Abstract
Epigenetic mechanisms refer to the complex and interrelated molecular processes that dynamically modulate gene expression and function within every cell in the body. These regulatory systems represent the long-sought-after molecular interfaces that mediate gene × environment interactions. Changes in the epigenome throughout life are responsible not only for controlling normal development, adult homeostasis, and aging but also for mediating responses to injury. Emerging evidence implicates a spectrum of epigenetic processes in the pathophysiology of stroke. In this review, we describe conventional epigenetic mechanisms (including DNA methylation, histone code modifications, nucleosome remodeling, and higher-order chromatin formation) and highlight the emerging roles each of these processes play in the pathobiology of stroke. We suggest that understanding these mechanisms may be important for discovering more sensitive and specific biomarkers for risk, onset, and progression of stroke. In addition, we highlight epigenetic approaches for stroke therapy, including the inhibition of DNA methyltransferase and histone deacetylase enzyme activities. These therapeutic approaches are still in their infancy, but preliminary results suggest that contemporary agents targeting these pathways can regulate the deployment of stress responses that modulate neural cell viability and promote brain repair and functional reorganization. Indeed, these agents even appear to orchestrate sophisticated cognitive functions, including learning and memory.
Dynamic interactions between a diverse array of environmental, vascular, systemic, and central nervous system (CNS) factors underlie the pathogenesis of stroke and are responsible for mediating functional recovery. However, current strategies for prevention and treatment of stroke focus primarily on the modification of environmental risks, the acute and chronic management of vascular and hematological factors, and physical rehabilitation. These important interventions are effective, continuously being refined, and, particularly in the case of endovascular treatments, increasingly being adopted in practice. Nevertheless, stroke remains one of the leading causes of serious, long-term disability and death in the United States.1 Emerging therapeutic strategies have, therefore, focused on the development of neuroprotective and neural regenerative approaches to modifying the extent of CNS injury and restoring neurological function by preventing neuronal and glial injury and cell death, preserving the structural and functional integrity of neural networks, as well as enhancing tissue remodeling and repair. Although a spectrum of neuroprotective and neural regenerative treatments have appeared promising in preclinical studies, the clinical trials evaluating them have largely been equivocal or unsuccessful, which suggests that novel approaches are required for developing therapeutic strategies with greater efficacy.2
In the postgenomic era, the field of epigenetics is now poised to revolutionize modern medicine. Epigenetics is the study of molecular and cellular processes responsible for specifically modulating single gene expression and functional gene networks and also encompasses the long-sought-after molecular interface that mediates gene × environment interactions.3 Although the genome in each cell within the body is identical, cell- and tissue-specific profiles of gene transcription, posttranscriptional-RNA-processing (eg, RNA modifications, quality control, and transport), and translation are selectively regulated by multiple layers of interlaced epigenetic mechanisms that include DNA methylation; histone code modifications, nucleosome remodeling, and higher-order chromatin formation; noncoding RNA; and RNA editing. Dynamic changes occur in the epigenome throughout life. These changes control normal development, adult homeostasis, and aging and mediate responses to environmental stimuli (including diet, physical and chemical exposures, and behavioral and social factors).3 Recent studies have started to elucidate the key roles played by epigenetic mechanisms in the susceptibility to and the pathogenesis of complex diseases such as cancer and have demonstrated that identifying epigenetic biomarkers is important for risk stratification and molecular diagnosis.3 In addition, preliminary studies utilizing agents that target epigenetic pathways have suggested that these agents may be useful for treating a wide range of diseases, and a number of drugs already approved by the US Food and Drug Administration have direct or indirect effects on epigenetic mechanisms.4,5 Furthermore, a number of epigenetic agents are currently being evaluated in preclinical and clinical trials for CNS disorders.3
In the CNS, epigenetic mechanisms serve as key regulators of development, homeostasis, and plasticity, all of which are highly sensitive to local and more global environmental, vascular, systemic, and intrinsic CNS factors.3 Not surprisingly, epigenetic processes are involved in the molecular and cellular mechanisms underlying stroke pathogenesis and recovery, including the deployment of stress responses that modulate cell viability and promote tissue repair and functional reorganization. In this review, we highlight emerging evidence elucidating the role of epigenetic factors in stroke and suggest that understanding these processes may be critical for enhancing assessment of patient risk, early diagnosis, and characterization of clinically relevant molecular mechanisms associated with various stroke subtypes. Moreover, because epigenetic mechanisms are critical for brain patterning, neural stem cell maintenance, neurogenesis and gliogenesis, and synaptic and neural network plasticity and because they are also implicated in sophisticated cognitive functions (including learning and memory), we further suggest that systemic or even more local delivery of epigenetic therapeutic agents may permit the targeted activation of neural stem cells and other cell types present within the brain and promote the development of more effective neuroprotective and neural regenerative treatments for safeguarding and even restoring CNS function.
This is the first of a 3-part series describing the emerging role of epigenetics in stroke: part 1 covers DNA methylation and chromatin modifications; part 2 covers RNA regulatory circuitry; and part 3 covers neural stem cell biology and regenerative medicine.
EPIGENETIC MECHANISMS IMPLICATED IN STROKE
DNA Methylation
DNA methylation, the most well-characterized epigenetic mechanism, plays a critical role in the regulation of global and specific gene expression profiles and in the promotion of important cellular processes, such as the maintenance of genomic stability, X chromosome inactivation, and genomic imprinting (Table 1).6 Abnormal DNA methylation profiles have been associated with a broad spectrum of disorders, including stroke, atherosclerosis, obesity, insulin resistance, kidney disease, cancer, and autoimmunity.6,7 Mechanistically, DNA methylation inhibits the process of transcription and promotes binding of methyl-CpG-binding domain proteins, which recruit regulatory complexes containing epigenetic factors to methylated genomic loci in order to coordinately orchestrate reversible as well as long-term gene-silencing events. DNA methyltransferases (DNMTs) mediate DNA methylation by transferring methyl groups from S-adenosylmethionine to cytosine residues in various genomic regions. Members of this enzymatic family include DNMT3a and DNMT3b, which stimulate de novo methylation, and DNMT1, which actively maintains methylation. The expression levels and functions of these factors in neural cells are exquisitely regulated in an activity-dependent manner throughout development and adult life and are responsible for modulating neural subtype specification, maturation, and survival.8
Table 1.
Epigenetic Regulatory Mechanisms in Stroke
| Epigenetic Mechanisms | Description | Relevance to Stroke |
|---|---|---|
| DNA methylation | Refers to the transfer of methyl groups from SAM to cytosine residues in various genomic regions | Levels are increased in the ischemic brain and may be responsible for promoting neural cell death |
| Mediated by DNA methyltransferase enzymes | Deficiency of methylenetetrahydrofolate reductase, which is involved in the formation of SAM, causes hyperhomocysteinemia and an increased risk of stroke | |
| Regulates gene expression as well as diverse cellular processes, including maintenance of genomic stability, XCI, and genomic imprinting | Extent of XCI in female heterozygotes with Fabry disease determines clinical involvement, including risk of stroke | |
| Imprinted GNAS genomic locus is important for glucose and lipid metabolism and platelet function | ||
| Abnormal DNA methylation is associated with atherosclerosis, obesity, insulin resistance, kidney disease, cancer, and autoimmunity | ||
| Histone code modifications, nucleosome remodeling, and higher-order chromatin formation | Refer to highly integrated epigenetic mechanisms that modulate chromatin structure and function at single nucleotides (histone code modification), specific gene loci (nucleosome remodeling), and more extensive genomic regions (higher-order chromatin formation) | Histone acetylation levels are perturbed in the ischemic brain and may be associated with mediating neural cell death and protective responses, including excitotoxicity, oxidative stress, inflammation, cell cycle regulation, DNA repair, and apoptosis |
| Mediated by histone-, nucleosome-, and chromatin-modifying enzymes that are often components of large, multifunctional epigenetic macromolecular complexes | Abnormal chromatin is a key feature of necrotic cell death and apoptotic cell death, which are both associated with neural injury in stroke | |
| Play vital roles in executing genomic programs such as gene activation and silencing | Schimke immunoosseous dysplasia is a disease characterized by increased risk of stroke, which is caused by mutation of a nucleosome remodeling enzyme (ie, SMARCAL1) | |
| Antichromatin and antihistone antibodies are found in systemic lupus erythematosus, which is associated with an increased risk of stroke due to multiple factors | ||
| Chromatin dynamics are important for modulating cholesterol synthesis, transport, and metabolic pathways |
Abbreviations: SAM, S-adenosylmethionine; SMARCAL1, SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily A-like 1; XCI, X chromosome inactivation.
The role of DNA methylation in cerebral ischemia is multifaceted, with genome-wide and gene-specific effects that influence the vulnerability of the CNS to injury. Following middle cerebral artery occlusion (MCAO) in mice, DNA methylation levels are increased in ischemic brain tissue and may be responsible for promoting cell death.9,10 In fact, treatment with an inhibitor of DNA methylation reduces the extent of ischemic injury following MCAO. Furthermore, transgenic mice with reduced levels of neuronal DNMT1 exhibit significantly smaller infarcts following MCAO, compared with control animals. In contrast, mice without neuronal DNMT1 are not protected from cerebral ischemia. These observations suggest that the dynamic modulation of DNMT expression and the status of DNA methylation represent important mechanisms for preventing cell death in cerebral ischemia.
Another factor important for DNA methylation is methylenetetrahydrofolate reductase (MTHFR), which is involved in folate metabolism and in the formation of cellular reservoirs of the methyl group donor, S-adenosylmethionine. Intriguingly, MTHFR deficiency causes hyperhomocysteinemia and results in an increased risk of stroke and cardiovascular disease.11,12 Specific MTHFR gene polymorphisms (eg, C677T) are similarly associated with hyperhomocysteinemia and an increased risk of stroke, cardiovascular disease, neural developmental disorders, and a variety of other disease entities. The mechanisms that underlie this increased risk of stroke have not been clearly delineated, but differential MTHFR activity is associated with variations in global DNA methylation levels, which suggests that the risk may, in part, be linked to the effects of DNA methylation status on the vulnerability of the brain to ischemic injury.
Furthermore, other inherited causes of stroke are associated with cellular processes such as X chromosome inactivation and genomic imprinting that are mediated by DNA methylation. For example, Fabry disease is an X-linked disorder caused by deficiency of the lysosomal enzyme α galactosidase A, which results in glycosphingolipid accumulation in the vascular endothelium and can lead to stroke. Male hemizygotes are generally affected, but in female heterozygotes, the extent of X chromosome inactivation is the major factor that determines the severity of clinical involvement.13,14 Disorders of imprinted genomic loci may also be linked with stroke. For example, Prader-Willi syndrome, the archetypal disorder characterized by genomic imprinting, is associated with the moyamoya phenomenon.15 In addition, the highly complex imprinted GNAS genomic locus is important for the mediation of critical processes, including glucose and lipid metabolism and platelet function.16,17 Genetic variation at the GNAS locus is associated with a variety of disorders, including platelet dysfunction leading to coagulopathy and bleeding diathesis.18,19 Finally, studies of ischemic stroke heritability demonstrate that women with stroke are more likely than men with stroke to have a family history of stroke from their mothers than from their fathers that is independent of traditional vascular risk factors. This observation may, in part, be due to the effects of genomic imprinting.20
DNA methylation–mediated regulation of specific genes also plays a role in the pathophysiology of stroke. For example, DNA methylation influences expression of the pleiotropic factor thrombospondin 1 (THBS1), which exhibits complex temporal and cellular expression profiles in response to cerebral ischemia and intracerebral hemorrhage.21,22 Specifically, in murine cerebral endothelial cells, in vitro oxygen glucose deprivation is associated with an increase in THBS1 promoter methylation and a concurrent decrease in THBS1 expression. Conversely, reoxygenation after oxygen glucose deprivation is associated with a decrease in THBS1 promoter methylation and a concurrent increase in THBS1 expression. These findings imply that DNA methylation plays a role in the dynamic regulation of THBS1 in response to cerebral ischemia, which results in activation of signaling pathways that promote inflammation and cell death, suppression of angiogenesis, as well as enhancement of synaptic plasticity and functional recovery.23 Furthermore, differential DNA methylation profiles mediate sex differences in the endogenous neuroprotective response to MCAO. Dramatic upregulation of the estrogen receptor α (ERα) is an endogenous response to MCAO that attenuates ischemic cell death in young female rats.24 Although ERα is primarily expressed during neonatal development, MCAO induces selective demethylation of the ERα gene promoter in females, leading to the increase in ERα expression.25 Intriguingly, the neuroprotective effect of the ERα ligand, estrogen, is attenuated in aged animals, which may be the result of age-dependent changes in DNA methylation.26
We have selectively highlighted the contributions made by DNA methylation to stroke pathobiology. However, DNA methylation is increasingly being implicated in the modulation of homeostasis, cell cycle dynamics, cell viability, and stress responses impacting vascular, systemic, and intrinsic CNS factors that are also related to stroke but are beyond the scope of this review.
Histone Code Modifications, Nucleosome Remodeling, and Higher-Order Chromatin Formation
Chromatin is not simply a passive structure that packages DNA within the cell nucleus; rather, it mediates nuclear processes through local and more global structural and functional dynamics. Homeostatic and stress pathways that modulate cell viability are associated with active regulation of chromatin. Moreover, abnormal chromatin is a key feature of necrotic cell death and apoptotic cell death, which are both associated with neural injury in stroke. Although the different roles played by chromatin regulation in the pathophysiology of cerebral ischemia are not well characterized, emerging evidence suggests that these functions are extremely important and, furthermore, that chromatin-modifying agents may be neuroprotective.27
A complex series of highly integrated epigenetic mechanisms mediate the architecture of chromatin and play a vital role in executing genomic programs such as transcriptional activation and gene silencing.3,28–30 Histone code modifications are responsible for modulating chromatin structure and function at single nucleotides, nucleosome remodeling is responsible for modulating chromatin structure and function at specific gene loci, and higher-order chromatin formation is responsible for modulating chromatin structure and function at more extensive genomic regions.28–30 The histone code refers to profiles of posttranslational modifications of histone proteins (eg, acetylation, methylation, phosphorylation, ubiquitylation, SUMOylation, and adenosine diphosphate–ribosylation) that are catalyzed by specific enzymes (eg, histone acetyltransferases and histone deacetylases [HDACs]).28 Individual histone modifications have particular effects, such as the ability to activate or repress transcription, and the code formed by combinations of histone modifications defines the chromatin architecture and establishes the functional microdomains in the nucleus.28 Nucleosomes are the basic units of chromatin and are composed of DNA wrapped around a core of classic histones (eg, H2A, H2B, H3, and H4), linker histones (eg, H1), and variant histones (eg, H2A.Z). Nucleosome remodeling enzymes modify the conformation of DNA and these histone proteins, which promotes the local repositioning of nucleosomes that selectively alters accessibility of genomic regulatory regions to transcription factors and other proteins, and which may also lead to more widespread chromatin remodeling.30 Chromatin also forms higher-order structures, including loosely packaged euchromatin that is open and functionally active as well as more densely packaged and inactive heterochromatin that maintains genomic integrity.29 Histone-, nucleosome-, and chromatin-modifying enzymes are often components of large multifunctional epigenetic macromolecular complexes that integrate intracellular and intercellular cues from diverse signaling pathways and that have dynamic and wide-ranging regulatory effects on genomic structure and function.29
These epigenetic mechanisms are relevant for understanding the molecular pathophysiology of stroke. In fact, chromatin states and chromatin-modifying enzyme complexes modulate cholesterol synthesis, transport, and metabolic pathways, thus implicating epigenetic mechanisms in the development of atherosclerosis and in the determination of stroke risk.31–33 Also, sirtuins (SIRTs) are a family of HDAC enzymes that are implicated in mediating a range of functions (including cellular stress resistance, genomic stability, and energy metabolism) important in stroke.34 Indeed, the protective agent found in red wine (ie, resveratrol) activates SIRT1 and mimics the neuroprotective effects of “subthreshold” ischemic preconditioning in the brain.35 Furthermore, an abnormality of a chromatin-remodeling enzyme is directly linked to increased risk of stroke. Schimke immunoosseous dysplasia is an autosomal recessive multisystem disease caused by mutation of a gene that encodes the nucleosome remodeling enzyme SMARCAL1 (SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily A-like 1).36 Neurovascular manifestations, including stroke and moyamoya phenomenon, commonly occur in this disease. Moreover, nucleosomes are released from cells that are stressed and dying, and their levels in the peripheral circulation may be elevated in various pathological conditions (including stroke).37,38 Although it is a controversial question whether nucleosomes are markers of nonspecific cell death or more specific pathogenic mechanisms, the serum levels of nucleosomes measured after moderate to severe strokes correlate with infarct volume and also provide independent prognostic information.37,38 In addition, global DNA hypomethylation and antichromatin and antihistone antibodies are commonly found in systemic lupus erythematosus, which is associated with an increased risk of stroke as a result of multiple factors, including but not limited to the presence of antiphospholipid antibodies.39
Dynamic modulation of chromatin states is increasingly recognized as being associated with the molecular mechanisms (including excitotoxicity, oxidative stress, inflammation, cell cycle regulation, DNA repair, and apoptosis) that mediate neural cell death and protective responses in stroke. In fact, brain tissue subjected to MCAO exhibits significant changes in histone acetylation levels.40Furthermore, the heat shock response provoked by cerebral ischemia is mediated by factors such as Hsp70 (70-kDa heat shock protein), and histone acetylation in the Hsp70 promoter region is associated with transcriptional activation and subsequent initiation of pathways preventing neural cell death.41,42 Intriguingly, several preclinical studies have shown that the administration of various HDAC inhibitors in animal models of cerebral ischemia decreases the extent of neuronal injury and improves functional outcomes, partly as a result of the effects that these HDAC inhibitors have on Hsp70 promoter acetylation.40 Moreover, histone acetylation also plays a role in the protection of neurons against oxidative stress by indirectly promoting the function of neuroprotective antioxidant enzymes (ie, peroxiredoxins).43 These and other studies using HDAC inhibitors for stroke have recently been reviewed, and the cumulative evidence suggests that these agents are generally promising for promoting neuroprotection.27 In contrast, inhibiting HDAC1 enzyme activity can lead to DNA damage, cell-cycle deregulation, and neuronal death, whereas promoting HDAC1 enzyme activity has been shown to protect against DNA damage and neurotoxicity in cultured neurons and in vivo models of cerebral ischemia.44
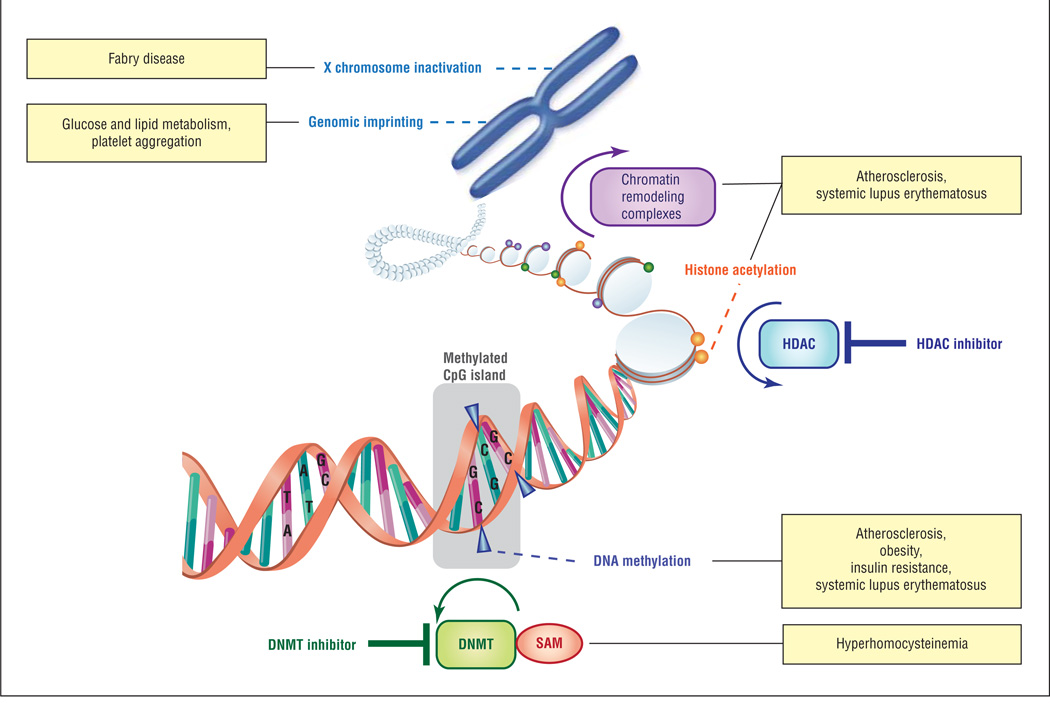
These observations highlight the complexity of chromatin structure and function and suggest that a detailed understanding of these epigenetic mechanisms, including the factors that permit gene-selective effects, is necessary for developing targeted epigenetic agents to prevent stroke and to treat stroke patients (Figure).
Figure.
Classic epigenetic mechanisms related to the pathogenesis of stroke. Schematic representing major classes of epigenetic mechanisms, sites of action for epigenetic therapeutic agents, and clinical conditions associated with stroke that result from deregulation of these epigenetic processes. DNMT indicates DNA methyltransferase; HDAC, histone deacetylase; SAM, S-adenosylmethionine.
THE ERA OF EPIGENOMIC MEDICINE: I
Contemporary Therapeutic Approaches
Drugs that target DNA methylation and histone modification pathways are already approved by the US Food and Drug Administration for the treatment of cancer and appear to be promising for other disorders (including hematological, immunological, and neuropsychiatric diseases). Pharmacological agents that affect these epigenetic mechanisms can reprogram cells and tissues with aberrant gene expression and function associated with various disease states.4 For example, these therapies act partly by correcting pathological changes in the balance between tumor suppressor genes and oncogenes in cancer and by reactivating the expression of an embryonic gene to compensate for a mutated adult gene in sickle cell disease.
Epigenetic agents are actively being evaluated in preclinical studies targeting stroke syndromes; however, these therapeutic approaches are still in their infancy (Table2). Among the most well-characterized epigenetic drugs are 5-azacytidine, 5-aza-2-deoxycytidine (or decitabine), and zebularine, which act as analogs of the nucleoside cytosine and nonspecifically inhibit the function of DNMT enzymes. Studies of cellular therapies for stroke have used a series of factors (including 5-azacytidine) to induce adipose stromal cells to differentiate into neuron-like cells and have transplanted these epigenetically engineered cells into animal models of stroke. Some of these exogenous cells survive, and neurological outcomes are improved; however, infarct volumes are not significantly reduced.45,46 In contrast, other in vivo studies have used 5-aza-2-deoxycytidine to treat animals with MCAO and have shown improved neurological outcomes with a reduction in the extent of ischemic injury, which suggests that epigenetic reprogramming of endogenous cells is a more effective treatment strategy than transplantation of exogenous cells.9 Zebularine has not been studied directly in stroke, but indirect evidence suggests that it can also play a role in CNS functions.47 These first-generation DNMT inhibitors are nonspecific and are associated with significant toxicity; therefore, more selective second-generation agents that affect DNA methylation are being developed.48 For example, MG98 is an antisense oligonucleotide designed to specifically target DNMT1 messenger RNA and inhibit its translation.48 Moreover, a number of commonly used drugs also have the ability to alter DNA methylation profiles. Agents such as hydralazine, procainamide, and valproic acid are associated with effects on DNA methylation that may be important for their clinical efficacy and toxicity.5 For example, the use of these epigenetic drugs can cause DNA hypomethylation and can also provoke adverse consequences such as the development of a syndrome similar to systemic lupus erythematosus, which is itself associated with global DNA hypomethylation as well as cerebrovascular complications.49
Table 2.
Contemporary Epigenetic Therapeutic Approaches for Stroke
| Epigenetic Mechanisms of Action | Agents | Relevance to Stroke |
|---|---|---|
| Inhibition of DNMT enzyme activity | 5-Azacytidine | Treatment with an inhibitor of DNA methylation reduces the extent of ischemic injury following MCAO |
| 5-Aza-2-deoxycytidine (or decitabine), zebularine, and MG98 | Mice with reduced levels of DNMT1 exhibit significantly smaller infarcts following MCAO, compared with control animals | |
| Inhibition of HDAC enzyme activity | Trichostatin A | Neuroprotective mechanisms affected by HDAC inhibition include the critical cellular processes that control growth and viability and stress responses |
| Suberoylanilide hydroxamic acid, sodium butyrate, sodium 4-phenylbutyrate, valproic acid, and curcumin | Paradigm for the restoration of impaired neural network connections and the recovery of lost neurological functions, including learning and memory |
Abbreviations: DNMT, DNA methyltransferase; HDAC, histone deacetylase; MCAO, middle cerebral artery occlusion.
In addition to agents that affect DNA methylation, a number of drugs that target histone modification pathways are also available. The majority of these agents are HDAC inhibitors, and preclinical studies in stroke models have used a range of HDAC inhibitors (including trichostatin A, suberoylanilide hydroxamic acid, sodium butyrate, sodium 4-phenylbutyrate, and valproic acid) to demonstrate that these agents can decrease the extent of neuronal injury and improve functional outcomes.27 Each of these HDAC inhibitors exhibits a varying degree of activity against different classes of HDAC enzymes, and several novel inhibitors are being developed for different classes and isotypes of HDACs. The precise neuroprotective mechanisms induced by HDAC inhibition are complex and include a broad array of factors implicated in mediating critical cellular processes, such as growth and viability (eg, p53) and stress responses (eg, Hsp70, inducible nitric oxide synthase, and tumor necrosis factor α).27 In addition to neuroprotective effects, HDAC inhibition also offers a paradigm for the restoration of impaired neural network connections and the recovery of seemingly lost neurological functions, including learning and memory.50 Indeed, histone acetylation has been implicated in mediating higher-order cognitive functions, and HDAC2 is specifically responsible for modulating the dendritic spine density, the number of synapses, and synaptic plasticity, all of which underlie learning and memory. Histone deacetylase inhibition and genetic manipulation of HDAC2 promote synaptic and neural network plasticity, which suggests that HDAC2-selective inhibitors may be particularly valuable for treating cognitive impairment.50 These studies have recently been discussed in an excellent review detailing the emerging role of HDAC inhibitors in the treatment of stroke.27
Future Therapeutic Strategies
Contemporary therapeutic approaches have shown the potential to prevent neural cell death and rescue CNS functions after injury; however, their range of actions are limited, and more sophisticated and combinatorial strategies will likely be necessary to combat complex disorders like stroke using dynamic epigenetic reprogramming. Existing agents exert relatively nonspecific effects directed toward a restricted number of epigenetic processes. In stark contrast, the epigenome is responsible for orchestrating and exquisitely calibrating the cell- and tissue-specific deployment of genes and functional gene networks through a much wider array of interrelated molecular mechanisms. These epigenetic processes may be globally or selectively deregulated in various stroke syndromes, as highlighted throughout this review. We suggest, therefore, that future therapeutic strategies for stroke, including those with the capacity to selectively activate subpopulations of endogenous regional neural stem cells and to promote neural regenerative programs, must encompass a spectrum of technologies with the potential to modulate the entire epigenome with high degrees of specificity, flexibility, efficacy, and tolerability through both systemic delivery and more localized endovascular approaches. These next-generation agents may include novel oligonucleotides and RNA-based therapies engineered in a sequence-specific manner to promote epigenetic reprogramming at precise genomic loci. These innovative strategies will help us to overcome the limitations of current, nonselective approaches for targeting DNA methylation and histone modification pathways. Furthermore, these strategies have the potential to enhance the activation of neuroprotective and neural regenerative genes and suppress genes that promote neural cell death.
Acknowledgments
Funding/Support: Dr Mehler is supported by grants NS38902 and MH66290 from the National Institutes of Health as well as by the Roslyn and Leslie Goldstein, the Mildred and Bernard H. Kayden, the F. M. Kirby, and the Alpern Family foundations.
Footnotes
Author Contributions: Study concept and design: Qureshi and Mehler. Acquisition of data: Qureshi and Mehler. Analysis and interpretation of data: Qureshi and Mehler. Drafting of the manuscript: Qureshi and Mehler. Critical revision of the manuscript for important intellectual content: Qureshi and Mehler. Obtained funding: Mehler. Administrative, technical, and material support: Qureshi and Mehler. Study supervision: Qureshi and Mehler.
Financial Disclosure: None reported.
REFERENCES
- 1.Lloyd-Jones D, Adams R, Carnethon M, et al. American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Heart disease and stroke statistics—2009 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 2009;119(3):480–486. doi: 10.1161/CIRCULATIONAHA.108.191259. [DOI] [PubMed] [Google Scholar]
- 2.Ginsberg MD. Current status of neuroprotection for cerebral ischemia: synoptic overview. Stroke. 2009;40(3) Suppl:S111–S114. doi: 10.1161/STROKEAHA.108.528877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mehler MF. Epigenetic principles and mechanisms underlying nervous system functions in health and disease. Prog Neurobiol. 2008;86(4):305–341. doi: 10.1016/j.pneurobio.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Szyf M. Epigenetics, DNA methylation, and chromatin modifying drugs. Annu Rev Pharmacol Toxicol. 2009;49:243–263. doi: 10.1146/annurev-pharmtox-061008-103102. [DOI] [PubMed] [Google Scholar]
- 5.Csoka AB, Szyf M. Epigenetic side-effects of common pharmaceuticals: a potential new field in medicine and pharmacology. Med Hypotheses. 2009;73(5):770–780. doi: 10.1016/j.mehy.2008.10.039. [DOI] [PubMed] [Google Scholar]
- 6.Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6(8):597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 7.Gluckman PD, Hanson MA, Buklijas T, Low FM, Beedle AS. Epigenetic mechanisms that underpin metabolic and cardiovascular diseases. Nat Rev Endocrinol. 2009;5(7):401–408. doi: 10.1038/nrendo.2009.102. [DOI] [PubMed] [Google Scholar]
- 8.Sharma RP, Tun N, Grayson DR. Depolarization induces downregulation of DNMT1 and DNMT3a in primary cortical cultures. Epigenetics. 2008;3(2):74–80. doi: 10.4161/epi.3.2.6103. [DOI] [PubMed] [Google Scholar]
- 9.Endres M, Meisel A, Biniszkiewicz D, et al. DNA methyltransferase contributes to delayed ischemic brain injury. J Neurosci. 2000;20(9):3175–3181. doi: 10.1523/JNEUROSCI.20-09-03175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Endres M, Fan G, Meisel A, Dirnagl U, Jaenisch R. Effects of cerebral ischemia in mice lacking DNA methyltransferase 1 in post-mitotic neurons. Neuroreport. 2001;12(17):3763–3766. doi: 10.1097/00001756-200112040-00032. [DOI] [PubMed] [Google Scholar]
- 11.Casas JP, Hingorani AD, Bautista LE, Sharma P. Meta-analysis of genetic studies in ischemic stroke: thirty-two genes involving approximately 18,000 cases and 58,000 controls. Arch Neurol. 2004;61(11):1652–1661. doi: 10.1001/archneur.61.11.1652. [DOI] [PubMed] [Google Scholar]
- 12.Kelly PJ, Rosand J, Kistler JP, et al. Homocysteine, MTHFR 677C-->T polymorphism, and risk of ischemic stroke: results of a meta-analysis. Neurology. 2002;59(4):529–536. doi: 10.1212/wnl.59.4.529. [DOI] [PubMed] [Google Scholar]
- 13.Giacomini PS, Shannon PT, Clarke JT, Jaigobin C. Fabry’s disease presenting as stroke in a young female. Can J Neurol Sci. 2004;31(1):112–114. doi: 10.1017/s0317167100002936. [DOI] [PubMed] [Google Scholar]
- 14.Dobrovolny R, Dvorakova L, Ledvinova J, et al. Relationship between X-inactivation and clinical involvement in Fabry heterozygotes: eleven novel mutations in the alpha-galactosidase A gene in the Czech and Slovak population. J Mol Med. 2005;83(8):647–654. doi: 10.1007/s00109-005-0656-2. [DOI] [PubMed] [Google Scholar]
- 15.Kusuhara T, Ayabe M, Hino H, Shoji H, Neshige R. A case of Prader-Willi syndrome with bilateral middle cerebral artery occlusion and moyamoya phenomenon [in Japanese] Rinsho Shinkeigaku. 1996;36(6):770–773. [PubMed] [Google Scholar]
- 16.Chen M, Gavrilova O, Liu J, et al. Alternative Gnas gene products have opposite effects on glucose and lipid metabolism. Proc Natl Acad Sci U S A. 2005;102(20):7386–7391. doi: 10.1073/pnas.0408268102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freson K, Izzi B, Labarque V, et al. GNAS defects identified by stimulatory G protein alpha-subunit signalling studies in platelets. J Clin Endocrinol Metab. 2008;93(12):4851–4859. doi: 10.1210/jc.2008-0883. [DOI] [PubMed] [Google Scholar]
- 18.Freson K, Jaeken J, Van Helvoirt M, et al. Functional polymorphisms in the paternally expressed XLalphas and its cofactor ALEX decrease their mutual interaction and enhance receptor-mediated cAMP formation. Hum Mol Genet. 2003;12(10):1121–1130. doi: 10.1093/hmg/ddg130. [DOI] [PubMed] [Google Scholar]
- 19.Freson K, Izzi B, Jaeken J, et al. Compound heterozygous mutations in the GNAS gene of a boy with morbid obesity, thyroid-stimulating hormone resistance, pseudohypoparathyroidism, and a prothrombotic state. J Clin Endocrinol Metab. 2008;93(12):4844–4849. doi: 10.1210/jc.2008-0233. [DOI] [PubMed] [Google Scholar]
- 20.Touzé E, Rothwell PM. Sex differences in heritability of ischemic stroke: a systematic review and meta-analysis. Stroke. 2008;39(1):16–23. doi: 10.1161/STROKEAHA.107.484618. [DOI] [PubMed] [Google Scholar]
- 21.Hu CJ, Chen SD, Yang DI, et al. Promoter region methylation and reduced expression of thrombospondin-1 after oxygen-glucose deprivation in murine cerebral endothelial cells. J Cereb Blood Flow Metab. 2006;26(12):1519–1526. doi: 10.1038/sj.jcbfm.9600304. [DOI] [PubMed] [Google Scholar]
- 22.Zhou HJ, Zhang HN, Tang T, et al. Alteration of thrombospondin-1 and -2 in rat brains following experimental intracerebral hemorrhage. J Neurosurg. doi: 10.3171/2010.1.JNS09637. [published online February 5, 2010] [DOI] [PubMed] [Google Scholar]
- 23.Xing C, Lee S, Kim WJ, et al. Neurovascular effects of CD47 signaling: promotion of cell death, inflammation, and suppression of angiogenesis in brain endothelial cells in vitro. J Neurosci Res. 2009;87(11):2571–2577. doi: 10.1002/jnr.22076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson ME, Westberry JM. Regulation of oestrogen receptor gene expression: new insights and novel mechanisms. J Neuroendocrinol. 2009;21(4):238–242. doi: 10.1111/j.1365-2826.2009.01830.x. [DOI] [PubMed] [Google Scholar]
- 25.Westberry JM, Prewitt AK, Wilson ME. Epigenetic regulation of the estrogen receptor alpha promoter in the cerebral cortex following ischemia in male and female rats. Neuroscience. 2008;152(4):982–989. doi: 10.1016/j.neuroscience.2008.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sunday L, Osuna C, Krause DN, Duckles SP. Age alters cerebrovascular inflammation and effects of estrogen. Am J Physiol Heart Circ Physiol. 2007;292(5):H2333–H2340. doi: 10.1152/ajpheart.01057.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langley B, Brochier C, Rivieccio MA. Targeting histone deacetylases as a multifaceted approach to treat the diverse outcomes of stroke. Stroke. 2009;40(8):2899–2905. doi: 10.1161/STROKEAHA.108.540229. [DOI] [PubMed] [Google Scholar]
- 28.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 29.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 30.Cairns BR. The logic of chromatin architecture and remodelling at promoters. Nature. 2009;461(7261):193–198. doi: 10.1038/nature08450. [DOI] [PubMed] [Google Scholar]
- 31.Gilardi F, Mitro N, Godio C, et al. The pharmacological exploitation of cholesterol 7alpha-hydroxylase, the key enzyme in bile acid synthesis: from binding resins to chromatin remodelling to reduce plasma cholesterol. Pharmacol Ther. 2007;116(3):449–472. doi: 10.1016/j.pharmthera.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 32.Shafaati M, O’Driscoll R, Björkhem I, Meaney S. Transcriptional regulation of cholesterol 24-hydroxylase by histone deacetylase inhibitors. Biochem Biophys Res Commun. 2009;378(4):689–694. doi: 10.1016/j.bbrc.2008.11.103. [DOI] [PubMed] [Google Scholar]
- 33.Fang S, Miao J, Xiang L, Ponugoti B, Treuter E, Kemper JK. Coordinated recruitment of histone methyltransferase G9a and other chromatin-modifying enzymes in SHP-mediated regulation of hepatic bile acid metabolism. Mol Cell Biol. 2007;27(4):1407–1424. doi: 10.1128/MCB.00944-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460(7255):587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raval AP, Dave KR, Pérez-Pinzón MA. Resveratrol mimics ischemic preconditioning in the brain. J Cereb Blood Flow Metab. 2006;26(9):1141–1147. doi: 10.1038/sj.jcbfm.9600262. [DOI] [PubMed] [Google Scholar]
- 36.Deguchi K, Clewing JM, Elizondo LI, et al. Neurologic phenotype of Schimke immunoosseous dysplasia and neurodevelopmental expression of SMARCAL1. J Neuropathol Exp Neurol. 2008;67(6):565–577. doi: 10.1097/NEN.0b013e3181772777. [DOI] [PubMed] [Google Scholar]
- 37.Geiger S, Holdenrieder S, Stieber P, et al. Nucleosomes in serum of patients with early cerebral stroke. Cerebrovasc Dis. 2006;21(1–2):32–37. doi: 10.1159/000089591. [DOI] [PubMed] [Google Scholar]
- 38.Geiger S, Holdenrieder S, Stieber P, et al. Nucleosomes as a new prognostic marker in early cerebral stroke. J Neurol. 2007;254(5):617–623. doi: 10.1007/s00415-006-0407-5. [DOI] [PubMed] [Google Scholar]
- 39.Muller S, Dieker J, Tincani A, Meroni PL. Pathogenic anti-nucleosome antibodies. Lupus. 2008;17(5):431–436. doi: 10.1177/0961203308090030. [DOI] [PubMed] [Google Scholar]
- 40.Faraco G, Pancani T, Formentini L, et al. Pharmacological inhibition of histone deacetylases by suberoylanilide hydroxamic acid specifically alters gene expression and reduces ischemic injury in the mouse brain. Mol Pharmacol. 2006;70(6):1876–1884. doi: 10.1124/mol.106.027912. [DOI] [PubMed] [Google Scholar]
- 41.Giffard RG, Han RQ, Emery JF, Duan M, Pittet JF. Regulation of apoptotic and inflammatory cell signaling in cerebral ischemia: the complex roles of heat shock protein 70. Anesthesiology. 2008;109(2):339–348. doi: 10.1097/ALN.0b013e31817f4ce0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao YM, Chen X, Sun H, et al. Effects of histone deacetylase inhibitors on transcriptional regulation of the hsp70 gene in Drosophila. Cell Res. 2006;16(6):566–576. doi: 10.1038/sj.cr.7310074. [DOI] [PubMed] [Google Scholar]
- 43.Soriano FX, Papadia S, Bell KF, Hardingham GE. Role of histone acetylation in the activity-dependent regulation of sulfiredoxin and sestrin 2. Epigenetics. 2009;4(3):152–158. doi: 10.4161/epi.4.3.8753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim D, Frank CL, Dobbin MM, et al. Deregulation of HDAC1 by p25/Cdk5 in neurotoxicity. Neuron. 2008;60(5):803–817. doi: 10.1016/j.neuron.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kang SK, Lee DH, Bae YC, Kim HK, Baik SY, Jung JS. Improvement of neurological deficits by intracerebral transplantation of human adipose tissue-derived stromal cells after cerebral ischemia in rats. Exp Neurol. 2003;183(2):355–366. doi: 10.1016/s0014-4886(03)00089-x. [DOI] [PubMed] [Google Scholar]
- 46.Lee TH, Yoon JG. Intracerebral transplantation of human adipose tissue stromal cells after middle cerebral artery occlusion in rats. J Clin Neurosci. 2008;15(8):907–912. doi: 10.1016/j.jocn.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 47.Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53(6):857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 48.Plummer R, Vidal L, Griffin M, et al. Phase I study of MG98, an oligonucleotide antisense inhibitor of human DNA methyltransferase 1, given as a 7-day infusion in patients with advanced solid tumors. Clin Cancer Res. 2009;15(9):3177–3183. doi: 10.1158/1078-0432.CCR-08-2859. [DOI] [PubMed] [Google Scholar]
- 49.Szyf M. Epigenetic therapeutics in autoimmune disease. Clin Rev Allergy Immunol. 2010;39(1):62–77. doi: 10.1007/s12016-009-8172-8. [DOI] [PubMed] [Google Scholar]
- 50.Guan JS, Haggarty SJ, Giacometti E, et al. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459(7243):55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]