Abstract
The dynamics of mitochondrial biogenesis and function is a complex interplay of cellular and molecular processes that ultimately shape bioenergetics capacity. Mitochondrial mass, by itself, represents the net balance between rates of biogenesis and degradation. Mitochondrial biogenesis is dependent on different signaling cascades and transcriptional complexes that promote the formation and assembly of mitochondria—a process that is heavily dependent on timely and coordinated transcriptional control of genes encoding for mitochondrial proteins. In this article, we discuss the major signals and transcriptional complexes, programming mitochondrial biogenesis, and bioenergetic activity. This regulatory network represents a new therapeutic window into the treatment of the wide spectrum of mitochondrial and neurodegenerative diseases characterized by dysregulation of mitochondrial dynamics and bioenergetic deficiencies.
Mitochondria contain >1000 proteins, but encode for only 13 of them. Thus, mitochondrial formation and function heavily depend on the coordinated transcriptional control of nuclear genes that encode the other mitochondrial proteins.
Mitochondria are dense, double membrane-enclosed organelles that are present in all mammalian cells except mature red blood corpuscles. They uniquely possess their own genome in a circular DNA molecule. Although this genome encodes for only a small fraction of the total genes needed for mitochondrial organelle assembly and function, it is thus necessarily complemented by nuclear-encoded genes. Mitochondria provide a wide variety of biochemical services to the cell. Although it is conventionally accepted that energy production through the aerobic oxidation of carbon substrates and the generation of ATP is the principal function of mitochondria, this narrow view overlooks the many other impressive biosynthetic and regulatory capacities of this versatile organelle. Mitochondria, for instance, are essential for the synthesis of pyrimidines and purines, contribute to the synthesis of heme, regulate nitrogen balance through the urea cycle, produce ketone bodies, are necessary for sex hormone production, are involved in the processing of xenobiotics, play a critical role in redox balance, and are key regulators of the apoptotic program in mammals. Given the centrality of mitochondria to the maintenance of so many pathways in mammalian cells, it is not surprising that there exist multiple layers of control that enable a cell to coordinate its net mitochondrial activity with fuel sources, biosynthetic demands, proliferation rates, and external stimuli. One important layer in this signaling control is the expression of mitochondrial nuclear-derived genes that initiate the cellular program of mitochondrial biogenesis. Defects and failure in the integrity of this signaling system can cause serious perturbations to cellular metabolism and give rise to a broad range of tissue specific as well as systemic pathologies in humans. The same signaling system, on the other hand, also opens many therapeutic opportunities for the correction of mitochondrial defects. In this article, we discuss some of the major signaling factors that serve to regulate mitochondrial biogenesis and their implications for the mitigation of mitochondrial disorders.
REGULATION OF MITOCHONDRIAL BIOGENESIS AND FUNCTION THROUGH NUCLEAR-BASED GENE EXPRESSION
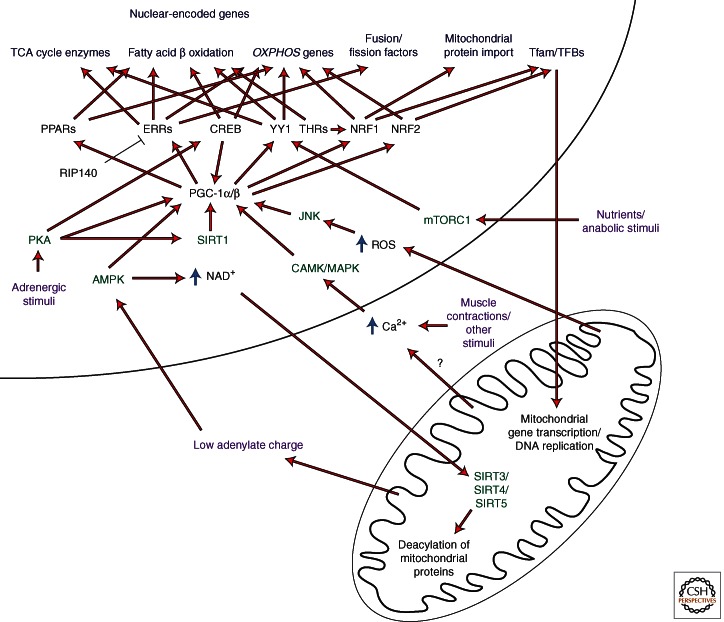
Although mitochondria contain in excess of 1000 proteins, its tiny circular genome encodes for only 13 proteins, 22 tRNAs, and two rRNAs; the balance of this genetic shortage is made up by the nuclear genome (Calvo and Mootha 2010). Because the vast majority of mitochondrial genes are situated in the nucleus, transcription complexes at promoters of these genes control their expression. This affords the cell the opportunity to coordinately regulate the expression of both mitochondrial and nonmitochondrial gene sets to affect a unified specific cellular response. Below we highlight some of the major nuclear transcriptional complexes regulating mitochondrial gene expression and the diverse signaling pathways to which they respond (Fig. 1).
Figure 1.
An overview of the transcriptional complexes regulating mitochdondrial gene expression and the signaling pathways converging upon them. Illustrated are the major transcriptional complexes/chromatin remodeling factors directly associated with changes in mitochondrial gene expression (see text for additional information).
Nuclear Respiratory Factors
Sequence analysis of the electron transport gene cytochrome c promoter initially identified nuclear respiratory factor 1 (NRF-1) as an important regulator of gene expression (Evans and Scarpulla 1989). NRF-1 controls the expression of a significant number of the proteins that make up the five respiratory complexes, as well as proteins integral to mitochondrial import and heme biosynthesis (Scarpulla 2008). NRF-1 is also able to integrate nuclear control of the transcriptional and replicative activity of the mitochondrial genome through the direct modulation of transcription factor A mitochondrial (TFAM) and transcription factor B proteins (TFBs) gene expression; TFAM and TFBs are major regulators of mitochondrial DNA transcription and replication (Gleyzer et al. 2005). NRF-1 may play an integral role in coupling mitochondrial replication with nuclear replication over the course of the cell cycle through regulation of E2F transcription factor targets and mitochondrial genes, indicating a link between cell division and mitochondrial biogenesis (Cam et al. 2004).
Studies involving the promoter region of cytochrome c oxidase complex subunit IV (COXIV) revealed the presence of another nuclear respiratory factor, NRF-2, which was able to regulate the expression of proteins in the electron transport chain (Carter et al. 1992). NRF-2 promotes the expression of genes that encode for mitochondrial complex IV cytochrome c oxidase (Gugneja et al. 1995). Like NRF1, NRF2 also controls the expression of TFAM and TFBs. The differential regulation of NRF1 and NRF2 is not completely understood but phosphorylation of these factors can alter their transcriptional activities (Scarpulla 2008). In addition, specific coactivators of the PGC-1 family sculpt NRFs-dependent control of mitochondrial gene expression in response to different signaling pathways (Wu et al. 1999).
Nuclear Hormone Receptors
Within the superfamily of nuclear hormone receptors, there are several subclasses that control mitochondrial function and biogenesis. The PPAR group of receptors is one such set. PPARα, which can be activated by long-chain fatty acids and eicosanoids, can potently promote the expression of genes involved in mitochondrial β-oxidation (Gulick et al. 1994). The activation of PPARγ activation, on the other hand, triggers mitochondrial biogenesis in white adipose tissue (Wilson-Fritch et al. 2004); PPARγ activation is also necessary to induce the expression of mitochondrial genes involved in the brown fat-mediated thermogenic program (Puigserver et al. 1998). PPARδ, in skeletal muscle, is able to up-regulate mitochondrial genes associated with fatty acid oxidation and specify slow twitch fiber type (Narkar et al. 2008; Kleiner et al. 2009).
Thyroid hormone receptors (THRs) can also promote mitochondrial biogenesis and tissue-specific function. This includes mitochondrial-driven thermogenesis that occurs in brown fat during the adaptation to lower temperatures (Silva 1995). In some instances, THRs directly drive the transcription of nuclear-encoded genes, whereas, in others, the effects can occur indirectly through the thyroid hormone-mediated up-regulation of NRF-1 (Venditti et al. 2009). A truncated form of thyroid hormone receptor α has been reported to be localized to the mitochondrion and may serve to directly activate the transcription of genes in the mitochondrial genome (Casas et al. 1999).
Estrogen-related receptors ERR-α, ERR-β, and ERR-γ are yet another set of nuclear hormone receptors capable of promoting mitochondrial biogenesis. These receptors have no known endogenous ligands and are primary expressed in tissues with high oxidative metabolism capacities (Eichner and Giguere 2011). DNA-binding sites for ERR-α have been mapped in a large number of nuclear-encoded mitochondrial genes, including those involved in oxidative phosphorylation, fatty acid oxidation, TCA cycle, and factors regulating mitochondrial fusion/fission. Although ERRs are considered orphan nuclear receptors, their activity can certainly be modulated through recruitment of coactivators/corepressor complexes. Indeed, the transcriptional coactivators PGC-1α/β are efficacious activators of ERRs and promote the expression of mitochondrial genes (Mootha et al. 2004; Schreiber et al. 2004). The transcriptional activity of this transcription factor/activator module is controlled through deacetylation by sirtuin 1, which allows for an important regulatory mechanism that connects nutrient/hormonal signaling to the control of mitochondrial function (Wilson et al. 2010). From the standpoint of repression of ERR activity, the RIP140 nuclear hormone corepressor seems to be important (Wilson et al. 2010).
Other Transcription Factors: CREB and YY1
CREB, a cAMP activated transcription factor, can promote the expression of several mitochondrial genes, including the proteins which make up complex IV and enzymes involved in the β-oxidation pathway (Gopalakrishnan and Scarpulla 1994). Many mitochondrial genes contain YY1 binding sites within their promoter regions and this transcription factor has been found to work in conjunction with PGC-1α to regulate their expression (Cunningham et al. 2007). Loss of YY1 expression in skeletal muscle causes exercise intolerance and morphological signs of mitochondrial myopathy in mice (Blattler et al. 2012b).
Transcriptional Coactivators
Transcriptional coactivators do not bind to DNA but can nonetheless promote gene transcription by enhancing the activity of true DNA-binding transcription factors. Biologically, these proteins can set into motion genome-wide transcriptional changes by coactivating many different transcription factors. In mammalian cells, the PGC-1 family of coactivators (PGC-1α, PGC-1β, and PRC) potentiate the activity of several transcription factors involved in the basic functions of the mitochondrion as well as its rate of biogenesis (Puigserver and Spiegelman 2003; Kelly and Scarpulla 2004). Indeed, PGC-1α and PGC-1β are sufficient to increase total mitochondrial mass, reactive oxygen species scavenging enzymes, oxidative phosphorylation components, mitochondrial metabolic pathways, protein import complexes, proteins involved in fission and fusion, and the levels of mitochondrial sirtuins (Mootha et al. 2003; Uldry et al. 2006; Cunningham et al. 2007; Rasbach et al. 2010; Handschin and Spiegelman 2011). This process occurs in part through the ability of PGC-1 proteins to potentiate the function of the NRFs, ERRs, and YY1.
In skeletal muscle, the increased mitochondrial biogenesis caused by transgenic overexpression of PGC-1α can attenuate the development of mitochondrial disease in mouse models of respiratory complex IV/cytochrome c oxidase deficiencies (Lin et al. 2002; Wenz et al. 2008; Viscomi et al. 2011). It has also been shown that PGC-1α/β expression improves mitochondrial respiration capacity in cells from human patients with a complex III or IV deficiency (Srivastava et al. 2009). It should also be noted that age-related declines in mitochondrial function are also mitigated by enhanced activity of PGC-1α. Skeletal muscle specific PGC-1α transgenic mice are less susceptible to exercise-induced fatigue and are protected against age-related diseases, such as sarcopenia and metabolic diseases, and as a consequence, these mice show an increased life span (Wenz et al. 2009). Nevertheless, it has yet to be determined whether the beneficial health effects of PGC-1α in skeletal muscle are attributable to its promotion of mitochondrial function or its effects on nonmitochondrial gene expression.
Sirtuin Proteins
Sirtuins are NAD+-dependent protein deacylases that are homologous to yeast Sir2p, which is one of the genes involved in transcriptional silencing in that organism (Klar et al. 1979). In mammals there are seven sirtuin paralogs. SIRT1, SIRT6, and SIRT7 are nuclear proteins, SIRT3, SIRT4, and SIRT5 are imported into mitochondria, and SIRT2 is principally cytoplasmic (Finkel et al. 2009; Haigis and Sinclair 2010; Verdin et al. 2010; Guarente 2011). Through their deacylation activities, sirtuins modulate the biological activities of a wide array of target pathways. From the standpoint of mitochondrial biology, SIRT1, SIRT3, SIRT4, and SIRT5 have profound effects on the function of this organelle. SIRT1 deacetylates several key transcription factors that result in the up-regulation of numerous genes involved in mitochondrial respiration (Rodgers et al. 2005; Purushotham et al. 2009) and may (Price et al. 2012) or may not (Park et al. 2012) be necessary for the promitochondrial effects of polyphenolic stilbenoid compounds like resveratrol. In addition, sirtuins deacetylate numerous metabolic enzymes to govern their specific activity. Changes in SIRT3 activity have been shown to be an important determinant in the acetylation state of mitochondrial in response to nutrient availability (Hirschey et al. 2011; Hebert et al. 2013). The acetylation of many mitochondrial proteins, such as isocitrate dehydrogenase 2 (Yu et al. 2012), can alter their catalytic/biological function. As such, loss of SIRT3 activity results in profound aberrations in mitochondrial function and exacerbates the metabolic pathologies associated with chronic nutrient excess. Although no consistent deacylase activities have been shown for SIRT4, it has been shown that SIRT4 antagonizes mitochondrial capacity for fatty acid oxidation in hepatic and skeletal muscle cells (Nasrin et al. 2010). SIRT5, which has been shown to display desuccinylase, demalonylase, and deacetylase activities, targets carbamoyl phosphate synthetase 1, and can regulate mitochondrial urea cycle flux (Nakagawa et al. 2009; Du et al. 2011).
SIGNALING MODULES THAT CONTROL MITOCHONDRIAL BIOGENESIS AND FUNCTION
Cells are constantly engaged in both temporal and spatial control of mitochondrial biogenesis in response to nutrient availability, hormonal cues, and changes in temperature. Integration of these variables frequently involves the use of signaling pathways that ultimately converge upon the transcriptional regulators described above. In this section, we discuss the key signaling modules that control mitochondrial genes and lead to increased mitochondrial mass.
AMP-Activated Protein Kinase (AMPK)
AMPK is a phylogenetically ancient cellular sensor that is triggered by high cellular energy demands (Hardie 2007). As such, it is not surprising that AMPK is strongly connected to mitochondrial bioenergetics. When ATP synthesis is impaired or ATP is being consumed and generating AMP at a high rate, AMPK becomes active, which, in turn, phosphorylates several enzymes involved in the stimulation of catabolic pathways, such as glucose transport and fatty acid oxidation and the inhibition of anabolic pathways, such as glycogen synthesis and lipogenesis (Kahn et al. 2005). Chronic activation of AMPK is also able to induce mitochondrial biogenesis through the control of nuclear transcription (Fig. 2). Essential to this process are the AMPK-mediated activation of PGC-1α and SIRT1. AMPK can directly phosphorylate and activate PGC-1α (Jager et al. 2007); other pathways that activate AMPK can result in a subsequent increase in PGC-1α protein expression and mitochondrial biogenesis (Birkenfeld et al. 2011). AMPK through Nampt-dependent and -independent mechanisms is also able to increase NAD+ levels, which promotes SIRT1 activity and the deacetylation/activation of PGC-1α (Canto et al. 2009). Thus, the AMPK/SIRT1/PGC1α signaling axis is thought to play a key role in the adaptive metabolic programming that occurs during caloric restriction and exercise (Canto and Auwerx 2009). Nevertheless, it has been alternatively suggested that caloric restriction-mediated activation of AMPK serves to maintain mitochondrial protein biosynthesis independently of transcription (Miller et al. 2012).
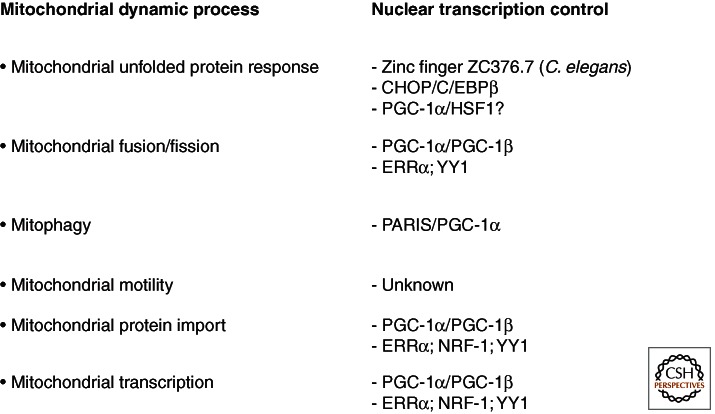
Figure 2.
Transcriptional control of processes involved in the regulation of mitochondrial dynamics. Nuclear transcription factors regulate the transcription of specific proteins that are basic components of specific mitochondrial processes associated with mitochondrial dynamics (see text for further information).
Calcium Signaling
Intracellular calcium dynamics are affected by a wide array of signaling pathways and calcium, as a second messenger molecule, and constitutes an important mechanism for initiating changes in mitochondrial gene expression. This phenomenon is most evident in skeletal muscle, a tissue that displays coordinated changes in calcium homeostasis as part of its basic contractile mechanism. In this tissue, calcium-sensitive protein networks, such as calcium/calmodulin-dependent kinase and p38 mitogen-activated kinase, are critical actors in the mobilization of the transcriptional machinery that promotes mitochondrial proliferation and fatty acid oxidation (Wu et al. 2002; Wright et al. 2007).
cAMP Pathway
Yet another well-conserved second messenger system capable of altering mitochondrial function is the cAMP-PKA pathway. The principal transcriptional effector of this pathway is the CREB transcription factor, whose phosphorylation by PKA promotes CREB activity and target gene transcription (Mayr and Montminy 2001). PKA activation can also enhance the expression of PGC-1α, through the intermediacy of CREB (Herzig et al. 2001), as well as directly activate the catalytic function of SIRT1 (Gerhart-Hines et al. 2011). Biologically, one of the most salient examples of the power of cAMP-mediated effects on mitochondrial function is found in the adaptive nonshivering thermogenic response to lower temperatures (Cannon and Nedergaard 2004). This heat-generating process is highly dependent on induction of uncoupled mitochondrial respiration in adipose depots—primarily brown fat tissue—through the up-regulation of uncoupling proteins such as UCP1 and the up-regulation of oxidative pathways (Cannon and Nedergaard 2004). Sympathetic stimulation of brown and “beige” fat tissues increases cAMP-PKA tone in these tissues and precipitates the initial uncoupling response. Whereas the specific determination of the brown fat cell lineage is transcriptionally specified through PRDM16 (Seale et al. 2007), the mitochondrial thermogenic response is defined by the PGC-1 coactivators, which account for approximately 30% of the cAMP response in brown fat cells (Uldry et al. 2006).
mTOR Pathway
The mechanistic target of rapamycin (mTOR) pathway is a logic gate for integrating cell growth and cell size with the availability of growth factors, energy levels, and certain nutrients, such as the branched-chain amino acids. These processes require a net increase in cellular anabolic/biosynthetic capacities, which will ultimately depend on enhanced mitochondrial biogenesis and bioenergetics. The effects of mTOR on mitochondrial biology occur through several mechanisms. One of the mechanisms seems to be independent of transcription through direct effects on mitochondrial organelles and stimulation of respiration (Schieke et al. 2006). In a second mechanism, mTOR modulates mitochondrial gene expression through the regulation of different transcription factors. In liver, mTOR affects genes of fatty acid oxidation and ketone body production through its ability to activate NCoR and inactivate PPARα (Sengupta et al. 2010). In skeletal muscle, mTOR binds to the transcription factor YY1 and recruits PGC1α, increasing the ability of YY1 to activate mitochondrial gene expression. mTOR inhibition by using rapamycin, for example, results in a dissociation of PGC1α and decreases mitochondrial gene expression (Cunningham et al. 2007; Blattler et al. 2012a).
Retrograde Signaling from Mitochondria to the Nucleus
The retrograde transmission of regulatory signals from the mitochondria to the nucleus, from a teleological standpoint, makes sense in that deficits or excesses in mitochondrial activity would need to be ultimately corrected by adjustments in nuclear gene expression. This phenomenon has been actively explored in yeast (Liu and Butow 2006). In mammalian cells, however, the nature of these signals and the mechanisms that they use are less well understood. Nevertheless, evidence of this regulatory phenomenon can be seen in the skeletal muscle of patients with mitochondrial diseases, such as those with genetic mutations of oxidative respiration proteins, which display increased mitochondrial proliferation—perhaps as an effort to compensate for impaired mitochondrial activity (DiMauro and Hirano 2009; Wallace and Fan 2009). The nature of this communication is unknown, but calcium and ROS signaling have been connected to this retrograde control.
Calcium Signaling
Much like the endoplasmic reticulum, mitochondria are quantitatively significant stores of calcium in the cell. As such, they can provide a potential signaling avenue for the regulation of cellular processes—indeed, overloading the mitochondrial calcium pool is a signal for the induction of apoptosis. At this point, although mechanisms of calcium uptake by the mitochondria have been characterized (Baughman et al. 2011; De Stefani et al. 2011), there is no clearly delineated pathway by which mitochondrial calcium can be released to regulate the calcium sensitive transcription/chromatin factor pathways discussed above. Nonetheless, PGC-1α increases mitochondrial calcium release, which establishes a positive feedback loop for the maintenance of a mutual connection between mitochondrial and nuclear gene expression (Bianchi et al. 2006).
Reactive Oxygen Species (ROS)
ROS are produced at respiratory chain complexes located in the inner mitochondrial membrane. In Drosophila, disruption of respiratory complex 1 signals to components of the G1 to S cell cycle machinery trough JNK and FoxO (Owusu-Ansah et al. 2008). In mammalian cells, a similar retrograde signaling has been uncovered through a systematic analysis of different mitochondrial dysfunction mutations; this pathway increases OXPHOS genes and involves ROS signaling through JNK and the RXRα/PGC1α pathway (Chae et al. 2013).
CONTROL OF MITOCHONDRIAL PROCESSES THAT CONTRIBUTE TO THE BIOGENESIS OF MITOCHONDRIA
Mitochondrial Unfolded Protein Response and Quality Control
The mitochondria contains an internal protease system that monitors accumulation of unfolded or misfolded proteins (encoded by the nuclear or mitochondrial genomes) in the different compartments, including the outer membrane, intermembrane space, inner membrane, and matrix. Thus, during mitochondrial biogenesis or stress, chaperones and quality-control proteases recognize hydrophobic amino acids that are in the interior structure of normal folded proteins (Baker and Haynes 2011). In addition to this organelle’s internal quality control, there is a retrograde signaling to the nucleus via translocation of several transcription factors. In Caenorhabditis elegans, mitochondrial chaperones such as mtHSP70 proteins bind to unfolded mitochondrial proteins; however, if the amount of unfolded proteins exceeds the chaperone capacity, a protease called ClpXP degrades these proteins into peptides. Mitochondrial efflux of these peptides activates a transcription factor, ZC376.7, which translocates to the nucleus to activate expression of mitochondrial chaperone genes (Haynes et al. 2010). In mammalian cells, accumulation of unfolded proteins in the mitochondrial matrix increases CHOP/C/EBPβ transcriptional activities on promoters encoding for mitochondrial chaperone genes, including chaperonins 10 and 60, mtDNAJ, and ClpP (Zhao et al. 2002). In hepatocyte cells, PGC-1α interacts with HSF1; however, it is unknown whether this pathway controls the mitochondrial unfolded protein response (Charos et al. 2012).
Mitochondrial Fusion/Fission
Mitochondria are dynamic organelles that undergo constant division or fission and fusion (Twig et al. 2008; Westermann 2010; Corrado et al. 2012). The mitochondrial mass undergoes different shapes depending on the cellular state. Fusion of the outer mitochondrial membrane is executed by the activity of two dynamin-related GTPases, MFN1 and MFN2, that form homo- or heterodimers. In addition, OPA1—another dynamin-related GTPase—controls mitochondrial fusion and has also been involved in cytochrome release and apoptosis. The mitochondrial fission machinery is made up of DRP1, FIS1, MFF, and MIEF1. DRP1 is a GTPase protein that is localized in the cytoplasm, but after calcineurin-mediated dephosphorylation is recruited to the mitochondria. Once at the mitochondria DRP1 oligomerizes and forms ring structures that will constrict and produce fission. DRP1 has adaptor proteins that either activate fission (MFF) or suppress fission (MIEF) modulating the DRP1 GTPase activity. FIS1 is the DRP1 receptor located in the outer mitochondrial membrane and also interacts and sequesters MIEF1. In cells, fusion makeup of interconnected mitochondria is essential to maintain a highly metabolic and energetic state. In contrast, quiescent or resting cells have fragmented mitochondria with small spherical shapes with low metabolic activities (Chen and Chan 2010).
Thus, it seems plausible that the mechanisms that control mitochondrial biogenesis are tightly associated with the fusion machinery. Along these lines, the transcriptional coactivators PGC-1α and PGC1-β are positive activators of MFN2 (Cartoni et al. 2005; Soriano et al. 2006). Whether there is independent transcriptional control of the fission machinery, or if their regulation is simply bundled with the rest of the nuclear-encoded mitochondrial proteins is currently unknown.
Mitophagy
Mitochondrial mass is defined by the balances of mitochondrial formation and degradation. This latter process largely occurs through a particular type of cellular autophagy, termed mitophagy, that clears defective mitochondria. Mitophagy is a cellular degradation process that maintains “healthy” mitochondrial mass (Ashrafi and Schwarz 2013). Elongated and fused mitochondrial are resistant to mitophagy, whereas fragmented mitochondria generated by fission are more accessible to mitophagy. The elimination of damaged mitochondria is mediated by the activities of PINK1 (PTEN-induced putative protein kinase 1) and the E3 ubiquitin ligase, Parkin. PINK1 and Parkin accumulate at the mitochondrial surface of damaged organelles through the loss of mitochondrial membrane potential (Youle and van der Bliek 2012). In turn, this promotes separation from the functional mitochondrial network followed by targeting defective mitochondria for degradation. This latter process requires Parkin-dependent ubiquitination of mitochondrial proteins, but the critical Parkin substrates necessary for mitophagy are unknown. MFN1 and MFN2 are ubiquitinated by Parkin, although they are not required for mitophagy. Mitochondria are selectively removed upon treatment with chemical uncouplers. In this process, opening of the mitochondrial permeability transition pore (PTP) also triggers selective mitophagy. In addition, starvation signals such as glucagon can also induce mitophagy but require opening of the PTP. Similar to mitochondrial fusion/fission, little is known about the transcriptional control of mitophagy. Interestingly, mutations that occur in Parkinson’s disease and inactivate the E3 ligase activity of Parkin, lead to the accumulation of Parkin substrates (Narendra et al. 2012). Among them is PARIS, a zinc finger corepressor, which can suppress the transcriptional coactivator PGC1α (Shin et al. 2011). Consistent with this, PGC-1α responsive genes are underexpressed in microdissected dopaminergic neurons of Parkinson’s disease mouse models, suggesting that this pathway could be causative in this disease (Beal 2009; Zheng et al. 2010). Based on these data, it is possible to speculate that during mitophagy Parkin activation can signal to the nucleus through Paris degradation, resulting in an increase in PGC-1α and will subsequently promote mitochondrial biogenesis.
Mitochondrial Motility
Depending on the cell type, an increase in mitochondrial mass needs to be coordinated with a precise intracellular localization of mitochondrial organelles to meet local energetic cellular demands. Mitochondrial motility is influenced by the organelle’s size, which is in turn controlled by fusion/fission processes. Failure in these processes prevents efficient intracellular movement of mitochondria, as is most evident in neurons, which critically depend on this pathway to localize mitochondria close to synaptic sites (Chen and Chan 2009).
Mitochondrial transport is based on microtubule dynamics. Kinesin-1 type motors are recruited to mitochondria through outer mitochondrial membrane proteins Miro1 and Miro2, involving an adaptor protein termed Milton. This is a calcium-dependent process: High concentrations of calcium halt mitochondria through the EF hands of Miro, anchoring mitochondria to sites of high ATP requirements. It is unclear whether calcium is the only regulator of this movement or there are additional sites of control including establishment of sensing gradients or transcriptional/translational regulation (Stowers et al. 2002; Wang et al. 2011).
Mitochondrial Protein Import
Biogenesis of functional and competent mitochondria requires the import and assembly of proteins synthesized in the cytoplasm. The mitochondria contain more than 1000 proteins that are mostly imported from the cytoplasm through the translocase of the outer mitochondrial membrane (TOM complex) (Endo et al. 2011; Gebert et al. 2011).
Once the mitochondrial protein has passed the TOM channel, it is sorted by interactions with different machineries located in the intermembrane space and inner membrane. Proteins that contain a presequence target signaling to the matrix are imported by the translocase of the inner membrane (TIM23) complex that is associated with the presequence translocase-associated motor (PAM). The TIM23 complex is also involved in laterally releasing proteins to the inner mitochondrial membrane. Several complexes in the intermembrane space participate in sorting mitochondrial proteins. For example, the MIA (mitochondrial intermembrane assembly) complex promotes oxidative folding of intermembrane proteins. The Tim9–Tim10 chaperone complex transfers hydrophobic proteins to the mitochondrial outer membrane through the SAM complex (e.g., β-barrel proteins) or the mitochondrial inner membrane through the TIM22 carrier complex (e.g., inner membrane carrier proteins). Additional import pathways include the oxidase assembly (OXA) machinery that transfers mitochondrial proteins from the matrix ribosomes to the inner membrane, or the mitochondrial import 1 (Mim1) that imports α-helical outer membrane proteins (Endo and Yamano 2009; Schmidt et al. 2010).
The dynamics of this complex mitochondrial protein import process will depend not only on the number of existing import complexes but also on the amounts of mitochondrial protein translated in the cytoplasm. Little is known about the protein stability or activities of these mitochondrial proteins, for example, control by ubiquitination or other posttranslational modifications (phosphorylation, acetylation, etc.) that control the import processes. However, an important regulatory control is the coordinated transcription of genes encoding for these proteins that will determine the rate of synthesis and cellular concentrations that will promote and contribute to mitochondrial mass. In mammalian cells, this transcriptional control involves the PGC-1 pathway and its interactions with transcription factors including NRFs and YY1 that are bound to promoters of mitochondrial import genes (Blattler et al. 2012b; Scarpulla et al. 2012). Whether there is selectivity or specificity among the different import pathways described here is currently unknown.
Mitochondrial Transcription
Balanced and efficient mitochondrial biogenesis requires the activation of mitochondrial transcription (Peralta et al. 2012). The mammalian mitochondrial DNA is a circular double-stranded genome (∼16.5 kb) that encodes 37 genes. Transcription of the mitochondrial DNA (mtDNA) is tightly coordinated with nuclear genes (particularly involved in OXPHOS) to achieve competent bieoenergetic mitochondrial organelles. The mtDNA contains two strands: the heavy (H) and the light (L) strands. The H strand contains most of the genes. In addition, there are two noncoding regions designated as the D-loop that harbors the promoter of both strands and the origin of replication. Mitochondrial transcription is bidirectional and starts in the D-loop region through the assembly of a complex composed by TFAM, TFB2M, and the RNA polymerase POLRMT. MTERF proteins also bind to promoters and control mitochondrial transcription (Rebelo et al. 2011). How the transcription of mitochondrial-encoded genes is precisely controlled is not completely understood. Several of the proteins that control mitochondrial DNA transcription have posttranslational modifications, including phosphorylation and acetylation. For example, TFAM is specifically phosphorylated (Lu et al. 2013) and acetylated (Dinardo et al. 2003), suggesting that its activity could be modified through cycles of acetylation/deacetylation in response to energetic signals. Another level of control of this mitochondrial transcriptional machinery is by inducing amounts of its components. Along these lines elements essential to mitochondrial transcription, such as TFAM, TFB2M, MTERF, and POLRMT, are regulated through the transcriptional coactivators PGC-1α and PGC-1β through their interaction with NRFs, ERRs, and YY1 (Mootha et al. 2004; Blattler et al. 2012b; Scarpulla et al. 2012).
THERAPEUTIC POTENTIAL OF SIGNALING PATHWAYS FOR THE ATTENUATION OF MITOCHONDRIAL-RELATED PATHOLOGIES
Mitochondrial dysfunction, whether it is attributable to deleterious mutations in nuclear or mtDNA genes (DiMauro and Hirano 2009; Wallace and Fan 2009) or to secondary systemic changes caused by factors such as aging/senescence, significantly impairs cell function and can result in a spectrum of pathologies. In certain circumstances, such as aging, the mechanisms for why or how mitochondrial function and dynamics become abnormal are unknown (Petersen et al. 2003; Reznick et al. 2007). Regardless of the mechanism, however, data gleaned from signaling studies suggest that certain elements of these pathways can be leveraged to either prevent or attenuate a wide array of mitochondrial dysfunctions.
Genetically Based Mitochondrial Disorders
Despite the fact that significant advances have been made in understanding the causative molecular mechanisms underlying many genetically based human mitochondrial disorders, there is a frightening paucity of demonstrable treatments (Hirano et al. 2012). In theory, however, interventions that boost the net mitochondrial activity in these patients to restore energy production and antioxidant capacities to some minimum threshold should be sufficient to attenuate afflicted organ dysfunction and the subsequent pathologies. As such, there are several druggable signaling avenues to pursue.
The first is the PGC-1α signaling axis. As mentioned earlier, transgenic overexpression of PGC-1α has been shown to significantly blunt the pathological effects of cytochrome c oxidase (Viscomi et al. 2011), COXIV deficiency in mice (Lin et al. 2002; Wenz et al. 2008), and COXIII/IV deficiency in human patient-derived cell lines. Thus, small molecule therapies that potentiate PGC-1α function or the function of one of its cognate transcription factors could yield tangible benefits. To date, efforts to specifically exploit this pathway have been largely limited to the testing of PPAR panagonists such as the fibrate family of compounds, which are already in clinical use as antihyperlipidemic drugs. Studies with bezofibrate suggest that it is of moderate use in humans suffering from mutations in carnitine palmitoyltransferase II deficiency (Bonnefont et al. 2009). Data on bezofibrate’s ability to correct the phenotypes associated with mutations in respiratory complex proteins, in contrast, have been confined largely to mouse models and have yielded very mixed results (Wenz et al. 2008; Viscomi et al. 2011; Yatsuga and Suomalainen 2012). The major limitation of fibrates is that they target only a single cognate transcription factor of PGC-1α, namely, PPARα, which is involved in up-regulation of the mitochondrial proteome. Small molecules that specifically up-regulate PGC-1α expression or promote its inherent biological activity, such as through deacetylation, are no doubt needed for a full effect.
The second signaling axis is that mediated by AMPK activation. High throughput screenings for compounds that improve the growth and ATP levels of fibroblasts from humans with a complex I deficiency revealed the AMPK activator 5-aminoimidazole-4-carboxamide ribonucleoside (AICAR) as one of the most efficacious molecules (Golubitzky et al. 2011). In several mouse models of cytochrome c oxidase deficiency, AICAR treatment was also shown to lead to a partial rescue of COXIV activity and some measure of improvement in motor performance (Viscomi et al. 2011).
The third most therapeutically attractive targets for mitochondrial diseases are the sirtuin family of proteins. Within this family, SIRT1 and SIRT3 offer the strongest connection with improvements in mitochondrial health. Small molecule activation of SIRT1 has been shown in vivo to improve mitochondrial respiratory capacity in mice on a high-fat diet (Minor et al. 2011). In healthy human volunteers, a next-generation SIRT1 activator (SRT2104) has been shown to decrease serum lipid levels and increase the recovery of adenosine diphosphate and phosphocreatine after exercise, which is suggestive of an increase in mitochondrial oxidative phosphorylation (Libri et al. 2012). Although there are no human in vivo studies of the effects of SIRT1 activation on mitochondrial diseases, in vitro studies with the putative SIRT1 activator resveratrol have shown improvements in mitochondrial fatty acid oxidation in fibroblasts derived from patients with carnitine palmitoyltransferase II or very long-chain acyl-CoA dehydrogenase deficiencies (Bastin et al. 2011). In the case of SIRT3, up-regulation of activity has been shown to improve mitochondrial homeostasis in mice (Brown et al. 2013), although no data are available in humans. Ideally, therapies that simultaneously increase SIRT1 and SIRT3 activities, such as supplementation with the precursor nicotinamide riboside to increase the levels of their NAD+ cosubstrate (Canto et al. 2012), would be the most desirable for promoting mitochondrial function.
Prevention of Mitochondrial Dysregulation Associated with Aging and Metabolic Disorders
Many of the therapeutic strategies described for the amelioration of genetically based mitochondrial disorders are applicable to the decline in mitochondrial function that accompanies age-related senescence or certain metabolic disease states such as Type-2 diabetes. Other therapeutic options are available to the nongenetic mitochondrial disorders, because of their generally reduced phenotypic severity. These options include dietary manipulation and physical exercise. Restriction of caloric intake can result in profound alterations in mitochondrial physiology, including increased proliferation, enhanced bioenergetic efficiency, reduced membrane potential, and reduced production of oxidative species in mice (Lopez-Lluch et al. 2006) and in humans (Civitarese et al. 2007). These mitochondrial changes are thought to be a major contributor to the ability of caloric restriction to prevent the development of diseases linked to aging. Indeed, calorie restriction extends life span in different organisms but the molecular mechanisms are not completely understood (Sohal and Weindruch 1996; Spindler 2001). Proposed mechanisms for the longevity effects include changes in the signaling pathways associated with nutrient availability and insulin levels that converge to regulate mTOR (Kaeberlein et al. 2005; Zoncu et al. 2011) and FoxOs (Salih and Brunet 2008) protein activities. In terms of mitochondrial function, however, other regulatory pathways appear to be connected with the effects of caloric restriction, namely, the AMPK, sirtuins, and PGC1α regulatory network.
Exercise is an extremely powerful inducer of mitochondrial activity and proliferation—particularly in skeletal muscle (Yan et al. 2011). Coincident with this change, exercise is also able to strongly protect against many types of age-related metabolic disorders. Less clear, however, is whether exercise can mimic the longevity effects of caloric restriction. An important point, however, is whether the beneficial health effects of exercise are to an increase in the mass of mitochondrial and/or the myriad other cellular and systemic changes that occur with exercise.
CONCLUDING REMARKS
Researchers across multiple disciplines have made considerable progress by uncovering many important signaling pathways that control mitochondrial function at the level of nuclear transcription and discerning how these pathways are targeted through extrinsic signals to ultimately govern mitochondrial activities. Collectively, these signaling pathways can effectively boost the cell’s mitochondrial mass, improve the mitochondrial bioenergetics profile, and protect it against oxidative stress. As such, they represent a very fertile ground for the development of therapies for the rectification of diseases characterized by mitochondrial dysfunction.
ACKNOWLEDGMENTS
Mitochondrial-related research in the Puigserver laboratory is supported from the Dana-Farber Cancer Institute funds and grants from the NIH/NIDDK, American Diabetes Association, American Heart Association, and Muscle Dystrophy Association.
Footnotes
Editors: Douglas C. Wallace and Richard J. Youle
Additional Perspectives on Mitochondria available at www.cshperspectives.org
REFERENCES
- Ashrafi G, Schwarz TL 2013. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ 20: 31–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker BM, Haynes CM 2011. Mitochondrial protein quality control during biogenesis and aging. Trends Biochem Sci 36: 254–261 [DOI] [PubMed] [Google Scholar]
- Bastin J, Lopes-Costa A, Djouadi F 2011. Exposure to resveratrol triggers pharmacological correction of fatty acid utilization in human fatty acid oxidation-deficient fibroblasts. Hum Mol Genet 20: 2048–2057 [DOI] [PubMed] [Google Scholar]
- Baughman JM, Perocchi F, Girgis HS, Plovanich M, Belcher-Timme CA, Sancak Y, Bao XR, Strittmatter L, Goldberger O, Bogorad RL, et al. 2011. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature 476: 341–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal MF 2009. Therapeutic approaches to mitochondrial dysfunction in Parkinson’s disease. Parkinsonism Relat Disord 15 (Suppl 3): S189–S194 [DOI] [PubMed] [Google Scholar]
- Bianchi K, Vandecasteele G, Carli C, Romagnoli A, Szabadkai G, Rizzuto R 2006. Regulation of Ca2+ signalling and Ca2+-mediated cell death by the transcriptional coactivator PGC-1α. Cell Death Differ 13: 586–596 [DOI] [PubMed] [Google Scholar]
- Birkenfeld AL, Lee HY, Guebre-Egziabher F, Alves TC, Jurczak MJ, Jornayvaz FR, Zhang D, Hsiao JJ, Martin-Montalvo A, Fischer-Rosinsky A, et al. 2011. Deletion of the mammalian INDY homolog mimics aspects of dietary restriction and protects against adiposity and insulin resistance in mice. Cell Metab 14: 184–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattler SM, Cunningham JT, Verdeguer F, Chim H, Haas W, Liu H, Romanino K, Ruegg MA, Gygi S, Shi Y, et al. 2012a. Yin Yang 1 deficiency in skeletal muscle protects against rapamycin-induced diabetic-like symptoms through activation of insulin/IGF signaling. Cell Metab 15: 505–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattler SM, Verdeguer F, Liesa M, Cunningham JT, Vogel RO, Chim H, Liu H, Romanino K, Shirihai OS, Vazquez F, et al. 2012b. Defective mitochondrial morphology and bioenergetic function in mice lacking the transcription factor Yin Yang 1 in skeletal muscle. Mol Cell Biol 32: 3333–3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnefont JP, Bastin J, Behin A, Djouadi F 2009. Bezafibrate for an inborn mitochondrial β-oxidation defect. N Engl J Med 360: 838–840 [DOI] [PubMed] [Google Scholar]
- Brown K, Xie S, Qiu X, Mohrin M, Shin J, Liu Y, Zhang D, Scadden DT, Chen D 2013. SIRT3 reverses aging-associated degeneration. Cell Rep 3: 319–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo SE, Mootha VK 2010. The mitochondrial proteome and human disease. Annu Rev Genomics Hum Genet 11: 25–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam H, Balciunaite E, Blais A, Spektor A, Scarpulla RC, Young R, Kluger Y, Dynlacht BD 2004. A common set of gene regulatory networks links metabolism and growth inhibition. Mol Cell 16: 399–411 [DOI] [PubMed] [Google Scholar]
- Cannon B, Nedergaard J 2004. Brown adipose tissue: Function and physiological significance. Physiol Rev 84: 277–359 [DOI] [PubMed] [Google Scholar]
- Canto C, Auwerx J 2009. PGC-1α, SIRT1 and AMPK, an energy sensing network that controls energy expenditure. Curr Opin Lipidol 20: 98–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J 2009. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 458: 1056–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Houtkooper RH, Pirinen E, Youn DY, Oosterveer MH, Cen Y, Fernandez-Marcos PH, Yamamoto H, Andreux PA, Cettour-Rose P, et al. 2012. The NAD+ precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metab 15: 838–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter RS, Bhat NK, Basu A, Avadhani NG 1992. The basal promoter elements of murine cytochrome c oxidase subunit IV gene consist of tandemly duplicated ets motifs that bind to GABP-related transcription factors. J Biol Chem 267: 23418–23426 [PubMed] [Google Scholar]
- Cartoni R, Leger B, Hock MB, Praz M, Crettenand A, Pich S, Ziltener JL, Luthi F, Deriaz O, Zorzano A, et al. 2005. Mitofusins 1/2 and ERRα expression are increased in human skeletal muscle after physical exercise. J Physiol 567: 349–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas F, Rochard P, Rodier A, Cassar-Malek I, Marchal-Victorion S, Wiesner RJ, Cabello G, Wrutniak C 1999. A variant form of the nuclear triiodothyronine receptor c-ErbAα1 plays a direct role in regulation of mitochondrial RNA synthesis. Mol Cell Biol 19: 7913–7924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae S, Ahn BY, Byun K, Cho YM, Yu MH, Lee B, Hwang D, Park KS 2013. A systems approach for decoding mitochondrial retrograde signaling pathways. Science Signal 6: rs4. [DOI] [PubMed] [Google Scholar]
- Charos AE, Reed BD, Raha D, Szekely AM, Weissman SM, Snyder M 2012. A highly integrated and complex PPARGC1A transcription factor binding network in HepG2 cells. Genome Res 22: 1668–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Chan DC 2009. Mitochondrial dynamics—fusion, fission, movement, and mitophagy—in neurodegenerative diseases. Hum Mol Genet 18: R169–R176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Chan DC 2010. Physiological functions of mitochondrial fusion. Ann NY Acad Sci 1201: 21–25 [DOI] [PubMed] [Google Scholar]
- Civitarese AE, Carling S, Heilbronn LK, Hulver MH, Ukropcova B, Deutsch WA, Smith SR, Ravussin E 2007. Calorie restriction increases muscle mitochondrial biogenesis in healthy humans. PLoS Med 4: e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrado M, Scorrano L, Campello S 2012. Mitochondrial dynamics in cancer and neurodegenerative and neuroinflammatory diseases. Int J Cell Biol 2012: 729290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P 2007. mTOR controls mitochondrial oxidative function through a YY1-PGC-1α transcriptional complex. Nature 450: 736–740 [DOI] [PubMed] [Google Scholar]
- De Stefani D, Raffaello A, Teardo E, Szabo I, Rizzuto R 2011. A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature 476: 336–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMauro S, Hirano M 2009. Pathogenesis and treatment of mitochondrial disorders. Adv Exp Med Biol 652: 139–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinardo MM, Musicco C, Fracasso F, Milella F, Gadaleta MN, Gadaleta G, Cantatore P 2003. Acetylation and level of mitochondrial transcription factor A in several organs of young and old rats. Biochem Biophys Res Commun 301: 187–191 [DOI] [PubMed] [Google Scholar]
- Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, Kim J, Woo J, Kim JH, Choi BH, et al. 2011. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science 334: 806–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichner LJ, Giguere V 2011. Estrogen related receptors (ERRs): A new dawn in transcriptional control of mitochondrial gene networks. Mitochondrion 11: 544–552 [DOI] [PubMed] [Google Scholar]
- Endo T, Yamano K 2009. Multiple pathways for mitochondrial protein traffic. Biol Chem 390: 723–730 [DOI] [PubMed] [Google Scholar]
- Endo T, Yamano K, Kawano S 2011. Structural insight into the mitochondrial protein import system. Biochim Biophys Acta 1808: 955–970 [DOI] [PubMed] [Google Scholar]
- Evans MJ, Scarpulla RC 1989. Interaction of nuclear factors with multiple sites in the somatic cytochrome c promoter. Characterization of upstream NRF-1, ATF, and intron Sp1 recognition sequences. J Biol Chem 264: 14361–14368 [PubMed] [Google Scholar]
- Finkel T, Deng CX, Mostoslavsky R 2009. Recent progress in the biology and physiology of sirtuins. Nature 460: 587–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebert N, Ryan MT, Pfanner N, Wiedemann N, Stojanovski D 2011. Mitochondrial protein import machineries and lipids: A functional connection. Biochim Biophys Acta 1808: 1002–1011 [DOI] [PubMed] [Google Scholar]
- Gerhart-Hines Z, Dominy JE Jr, Blattler SM, Jedrychowski MP, Banks AS, Lim JH, Chim H, Gygi SP, Puigserver P 2011. The cAMP/PKA pathway rapidly activates SIRT1 to promote fatty acid oxidation independently of changes in NAD+. Mol Cell 44: 851–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleyzer N, Vercauteren K, Scarpulla RC 2005. Control of mitochondrial transcription specificity factors (TFB1M and TFB2M) by nuclear respiratory factors (NRF-1 and NRF-2) and PGC-1 family coactivators. Mol Cell Biol 25: 1354–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golubitzky A, Dan P, Weissman S, Link G, Wikstrom JD, Saada A 2011. Screening for active small molecules in mitochondrial complex I deficient patient’s fibroblasts, reveals AICAR as the most beneficial compound. PloS ONE 6: e26883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan L, Scarpulla RC 1994. Differential regulation of respiratory chain subunits by a CREB-dependent signal transduction pathway. Role of cyclic AMP in cytochrome c and COXIV gene expression. J Biol Chem 269: 105–113 [PubMed] [Google Scholar]
- Guarente L 2011. Franklin H. Epstein Lecture: Sirtuins, aging, and medicine. N Engl J Med 364: 2235–2244 [DOI] [PubMed] [Google Scholar]
- Gugneja S, Virbasius JV, Scarpulla RC 1995. Four structurally distinct, non-DNA-binding subunits of human nuclear respiratory factor 2 share a conserved transcriptional activation domain. Mol Cell Biol 15: 102–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulick T, Cresci S, Caira T, Moore DD, Kelly DP 1994. The peroxisome proliferator-activated receptor regulates mitochondrial fatty acid oxidative enzyme gene expression. Proc Natl Acad Sci 91: 11012–11016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis MC, Sinclair DA 2010. Mammalian sirtuins: Biological insights and disease relevance. Annu Rev Pathol 5: 253–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handschin C, Spiegelman BM 2011. PGC-1 coactivators and the regulation of skeletal muscle fiber-type determination. Cell Metab 13: 351; author reply 352 [DOI] [PubMed] [Google Scholar]
- Hardie DG 2007. AMP-activated/SNF1 protein kinases: Conserved guardians of cellular energy. Nat Rev Mol Cell Biol 8: 774–785 [DOI] [PubMed] [Google Scholar]
- Haynes CM, Yang Y, Blais SP, Neubert TA, Ron D 2010. The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans. Mol Cell 37: 529–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert AS, Dittenhafer-Reed KE, Yu W, Bailey DJ, Selen ES, Boersma MD, Carson JJ, Tonelli M, Balloon AJ, Higbee AJ, et al. 2013. Calorie restriction and SIRT3 trigger global reprogramming of the mitochondrial protein acetylome. Mol Cell 49: 186–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzig S, Long F, Jhala US, Hedrick S, Quinn F, Bauer A, Rudolph D, Schutz G, Yoon C, Puigserver P, et al. 2001. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature 413: 179–183 [DOI] [PubMed] [Google Scholar]
- Hirano M, Garone C, Quinzii CM 2012. CoQ10 deficiencies and MNGIE: Two treatable mitochondrial disorders. Biochim Biophys Acta 1820: 625–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschey MD, Shimazu T, Jing E, Grueter CA, Collins AM, Aouizerat B, Stancakova A, Goetzman E, Lam MM, Schwer B, et al. 2011. SIRT3 deficiency and mitochondrial protein hyperacetylation accelerate the development of the metabolic syndrome. Mol Cell 44: 177–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager S, Handschin C, St-Pierre J, Spiegelman BM 2007. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1α. Proc Natl Acad Sci 104: 12017–12022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW III, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK 2005. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science 310: 1193–1196 [DOI] [PubMed] [Google Scholar]
- Kahn BB, Alquier T, Carling D, Hardie DG 2005. AMP-activated protein kinase: Ancient energy gauge provides clues to modern understanding of metabolism. Cell Metab 1: 15–25 [DOI] [PubMed] [Google Scholar]
- Kelly DP, Scarpulla RC 2004. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev 18: 357–368 [DOI] [PubMed] [Google Scholar]
- Klar AJ, Fogel S, Macleod K 1979. MAR1—A regulator of the HMa and HMα loci in Saccharomyces cerevisiae. Genetics 93: 37–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiner S, Nguyen-Tran V, Bare O, Huang X, Spiegelman B, Wu Z 2009. PPARδ agonism activates fatty acid oxidation via PGC-1α but does not increase mitochondrial gene expression and function. J Biol Chem 284: 18624–18633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri V, Brown AP, Gambarota G, Haddad J, Shields GS, Dawes H, Pinato DJ, Hoffman E, Elliot PJ, Vlasuk GP, et al. 2012. A pilot randomized, placebo controlled, double blind phase I trial of the novel SIRT1 activator SRT2104 in elderly volunteers. PloS ONE 7: e51395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN, et al. 2002. Transcriptional co-activator PGC-1 α drives the formation of slow-twitch muscle fibres. Nature 418: 797–801 [DOI] [PubMed] [Google Scholar]
- Liu Z, Butow RA 2006. Mitochondrial retrograde signaling. Annu Rev Genet 40: 159–185 [DOI] [PubMed] [Google Scholar]
- Lopez-Lluch G, Hunt N, Jones B, Zhu M, Jamieson H, Hilmer S, Cascajo MV, Allard J, Ingram DK, Navas P, et al. 2006. Calorie restriction induces mitochondrial biogenesis and bioenergetic efficiency. Proc Natl Acad Sci 103: 1768–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, Lee J, Nie X, Li M, Morozov YI, Venkatesh S, Bogenhagen DF, Temiakov D, Suzuki CK 2013. Phosphorylation of human TFAM in mitochondria impairs DNA binding and promotes degradation by the AAA+ Lon protease. Mol Cell 49: 121–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr B, Montminy M 2001. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol 2: 599–609 [DOI] [PubMed] [Google Scholar]
- Miller BF, Robinson MM, Bruss MD, Hellerstein M, Hamilton KL 2012. A comprehensive assessment of mitochondrial protein synthesis and cellular proliferation with age and caloric restriction. Aging Cell 11: 150–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor RK, Baur JA, Gomes AP, Ward TM, Csiszar A, Mercken EM, Abdelmohsen K, Shin YK, Canto C, Scheibye-Knudsen M, et al. 2011. SRT1720 improves survival and healthspan of obese mice. Scientific Rep 1: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, et al. 2003. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34: 267–273 [DOI] [PubMed] [Google Scholar]
- Mootha VK, Handschin C, Arlow D, Xie X, St Pierre J, Sihag S, Yang W, Altshuler D, Puigserver P, Patterson N, et al. 2004. Errα and Gabpa/b specify PGC-1α-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc Natl Acad Sci 101: 6570–6575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Lomb DJ, Haigis MC, Guarente L 2009. SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell 137: 560–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D, Walker JE, Youle R 2012. Mitochondrial quality control mediated by PINK1 and Parkin: Links to parkinsonism. Cold Spring Harb Perspect Biol 4: a011338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narkar VA, Downes M, Yu RT, Embler E, Wang YX, Banayo E, Mihaylova MM, Nelson MC, Zou Y, Juguilon H, et al. 2008. AMPK and PPARδ agonists are exercise mimetics. Cell 134: 405–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrin N, Wu X, Fortier E, Feng Y, Bare OC, Chen S, Ren X, Wu Z, Streeper RS, Bordone L 2010. SIRT4 regulates fatty acid oxidation and mitochondrial gene expression in liver and muscle cells. J Biol Chem 285: 31995–32002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owusu-Ansah E, Yavari A, Mandal S, Banerjee U 2008. Distinct mitochondrial retrograde signals control the G1-S cell cycle checkpoint. Nat Genet 40: 356–361 [DOI] [PubMed] [Google Scholar]
- Park SJ, Ahmad F, Philp A, Baar K, Williams T, Luo H, Ke H, Rehmann H, Taussig R, Brown AL, et al. 2012. Resveratrol ameliorates aging-related metabolic phenotypes by inhibiting cAMP phosphodiesterases. Cell 148: 421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peralta S, Wang X, Morales CT 2012. Mitochondrial transcription: Lessons from mouse models. Biochim Biophys Acta 1819: 961–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen KF, Befroy D, Dufour S, Dziura J, Ariyan C, Rothman DL, DiPietro L, Cline GW, Shulman GI 2003. Mitochondrial dysfunction in the elderly: Possible role in insulin resistance. Science 300: 1140–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price NL, Gomes AP, Ling AJ, Duarte FV, Martin-Montalvo A, North BJ, Agarwal B, Ye L, Ramadori G, Teodoro JS, et al. 2012. SIRT1 is required for AMPK activation and the beneficial effects of resveratrol on mitochondrial function. Cell Metab 15: 675–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver P, Spiegelman BM 2003. Peroxisome proliferator-activated receptor-γ coactivator 1α (PGC-1α): Transcriptional coactivator and metabolic regulator. Endocr Rev 24: 78–90 [DOI] [PubMed] [Google Scholar]
- Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM 1998. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92: 829–839 [DOI] [PubMed] [Google Scholar]
- Purushotham A, Schug TT, Xu Q, Surapureddi S, Guo X, Li X 2009. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab 9: 327–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasbach KA, Gupta RK, Ruas JL, Wu J, Naseri E, Estall JL, Spiegelman BM 2010. PGC-1α regulates a HIF2α-dependent switch in skeletal muscle fiber types. Proc Natl Acad Sci 107: 21866–21871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebelo AP, Dillon LM, Moraes CT 2011. Mitochondrial DNA transcription regulation and nucleoid organization. J Inherit Metabol Dis 34: 941–951 [DOI] [PubMed] [Google Scholar]
- Reznick RM, Zong H, Li J, Morino K, Moore IK, Yu HJ, Liu ZX, Dong J, Mustard KJ, Hawley SA, et al. 2007. Aging-associated reductions in AMP-activated protein kinase activity and mitochondrial biogenesis. Cell Metab 5: 151–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P 2005. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature 434: 113–118 [DOI] [PubMed] [Google Scholar]
- Salih DA, Brunet A 2008. FoxO transcription factors in the maintenance of cellular homeostasis during aging. Curr Opin Cell Biol 20: 126–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla RC 2008. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev 88: 611–638 [DOI] [PubMed] [Google Scholar]
- Scarpulla RC, Vega RB, Kelly DP 2012. Transcriptional integration of mitochondrial biogenesis. Trends Endocrinol Metab: 23: 459–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schieke SM, Phillips D, McCoy JP Jr, Aponte AM, Shen RF, Balaban RS, Finkel T 2006. The mammalian target of rapamycin (mTOR) pathway regulates mitochondrial oxygen consumption and oxidative capacity. J Biol Chem 281: 27643–27652 [DOI] [PubMed] [Google Scholar]
- Schmidt O, Pfanner N, Meisinger C 2010. Mitochondrial protein import: From proteomics to functional mechanisms. Nat Rev Mol Cell Biol 11: 655–667 [DOI] [PubMed] [Google Scholar]
- Schreiber SN, Emter R, Hock MB, Knutti D, Cardenas J, Podvinec M, Oakeley EJ, Kralli A 2004. The estrogen-related receptor α (ERRα) functions in PPARγ coactivator 1α (PGC-1α)-induced mitochondrial biogenesis. Proc Natl Acad Sci 101: 6472–6477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Kajimura S, Yang W, Chin S, Rohas LM, Uldry M, Tavernier G, Langin D, Spiegelman BM 2007. Transcriptional control of brown fat determination by PRDM16. Cell Metab 6: 38–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Peterson TR, Laplante M, Oh S, Sabatini DM 2010. mTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Nature 468: 1100–1104 [DOI] [PubMed] [Google Scholar]
- Shin JH, Ko HS, Kang H, Lee Y, Lee YI, Pletinkova O, Troconso JC, Dawson VL, Dawson TM 2011. PARIS (ZNF746) repression of PGC-1α contributes to neurodegeneration in Parkinson’s disease. Cell 144: 689–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JE 1995. Thyroid hormone control of thermogenesis and energy balance. Thyroid 5: 481–492 [DOI] [PubMed] [Google Scholar]
- Sohal RS, Weindruch R 1996. Oxidative stress, caloric restriction, and aging. Science 273: 59–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano FX, Liesa M, Bach D, Chan DC, Palacin M, Zorzano A 2006. Evidence for a mitochondrial regulatory pathway defined by peroxisome proliferator-activated receptor-gamma coactivator-1α, estrogen-related receptor-α, and mitofusin 2. Diabetes 55: 1783–1791 [DOI] [PubMed] [Google Scholar]
- Spindler SR 2001. Calorie restriction enhances the expression of key metabolic enzymes associated with protein renewal during aging. Ann NY Acad Sci 928: 296–304 [DOI] [PubMed] [Google Scholar]
- Srivastava S, Diaz F, Iommarini L, Aure K, Lombes A, Moraes CT 2009. PGC-1α/β induced expression partially compensates for respiratory chain defects in cells from patients with mitochondrial disorders. Hum Mol Genet 18: 1805–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowers RS, Megeath LJ, Gorska-Andrzejak J, Meinertzhagen IA, Schwarz TL 2002. Axonal transport of mitochondria to synapses depends on milton, a novel Drosophila protein. Neuron 36: 1063–1077 [DOI] [PubMed] [Google Scholar]
- Twig G, Hyde B, Shirihai OS 2008. Mitochondrial fusion, fission and autophagy as a quality control axis: The bioenergetic view. Biochim Biophys Acta 1777: 1092–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uldry M, Yang W, St-Pierre J, Lin J, Seale P, Spiegelman BM 2006. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab 3: 333–341 [DOI] [PubMed] [Google Scholar]
- Venditti P, Bari A, Di Stefano L, Cardone A, Della Ragione F, D’Esposito M, Di Meo S 2009. Involvement of PGC-1, NRF-1, and NRF-2 in metabolic response by rat liver to hormonal and environmental signals. Mol Cell Endocrinol 305: 22–29 [DOI] [PubMed] [Google Scholar]
- Verdin E, Hirschey MD, Finley LW, Haigis MC 2010. Sirtuin regulation of mitochondria: Energy production, apoptosis, and signaling. Trends Biochem Sci 35: 669–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viscomi C, Bottani E, Civiletto G, Cerutti R, Moggio M, Fagiolari G, Schon EA, Lamperti C, Zeviani M 2011. In vivo correction of COX deficiency by activation of the AMPK/PGC-1α axis. Cell Metab 14: 80–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC, Fan W 2009. The pathophysiology of mitochondrial disease as modeled in the mouse. Genes Dev 23: 1714–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Winter D, Ashrafi G, Schlehe J, Wong YL, Selkoe D, Rice S, Steen J, LaVoie MJ, Schwarz TL 2011. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell 147: 893–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenz T, Diaz F, Spiegelman BM, Moraes CT 2008. Activation of the PPAR/PGC-1α pathway prevents a bioenergetic deficit and effectively improves a mitochondrial myopathy phenotype. Cell Metab 8: 249–256 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wenz T, Rossi SG, Rotundo RL, Spiegelman BM, Moraes CT 2009. Increased muscle PGC-1α expression protects from sarcopenia and metabolic disease during aging. Proc Natl Acad Sci 106: 20405–20410 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Westermann B 2010. Mitochondrial fusion and fission in cell life and death. Nat Rev Mol Cell Biol 11: 872–884 [DOI] [PubMed] [Google Scholar]
- Wilson BJ, Tremblay AM, Deblois G, Sylvain-Drolet G, Giguere V 2010. An acetylation switch modulates the transcriptional activity of estrogen-related receptor α. Mol Endocrinol 24: 1349–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson-Fritch L, Nicoloro S, Chouinard M, Lazar MA, Chui PC, Leszyk J, Straubhaar J, Czech MP, Corvera S 2004. Mitochondrial remodeling in adipose tissue associated with obesity and treatment with rosiglitazone. J Clin Invest 114: 1281–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright DC, Geiger PC, Han DH, Jones TE, Holloszy JO 2007. Calcium induces increases in peroxisome proliferator-activated receptor γ coactivator-1α and mitochondrial biogenesis by a pathway leading to p38 mitogen-activated protein kinase activation. J Biol Chem 282: 18793–18799 [DOI] [PubMed] [Google Scholar]
- Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, et al. 1999. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 98: 115–124 [DOI] [PubMed] [Google Scholar]
- Wu H, Kanatous SB, Thurmond FA, Gallardo T, Isotani E, Bassel-Duby R, Williams RS 2002. Regulation of mitochondrial biogenesis in skeletal muscle by CaMK. Science 296: 349–352 [DOI] [PubMed] [Google Scholar]
- Yan Z, Okutsu M, Akhtar YN, Lira VA 2011. Regulation of exercise-induced fiber type transformation, mitochondrial biogenesis, and angiogenesis in skeletal muscle. J Appl Physiol 110: 264–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsuga S, Suomalainen A 2012. Effect of bezafibrate treatment on late-onset mitochondrial myopathy in mice. Hum Mol Genet 21: 526–535 [DOI] [PubMed] [Google Scholar]
- Youle RJ, van der Bliek AM 2012. Mitochondrial fission, fusion, and stress. Science 337: 1062–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Dittenhafer-Reed KE, Denu JM 2012. SIRT3 protein deacetylates isocitrate dehydrogenase 2 (IDH2) and regulates mitochondrial redox status. J Biol Chem 287: 14078–14086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Wang J, Levichkin IV, Stasinopoulos S, Ryan MT, Hoogenraad NJ 2002. A mitochondrial specific stress response in mammalian cells. EMBO J 21: 4411–4419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Liao Z, Locascio JJ, Lesniak KA, Roderick SS, Watt ML, Eklund AC, Zhang-James Y, Kim PD, Hauser MA, et al. 2010. PGC-1α, a potential therapeutic target for early intervention in Parkinson’s disease. Sci Transl Med 2: 52ra73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM 2011. mTOR: From growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol 12: 21–35 [DOI] [PMC free article] [PubMed] [Google Scholar]