Abstract
The accurate copying of genetic information in the double helix of DNA is essential for inheritance of traits that define the phenotype of cells and the organism. The core machineries that copy DNA are conserved in all three domains of life: bacteria, archaea, and eukaryotes. This article outlines the general nature of the DNA replication machinery, but also points out important and key differences. The most complex organisms, eukaryotes, have to coordinate the initiation of DNA replication from many origins in each genome and impose regulation that maintains genomic integrity, not only for the sake of each cell, but for the organism as a whole. In addition, DNA replication in eukaryotes needs to be coordinated with inheritance of chromatin, developmental patterning of tissues, and cell division to ensure that the genome replicates once per cell division cycle.
DNA replication involves an unimaginable amount of information transfer, especially in eukaryotes. It requires machinery that repairs errors, ensures chromatin inheritance, and coordinates with cell division.
The genetic information within the cells of our body is stored in the double helix of DNA, a long cylinderlike structure with a radius that is only 10 Å or one billionth of a meter but can be of considerable length. A single DNA molecule within a bacterium that grows in our gut flora is approximately 5 million base pairs in length and when stretched out, is about 1.6 mm in length, roughly the diameter of a pinhead. In contrast, the single DNA molecule in the largest human chromosome is 245,203,898 base pairs or about 8.33 cm long. The entire human genome, consisting of its 24 different chromosomes in a male is about 3 billion base pairs or 1 m long. Each cell in our body, with rare exceptions, contains two copies of the genome and thus 2 m of total DNA. Thus the scale and complexity of duplicating genomes is remarkable. For example, ∼2200 human cells can sit on the top of a 1.5 mm pinhead and when extracted and laid out in a line, the DNA from these cells would be ∼4.5 km (2.8 miles) long. In our body, about 500–700 million new blood cells are born every minute in the bone marrow (Doulatov et al. 2012), containing a total of about 1 million km of DNA, or enough DNA to wrap around the equator of the earth 25 times. Thus DNA replication is a serious business in our body, occurring from the time that a fertilized egg first begins duplicating DNA to yield the many trillions of cells that make up an adult body and continuing in all tissues of the adult body throughout our life. The amount of DNA duplicated in an entire human body represents an unimaginable amount of information transfer. Moreover, each round of duplication needs to be highly accurate, making one mistake in less than 100 million bases copied per cell division. How copying of the double helix occurs and how it is so highly accurate is the topic of this collection. Inevitably the processes of accurate copying of the genome can go awry, yielding mutations that affect our lives, and thus the collection outlines the disorders that accelerate human disease.
However, the problem of copying DNA is much more complicated than indicated above. The 2 m of DNA in each human cell is wrapped up with histone proteins within the cell’s nucleus that is only about 5 μm wide, presenting a compaction in DNA length of about 2 million-fold. How can the copying process deal with the fact that the DNA is wrapped around proteins and scrunched into a volume that creates a spatial organization problem of enormous magnitude? Not only is the DNA copied, but the proteins associated with the DNA need to be duplicated, along with all the chemical modifications attached to DNA and histones that greatly influence developmental patterning of gene expression. The protein machineries that replicate DNA and duplicate proteins within the chromosomes are some of the most complex and intriguing machineries known. Furthermore, the regulations of the processes are some of the most complex because they need to ensure that each DNA molecule in each chromosome is copied once, and only once each time before a cell divides. Errors in the regulation of DNA replication lead to accelerated mutation rates, often associated with increased rates of cancer and other diseases.
The process of accurately copying a genome can be broken down into various subprocesses that combine to provide efficient genome duplication. Central to the entire process is the machinery that actually copies the DNA with high fidelity, including proteins that start the entire process and the proteins that actually copy one helix to produce two. Superimposed on this fundamental process are mechanisms that detect and repair errors and damage to the DNA. Also associated with the DNA replication apparatus are the proteins that ensure that the histone proteins and their modifications in chromatin are inherited along with the DNA. Finally, other machineries cooperate with the DNA replication apparatus to ensure that the resulting two DNA molecules, the sister chromatids, are tethered together until the cell completes duplicating all of its DNA and segregates the sister chromatids evenly to the two daughter cells. Only by combining all of these processes can genetic inheritance ensure that each cell has a faithful copy of its parent’s genome.
WHERE TO START
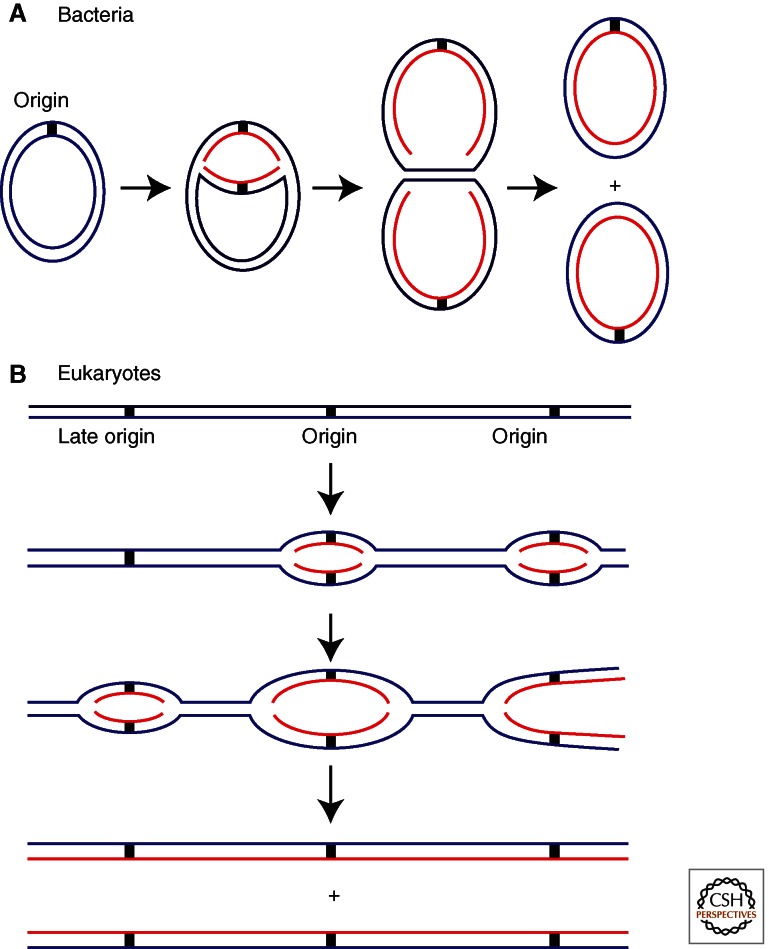
Replication begins at particular positions in chromosomes called “origins” where designated initiator proteins bind to DNA to start the process of replication. There are important differences among bacteria, archaea, and eukaryotes in this process, but there are also many striking similarities that suggest the process dates back to the last universal cellular ancestor (Stillman 2005; Kaguni 2011). Bacteria often contain only one chromosome with one origin at which two replication forks assemble and move in opposite directions (Fig. 1A). Although not all bacteria follow this paradigm, this is the case for the Escherichia coli circular 4.4 Mb genome, forming a single replicon or unit of replication from a single origin. At a rate of 1 Kb/s for each fork, this genome is replicated within 30 min. In contrast, eukaryotes typically have multiple linear chromosomes, each with many origins. Multiple origins are a necessity for eukaryotes as they have much larger genomes than bacteria and eukaryotic replication forks move about 20 times more slowly than bacterial replication forks. As an example, the largest human chromosome (chromosome #1) is 250 Mb and if it had only one origin, it would require more than 50 days to replicate compared to the typical 24 h division time of a eukaryotic cell and approximately 8 h for copying DNA in S phase. Initiation at each origin produces two divergent DNA replication forks along the chromosome to create a replicon that is duplicated only once per cell division. The duplication of many replicons eventually yields two daughter chromosomes called sister chromatids that are tethered together until they separate during mitosis (Fig. 1B). Although few archaea species have been characterized, they appear to be evolutionary hybrids between bacteria and eukaryotes, because some species have a single chromosome with a single origin, whereas other species have multiple origins per chromosome (Samson and Bell 2011). Moreover, the ploidy of the genome in archaea varies considerably, with some species having a 1C–2C distribution throughout their cell cycle, whereas others have up to 25 copies of their genome in proliferating cells. The rate of DNA replication fork progression also appears to be in between that in bacteria and eukaryotes, at about 20 kB/min, ∼10 times faster than that in eukaryotes. Although some bacteria like E. coli replicate their genomes much faster, others such as Caulobacter crescentus replicate at roughly the same rate as some archaea, such as Pyrococcus abyssi (Dingwall and Shapiro 1989; Myllykallio et al. 2000).
Figure 1.
Replication initiation in bacteria and eukaryotes. (A) Most bacteria have a circular chromosome with one origin, although there are exceptions to this. Illustrated here is the E. coli chromosome that has one origin from which two replication forks proceed in opposite directions. (B) Eukaryotes have long linear chromosomes. Bidirectional replication is initiated at multiple origins along each chromosome.
Bacterial origins are well-defined sequences to which the replication initiator proteins bind. In contrast, eukaryotic origins are not typically defined at the level of DNA sequence (with the important exception of the budding yeast Saccharomyces cerevisiae). Significant recent progress has indicated that eukaryotic origins are defined less by DNA sequence than by chromatin organization, with many origins corresponding to regions of DNA with transcriptional activity or other features that allow access to origin-binding proteins (Masai et al. 2010). Many human cell origins occur in sequences that are evolutionarily conserved among mammals, suggesting that they are far from arbitrary (Cadoret et al. 2008). In most eukaryotes, a small subset of potential origins is used in a typical cell cycle in individual cells, but origin utilization can be greatly increased to facilitate astonishingly rapid cell division, as seen in the fertilized eggs of many animals (Rhind 2006). Whether an origin is used or not is a stochastic process that depends on the chromatin context and in some cases the developmental state of cells in multicellular organisms. Furthermore the multiple origins in eukaryotic chromosomes are organized into clusters that are activated at specific times during S phase of the cell cycle, and the temporal patterning varies again with developmental patterning of cells (Gilbert et al. 2010). Origins of replication are discussed in Leonard and Méchali (2013).
To begin the process of activating an origin for replication, bacterial, archaeal, and eukaryotic cells use origin-binding proteins composed of AAA+ family subunit(s) (Erzberger and Berger 2006). AAA+ proteins generally function as multimeric machines. For example in bacteria, multiple copies of the DnaA origin-binding protein form a helical filament that binds the origin (Kaguni 2011). The DnaA filament binds ATP to unwind an A/T-rich region of the origin, resulting in a single-strand DNA (ssDNA) “bubble” onto which the replicative helicase loads (described in the next section).
Eukaryotes contain a six subunit origin-binding protein referred to as ORC (origin recognition complex) (Stillman 2005). Five of the ORC subunits are related to AAA+ proteins and together with another AAA+ protein called Cdc6 that is highly related in sequence to the largest ORC subunit, Orc1, they form a ring-shaped hexamer that binds DNA (Sun et al. 2012). However, unlike bacterial DnaA, ORC does not unwind DNA at regions to which it binds. Archaeal cells also use AAA+ proteins that are related to the largest subunit of ORC, Orc1 and to Cdc6, but the number of these subunits varies depending on the particular type of archaeal cell (Barry and Bell 2006). Both DnaA and ORC are used in other processes besides replication (see Bell and Kaguni 2013).
LOADING THE HELICASE
The objective of origin-binding proteins in bacteria, archaea, and eukaryotes is the loading of two helicases onto DNA, which eventually give rise to two DNA replication forks that move in opposite directions from each origin. In all three domains of life, the helicase is a six subunit complex that unwinds DNA by encircling one strand of the parental duplex (Gai et al. 2010). Each helicase uses ATP hydrolysis to translocate along the single strand, acting as a moving wedge to force the parental duplex apart. The cellular DNA helicases are similar to hexameric enzymes present in several eukaryotic cell viruses such as the simian virus 40 T antigen and the human papillomavirus E1 helicase. Beyond these important similarities lie many differences among the replicative helicases of bacteria, archaea, and eukaryotic cells and their viruses. The bacterial helicase is a homohexamer that is placed around ssDNA generated by the DnaA protein at the origin; it travels 5′–3′ along the strand onto which it is bound. This directionality places the bacterial helicase around the lagging-strand template at a replication fork. In contrast, the eukaryotic helicase is a heterohexamer known as the MCM2-7 complex. Each of the six MCM subunits is encoded by a separate gene but they are related in sequence and are AAA+ protein ATPases, whereas the bacterial helicase ATPase architecture is based on a RecA-like fold (Enemark and Joshua-Tor 2008). Eukaryotic MCM encircles the leading strand template at a replication fork and tracks along ssDNA 3′–5′, the opposite polarity of the bacterial helicase. Another distinctive feature of the eukaryotic MCM2-7 helicase is that it is initially loaded onto the origin as a head-to-head double hexamer with the double-strand DNA (dsDNA) passing through the hexamer channel and therefore must transition to ssDNA to function as a helicase (Masai et al. 2010). This transition, although not well understood, is an important feature regulating replication initiation and involves addition of the Cdc45 and GINS proteins to form an active helicase called the CMG (Cdc45-Mcm2-7-GINS) (Ilves et al. 2010). Without these accessory proteins, the MCM2-7 is inactive as a helicase. Interestingly, the archaeal helicase is also a double hexamer, but in this case made up from a single protein called MCM that is related to the eukaryotic cell helicase. It also travels on ssDNA in the 3′–5′ direction and hence on the leading strand template and does not require accessory proteins for its helicase activity (Barry and Bell 2006).
All cells require other factors in addition to the origin-binding protein to load the helicase onto DNA. Before loading, bacterial DnaB and eukaryotic MCM are bound by DnaC and Cdt1, respectively, which facilitate delivery of the helicase complexes to the origin. DnaC is an AAA+ protein that uses ATP to bind the DnaB helicase in an inactive form and it cooperates with DnaA to load DnaB onto the ssDNA bubble formed at the origin by DnaA. ATP hydrolysis ejects DnaC after the loading step, enabling the helicase to become active in DNA unwinding (Kaguni 2011). In eukaryotes, Cdt1 brings the MCM2-7 helicase to the ORC-Cdc6 complex that is bound to the origin DNA (Masai et al. 2010). MCM loading triggers ATP hydrolysis by Cdc6, ejecting it from the DNA and promoting release of Cdt1. Archaea have the AAA+ Orc1/Cdc6 origin-binding protein, but to date no archaeal Cdt1 homolog has been identified, so the MCM hexamer may bind directly to the initiator protein (Barry and Bell 2006). The precise mechanism by which these proteins load the helicase is unknown in any system. MCM2-7 loading by ORC, Cdc6, and Cdt1 forms a prereplicative complex (the Pre-RC) in which MCM2-7 surrounds duplex DNA, but it remains inactive for DNA unwinding until cells commit to enter S phase of the cell cycle (Masai et al. 2010).
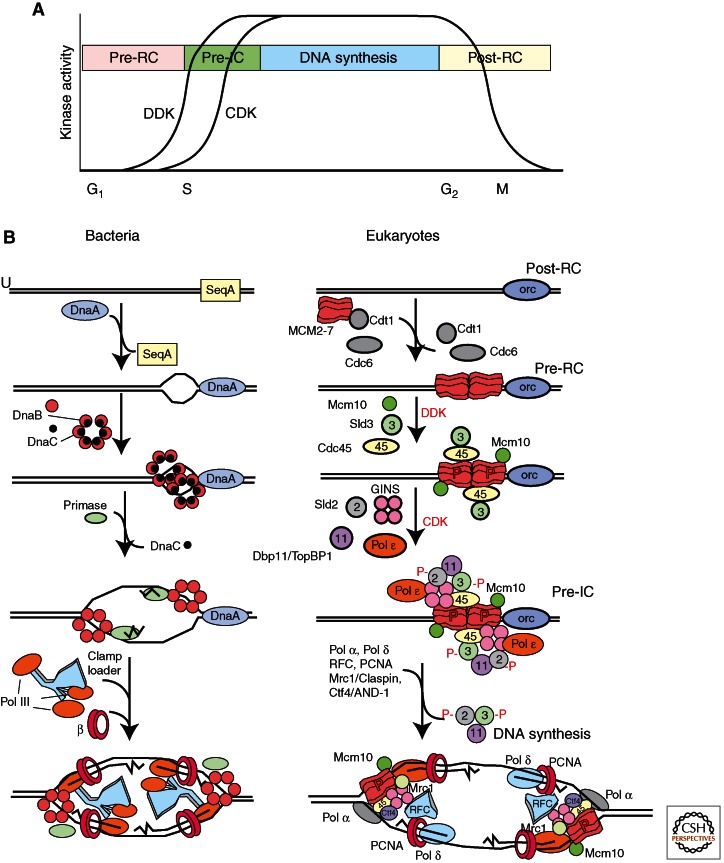
Loading the helicase and activating it to unwind DNA are central replication control points in all cell types, but the way bacteria and eukaryotes regulate this process is fundamentally different. E. coli DnaA binding at the origin is regulated by SeqA, which sequesters the origin and prevents access to DnaA (Dame et al. 2011). Sequestration is dependent on the methylation state of the origin DNA (SeqA can only bind newly replicated, hemimethylated DNA). Under optimal growth conditions, E. coli reinitiates DNA synthesis at the origin before completing the previous round of replication, yielding multiple chromosomes in one cell; the chromosomes eventually segregate into individual cells. Eukaryotes cannot afford the luxury of rereplication because of their requirement for multiple origins on each chromosome. Reinitiation at some origins and not others would lead to copy number differences within regions of a chromosome and problems with chromosome segregation during mitosis. Hence, under most circumstances, eukaryotic origin initiation is tightly regulated so that origins initiate once, and only once, per cell division. Eukaryotes achieve this exquisite level of control by separating initiation events into different phases of the cell cycle and imposing multiple regulatory processes on the mechanism of initiation of DNA replication (Diffley 2011), whereas bacteria lack a well-defined cell cycle (see Fig. 2) (Morgan 2007). Progression from one eukaryotic cell phase to the next is driven by many regulated events including the synthesis of new proteins, the destruction of others, and protein modification such as phosphorylation by kinases, a modification that is largely absent among bacterial replication proteins. An additional level of control, also distinct from bacteria, is that eukaryotic replication occurs in the nucleus and this compartmentalization allows for tight regulation by excluding key proteins from the nucleus when their activity is not required or when it might be detrimental.
Figure 2.
Origin activation and replisome assembly in bacteria and eukaryotes. (A) Origin activation in eukaryotes is regulated by DDK (Dbf4-Cdc7 kinase) and CDK (cyclin-dependent protein kinase) kinases that have low activity in G1 phase, and high activity in S phase. (B) Steps in origin activation and replisome assembly in bacteria and eukaryotes. The relatively more complex process of DNA replication in eukaryotes is reflected in the larger number of proteins required to initiate and elongate DNA synthesis from each origin. See text for details.
Formation of the pre-RC occurs during mitotic exit in rapidly proliferating eukaryotic cells or during the G1 phase of the cell cycle, but the MCM2-7 helicase remains inactive after it is loaded onto the dsDNA. The establishment of active replication forks is regulated by kinases that drive the cell into S phase by directing the chromatin association of many factors, most importantly Cdc45 and GINS, which are now known to activate the MCM2-7 helicase activity (Ilves et al. 2010). During S phase, the Cdc6 and Cdt1 proteins are eliminated by either selective protein degradation or nuclear exclusion, thereby preventing further MCM2-7 loading and reinitiation in S phase. These events underlie the phenomenon known as “licensing” of origins (Masai et al. 2010). Origins are licensed by MCM2-7 loading during mitotic exit or during G1, and then activated in S phase. By separating the helicase loading and DNA synthesis steps into two different phases of the cell cycle, eukaryotes prevent origins from initiating more than once (Diffley 2011). After replication, cells enter G2 phase and then M (mitosis) phase, in which the duplicated chromosomes are segregated into new daughter cells (see Fig. 2). Cell-cycle phases and their regulation are explained in McIntosh and Blow (2012), Siddiqui et al. (2013), Skarstad and Katayama (2013), and Zielke et al. (2013).
REACTIONS LEADING TO PRIMING
In all cell types studied so far, DNA polymerases cannot initiate new chains of nucleic acids and thus the synthesis of a primer by a DNA-dependent RNA polymerase is needed to begin cellular DNA replication (Frick and Richardson 2001). Priming occurs only on ssDNA, which requires prior helicase loading and unwinding activity. In E. coli, the primase is a single-subunit enzyme, DnaG, which transiently binds DnaB helicase to synthesize an RNA primer of ∼12 nucleotides (nt). Binding of DnaG primase to DnaB also stimulates release of the regulatory protein DnaC from DnaB, indicating that initial priming and unwinding are tightly coordinated (Kaguni 2011). Priming in eukaryotes is performed by the four-subunit DNA polymerase α-primase complex (Pol α/primase) that synthesizes an RNA of ∼12 nt and extends this primer RNA with ∼25 nt of DNA to form an RNA/DNA hybrid primer. Priming in eukaryotes occurs only after G1 phase transits to the S phase (see Fig. 2). How the Pol α/primase is recruited is not known, but in the case of SV40 DNA replication it binds directly to the helicase. In eukaryotic cells, recruitment of Pol α/primase may be mediated by the MCM10 protein that is required for initiation of DNA replication, but MCM10 then dissociates so that it cannot keep Pol α/primase on the lagging strand (Waga and Stillman 1998; van Deursen et al. 2012). The G1/S transition in eukaryotes, and eventual priming of DNA, involves activation of the S-phase CDK (cyclin-dependent protein kinase) and DDK (Dbf4-Cdc7 kinase) kinases (Diffley 2010). In all cells, elongation of the first primer becomes the leading strand. Subsequent priming events occur on the lagging strand to form Okazaki fragments. Okazaki fragments are 1–2 kb in bacteria, and only 100–200 bp in eukaryotes (Balakrishnan and Bambara 2013).
The reactions leading to priming in eukaryotes are still rather mysterious, and several proteins are involved that have no clear homologs in bacteria including Sld2, Sld3, Sld7, Cdc45, the four-subunit GINS complex, and Dpb11/TopBP1 (Masai et al. 2010). An S-phase-specific CDK phosphorylates Sld2 and Sld3 that allows them to bind to Dbp11, whereas the DDK phosphorylation of the MCM4 subunit is necessary for initiation of DNA replication (Diffley 2010; Labib 2010; Sheu and Stillman 2010). Additional details on the complex transactions that activate a licensed origin in eukaryotes are in Tanaka and Araki (2013). Current studies indicate that DNA polymerase ε (Pol ε), a replicative DNA polymerase, associates with the origin before Pol α/primase in a complex with GINS, Dpb11, and CDK-phosphorylated Sld2 (Araki 2010). Thus the initial priming event may be performed by some protein other than Pol α/primase. Alternatively, Pol ε may act as a structural component that helps recruit Pol α/primase. At an undefined point in the process, the MCM2-7 complex transitions from encircling dsDNA to encircling ssDNA. Only when this transition is complete can DNA be unwound by the CMG and provide an ssDNA template for primase activity to initiate replication.
THE REPLICATION MACHINE
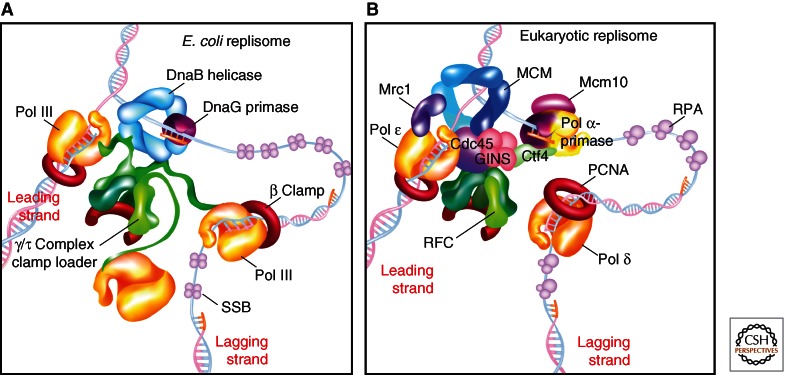
Replication is performed by a multiprotein replication machine that synthesizes both daughter duplexes simultaneously. Replication machines have the same core components in all cells: DNA polymerases, circular sliding clamps, a pentameric clamp loader, helicase, primase, and SSB (single-strand binding protein) (Waga and Stillman 1998; Garg and Burgers 2005; Johnson and O’Donnell 2005; Barry and Bell 2006). The way in which these proteins are arranged and connect to one another differs among cell types. The replication machine is often referred to as a “replisome.” The bacterial replisome, illustrated in Figure 3A, is organized by the clamp loader, which contains three identical τ subunits that bind three C-family DNA polymerase III (Pol III) polymerases (see Fig. 3). The τ subunit also binds the homohexameric helicase (i.e., E. coli DnaB). As primase forms RNA primers, the clamp loader repeatedly loads new circular β clamps onto the primer/template for use by the lagging-strand Pol III(s). This primase/polymerase switch is facilitated by SSB, which binds ssDNA and enables the clamp loader to dislodge primase from the primed site. SSB protects the ssDNA from nucleases and facilitates elongation by Pol III-β on ssDNA. The bacterial replisome is highly processive, meaning that in E. coli it can synthesize ∼86 kb on the leading strand without dissociating from the template (Georgescu et al. 2010). However, high processivity is a disadvantage on the lagging strand, which is synthesized as numerous short ∼1000 nt Okazaki fragments and requires the polymerase to dissociate from the template DNA and reassociate at a new primed site for each Okazaki fragment. To overcome this “processivity barrier,” specific mechanisms have evolved that pry the polymerase from the clamp to release an Okazaki fragment, on which the polymerase reassociates with a new clamp at the next RNA primed site. The three Pol IIIs in the bacterial replisome aid the production of multiple Okazaki fragments. These mechanisms will be further discussed in Hedglin et al. (2013).
Figure 3.
Organization of bacterial and eukaryotic replisome machines. (A) Replisome architecture in E. coli. The helicase (DnaB) encircles the lagging strand. Three molecules of Pol III are attached to one clamp loader. The clamp loader binds the helicase and repeatedly assembles β clamps onto primed sites as they are formed by primase. (B) Proposed architecture of a eukaryotic replisome. The MCM2-7 helicase encircles the leading strand; unwinding is aided by association of GINS and Cdc45 with MCM2-7 to form the CMG complex. The RFC clamp loader repeatedly loads PCNA (proliferating cell nuclear antigen) clamps onto lagging-strand primers formed by Pol α-primase. Unlike E. coli, the clamp loader may not form stabile attachments to the replisome. The leading-strand polymerase (Pol ε) is stabilized on DNA by Mrc1. Pol δ replicates the lagging strand. Contacts between Pol δ and other components of the replisome are not yet defined. Mcm10 and Ctf4 contact Pol α-primase.
Although eukaryotic and archaeal replisomes have similar components, the connections among the components are quite different from bacterial replisomes (Fig. 3B). The hexameric MCM2-7 encircles the leading strand, not the lagging strand and needs the Cdc45 and GINS proteins for it to function in unwinding DNA. Also, the eukaryotic replisome contains two different B-family polymerases that function separately for the leading and lagging strands, Pol ε and Pol δ, respectively, in addition to Pol α/primase, which starts each Okazaki fragment. The lagging strand is coated by replication protein A (RPA), a heterotrimer SSB that is structurally and functionally analogous to the bacterial SSB tetramer. As in bacteria, a primase/polymerase switch is mediated by RPA. Both Pol ε and Pol δ function with a ring-shaped proliferating cell nuclear antigen (PCNA) clamp of similar structure to E. coli β. PCNA is assembled on DNA by the pentameric replication factor C (RFC) clamp loader, composed of subunits with sequence homology and similar structure to the bacterial clamp loader (Garg and Burgers 2005). Unlike bacteria, RFC does not appear to contact the polymerases or the helicase, and the connections among replisome components remain unclear. There are numerous candidates among the many proteins known to be required for efficient eukaryotic replication that have no homolog in bacteria. For example, the GINS heterotetramer and the Cdc45 protein that form a complex with the MCM2-7 helicase to yield the CMG complex may bind Pol α/primase. GINS and Cdc45 are required for efficient DNA unwinding activity and may bind other factors in addition to the MCM2-7 complex to organize the replisome. In budding yeast, a replication progression complex has been identified containing potential links between the CMG and Pol α/primase via Ctf4/AND-1, and between CMG and Pol ε via Mrc1/Claspin (Masai et al. 2010). Detailed evidence for these and other possible connections, the functions of analogous proteins in other eukaryotes, and their importance for fork progression can be found in Bell and Botchan (2013).
The oligomeric structures of sliding clamps enable multiple proteins to bind one clamp simultaneously, referred to as the “toolbelt” hypothesis (Pages and Fuchs 2002). An extreme case is an archaeal PCNA heterotrimer in which each different subunit binds a different partner protein (i.e., polymerase, ligase, and Fen1) that is involved in the synthesis and maturation of Okazaki fragments (Barry and Bell 2006). These features of sliding clamps are discussed in Hedglin et al. (2013), but one aspect will be mentioned here as it has important implications for replisome structure and function. In particular, all cells contain a variety of low-fidelity DNA polymerases that can bypass lesions in the DNA, referred to as “TLS” Pols (translesion synthesis polymerases). Different Pols can bind the clamp simultaneously and trade places with one another, making the replisome a much more dynamic machine than originally thought (Langston et al. 2009). E. coli contains three “translesion” DNA polymerases (TLS Pols), which are induced on DNA damage. Studies have shown that at high concentrations such as those induced on DNA damage, the TLS Pols bind the β clamp and trade places with Pol III at a moving fork, yet retain the helicase to form a “TLS replisome.” TLS replisomes move much more slowly than Pol III and dictate the speed of the helicase. An obvious advantage of forming TLS replisomes on DNA damage is to slow the replication fork, giving time for DNA repair before a replication fork encounters a lesion. In the event a lesion is encountered, it can be bypassed by the TLS polymerase.
DEALING WITH DNA DAMAGE DURING REPLICATION
In bacteria and eukaryotes there are mechanisms to activate a stalled DNA replication fork that may be caused by the replisome running into DNA damage, a double-strand break, or a protein block. There are many repair mechanisms, such as error-free and error-prone DNA synthesis at the DNA replication fork, or post- replicative repair by nucleotide excision repair or base excision repair (Lazzaro et al. 2009; Hubscher and Maga 2011; Lehmann 2011). The multiple DNA polymerases involved in DNA repair are discussed in Goodman and Woodgate (2013) and Yeeles et al. (2013). Alternatively, branch migration followed by DNA synthesis or intersister chromatid recombination can be used to allow replicative bypass of DNA lesions. If these repair events occur while DNA replication is still occurring, stalled DNA replication forks must either be reactivated or bypassed, events that are discussed in McIntosh and Blow (2012) and Yeeles et al. (2013). Unlike bacteria, eukaryotes have multiple origins and thus failure to replicate a complete replicon can be compensated by activation of adjacent origins of DNA replication, another way of bypassing a stalled DNA replication fork (Blow et al. 2011).
If significant DNA damage occurs either before or during DNA replication in eukaryotes, so-called checkpoint mechanisms signal to the cell-cycle regulatory machinery, principally the CDK and DDK protein kinases, that subsequent events in the cell cycle should wait until DNA damage is repaired. The biochemistry of these varied signaling events is still being worked out, but one common signal is the stable presence of RPA-coated ssDNA. Normally ssDNA should not exist in a cell, but its presence signals that a stalled replication fork exists, that resection of damaged DNA has occurred, or that recombination is taking place. In any event, the cell does not want to progress until the damage has been repaired. In both bacteria and eukaryotes, restarting a stalled DNA replication fork is a key process that involves DNA helicases and priming events that are unique to stalled fork recovery. For example, in eukaryotes cancer-prone disorders called Bloom’s syndrome and Warner’s syndrome have defects in specialized DNA helicases that are involved in dealing with DNA damage at stalled replication forks. Another cancer-prone syndrome called Fanconi anemia has revealed defects in a family of proteins that handle cross-links in DNA. These pathways, as well as the recovery of stalled forks are discussed in Abbas et al. (2013), Jackson et al. (2013), and Yeeles et al. (2013).
SPATIAL AND TEMPORAL ORGANIZATION OF DNA REPLICATION
In bacteria, the DNA replication machinery is assembled at the single origin of DNA replication in a characteristic location. In Caulobacter, the replisome is located at one end of the rod-shaped bacterium but in E. coli, it is located in the middle of the cell (Toro and Shapiro 2010). After initiation of DNA replication, the new replicated origins spool out of the replisome, which stays in place, and the origins move to predetermined locations. In E. coli, the origins move to the quarters of the cell but in Caulobacter, the origins move to the other end of the cell. Thus the origins of DNA replication are associated with DNA elements that can move the DNA and prepare the cell for separating the two daughter chromosomes during cell division, thereby remotely resembling centromeres in eukaryotic cells. Interestingly, pre-RC proteins in eukaryotes play a role at centromeres and centrosomes, perhaps reflecting an evolutionary link between chromosome replication and segregation (Saffery et al. 2000; Prasanth et al. 2004; Hemerly et al. 2009; Hossain and Stillman 2012; Varma et al. 2012).
As the two DNA bacterial forks move in opposite directions away from the origin, they eventually meet at a termination site that locates to the center of the cell before cell division. In rapidly growing bacteria, it is possible to reinitiate DNA replication from the origin before cell division actually takes place and this is made possible because of the spatial separation of the origin and termination regions of the genome and their placement relative to the plane of cell division. Such reinitiation is lethal in eukaryotic cells because it causes aneuploidy and genome instability.
In contrast to bacteria, eukaryotes have to deal with multiple chromosomes and numerous origins. The fastest way to replicate the multiple chromosomes in eukaryotes is for all origins to fire at the same time, but this is rare and occurs in the early and rapid cell divisions of Xenopus embryos and during the replication of the syncytial nuclei in the Drosophila embryo. In somatic cells origins of DNA replication fire at specific times during S phase, with some firing early and others late in a specific temporal pattern that is characteristic for each cell type (Fig. 1B) (Toro and Shapiro 2010). Within a chromosome, clusters of origins of DNA replication that are located adjacent to each other initiate DNA replication at the same time, creating megabase-pair-sized domains of chromosomes that are copied synchronously. Remarkably, whole genome analysis of these domains of chromosomes that are replicated at the same time has revealed that they correspond to regions of the genome that are spatially associated with each other in the nucleus (Ryba et al. 2010). Thus, the genome within the nucleus is organized into domains that are physically associated with each other and these domains replicate at characteristic times during S phase. Such an arrangement explains why replication proteins such as the DNA polymerase clamp PCNA form temporally regulated patterns in the nucleus during S phase that correspond to sites of DNA synthesis of large regions of the genome (Kill et al. 1991; O’Keefe et al. 1992; Leonhardt et al. 2000).
Pre-RCs are assembled at all origins of DNA replication before the start of S phase and the temporal patterning of actual DNA synthesis from each origin is governed by the chromatin context, including the nature of histone modifications, and by rate-limiting proteins that establish the preinitiation complex (Douglas and Diffley 2012). The activation of pre-RCs is regulated by at least two independent protein kinase signaling systems, the CDK and the DDK (Labib 2010; Tanaka and Araki 2013). Once the pre-RC is used or once passive DNA replication passes through a pre-RC, the pre-RC is destroyed and cannot be re-formed during the time in the cell cycle that CDKs are active. Thus pre-RCs cannot be established until the cells pass through the metaphase to anaphase transition when the cyclin moieties of CDKs are destroyed. Such a mechanism limits DNA replication from all origins to once per cell division cycle (Diffley 2011). So, unlike bacterial chromosome replication, it is clear that the complex genomes of eukaryotes have demanded more complex regulatory systems to maintain genome integrity. These issues are discussed in Siddiqui et al. (2013) and Zielke et al. (2013).
WHAT NEEDS TO BE DONE?
Although the broad outline and many important details of DNA replication have been identified, many important aspects of this central process remain to be discovered. In large part, we still do not know how origins function. How do origin-binding proteins organize the DNA? Exactly how do helicase loaders function? How and when does the MCM helicase transition from encircling dsDNA to encircling ssDNA? The functions of several proteins required for origin activation and priming in eukaryotes are still shrouded in mystery. Priming and replisome assembly require numerous proteins that lack homologs in bacteria. What are the functions of Sld2, Sld3, Dbp11, Mcm10, GINS, and Cdc45 and how is their function influenced by phosphorylation? We lack an understanding of how the multiple origins in eukaryotes are coordinated and how the domain structure is established and maintained through multiple cell divisions. For example, just what are nuclear foci and how are replication foci organized within them? Are origins within one replicon clustered into one focus? Once replication forks are established, we know little about how they are regulated. If one replication fork in a focus were to stop, would it halt the other forks within that focus? How do replisomes move through nucleosomes, especially in highly condensed DNA and how are the parental nucleosomes inherited to the sister chromatids? How do replisomes deal with cohesin rings and how are these loaded? We have barely scratched the surface on questions surrounding the interface of replication with repair and recombination. For example, how can replication forks form during break-induced replication in S and G2 phase when the MCMs are thought to be loaded only in G1? How do checkpoint mechanisms act on moving replication forks? The newly revealed coordination of DNA metabolism with chromatin establishment, gene silencing, and epigenetic control is only beginning to be explored. Most of what we know about DNA replication has been learned in organisms with stable karyotypes and ploidy. However some organisms, particularly microbial eukaryotes, have extreme variations in ploidy and variable numbers of chromosomes. What mechanisms exist to facilitate this yet maintain order in this apparent chaos? Finally, and perhaps most important, some types of human disease, including certain cancers, have their basis in replication. Clearly many important questions remain, despite the enormous progress of recent years. We hold hope that understanding the mechanistic details of these processes may lead to cures, or at least treatment of human disease in the future.
ACKNOWLEDGMENTS
The authors’ research is supported by the Howard Hughes Medical Institute (M.O.) and the National Institutes of Health (GM38839 to M.O.; CA13016 and GM45436 to B.S.).
Footnotes
Editors: Stephen D. Bell, Marcel Méchali, and Melvin L. DePamphilis
Additional Perspectives on DNA Replication available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- *.Abbas T, Keaton MA, Dutta A 2013. Genomic instability in cancer. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a012914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki H 2010. Cyclin-dependent kinase-dependent initiation of chromosomal DNA replication. Curr Opin Cell Biol 22: 766–771 [DOI] [PubMed] [Google Scholar]
- *.Balakrishnan L, Bambara RA 2013. Okazaki fragment metabolism. Cold Spring Harb Perspect Biol 5: a010173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry ER, Bell SD 2006. DNA replication in the archaea. Microbiol Mol Biol Rev 70: 876–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Bell SD, Botchan MR 2013. The minichromosome maintenance replicative helicase. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a012807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Bell SP, Kaguni JM 2013. Helicase loading at chromosomal origins of replication. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a010124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Ge XQ, Jackson DA 2011. How dormant origins promote complete genome replication. Trends Biochem Sci 36: 405–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadoret JC., Meisch F, Hassan-Zadeh V, Luyten I, Guillet C, Duret L, Quesneville H, Prioleau MN 2008. Genome-wide studies highlight indirect links between human replication origins and gene regulation. Proc Natl Acad Sci 105: 15837–15842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dame RT, Kalmykowa OJ, Grainger DC 2011. Chromosomal macrodomains and associated proteins: Implications for DNA organization and replication in gram negative bacteria. PLoS Genet 7: e1002123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley JF 2010. The many faces of redundancy in DNA replication control. Cold Spring Harb Symp Quant Biol 75: 135–142 [DOI] [PubMed] [Google Scholar]
- Diffley JF 2011. Quality control in the initiation of eukaryotic DNA replication. Philos Trans R Soc Lond B Biol Sci 366: 3545–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall A, Shapiro L 1989. Rate, origin, and bidirectionality of Caulobacter chromosome replication as determined by pulsed-field gel electrophoresis. Proc Natl Acad Sci 86: 119–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas ME, Diffley JF 2012. Replication timing: The early bird catches the worm. Curr Biol 22: R81–R82 [DOI] [PubMed] [Google Scholar]
- Doulatov S, Notta F, Laurenti E, Dick JE 2012. Hematopoiesis: A human perspective. Cell Stem Cell 10: 120–136 [DOI] [PubMed] [Google Scholar]
- Enemark EJ, Joshua-Tor L 2008. On helicases and other motor proteins. Curr Opin Struct Biol 18: 243–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erzberger JP, Berger JM 2006. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu Rev Biophys Biomol Struct 35: 93–114 [DOI] [PubMed] [Google Scholar]
- Frick DN, Richardson CC 2001. DNA primases. Annu Rev Biochem 70: 39–80 [DOI] [PubMed] [Google Scholar]
- Gai D, Chang YP, Chen XS 2010. Origin DNA melting and unwinding in DNA replication. Curr Opin Struct Biol 20: 756–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg P, Burgers PM 2005. DNA polymerases that propagate the eukaryotic DNA replication fork. Crit Rev Biochem Mol Biol 40: 115–128 [DOI] [PubMed] [Google Scholar]
- Georgescu RE, Yao NY, O’Donnell M 2010. Single-molecule analysis of the Escherichia coli replisome and use of clamps to bypass replication barriers. FEBS Lett 584: 2596–2605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DM, Takebayashi SI, Ryba T, Lu J, Pope BD, Wilson KA, Hiratani I 2010. Space and time in the nucleus: Developmental control of replication timing and chromosome architecture. Cold Spring Harb Symp Quant Biol 75: 143–153 [DOI] [PubMed] [Google Scholar]
- *.Goodman MF, Woodgate R 2013. Translesion DNA polymerases. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a010363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Hedglin M, Kumar R, Benkovic SJ 2013. Replication clamps and clamp loaders. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a010165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemerly AS, Prasanth SG, Siddiqui K, Stillman B 2009. Orc1 controls centriole and centrosome copy number in human cells. Science 323: 789–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain M, Stillman B 2012. Meier-Gorlin syndrome mutations disrupt an Orc1 CDK inhibitory domain and cause centrosome re-duplication. Genes Dev 26: 1797–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubscher U, Maga G 2011. DNA replication and repair bypass machines. Curr Opin Chem Biol 15: 627–635 [DOI] [PubMed] [Google Scholar]
- Ilves I, Petojevic T, Pesavento JJ, Botchan MR 2010. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol Cell 37: 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Jackson AP, Laskey RA, Coleman N 2013. Replication proteins and human disease. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a013060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A, O’Donnell M 2005. Cellular DNA replicases: Components and dynamics at the replication fork. Annu Rev Biochem 74: 283–315 [DOI] [PubMed] [Google Scholar]
- Kaguni JM 2011. Replication initiation at the Escherichia coli chromosomal origin. Curr Opin Chem Biol 15: 606–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kill IR, Bridger JM, Campbell KH, Maldonado-Codina G, Hutchison CJ 1991. The timing of the formation and usage of replicase clusters in S-phase nuclei of human diploid fibroblasts. J Cell Sci 100: 869–876 [DOI] [PubMed] [Google Scholar]
- Labib K 2010. How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes Dev 24: 1208–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langston LD, Indiani C, O’Donnell M 2009. Whither the replisome: Emerging perspectives on the dynamic nature of the DNA replication machinery. Cell Cycle 8: 2686–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaro F, Giannattasio M, Puddu F, Granata M, Pellicioli A, Plevani P, Muzi-Falconi M 2009. Checkpoint mechanisms at the intersection between DNA damage and repair. DNA Repair (Amst) 8: 1055–1067 [DOI] [PubMed] [Google Scholar]
- Lehmann AR 2011. DNA polymerases and repair synthesis in NER in human cells. DNA Repair (Amst) 10: 730–733 [DOI] [PubMed] [Google Scholar]
- *.Leonard AC, Méchali M 2013. DNA replication origins. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a010116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt H, Rahn HP, Weinzierl P, Sporbert A, Cremer T, Zink D, Cardoso MC 2000. Dynamics of DNA replication factories in living cells. J Cell Biol 149: 271–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai H, Matsumoto S, You Z, Yoshizawa-Sugata N, Oda M 2010. Eukaryotic chromosome DNA replication: Where, when, and how? Annu Rev Biochem 79: 89–130 [DOI] [PubMed] [Google Scholar]
- *.McIntosh D, Blow JJ 2012. Dormant origins, the licensing checkpoint, and the response to replicative stresses. Cold Spring Harb Perspect Biol 4: a012955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan DO 2007. The cell cycle: Principles of control. Oxford University Press, London [Google Scholar]
- Myllykallio H, Lopez P, Lopez-Garcia P, Heilig R, Saurin W, Zivanovic Y, Philippe H, Forterre P 2000. Bacterial mode of replication with eukaryotic-like machinery in a hyperthermophilic archaeon. Science 288: 2212–2215 [DOI] [PubMed] [Google Scholar]
- O’Keefe RT, Henderson SC, Spector DL 1992. Dynamic organization of DNA replication in mammalian cell nuclei: Spatially and temporally defined replication of chromosome-specific α-satellite DNA sequences. J Cell Biol 116: 1095–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pages V, Fuchs RP 2002. How DNA lesions are turned into mutations within cells? Oncogene 21: 8957–8966 [DOI] [PubMed] [Google Scholar]
- Prasanth SG, Prasanth KV, Siddiqui K, Spector DL, Stillman B 2004. Human Orc2 localizes to centrosomes, centromeres and heterochromatin during chromosome inheritance. Embo J 23: 2651–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N 2006. DNA replication timing: Random thoughts about origin firing. Nat Cell Biol 8: 1313–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, Zhang J, Schulz TC, Robins AJ, Dalton S, Gilbert DM 2010. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res 20: 761–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffery R, Irvine DV, Griffiths B, Kalitsis P, Wordeman L, Choo KH 2000. Human centromeres and neocentromeres show identical distribution patterns of >20 functionally important kinetochore-associated proteins. Hum Mol Genet 9: 175–185 [DOI] [PubMed] [Google Scholar]
- Samson RY, Bell SD 2011. Cell cycles and cell division in the archaea. Curr Opin Microbiol 14: 350–356 [DOI] [PubMed] [Google Scholar]
- Sheu YJ, Stillman B 2010. The Dbf4-Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature 463: 113–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Siddiqui K, On KF, Diffley JFX 2013. Regulating DNA replication in eukarya. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a012930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Skarstad K, Katayama T 2013. Regulating DNA replication in bacteria. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a012922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman B 2005. Origin recognition and the chromosome cycle. FEBS Lett 579: 877–884 [DOI] [PubMed] [Google Scholar]
- Sun J, Kawakami H, Zech J, Speck C, Stillman B, Li H 2012. Cdc6-induced conformational changes in ORC bound to origin DNA revealed by cryo-electron microscopy. Structure 20: 534–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Araki H 2010. Regulation of the initiation step of DNA replication by cyclin-dependent kinases. Chromosoma 119: 565–574 [DOI] [PubMed] [Google Scholar]
- *.Tanaka S, Araki H 2013. Helicase activation and establishment of replication forks at chromosomal origins of replication. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a010371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro E, Shapiro L 2010. Bacterial chromosome organization and segregation. Cold Spring Harb Perspect Biol 2: a000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deursen F, Sengupta S, De Piccoli G, Sanchez-Diaz A, Labib K 2012. Mcm10 associates with the loaded DNA helicase at replication origins and defines a novel step in its activation. Embo J 31: 2195–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma D, Chandrasekaran S, Sundin LJ, Reidy KT, Wan X, Chasse DA, Nevis KR, Deluca JG, Salmon ED, Cook JG 2012. Recruitment of the human Cdt1 replication licensing protein by the loop domain of Hec1 is required for stable kinetochore-microtubule attachment. Nat Cell Biol 14: 593–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waga S, Stillman B 1998. The DNA replication fork in eukaryotic cells. Annu Rev Biochem 67: 721–751 [DOI] [PubMed] [Google Scholar]
- *.Yeeles JTP, Poli J, Marians KJ, Pasero P 2013. Rescuing stalled or damaged replication forks. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a012815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Zielke N, Edgar BA, DePamphilis ML 2013. Endoreplication. Cold Spring Harb Perspect Biol 5: a012948. [DOI] [PMC free article] [PubMed] [Google Scholar]