord-specific gene expression provided by embryos of the Ci-Bra mutant line and by transgenic embryos that phenocopy them. Furthermore, the results presented here highlight the existence of transcription factors whose expression in the notochord is less sensitive to changes in the level of Ci-Bra.
EXPERIMENTAL PROCEDURES
Embryo culture, fixation and electroporation
Adult Ciona intestinalis were purchased from Marine Research and Educational Products (M-REP; Carlsbad, CA, USA) and kept in an aquarium in recirculating artificial sea water at 17–18°C. Wild-type embryos for in situ experiments were obtained by in vitro fertilization and fixed at the desired stages in 4% paraformaldehyde, 0.1M MOPS (pH 7.5) and 0.5M NaCl at 4°C overnight. Ci-Bra mutant embryos were kindly provided by Drs. S. Chiba and W. Smith (University of California at Santa Barbara, USA). Electroporations were performed as previously described (Oda-Ishii and Di Gregorio, 2007).
Probe preparation
Digoxigenin-labeled RNA probes for each gene were generated from the following EST cDNA clones from the Ciona intestinalis Gene Collection release 1 (Satou et al., 2002): GC23n09 (Sall-a), GC45i22 (Lmx-like), GC31h17 (Lmx), GC40i13 (NFAT5), and from the following clones found in the Ciona intestinalis Gateway-compatible Unigene collection (Beckman Coulter Genomics, Grenoble, France): 63M13 (Fos-a), 85P09 (AFF), 83I17 (STAT5/6-b) and 104P16 (Klf15) (see also Table 1). Plasmid DNA for all clones was purified using the NucleoSpin Plasmid isolation kit (Macherey-Nagel, Bethlehem, PA, USA). Clones GC23n09, GC45i22, GC31h17 and GC40i13 were linearized by enzymatic digestion with NotI for the synthesis of the antisense probes (New England Biolabs, Ipswich, MA, USA). For 63M13, 85P09, 83I17 and 104P16, fragments were amplified by PCR using M13F and M13R reverse primers and Hi-Fi Taq DNA polymerase (Invitrogen, Carlsbad, CA, USA). DNA templates were purified by standard phenol-chloroform extraction and ethanol precipitation, and RNA probes were generated and purified as previously described (Kugler et al., 2008).
Whole-mount in situ hybridization (WMISH)
Whole-mount in situ hybridizations were carried out essentially as previously published (Oda-Ishii and Di Gregorio, 2007), using hybridization temperatures of either 42°C (Lmx-like and Sall-a) or 50°C (NFAT5, Fos-a, AFF, STAT5/6-b and Klf15).
Phylogenetic analyses
Multiple alignments of metazoan protein sequences were obtained using the MUSCLE 3.6 software (Edgar, 2004) and modified manually whenever necessary. Sequence alignments are available upon request. Neighbor-joining analyses and Bayesian inferences were performed as previously described (Kugler et al., 2011), except for the Bayesian inference analysis of Klf which required 500,000 generations of the four Markov Chain Monte Carlo (MCMC) chains run, hence requiring 12,500 sampled trees to be discarded as “burn-in”.
Acknowledgments
Grant information: NIH/NICHD: R01HD050704; March of Dimes Birth Defects Foundation: 1-FY08-430; Charles A. Frueauff Foundation (to A.D.G.); Norwegian Research Council: 133335/V40 and 183302/S10; (to D.J.); NIH: T32 GM008539 (to D.S.J.-E.)
We are indebted to Drs. S. Chiba and W. Smith (University of California at Santa Barbara) for kindly providing the Ci-Bra mutant embryos. We thank Drs. Nori Satoh and Yutaka Satou (Kyoto University) for the Gene Collection release 1 and Drs. U. Rothbacher and P. Lemaire (IBDML) and M. Gilchrist (Wellcome Trust/Cancer Research UK Gurdon Institute) for making the Unigene cDNA collection available. This work was supported by grant NIH/NICHD R01HD050704, grant 1-FY08-430 from the March of Dimes Birth Defects Foundation and a grant from the Charles A. Frueauff Foundation to A.D.G., and by grants 133335/V40 and 183302/S10 from the Norwegian Research Council to D.J. D.S.J.-E. was supported in part by NIH training grant T32 GM008539.
Abbreviations
- cDNA
complementary DNA
- CNS
central nervous system
- ENU
N-ethyl-N-nitrosourea
- EST
Expressed Sequence Tag
- kb
kilobase(s), or 1000 base pairs
- MOPS
3-(N-morpholino) propanesulfonic acid
- PCR
Polymerase Chain Reaction
- WMISH
whole-mount in situ hybridization
References
- Abdelkhalek HB, Beckers A, Schuster-Gossler K, Pavlova MN, Burkhardt H, Lickert H, Rossant J, Reinhardt R, Schalkwyk LC, Muller I, Herrmann BG, Ceolin M, Rivera-Pomar R, Gossler A. The mouse homeobox gene Not is required for caudal notochord development and affected by the truncate mutation. Genes Dev. 2004;18:1725–1736. doi: 10.1101/gad.303504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoutzias GD, Veron AS, Weiner J, 3rd, Robinson-Rechavi M, Bornberg-Bauer E, Oliver SG, Robertson DL. One billion years of bZIP transcription factor evolution: conservation and change in dimerization and DNA-binding site specificity. Mol Biol Evol. 2007;24:827–835. doi: 10.1093/molbev/msl211. [DOI] [PubMed] [Google Scholar]
- Aramburu J, Drews-Elger K, Estrada-Gelonch A, Minguillon J, Morancho B, Santiago V, Lopez-Rodriguez C. Regulation of the hypertonic stress response and other cellular functions by the Rel-like transcription factor NFAT5. Biochem Pharmacol. 2006;72:1597–1604. doi: 10.1016/j.bcp.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Asbreuk CH, Vogelaar CF, Hellemons A, Smidt MP, Burbach JP. CNS expression pattern of Lmx1b and coexpression with ptx genes suggest functional cooperativity in the development of forebrain motor control systems. Mol Cell Neurosci. 2002;21:410–420. doi: 10.1006/mcne.2002.1182. [DOI] [PubMed] [Google Scholar]
- Bergstrom DE, Gagnon LH, Eicher EM. Genetic and physical mapping of the dreher locus on mouse chromosome 1. Genomics. 1999;59:291–299. doi: 10.1006/geno.1999.5873. [DOI] [PubMed] [Google Scholar]
- Bitoun E, Davies KE. The robotic mouse: unravelling the function of AF4 in the cerebellum. Cerebellum. 2005;4:250–260. doi: 10.1080/14734220500325897. [DOI] [PubMed] [Google Scholar]
- Bitoun E, Oliver PL, Davies KE. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum Mol Genet. 2007;16:92–106. doi: 10.1093/hmg/ddl444. [DOI] [PubMed] [Google Scholar]
- Bork P, Dandekar T, Diaz-Lazcoz Y, Eisenhaber F, Huynen M, Yuan Y. Predicting function: from genes to genomes and back. J Mol Biol. 1998;283:707–725. doi: 10.1006/jmbi.1998.2144. [DOI] [PubMed] [Google Scholar]
- Britanova O, Lukyanov S, Gruss P, Tarabykin V. The mouse Laf4 gene: exon/intron organization, cDNA sequence, alternative splicing, and expression during central nervous system development. Genomics. 2002;80:31–37. doi: 10.1006/geno.2002.6796. [DOI] [PubMed] [Google Scholar]
- Camp E, Hope R, Kortschak RD, Cox TC, Lardelli M. Expression of three spalt (sal) gene homologues in zebrafish embryos. Dev Genes Evol. 2003;213:35–43. doi: 10.1007/s00427-002-0284-6. [DOI] [PubMed] [Google Scholar]
- Casey ES, Tada M, Fairclough L, Wylie CC, Heasman J, Smith JC. Bix4 is activated directly by VegT and mediates endoderm formation in Xenopus development. Development. 1999;126:4193–4200. doi: 10.1242/dev.126.19.4193. [DOI] [PubMed] [Google Scholar]
- Chiba S, Jiang D, Satoh N, Smith WC. Brachyury null mutant-induced defects in juvenile ascidian endodermal organs. Development. 2009;136:35–39. doi: 10.1242/dev.030981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinenov Y, Kerppola TK. Close encounters of many kinds: Fos-Jun interactions that mediate transcription regulatory specificity. Oncogene. 2001;20:2438–2452. doi: 10.1038/sj.onc.1204385. [DOI] [PubMed] [Google Scholar]
- Cleaver O, Krieg PA. Notochord patterning of the endoderm. Dev Biol. 2001;234:1–12. doi: 10.1006/dbio.2001.0214. [DOI] [PubMed] [Google Scholar]
- Corbo JC, Levine M, Zeller RW. Characterization of a notochord-specific enhancer from the Brachyury promoter region of the ascidian, Ciona intestinalis. Development. 1997;124:589–602. doi: 10.1242/dev.124.3.589. [DOI] [PubMed] [Google Scholar]
- de Celis JF, Barrio R. Regulation and function of Spalt proteins during animal development. Int J Dev Biol. 2009;53:1385–1398. doi: 10.1387/ijdb.072408jd. [DOI] [PubMed] [Google Scholar]
- Di Gregorio A, Levine M. Regulation of Ci-tropomyosin-like, a Brachyury target gene in the ascidian, Ciona intestinalis. Development. 1999;126:5599–5609. doi: 10.1242/dev.126.24.5599. [DOI] [PubMed] [Google Scholar]
- Dunn MP, Di Gregorio A. The evolutionarily conserved leprecan gene: its regulation by Brachyury and its role in the developing Ciona notochord. Dev Biol. 2009;328:561–574. doi: 10.1016/j.ydbio.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynan WS, Tjian R. The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell. 1983;35:79–87. doi: 10.1016/0092-8674(83)90210-6. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucl Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Failli V, Bachy I, Retaux S. Expression of the LIM-homeodomain gene Lmx1a (dreher) during development of the mouse nervous system. Mech Dev. 2002;118:225–228. doi: 10.1016/s0925-4773(02)00254-x. [DOI] [PubMed] [Google Scholar]
- Fisch S, Gray S, Heymans S, Haldar SM, Wang B, Pfister O, Cui L, Kumar A, Lin Z, Sen-Banerjee S, Das H, Petersen CA, Mende U, Burleigh BA, Zhu Y, Pinto YM, Liao R, Jain MK. Kruppel-like factor 15 is a regulator of cardiomyocyte hypertrophy. Proc Natl Acad Sci U S A. 2007;104:7074–7079. doi: 10.1073/pnas.0701981104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Nelson DL. FMR2 function: insight from a mouse knockout model. Cytogenet Genome Res. 2003;100:129–139. doi: 10.1159/000072847. [DOI] [PubMed] [Google Scholar]
- Hamada M, Shimozono N, Ohta N, Satou Y, Horie T, Kawada T, Satake H, Sasakura Y, Satoh N. Expression of neuropeptide- and hormone-encoding genes in the Ciona intestinalis larval brain. Dev Biol. 2011 doi: 10.1016/j.ydbio.2011.01.006. [DOI] [PubMed] [Google Scholar]
- Hennighausen L, Robinson GW. Interpretation of cytokine signaling through the transcription factors STAT5A and STAT5B. Genes Dev. 2008;22:711–721. doi: 10.1101/gad.1643908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hino K, Satou Y, Yagi K, Satoh N. A genomewide survey of developmentally relevant genes in Ciona intestinalis. VI. Genes for Wnt, TGFbeta, Hedgehog and JAK/STAT signaling pathways. Dev Genes Evol. 2003;213:264–272. doi: 10.1007/s00427-003-0318-8. [DOI] [PubMed] [Google Scholar]
- Hogan PG, Chen L, Nardone J, Rao A. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 2003;17:2205–2232. doi: 10.1101/gad.1102703. [DOI] [PubMed] [Google Scholar]
- Hotta K, Mitsuhara K, Takahashi H, Inaba K, Oka K, Gojobori T, Ikeo K. A web-based interactive developmental table for the ascidian Ciona intestinalis, including 3D real-image embryo reconstructions: I. From fertilized egg to hatching larva. Dev Dyn. 2007;236:1790–1805. doi: 10.1002/dvdy.21188. [DOI] [PubMed] [Google Scholar]
- Hotta K, Takahashi H, Asakura T, Saitoh B, Takatori N, Satou Y, Satoh N. Characterization of Brachyury-downstream notochord genes in the Ciona intestinalis embryo. Dev Biol. 2000;224:69–80. doi: 10.1006/dbio.2000.9765. [DOI] [PubMed] [Google Scholar]
- Hotta K, Takahashi H, Erives A, Levine M, Satoh N. Temporal expression patterns of 39 Brachyury-downstream genes associated with notochord formation in the Ciona intestinalis embryo. Dev Growth Differ. 1999;41:657–664. doi: 10.1046/j.1440-169x.1999.00467.x. [DOI] [PubMed] [Google Scholar]
- Hotta K, Takahashi H, Satoh N, Gojobori T. Brachyury-downstream gene sets in a chordate, Ciona intestinalis: integrating notochord specification, morphogenesis and chordate evolution. Evol Dev. 2008;10:37–51. doi: 10.1111/j.1525-142X.2007.00212.x. [DOI] [PubMed] [Google Scholar]
- Hotta K, Takahashi H, Ueno N, Gojobori T. A genome-wide survey of the genes for planar polarity signaling or convergent extension-related genes in Ciona intestinalis and phylogenetic comparisons of evolutionary conserved signaling components. Gene. 2003;317:165–185. doi: 10.1016/s0378-1119(03)00700-5. [DOI] [PubMed] [Google Scholar]
- Hou SX, Zheng Z, Chen X, Perrimon N. The Jak/STAT pathway in model organisms: emerging roles in cell movement. Dev Cell. 2002;3:765–778. doi: 10.1016/s1534-5807(02)00376-3. [DOI] [PubMed] [Google Scholar]
- Hunter CS, Rhodes SJ. LIM-homeodomain genes in mammalian development and human disease. Mol Biol Rep. 2005;32:67–77. doi: 10.1007/s11033-004-7657-z. [DOI] [PubMed] [Google Scholar]
- Imai KS, Hino K, Yagi K, Satoh N, Satou Y. Gene expression profiles of transcription factors and signaling molecules in the ascidian embryo: towards a comprehensive understanding of gene networks. Development. 2004;131:4047–4058. doi: 10.1242/dev.01270. [DOI] [PubMed] [Google Scholar]
- Imai KS, Levine M, Satoh N, Satou Y. Regulatory blueprint for a chordate embryo. Science. 2006;312:1183–1187. doi: 10.1126/science.1123404. [DOI] [PubMed] [Google Scholar]
- Imai KS, Stolfi A, Levine M, Satou Y. Gene regulatory networks underlying the compartmentalization of the Ciona central nervous system. Development. 2009;136:285–293. doi: 10.1242/dev.026419. [DOI] [PubMed] [Google Scholar]
- Ishibashi T, Usami T, Fujie M, Azumi K, Satoh N, Fujiwara S. Oligonucleotide-based microarray analysis of retinoic acid target genes in the protochordate, Ciona intestinalis. Dev Dyn. 2005;233:1571–1578. doi: 10.1002/dvdy.20486. [DOI] [PubMed] [Google Scholar]
- Jauliac S, Lopez-Rodriguez C, Shaw LM, Brown LF, Rao A, Toker A. The role of NFAT transcription factors in integrin-mediated carcinoma invasion. Nat Cell Biol. 2002;4:540–544. doi: 10.1038/ncb816. [DOI] [PubMed] [Google Scholar]
- Jiang D, Munro EM, Smith WC. Ascidian prickle regulates both mediolateral and anterior-posterior cell polarity of notochord cells. Curr Biol. 2005;15:79–85. doi: 10.1016/j.cub.2004.12.041. [DOI] [PubMed] [Google Scholar]
- Jiang D, Smith WC. Ascidian notochord morphogenesis. Dev Dyn. 2007;236:1748–1757. doi: 10.1002/dvdy.21184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsenty G. Transcriptional control of skeletogenesis. Annu Rev Genomics Hum Genet. 2008;9:183–196. doi: 10.1146/annurev.genom.9.081307.164437. [DOI] [PubMed] [Google Scholar]
- Keyser P, Borge-Renberg K, Hultmark D. The Drosophila NFAT homolog is involved in salt stress tolerance. Insect Biochem Mol Biol. 2007;37:356–362. doi: 10.1016/j.ibmb.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Kraemer J, Kolditz D, Gowin R. Water and electrolyte content of human intervertebral discs under variable load. Spine (Phila Pa 1976) 1985;10:69–71. doi: 10.1097/00007632-198501000-00011. [DOI] [PubMed] [Google Scholar]
- Kubo A, Suzuki N, Yuan X, Nakai K, Satoh N, Imai KS, Satou Y. Genomic cis-regulatory networks in the early Ciona intestinalis embryo. Development. 2010;137:1613–1623. doi: 10.1242/dev.046789. [DOI] [PubMed] [Google Scholar]
- Kugler JE, Gazdoiu S, Oda-Ishii I, Passamaneck YJ, Erives AJ, Di Gregorio A. Temporal regulation of the muscle gene cascade by Macho1 and Tbx6 transcription factors in Ciona intestinalis. J Cell Sci. 2010;123:2453–2463. doi: 10.1242/jcs.066910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugler JE, Kerner P, Bouquet JM, Jiang D, Di Gregorio A. Evolutionary changes in the notochord genetic toolkit: a comparative analysis of notochord genes in the ascidian Ciona and the larvacean Oikopleura. BMC Evol Biol. 2011;11:21. doi: 10.1186/1471-2148-11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugler JE, Passamaneck YJ, Feldman TG, Beh J, Regnier TW, Di Gregorio A. Evolutionary conservation of vertebrate notochord genes in the ascidian Ciona intestinalis. Genesis. 2008;46:697–710. doi: 10.1002/dvg.20403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CR, Sakai D, Nakai T, Toyama K, Mochida J, Alini M, Grad S. A phenotypic comparison of intervertebral disc and articular cartilage cells in the rat. Eur Spine J. 2007;16:2174–2185. doi: 10.1007/s00586-007-0475-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis RS, Ward AC. Conservation, duplication and divergence of the zebrafish stat5 genes. Gene. 2004;338:65–74. doi: 10.1016/j.gene.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Lopez-Rodriguez C, Antos CL, Shelton JM, Richardson JA, Lin F, Novobrantseva TI, Bronson RT, Igarashi P, Rao A, Olson EN. Loss of NFAT5 results in renal atrophy and lack of tonicity-responsive gene expression. Proc Natl Acad Sci U S A. 2004;101:2392–2397. doi: 10.1073/pnas.0308703100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschalek R. Mixed lineage leukemia: roles in human malignancies and potential therapy. FEBS J. 2010;277:1822–1831. doi: 10.1111/j.1742-4658.2010.07608.x. [DOI] [PubMed] [Google Scholar]
- Matsumoto J, Kumano G, Nishida H. Direct activation by Ets and Zic is required for initial expression of the Brachyury gene in the ascidian notochord. Dev Biol. 2007;306:870–882. doi: 10.1016/j.ydbio.2007.03.034. [DOI] [PubMed] [Google Scholar]
- Matthews CP, Colburn NH, Young MR. AP-1 a target for cancer prevention. Curr Cancer Drug Targets. 2007;7:317–324. doi: 10.2174/156800907780809723. [DOI] [PubMed] [Google Scholar]
- Mishima Y, Lindgren AG, Chizhikov VV, Johnson RL, Millen KJ. Overlapping function of Lmx1a and Lmx1b in anterior hindbrain roof plate formation and cerebellar growth. J Neurosci. 2009;29:11377–11384. doi: 10.1523/JNEUROSCI.0969-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagi C, Yamashita S, Ohba Y, Yoshizaki H, Matsuda M, Hirano T. STAT3 noncell-autonomously controls planar cell polarity during zebrafish convergence and extension. J Cell Biol. 2004;166:975–981. doi: 10.1083/jcb.200403110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto DM, Crowther RJ. Formation of the notochord in living ascidian embryos. J Embryol Exp Morphol. 1985;86:1–17. [PubMed] [Google Scholar]
- Morello R, Zhou G, Dreyer SD, Harvey SJ, Ninomiya Y, Thorner PS, Miner JH, Cole W, Winterpacht A, Zabel B, Oberg KC, Lee B. Regulation of glomerular basement membrane collagen expression by LMX1B contributes to renal disease in nail patella syndrome. Nat Genet. 2001;27:205–208. doi: 10.1038/84853. [DOI] [PubMed] [Google Scholar]
- Moret F, Christiaen L, Deyts C, Blin M, Vernier P, Joly JS. Regulatory gene expressions in the ascidian ventral sensory vesicle: evolutionary relationships with the vertebrate hypothalamus. Dev Biol. 2005;277:567–579. doi: 10.1016/j.ydbio.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Morley RH, Lachani K, Keefe D, Gilchrist MJ, Flicek P, Smith JC, Wardle FC. A gene regulatory network directed by zebrafish No tail accounts for its roles in mesoderm formation. Proc Natl Acad Sci U S A. 2009;106:3829–3834. doi: 10.1073/pnas.0808382106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro EM, Odell G. Morphogenetic pattern formation during ascidian notochord formation is regulative and highly robust. Development. 2002;129:1–12. doi: 10.1242/dev.129.1.1. [DOI] [PubMed] [Google Scholar]
- Oates AC, Wollberg P, Pratt SJ, Paw BH, Johnson SL, Ho RK, Postlethwait JH, Zon LI, Wilks AF. Zebrafish stat3 is expressed in restricted tissues during embryogenesis and stat1 rescues cytokine signaling in a STAT1-deficient human cell line. Dev Dyn. 1999;215:352–370. doi: 10.1002/(SICI)1097-0177(199908)215:4<352::AID-AJA7>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Oda-Ishii I, Di Gregorio A. Lineage-independent mosaic expression and regulation of the Ciona multidom gene in the ancestral notochord. Dev Dyn. 2007;236:1806–1819. doi: 10.1002/dvdy.21213. [DOI] [PubMed] [Google Scholar]
- Ott T, Parrish M, Bond K, Schwaeger-Nickolenko A, Monaghan AP. A new member of the spalt like zinc finger protein family, Msal-3, is expressed in the CNS and sites of epithelial/mesenchymal interaction. Mech Dev. 2001;101:203–207. doi: 10.1016/s0925-4773(00)00552-9. [DOI] [PubMed] [Google Scholar]
- Pascal A, Riou JF, Carron C, Boucaut JC, Umbhauer M. Cloning and developmental expression of STAT5 in Xenopus laevis. Mech Dev. 2001;106:171–174. doi: 10.1016/s0925-4773(01)00435-x. [DOI] [PubMed] [Google Scholar]
- Passamaneck YJ, Katikala L, Perrone L, Dunn MP, Oda-Ishii I, Di Gregorio A. Direct activation of a notochord cis-regulatory module by Brachyury and FoxA in the ascidian Ciona intestinalis. Development. 2009;136:3679–3689. doi: 10.1242/dev.038141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson R, Fleetwood J, Eaton S, Crossley M, Bao S. Kruppel-like transcription factors: a functional family. Int J Biochem Cell Biol. 2008;40:1996–2001. doi: 10.1016/j.biocel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- Pressman CL, Chen H, Johnson RL. LMX1B, a LIM homeodomain class transcription factor, is necessary for normal development of multiple tissues in the anterior segment of the murine eye. Genesis. 2000;26:15–25. [PubMed] [Google Scholar]
- Risbud MV, Schaer TP, Shapiro IM. Toward an understanding of the role of notochordal cells in the adult intervertebral disc: from discord to accord. Dev Dyn. 2010;239:2141–2148. doi: 10.1002/dvdy.22350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satou Y, Satoh N. Cataloging transcription factor and major signaling molecule genes for functional genomic studies in Ciona intestinalis. Dev Genes Evol. 2005;215:580–596. doi: 10.1007/s00427-005-0016-9. [DOI] [PubMed] [Google Scholar]
- Satou Y, Yamada L, Mochizuki Y, Takatori N, Kawashima T, Sasaki A, Hamaguchi M, Awazu S, Yagi K, Sasakura Y, Nakayama A, Ishikawa H, Inaba K, Satoh N. A cDNA resource from the basal chordate Ciona intestinalis. Genesis. 2002;33:153–154. doi: 10.1002/gene.10119. [DOI] [PubMed] [Google Scholar]
- Schwab JH, Boland PJ, Agaram NP, Socci ND, Guo T, O’Toole GC, Wang X, Ostroumov E, Hunter CJ, Block JA, Doty S, Ferrone S, Healey JH, Antonescu CR. Chordoma and chondrosarcoma gene profile: implications for immunotherapy. Cancer Immunol Immunother. 2009;58:339–349. doi: 10.1007/s00262-008-0557-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott A, Stemple DL. Zebrafish notochordal basement membrane: signaling and structure. Curr Top Dev Biol. 2005;65:229–253. doi: 10.1016/S0070-2153(04)65009-5. [DOI] [PubMed] [Google Scholar]
- Shapiro IM, Risbud MV. Transcriptional profiling of the nucleus pulposus: say yes to notochord. Arthritis Res Ther. 2010;12:117. doi: 10.1186/ar3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood DR, Butler JA, Kramer JM, Sternberg PW. FOS-1 promotes basement-membrane removal during anchor-cell invasion in C. elegans. Cell. 2005;121:951–962. doi: 10.1016/j.cell.2005.03.031. [DOI] [PubMed] [Google Scholar]
- Shi W, Peyrot SM, Munro E, Levine M. FGF3 in the floor plate directs notochord convergent extension in the Ciona tadpole. Development. 2009;136:23–28. doi: 10.1242/dev.029157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smidt MP, Asbreuk CH, Cox JJ, Chen H, Johnson RL, Burbach JP. A second independent pathway for development of mesencephalic dopaminergic neurons requires Lmx1b. Nat Neurosci. 2000;3:337–341. doi: 10.1038/73902. [DOI] [PubMed] [Google Scholar]
- Sohn P, Cox M, Chen D, Serra R. Molecular profiling of the developing mouse axial skeleton: a role for Tgfbr2 in the development of the intervertebral disc. BMC Dev Biol. 2010;10:29. doi: 10.1186/1471-213X-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, Larroux C, Lu DR, Mohanty K, Chapman J, Degnan BM, Rokhsar DS. Early evolution of the LIM homeobox gene family. BMC Biol. 2010;8:4. doi: 10.1186/1741-7007-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemple DL. Structure and function of the notochord: an essential organ for chordate development. Development. 2005;132:2503–2512. doi: 10.1242/dev.01812. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Aizawa K, Matsumura T, Nagai R. Vascular implications of the Kruppel-like family of transcription factors. Arterioscler Thromb Vasc Biol. 2005;25:1135–1141. doi: 10.1161/01.ATV.0000165656.65359.23. [DOI] [PubMed] [Google Scholar]
- Sweetman D, Munsterberg A. The vertebrate spalt genes in development and disease. Dev Biol. 2006;293:285–293. doi: 10.1016/j.ydbio.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Sweetman D, Smith TG, Farrell ER, Munsterberg A. Expression of csal1 in pre limb-bud chick embryos. Int J Dev Biol. 2005;49:427–430. doi: 10.1387/ijdb.051985ds. [DOI] [PubMed] [Google Scholar]
- Tada M, Casey ES, Fairclough L, Smith JC. Bix1, a direct target of Xenopus T-box genes, causes formation of ventral mesoderm and endoderm. Development. 1998;125:3997–4006. doi: 10.1242/dev.125.20.3997. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Hotta K, Erives A, Di Gregorio A, Zeller RW, Levine M, Satoh N. Brachyury downstream notochord differentiation in the ascidian embryo. Genes Dev. 1999;13:1519–1523. doi: 10.1101/gad.13.12.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassy O, Dauga D, Daian F, Sobral D, Robin F, Khoueiry P, Salgado D, Fox V, Caillol D, Schiappa R, Laporte B, Rios A, Luxardi G, Kusakabe T, Joly JS, Darras S, Christiaen L, Contensin M, Auger H, Lamy C, Hudson C, Rothbacher U, Gilchrist MJ, Makabe KW, Hotta K, Fujiwara S, Satoh N, Satou Y, Lemaire P. The ANISEED database: digital representation, formalization, and elucidation of a chordate developmental program. Genome Res. 2010;20:1459–1468. doi: 10.1101/gr.108175.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai TT, Danielson KG, Guttapalli A, Oguz E, Albert TJ, Shapiro IM, Risbud MV. TonEBP/OREBP is a regulator of nucleus pulposus cell function and survival in the intervertebral disc. J Biol Chem. 2006;281:25416–25424. doi: 10.1074/jbc.M601969200. [DOI] [PubMed] [Google Scholar]
- van der Windt AE, Haak E, Das RH, Kops N, Welting TJ, Caron MM, van Til NP, Verhaar JA, Weinans H, Jahr H. Physiological tonicity improves human chondrogenic marker expression through nuclear factor of activated T-cells 5 in vitro. Arthritis Res Ther. 2010;12:R100. doi: 10.1186/ar3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Zwaag B, Burbach JP, Scharfe C, Oefner PJ, Brunner HG, Padberg GW, van Bokhoven H. Identifying new candidate genes for hereditary facial paresis on chromosome 3q21-q22 by RNA in situ hybridization in mouse. Genomics. 2005;86:55–67. doi: 10.1016/j.ygeno.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Veeman MT, Nakatani Y, Hendrickson C, Ericson V, Lin C, Smith WC. Chongmague reveals an essential role for laminin-mediated boundary formation in chordate convergence and extension movements. Development. 2008;135:33–41. doi: 10.1242/dev.010892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vujovic S, Henderson S, Presneau N, Odell E, Jacques TS, Tirabosco R, Boshoff C, Flanagan AM. Brachyury, a crucial regulator of notochordal development, is a novel biomarker for chordomas. J Pathol. 2006;209:157–165. doi: 10.1002/path.1969. [DOI] [PubMed] [Google Scholar]
- Wada H. Origin and genetic evolution of the vertebrate skeleton. Zoolog Sci. 2010;27:119–123. doi: 10.2108/zsj.27.119. [DOI] [PubMed] [Google Scholar]
- Wada H, Okuyama M, Satoh N, Zhang S. Molecular evolution of fibrillar collagen in chordates, with implications for the evolution of vertebrate skeletons and chordate phylogeny. Evol Dev. 2006;8:370–377. doi: 10.1111/j.1525-142X.2006.00109.x. [DOI] [PubMed] [Google Scholar]
- Wada S, Tokuoka M, Shoguchi E, Kobayashi K, Di Gregorio A, Spagnuolo A, Branno M, Kohara Y, Rokhsar D, Levine M, Saiga H, Satoh N, Satou Y. A genomewide survey of developmentally relevant genes in Ciona intestinalis. II. Genes for homeobox transcription factors. Dev Genes Evol. 2003;213:222–234. doi: 10.1007/s00427-003-0321-0. [DOI] [PubMed] [Google Scholar]
- Wang Y, Ko BC, Yang JY, Lam TT, Jiang Z, Zhang J, Chung SK, Chung SS. Transgenic mice expressing dominant-negative osmotic-response element-binding protein (OREBP) in lens exhibit fiber cell elongation defect associated with increased DNA breaks. J Biol Chem. 2005;280:19986–19991. doi: 10.1074/jbc.M501689200. [DOI] [PubMed] [Google Scholar]
- Wilson L, Maden M. The mechanisms of dorsoventral patterning in the vertebrate neural tube. Dev Biol. 2005;282:1–13. doi: 10.1016/j.ydbio.2005.02.027. [DOI] [PubMed] [Google Scholar]
- Woodgett JR. Fos and jun: two into one will go. Semin Cancer Biol. 1990;1:389–397. [PubMed] [Google Scholar]
- Yagi K, Satou Y, Mazet F, Shimeld SM, Degnan B, Rokhsar D, Levine M, Kohara Y, Satoh N. A genomewide survey of developmentally relevant genes in Ciona intestinalis. III. Genes for Fox, ETS, nuclear receptors and NFkappaB. Dev Genes Evol. 2003;213:235–244. doi: 10.1007/s00427-003-0322-z. [DOI] [PubMed] [Google Scholar]
- Yagi K, Satou Y, Satoh N. A zinc finger transcription factor, ZicL, is a direct activator of Brachyury in the notochord specification of Ciona intestinalis. Development. 2004;131:1279–1288. doi: 10.1242/dev.01011. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Sakaguchi M, Medina RJ, Niida A, Sakaguchi Y, Miyazaki M, Kataoka K, Huh NH. Transcriptional regulation of a brown adipocyte-specific gene, UCP1, by KLF11 and KLF15. Biochem Biophys Res Commun. 2010;400:175–180. doi: 10.1016/j.bbrc.2010.08.039. [DOI] [PubMed] [Google Scholar]
- Yan R, Small S, Desplan C, Dearolf CR, Darnell JE., Jr Identification of a Stat gene that functions in Drosophila development. Cell. 1996;84:421–430. doi: 10.1016/s0092-8674(00)81287-8. [DOI] [PubMed] [Google Scholar]
- Yang XR, Ng D, Alcorta DA, Liebsch NJ, Sheridan E, Li S, Goldstein AM, Parry DM, Kelley MJ. T (brachyury) gene duplication confers major susceptibility to familial chordoma. Nat Genet. 2009;41:1176–1178. doi: 10.1038/ng.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger J, Kockel L, Peverali FA, Jackson DB, Mlodzik M, Bohmann D. Defective dorsal closure and loss of epidermal decapentaplegic expression in Drosophila fos mutants. EMBO J. 1997;16:7393–7401. doi: 10.1093/emboj/16.24.7393. [DOI] [PMC free article] [PubMed] [Google Scholar]