Abstract
Under favorable conditions DNA can survive for thousands of years in the remains of dead organisms. The DNA extracted from such remains is invariably degraded to a small average size by processes that at least partly involve depurination. It also contains large amounts of deaminated cytosine residues that are accumulated toward the ends of the molecules, as well as several other lesions that are less well characterized.
DNA fragments from ancient specimens are short (40–500 bp) and contain lesions that block DNA polymerases and cause replication errors. Degradation involves depurination and cytosine deamination, but other processes may be involved.
In living cells, DNA molecules continuously suffer chemical insults, which are countered by enzymatic repair mechanisms that maintain the integrity of the genome (Lindahl 1993). On death, these cellular repair mechanisms cease to function. As a consequence, the genome becomes exposed to the unmitigated effects of numerous factors that threaten its stability. These factors include intracellular nucleases, which are no longer sequestered in the cell and can thus gain access to DNA and degrade it, as well as microorganisms that spread in the decaying tissues. Together these factors may lead to the loss of all retrievable DNA. However, under favorable environmental conditions, for example when tissues are frozen or become desiccated quickly after death, these processes become inhibited before the complete destruction of all DNA endogenous to the organism. In these instances other destructive factors, particularly hydrolytic and oxidative processes, become limiting to the time that DNA survives in a tissue.
When DNA is extracted and analyzed from ancient samples these destructive factors manifest themselves in three different ways: (i) a reduction in DNA fragment size, (ii) lesions that block the replication of the DNA molecules by polymerases, thus impeding many forms of analysis, and (iii) lesions that cause incorrect nucleotides to be incorporated when the DNA is replicated. Here, we summarize what is known about each of these forms of damage in ancient DNA.
FRAGMENTATION
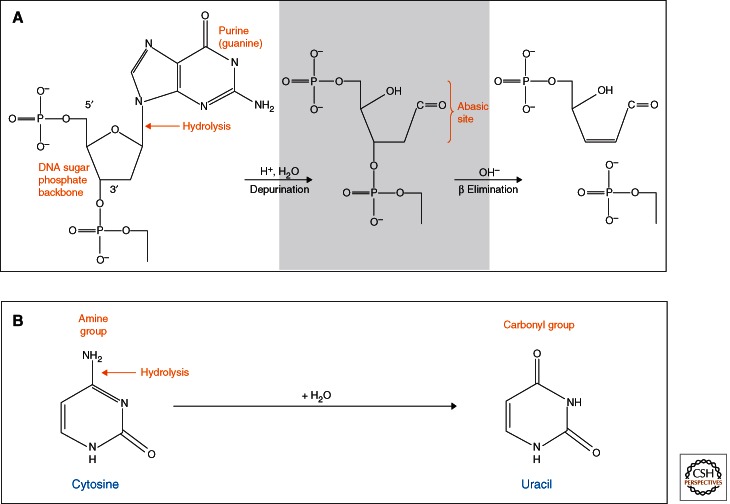
Already in the first systematic study of the properties of ancient DNA (Pääbo 1989), it was shown that almost all DNA extracted from samples varying in age between 4 and 13,000 years was degraded to fragments of 40–500 bp. Subsequent work has confirmed that this is a general feature of DNA extracted from almost all ancient remains. Based on in vitro experiments using modern DNA (Lindahl and Andersson 1972; Lindahl and Nyberg 1972), it has been suggested that fragmentation is owing to hydrolytic depurination and subsequent β elimination resulting in single-strand breaks (Fig. 1) (Pääbo and Wilson 1991; Lindahl 1993). Indeed, alkali treatment of ancient DNA extracts indicates the presence of terminal 5′-phosphate and aldehydic 3′ ends, which are products of β elimination (Jones et al. 1968).
Figure 1.
Fragmentation and deamination. (A) A likely cause of fragmentation in ancient DNA is depurination, in which the N-glycosyl bond between a sugar and an adenine or guanine residue is cleaved, resulting in an abasic site. The DNA strand is then fragmented through β elimination, leaving 3′-aldehydic and 5′-phosphate ends. (B) Deamination of cytosine to uracil is the major mechanism leading to miscoding lesions in ancient DNA. DNA polymerases will incorporate an A across from the U, and in turn a T across from the A, causing apparent G to A and C to T substitutions.
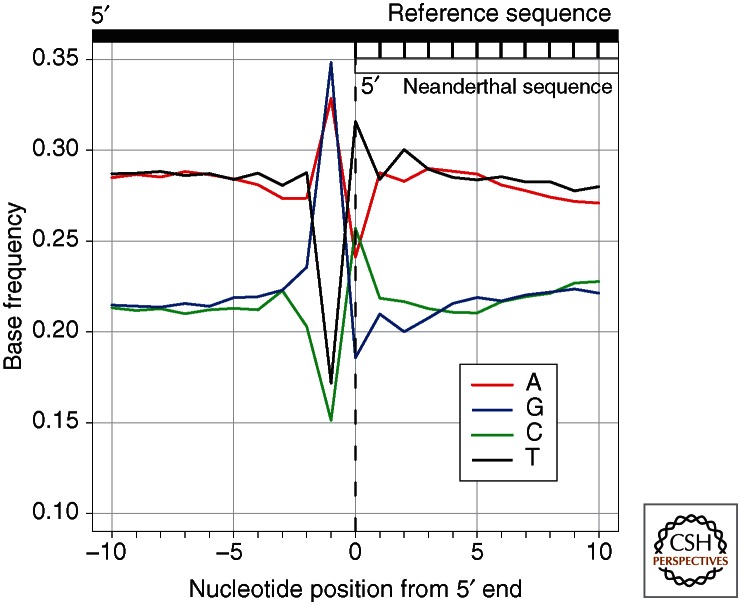
During the last decade, high-throughput sequencing methods have allowed a better understanding of fragmentation patterns in ancient DNA. This is because these methods rely on the ligation of DNA adaptors to the ends of DNA molecules to construct sequencing libraries (Margulies et al. 2005). Subsequent sequencing from the adaptors allows the location of strand breaks to be determined as long as the terminal nucleotides are amenable to ligation and not removed by enzymatic treatments used during library preparation. Using this approach and a reference genome to infer the bases immediately adjacent to the ancient DNA fragments, it was shown that the purines adenine (A) and guanine (G) are overrepresented next to the 5′ ends of DNA fragments extracted from the remains of Neanderthals, mammoths, and cave bears, which were all ∼40,000 years old (Fig. 2) (Briggs et al. 2007). In Pleistocene horse remains preserved in permafrost (Orlando et al. 2011) it was shown that G residues were even more overrepresented than A residues adjacent to the 5′ ends. A possible explanation for the preferential depurination of G over A residues is a resonance structure in guanine that may lower the activation energy required to break its N-glycosyl bond (Overballe-Petersen et al. 2012).
Figure 2.
Base frequencies of the human reference genome around 5′ ends of Neanderthal DNA fragments. An excess of A and G immediately outside of the Neanderthal sequence indicates that an elevated number of DNA fragments begin adjacent to purines, supporting the hypothesis that depurination is a major mechanism of fragmentation of ancient DNA.
Most methods for preparing DNA libraries for sequencing involve blunt-end repair, an enzymatic process that extends recessed and degrades overhanging 3′ ends of DNA fragments, thus precluding the determination of the exact location of 3′ ends (Briggs et al. 2007). A first attempt to determine the sequence context around 3′ ends was made using a sequencing technology that sequences from polyadenylated, but otherwise unmodified, 3′ ends (Orlando et al. 2011). For permafrost-preserved Pleistocene horse DNA this yielded no indication that strand breakage occurs preferentially at any particular nucleotide. However, this may be owing to inefficient poly(A)-tailing of aldehydic 3′ ends, which would exclude fragments generated by β elimination (Orlando et al. 2011). A recently developed DNA library preparation method in which adaptors are ligated to single-stranded DNA, preserves both the 5′ and 3′ ends and has confirmed that purines, especially guanine residues, are overrepresented immediately adjacent to both ends of ancient DNA fragments in DNA extracted from bones preserved for tens of thousands of years in caves (Meyer et al. 2012).
BLOCKING LESIONS
Some DNA modifications obstruct the movement of DNA polymerases along a template strand, preventing their amplification and sequencing. Such blocking lesions occur in the form of nucleotide modifications and cross-links, which can form either between DNA strands, between different DNA fragments, or between DNA and other molecules.
Several nucleotide modifications block polymerase-mediated DNA synthesis. In an analysis of DNA extracted from 11 samples representing various permafrost and nonpermafrost preservation conditions, it was shown that different amounts of 5-hydroxy-5-methylhydantoin and 5-hydroxyhydantoin, which are oxidation products of pyrimidines, were present in all samples. Amplification of mitochondrial DNA was only possible in five of the samples that contained threefold to 29-fold less hydantoins than samples in which amplification failed (Höss et al. 1996). Little is known about other blocking lesions in ancient DNA although primer extension experiments have shown an elevated termination frequency at guanine residues in ancient samples (Heyn et al. 2010), suggesting that a modification of guanine may also act as a blocking lesion.
Early work using electron microscopy identified intermolecular cross-links in ancient DNA (Pääbo 1989). In addition, it has been shown by gas chromatography/mass spectrometry (GC/MS) that furanones, furaldehydes, and alkylpyrazines—products of the Maillard reaction, which can produce cross-links between macromolecules like DNA and proteins—are present in 20,000-yr-old sloth coprolites (Poinar et al. 1998). Amplification of a short mitochondrial DNA fragment from the coprolites was possible only after treatment with N-phenacylthiazolium bromide (N-PTB), a compound that cleaves Maillard products. However, although N-PTB is helpful in some instances, subsequent studies have reported less success using N-PTB treatments to improve amplification yield (Rohland and Hofreiter 2007; Binladen and Willerslev 2010).
It has also been suggested, based on the refractivity of permafrost-derived ancient DNA to heat denaturation, that cross-links accumulate about 100 times faster than single-strand breaks in such DNA (Hansen et al. 2006). In contrast, primer extensions on ancient DNA fragments that were 27,000–48,000 years old and came from three bones preserved in permafrost and one bone from a cave site concluded that all blocking lesions, including cross-links, were present in no more than 40% of molecules (Heyn et al. 2010). Thus, there is currently conflicting evidence about the extent to which blocking lesions are present in ancient DNA.
MISCODING LESIONS
Nucleotide bases are susceptible to hydrolytic deamination, resulting in modifications that cause them to be misread by DNA polymerases. A primary target of deamination is cytosine. Its product, uracil (Fig. 1), will direct the incorporation of adenine (A) during DNA replication, resulting in apparent C to T or G to A substitutions (depending on the strand sequenced). Because ancient DNA is sensitive to uracil-N-glycosylase (UNG) it has been inferred that it contains uracil residues (Pääbo 1989). Furthermore, asymmetric PCR that amplifies only one DNA strand has shown that the large majority of nucleotide substitutions in ancient DNA sequences are from C to T and that treatment with UNG results in a substantial reduction of such substitutions (Hofreiter et al. 2001). Some investigators have proposed that adenine is deaminated to hypoxanthine in ancient DNA, causing apparent A to G changes during amplification (Hansen et al. 2001; Gilbert et al. 2003). Although this may occur at some level, many of these substitutions are likely to be the result of damage-independent misincorporations by the Taq polymerase used for PCR (Hofreiter et al. 2001; Pääbo et al. 2004; Binladen et al. 2006).
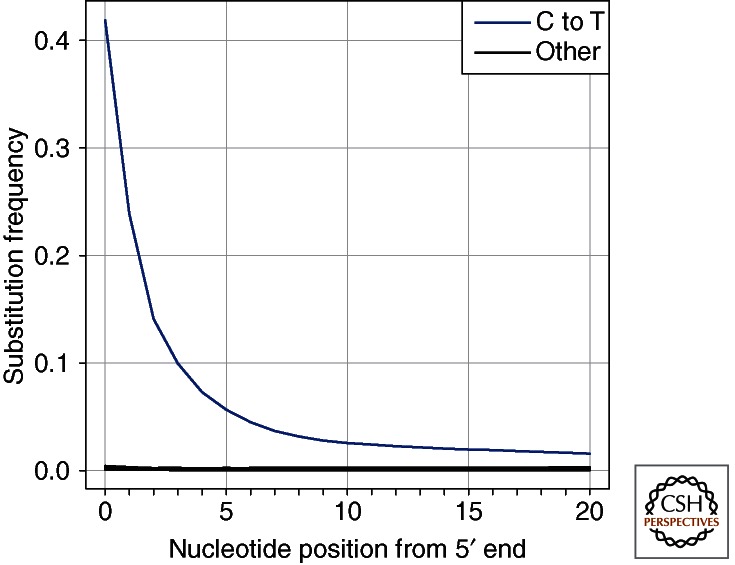
High-throughput sequencing of libraries constructed from ancient DNA has confirmed that C to T substitutions are greatly increased in number over other substitutions (Stiller et al. 2006; Gilbert et al. 2007). Moreover, it has been found that these substitutions are primarily localized to fragment ends where up to 40% of all cytosines appear as thymines, followed by an exponential decrease of such substitutions along the DNA molecule (Fig. 3) (Briggs et al. 2007; Brotherton et al. 2007). Notably, blunt-end repair by T4 DNA polymerase used to prepare the DNA sequencing libraries results in the elimination of overhanging 3′ ends, whereas overhanging 5′ ends are filled in. Consequently, toward the 3′ ends of the sequenced molecules, deaminated cytosine residues result in apparent G to A substitutions, explaining the occurrence of these substitutions in libraries prepared from ancient DNA (Briggs et al. 2007; Brotherton et al. 2007), which was initially mistaken for deamination of guanine residues (Stiller et al. 2006). Analysis of the 3′ end of ancient DNA fragments, made possible by both new sequencing technologies (Orlando et al. 2011) and the recent single-stranded library preparation method (Meyer et al. 2012), unequivocally shows that C to T changes occur at similar and high frequencies on both ends of ancient DNA fragments.
Figure 3.
Nucleotide substitution frequencies at the 5′ end of Neanderthal sequences. The frequencies of all possible differences to the human reference genome sequence are plotted in relation to their location in Neanderthal DNA sequences. Cytosine to thymine substitutions are the most frequent, and they occur most often at ends of molecule. All remaining nucleotide substitutions are indicated by the black line.
Because deaminated cytosines are localized primarily to the ends of DNA fragments and because the rate of cytosine deamination is about two orders of magnitude faster in single-stranded than double-stranded DNA (Lindahl 1993), it is likely that the accumulation of C to T substitutions at the ends of ancient DNA fragments reflects the occurrence of single-stranded overhanging ends in ancient DNA (Briggs et al. 2007; Orlando et al. 2011). Because uracils occur only rarely in the middle of ancient DNA strands they can be removed using uracil-N-glycosylase and Escherichia coli endonuclease VIII to improve sequence quality without substantially impairing the overall recovery of DNA sequence (Briggs et al. 2010).
DAMAGE THROUGH TIME
Although environmental conditions such as humidity, temperature, salinity, and pH will have a strong effect on DNA preservation, it has been estimated through extrapolation from in vitro experiments (Lindahl 1993) that DNA would survive no more than a few hundred thousand years. Indeed, it was recently estimated that the half-life of 242 bp mitochondrial DNA fragments in bird bones excavated in a small area in New Zealand is around 500 years and that only preservation in frozen conditions might allow DNA to survive for more than a million years (Allentoft et al. 2012). When several such detailed studies of DNA survival from different environments become available it will hopefully be possible to estimate the likelihood of DNA survival given the environmental conditions at an archaeological site. However, even then, conditions of relevance for DNA survival such as amount of water percolation, salinity, pH, and microbial growth are likely to vary within a site or an archaeological stratum and make preservation conditions variable on a “microscale.”
Nevertheless, it is clear that DNA will not survive over geological time scales. The oldest credible samples that have been sequenced stem from plant and insect DNA found in 450,000- to 800,000-yr-old ice cores from Greenland (Willerslev et al. 2007). In contrast, a number of publications of DNA sequences determined from remains that are several million years old exist in the literature, including inclusions of animals and plants enclosed in amber (DeSalle et al. 1992) and fossils of plants and dinosaurs (Golenberg et al. 1990; Woodward et al. 1994). These probably all represent DNA from present-day organisms that have contaminated the samples or experiments (Pääbo and Wilson 1991; Lindahl 1993; Pääbo et al. 2004).
DNA sequence features indicative of DNA damage have recently been assessed in animal remains varying in age from 18 to 60,000 years (Sawyer et al. 2012). No correlation between fragment length and age was found, and only a weak negative correlation between age and the occurrence of purines immediately adjacent to the 5′ ends of fragments was seen. Interestingly, in samples younger than 100 years of age, adenine residues predominated adjacent to the 5′ ends of fragments, whereas guanine residues predominated in samples older than 40,000 years. This suggests that some process other than depurination, which likely causes the elevated guanine signal in older samples, occurs in younger samples, perhaps degradation by enzymes shortly after death. Of the DNA sequence features studied, only cytosine deamination was strongly positively correlated with age, despite the fact that the samples came from different sites and a variety of burial conditions. Cytosine deamination, manifested by an elevated C to T substitution frequency, can therefore be used as an indication that DNA molecules are indeed ancient (Krause et al. 2010).
CONCLUDING REMARKS
Our understanding of postmortem DNA damage is still very fragmentary. For example, depurination is a well-characterized mechanism of DNA degradation but its contribution to ancient DNA fragmentation is based entirely on indirect evidence and it probably accounts for only 10%–40% of the fragmentation that occurs in ancient DNA (Sawyer et al. 2012). Furthermore, it is unclear how prevalent blocking lesions are in ancient DNA. Fortunately, technologies are likely to emerge that will allow nucleotide modifications and lesions to be directly detected and characterized without prior amplification or enzymatic modifications (e.g., Branton et al. 2008). An improved understanding of ancient DNA damage may allow the development of new repair strategies that might increase the number of DNA sequences that can be retrieved from ancient remains even further.
ACKNOWLEDGMENTS
We thank Susanna Sawyer and the Multimedia Department for assistance with figures. Our work is funded by the Max Planck Society.
Footnotes
Editors: Errol C. Friedberg, Stephen J. Elledge, Alan R. Lehmann, Tomas Lindahl, and Marco Muzi-Falconi
Additional Perspectives on DNA Repair, Mutagenesis, and Other Responses to DNA Damage available at www.cshperspectives.org
REFERENCES
- Allentoft ME, Collins M, Harker D, Haile J, Oskam CL, Hale ML, Campos PF, Samaniego JA, Gilbert MTP, Willerslev E, et al. 2012. The half-life of DNA in bone: Measuring decay kinetics in 158 dated fossils. Proc R Soc B 10.1098/rspb.2012.1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binladen J, Willerslev E 2010. Why study ancient DNA damage? J Nordic Archaeol Sci 14: 11–14 [Google Scholar]
- Binladen J, Wiuf C, Gilbert MTP, Bunce M, Barnett R, Larson G, Greenwood AD, Haile J, Ho SYW, Hansen AJ, et al. 2006. Assessing the fidelity of ancient DNA sequences amplified from nuclear genes. Genetics 172: 733–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branton D, Deamer DW, Marziali A, Bayley H, Benner SA, Butler T, Di Ventra M, Garaj S, Hibbs A, Huang X, et al. 2008. The potential and challenges of nanopore sequencing. Nat Biotechnol 26: 1146–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs AW, Stenzel U, Johnson PLF, Green RE, Kelso J, Prüfer K, Meyer M, Krause J, Ronan MT, Lachmann M, et al. 2007. Patterns of damage in genomic DNA sequences from a Neandertal. Proc Natl Acad Sci 104: 14616–14621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs AW, Stenzel U, Meyer M, Krause J, Kircher M, Pääbo S 2010. Removal of deaminated cytosines and detection of in vivo methylation in ancient DNA. Nucleic Acids Res 38: e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brotherton P, Endicott P, Sanchez JJ, Beaumont M, Barnett R, Austin J, Cooper A 2007. Novel high-resolution characterization of ancient DNA reveals C > U-type base modification events as the sole cause of post mortem miscoding lesions. Nucleic Acids Res 35: 5717–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSalle R, Gatesy J, Wheeler W, Grimaldi D 1992. DNA sequences from a fossil termite in Oligo-Miocene amber and their phylogenetic implications. Science (New York, NY) 257: 1933–1936 [DOI] [PubMed] [Google Scholar]
- Gilbert MTP, Willerslev E, Hansen AJ, Barnes I, Rudbeck L, Lynnerup N, Cooper A 2003. Distribution patterns of postmortem damage in human mitochondrial DNA. Am J Hum Genet 72: 32–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert MTP, Binladen J, Miller W, Wiuf C, Willerslev E, Poinar H, Carlson JE, Leebens-Mack JH, Schuster SC 2007. Recharacterization of ancient DNA miscoding lesions: Insights in the era of sequencing-by-synthesis. Nucleic Acids Res 35: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golenberg EM, Giannasi DE, Clegg MT, Smiley CJ, Durbin M, Henderson D, Zurawski G 1990. Chloroplast DNA sequence from a miocene Magnolia species. Nature 344: 656–658 [DOI] [PubMed] [Google Scholar]
- Hansen AJ, Willerslev E, Wiuf C, Mourier T, Arctander P 2001. Statistical evidence for miscoding lesions in ancient DNA templates. Mol Biol Evol 18: 262–265 [DOI] [PubMed] [Google Scholar]
- Hansen AJ, Mitchell DL, Wiuf C, Paniker L, Brand TB, Binladen J, Gilichinsky D A, Rønn R, Willerslev E 2006. Crosslinks rather than strand breaks determine access to ancient DNA sequences from frozen sediments. Genetics 173: 1175–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyn P, Stenzel U, Briggs AW, Kircher M, Hofreiter M, Meyer M 2010. Road blocks on paleogenomes–polymerase extension profiling reveals the frequency of blocking lesions in ancient DNA. Nucleic Acids Res 38: e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofreiter M, Jaenicke V, Serre D, von Haeseler A, Pääbo S 2001. DNA sequences from multiple amplifications reveal artifacts induced by cytosine deamination in ancient DNA. Nucleic Acids Res 29: 4793–4799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höss M, Jaruga P, Zastawny TH, Dizdaroglu M, Pääbo S 1996. DNA damage and DNA sequence retrieval from ancient tissues. Nucleic Acids Res 24: 1304–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AS, Mian AM, Walker RTJ 1968. Chem Soc C 2042–2044 [Google Scholar]
- Krause J, Briggs AW, Kircher M, Maricic T, Zwyns N, Derevianko A, Pääbo S 2010. A complete mtDNA genome of an early modern human from Kostenki, Russia. Curr Biol 20: 231–236 [DOI] [PubMed] [Google Scholar]
- Lindahl T 1993. Instability and decay of the primary structure of DNA. Nature 362: 709–715 [DOI] [PubMed] [Google Scholar]
- Lindahl T, Andersson A 1972. Rate of chain breakage at apurinic sites in double-stranded deoxyribonucleic acid. Biochemistry 11: 3618–3623 [DOI] [PubMed] [Google Scholar]
- Lindahl T, Nyberg B 1972. Rate of depurination of native deoxyribonucleic acid. Biochemistry 11: 3610–3618 [DOI] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen Y-J, Chen Z, et al. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437: 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, Kircher M, Gansauge M-T, Li H, Racimo F, Mallick S, Schraiber JG, Jay F, Prüfer K, De Filippo C, et al. 2012. A high-coverage genome sequence from an archaic Denisovan individual. Science (New York, NY) 338: 222–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando L, Ginolhac A, Raghavan M, Vilstrup J, Rasmussen M, Magnussen K, Steinmann KE, Kapranov P, Thompson JF, Zazula G, et al. 2011. True single-molecule DNA sequencing of a pleistocene horse bone. Genome Res 21: 1705–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overballe-Petersen S, Orlando L, Willerslev E 2012. Next-generation sequencing offers new insights into DNA degradation. Trends Biotechnol 30: 364–368 [DOI] [PubMed] [Google Scholar]
- Pääbo S 1989. Ancient DNA: Extraction, characterization, molecular cloning, and enzymatic amplification. Proc Natl Acad Sci 86: 1939–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pääbo S, Wilson AC 1991. Miocene DNA sequences—A dream come true? Curr Biol 1: 45–46 [DOI] [PubMed] [Google Scholar]
- Pääbo S, Poinar H, Serre D, Jaenicke-Despres V, Hebler J, Rohland N, Kuch M, Krause J, Vigilant L, Hofreiter M 2004. Genetic analyses from ancient DNA. Annu Rev Genet 38: 645–679 [DOI] [PubMed] [Google Scholar]
- Poinar HN, Hofreiter M, Spaulding GW, Martin PS, Stankiewicz BA, Bland H, Evershed RP, Possnert G, Pääbo S 1998. Molecular coproscopy: Dung and diet of the extinct ground sloth Nothrotheriops shastensis. Science 281: 402–406 [DOI] [PubMed] [Google Scholar]
- Rohland N, Hofreiter M 2007. Comparison and optimization of ancient DNA extraction. BioTechniques 42: 343–352 [DOI] [PubMed] [Google Scholar]
- Sawyer S, Krause J, Guschanski K, Savolainen V, Pääbo S 2012. Temporal patterns of nucleotide misincorporations and DNA fragmentation in ancient DNA. PloS ONE 7: e34131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiller M, Green RE, Ronan M, Simons JF, Du L, He W, Egholm M, Rothberg JM, Keates SG, Keats SG, et al. 2006. Patterns of nucleotide misincorporations during enzymatic amplification and direct large-scale sequencing of ancient DNA. Proc Natl Acad Sci 103: 13578–13584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willerslev E, Cappellini E, Boomsma W, Nielsen R, Hebsgaard MB, Brand TB, Hofreiter M, Bunce M, Poinar HN, Dahl-Jensen D, et al. 2007. Ancient biomolecules from deep ice cores reveal a forested southern Greenland. Science (New York, NY) 317: 111–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward SR, Weyand NJ, Bunnell M 1994. DNA sequence from Cretaceous period bone fragments. Science (New York, NY) 266: 1229–1232 [DOI] [PubMed] [Google Scholar]