Abstract
International drinking water quality monitoring programs have been established in order to prevent or to reduce the risk of contracting water-related infections. A survey was performed on groundwater-derived drinking water from 13 different hospitals in the Mahabubnagar District. A total of 55 bacterial strains were isolated which belonged to both gram-positive and gram-negative bacteria. All the taxa were identified based on the 16S rRNA gene sequence analysis based on which they are phylogenetically close to 27 different taxa. Many of the strains are closely related to their phylogenetic neighbors and exhibit from 98.4 to 100% sequence similarity at the 16S rRNA gene sequence level. The most common group was similar to Acinetobacter junii (21.8%) and Acinetobacter calcoaceticus (10.9%) which were shared by 7 and 5 water samples, respectively. Out of 55 isolates, only 3 isolates belonged to coliform group which are Citrobacter freundii and Pantoea anthophila. More than half (52.7%, 29 strains) of the phylogenetic neighbors which belonged to 12 groups were reported to be pathogenic and isolated from clinical specimens. Out of 27 representative taxa are affiliated have eight representative genera in drinking water except for those affiliated with the genera Exiguobacterium, Delftia, Kocuria, and Lysinibacillus.
1. Introduction
Human population growth wields several and various pressures on the quality and the quantity of drinking and fresh water resources and on the access to them. Safe drinking water remains inaccessible to several millions people in the globe. Safe drinking water, thus, reduces the load of waterborne diseases. Contamination of drinking water due to natural and manmade contaminants is frequently reported in developing countries where mainstream of the inhabitants survive in countryside and uptown areas with meager hygiene and waste clearance practices. In developing countries, poor water quality is the most important risk of child mortalities which are mainly through infectious diarrhea. In India, about 10% of the countryside and city populations do not have access to usual safe drinking water and several others are threatened [1].
Potable or drinking water is defined as having a satisfactory quality in terms of its physical, chemical, and bacteriological parameters so that it can be securely used for drinking and cooking. The most common and widespread health risks associated with drinking water in developing countries are of biological origin. The WHO estimates that about 1100 million people globally drink unsafe water, and the greater part of diarrheal disease in the globe (88%) is attributable to insecure water, sanitation, and hygiene [2]. Ten major waterborne diseases are responsible for over 28 billion disease episodes annually in developing countries. The quality of drinking water might be ascertained by its microbiological examination. The maximum threat from microbes (coliform) in water is related to consumption of drinking water, that is, contaminated with human or animal excreta. Coliform bacteria include members of the family Enterobacteriaceae, for example, Escherichia coli, Enterobacter aerogenes, Salmonella, and Klebsiella in water that are accountable for a variety of diseases like cholera, typhoid, dysenteries, bacillary dysentery, and so forth in human and livestock [3].
Recently, Suthar et al. [4, 5] reported a wide range of pathogenic bacteria in potable water samples from some rural habitations of Northern Rajasthan, India. A total of ten bacterial species, E. coli, Pseudomonas aeruginosa, E. aerogenes, Klebsiella sp., Proteus vulgaris, Alcaligenes faecalis, Bacillus cereus, Staphylococcus aureus, Streptococcus lactis, and Micrococcus luteus, were isolated and identified from the potable water samples from this region [4] M. luteus, S. lactis, Klebsiella sp., and E. coli were the dominant microflora (recorded from 73.1% of the villages/towns) in the water samples. The presence of coliforms shows the danger of fecal pollution and a consequent hazard of contracting a disease through pathogenic organisms. Nonetheless, the disease-causing organisms (pathogens) mostly transmitted via drinking water are predominantly of fecal origin.
The results of the recent study on the detection of potentially pathogenic bacteria in the drinking water distribution system of a hospital in Hungary by Felföldi et al. [5] had shown the presence of Legionella, Pseudomonas aeruginosa, and also several other opportunistic pathogenic bacteria, such as Escherichia albertii, Acinetobacter lwoffii and Corynebacterium tuberculostearicum, emphasizing that drinking water systems, particularly those with stagnant water sections, could be the source of nosocomial infections.
Hospitals drinking (potable) water systems are the most important and controllable as well as the most overlooked source of nosocomial pathogens. Conventional culture-based microbiological water quality monitoring techniques take a long time (several days), and usually a small volume of water is sampled (typically 100 mL), which gives rise to inadequate detection limits with regard to drinking water safety. Furthermore, the presence of some important waterborne pathogens (such as Pseudomonas aeruginosa or legionellae) shows no correlation with conventional indicator organism counts. Water-related pathogens can also find niches in water systems (i.e., an association with biofilms or free-living amoebae), rendering their observation with conventional techniques more difficult. Molecular techniques provide new and rapid facilities for the detection of pathogens involved in nosocomial infections. Five representative end-points (taps) in the drinking water system of a hospital (Budapest, Hungary) were sampled in October 2005.
2. Materials and Methods
2.1. Source of Samples
Thirteen drinking water samples were collected from different government hospitals, Mahabubnagar District, Andhra Pradesh, India, on 22nd and 23rd of November, 2009. Fifty five strains were isolated from these 13 drinking water sources on nutrient agar medium at 37°C. For isolation of bacteria, 100 μL of the water sample was plated on nutrient agar medium and incubated at 37°C for 2 days. Based on the different colony morphology from each sample, a total of 55 strains were selected and characterized in the present study.
2.2. 16S rRNA Gene Sequencing and Phylogenetic Analysis
For 16S rRNA gene sequencing, DNA was prepared using the Mo Bio microbial DNA isolation kit (Mo Bio Laboratories Inc., Solano Beach, CA, USA) and sequenced as described previously [6]. The resultant almost complete sequence of the 16S rRNA gene contained 1502 nucleotides. The 16S rRNA gene sequence of the isolate was subjected to BLAST sequence similarity search [7] and EzTaxon [8] to identify the nearest taxa. The entire related 16S rRNA gene sequences were downloaded from the database (http://www.ncbi.nlm.nih.gov/) aligned using the CLUSTAL_X program [9] and the alignment corrected manually. Phylogenetic tree was constructed using tree-making algorithm and the Maximum likelihood (ML) using the PhyML program [10].
2.3. Biochemical Tests
The biochemical tests were performed for all the strains obtained by using the HiMViCTM Biochemical Test Kit (HIMEDIA-KB001) and HiAssorted Biochemical Test Kit (HIMEDIA-KB002).
3. Results and Discussion
3.1. Characterization of the Bacterial Strains from Drinking Water
All these isolates were from different drinking water sources, and all are mesophilic and could grow in the temperature range of 20 to 40°C with optimum growth temperature of 37°C. All the strains could grow without NaCl in the medium, and the tolerance to NaCl varied from 1 to 2%. All strains could grow either in the pH range of 6–8.
3.2. Identification of Isolated Strains from Drinking Water
A total of 55 bacterial strains were recovered from the 13 drinking water sources (Table 1). Taxonomic analysis of all the 55 strains isolated from water samples indicated that 43 strains were Gram-negative and 12 Gram-positive. The nearest phylogenetic neighbor of all 55 isolated strains was identified following BLAST analysis of the 16S rRNA gene sequence. BLAST analysis indicated that twenty one strains belonged to the genus Acinetobacter, nine strains belonged to the genus Bacillus, eight strains belonged to the genus Pseudomonas, seven strains to the genus Aeromonas, two strains each belonged to the genera Methylobacterium and Pantoea, and one strain belonged to the genera Cupriavidus, Delftia, Exiguobacterium, Kocuria, and Lysinibacillus, respectively (Table 2). A total 27 different taxa were identified based on the BLAST analysis out of 55 strains. The most common group was similar to Acinetobacter junii (21.8%) and Acinetobacter calcoaceticus (10.9%) which were shared by 7 and 5 water samples, respectively. The phylogenetic trees constructed to determine their affiliations are shown in Figures 1 and 2. Out of 12 Gram-positive groups, 11 belonged to the phylum Firmicutes and 1 belonged to the phylum Actinobacteria (Table 2 and Figure 2). All 43 Gram-negative groups were belonged to the phylum Proteobacteria. Among the Proteobacteria, the Alphaproteobacteria, Betaproteobacteria, and Gammaproteobacteria were represented by 2, 2, and 39 groups, respectively (Table 2 and Figure 2). Phylogenetic analysis based on 16S rRNA gene sequences indicated that the 21 Acinetobacter strains belonged to 4 groups, 9 Bacillus strains belonged to 7 groups, 8 Pseudomonas strains belonged to 3 groups, 7 Aeromonas strains belonged to 5 groups, and 2 Methylobacterium strains formed two distinct groups (Figures 1 and 2). The affiliation of the strains to the nearest phylogenetic neighbor and the percentage of 16S rRNA gene sequence similarities are indicated in Table 2. Many of the strains are closely related to one another and exhibit 98.4 to 100% sequence similarity at the 16S rRNA gene sequence level like KL-1 and JD-2 which are closely related to one another (100%) and are affiliated to Acinetobacter calcoaceticus DSM 30006T (98.5%).
Table 1.
Bacterial abundance from the drinking water samples collected from government hospitals, Mahabubnagar.
| Serial number | Sample number | Place of sample collection | Number of isolates | Isolated strains |
|---|---|---|---|---|
| 1 | AL | Alampur | 3 | AL1, AL3, AL4 |
| 2 | AP | Achampet | 3 | AP1, AP2, AP4 |
| 3 | D | Shadnagar | 4 | D12, D22, D24, D25 |
| 4 | GD | Gadwal | 4 | GD2, GD4–D6 |
| 5 | JD | Jadcherla | 4 | JD1–JD4 |
| 6 | JU | Jurala | 4 | JU1–JU4 |
| 7 | KL | Kalwakurthy | 5 | KL1–KL5 |
| 8 | MB | Mahabubnagar-1 | 4 | MB1–MB4 |
| 9 | MK | Makthal | 3 | MK2–MK4 |
| 10 | NG | Nagar Kurnool | 2 | NG1, NG2 |
| 11 | NR | Narayanpet | 3 | NR2–NR4 |
| 12 | R | Mahabubnagar-2 | 12 | R1, R2, R5–R8, R21–R23, R26, R30, R31 |
| 13 | WN | Wanaparthy | 4 | WN1, WN3–WN5 |
Table 2.
Identification of the 55 bacterial strains isolated from the drinking water samples collected from different government hospitals, Mahabubnagar, based on BLAST analysis of the 16S rRNA gene sequences.
| Sl no. | Strain no. | Nearest phylogenetic neighbor | 16S rRNA gene sequence similarity % |
|---|---|---|---|
| Gram-negative bacterial strains | |||
|
| |||
| Proteobacteria | |||
| Alphaproteobacteria | |||
| 1 | D24 | Methylobacterium chloromethanicum CM4T CP001298 | 99.6 |
| 2 | AL1 | Methylobacterium zatmanii DSM 5688T AB175647 | 99.7 |
| Betaproteobacteria | |||
| 3 | AP1 | Cupriavidus pauculus LMG 3413T AF085226 | 100.0 |
| 4 | AL3 | Delftia lacustris DSM 21246T EU888308 | 100.0 |
| Gammaproteobacteria | |||
| 5 | JD2 | Acinetobacter calcoaceticus DSM 30006T X81661 | 98.5 |
| 6 | KL1 | Acinetobacter calcoaceticus DSM 30006T X81661 | 98.5 |
| 7 | KL5 | Acinetobacter calcoaceticus DSM 30006T X81661 | 98.5 |
| 8 | MB1 | Acinetobacter calcoaceticus DSM 30006T X81661 | 98.4 |
| 9 | MK4 | Acinetobacter calcoaceticus DSM 30006T X81661 | 98.5 |
| 10 | NR4 | Acinetobacter calcoaceticus DSM 30006T X81661 | 98.4 |
| 11 | GD5 | Acinetobacter johnsonii DSM 6963T X81663 | 99.9 |
| 12 | NG1 | Acinetobacter johnsonii DSM 6963T X81663 | 99.5 |
| 13 | AP2 | Acinetobacter junii LMG 998T AM410704 | 100.0 |
| 14 | AP4 | Acinetobacter junii LMG 998T AM410704 | 100.0 |
| 15 | GD2 | Acinetobacter junii LMG 998T AM410704 | 100.0 |
| 16 | JD4 | Acinetobacter junii LMG 998T AM410704 | 100.0 |
| 17 | KL3 | Acinetobacter junii LMG 998T AM410704 | 100.0 |
| 18 | KL4 | Acinetobacter junii LMG 998T AM410704 | 100.0 |
| 19 | MB3 | Acinetobacter junii LMG 998T AM410704 | 100.0 |
| 20 | R1 | Acinetobacter junii LMG 998T AM410704 | 99.9 |
| 21 | WN1 | Acinetobacter junii LMG 998T AM410704 | 99.9 |
| 22 | WN3 | Acinetobacter junii LMG 998T AM410704 | 100.0 |
| 23 | WN4 | Acinetobacter junii LMG 998T AM410704 | 100.0 |
| 24 | WN5 | Acinetobacter junii LMG 998T AM410704 | 99.9 |
| 25 | MB4 | Acinetobacter ursingii LUH 3792T AJ275038 | 99.6 |
| 26 | JD1 | Aeromonas aquariorum MDC47T EU085557 | 100.0 |
| 27 | R31 | Aeromonas aquariorum MDC47T EU085557 | 100.0 |
| 28 | R30 | Aeromonas aquariorum MDC47T EU085557 | 99.9 |
| 29 | GD4 | Aeromonas hydrophila subsp. hydrophila ATCC 7966T CP000462 | 100.0 |
| 30 | JD3 | Aeromonas ichthiosmia DSM 6393T X71120 | 99.8 |
| 31 | MK2 | Aeromonas ichthiosmia DSM 6393T X71120 | 99.8 |
| 32 | R26 | Aeromonas punctata subsp. punctata NCIMB 13016T X60408 | 99.2 |
| 33 | NG2 | Citrobacter freundii DSM 30039T AJ233408 | 98.3 |
| 34 | R21 | Pantoea anthophila LMG 2558T EF688010 | 99.8 |
| 35 | R22 | Pantoea anthophila LMG 2558T EF688010 | 99.9 |
| 36 | NR2 | Pseudomonas aeruginosa LMG 1242T Z76651 | 99.9 |
| 37 | R6 | Pseudomonas alcaligenes LMG 1224T Z76653 | 100.0 |
| 38 | R7 | Pseudomonas alcaligenes LMG 1224T Z76653 | 99.8 |
| 39 | R23 | Pseudomonas alcaligenes LMG 1224T Z76653 | 99.8 |
| 40 | D22 | Pseudomonas otitidis MCC10330T AY953147 | 99.8 |
| 41 | GD6 | Pseudomonas otitidis MCC10330T AY953147 | 99.8 |
| 42 | MB2 | Pseudomonas otitidis MCC10330T AY953147 | 99.6 |
| 43 | MK3 | Pseudomonas otitidis MCC10330T AY953147 | 99.7 |
|
| |||
| Gram-positive bacterial strains | |||
|
| |||
| Actinobacteria | |||
| 44 | R8 | Kocuria rhizophila DSM 11926T Y16264 | 100.0 |
| Firmicutes | |||
| Bacilli | |||
| 45 | D12 | Bacillus anthracis ATCC 14578T AB190217 | 100.0 |
| 46 | KL2 | Bacillus aryabhattai B8W22T EF114313 | 98.7 |
| 47 | NR3 | Bacillus aryabhattai B8W22T EF114313 | 99.7 |
| 48 | R5 | Bacillus cereus ATCC 14579T AE016877 | 100.0 |
| 49 | JU1 | Bacillus infantis SMC 4352-1T AY904032 | 99.5 |
| 50 | JU3 | Bacillus licheniformis ATCC 14580T CP000002 | 99.9 |
| 51 | JU4 | Bacillus niabensis 4T19T AY998119 | 98.9 |
| 52 | AL4 | Bacillus stratosphericus 41KF2aT AJ831841 | 99.9 |
| 53 | R2 | Bacillus stratosphericus 41KF2aT AJ831841 | 100.0 |
| 54 | D25 | Exiguobacterium indicum HHS31 T AJ846291 | 99.9 |
| 55 | JU2 | Lysinibacillus fusiformis NBRC 15717T AB271743 | 99.8 |
Figure 1.
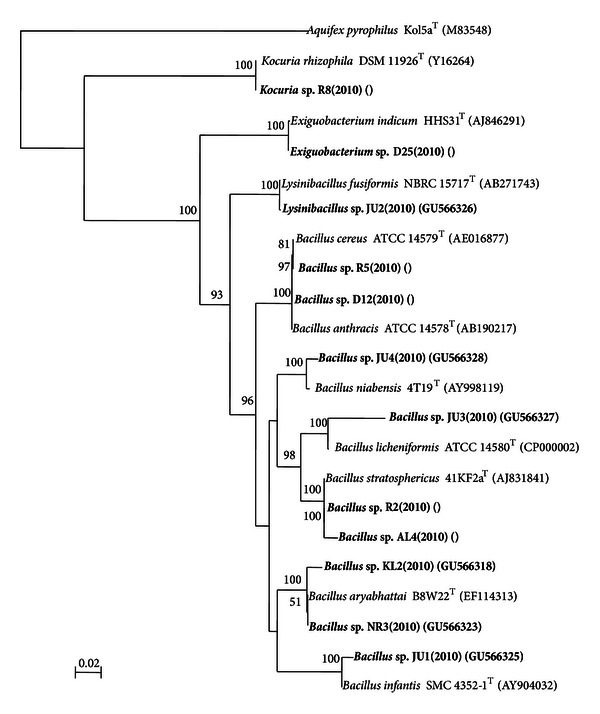
The affiliation of the strains to the nearest phylogenetic neighbor and the percentage of 16S rRNA gene sequence similarities.
Figure 2.

The affiliation of the strains to the nearest phylogenetic neighbor and the percentage of 16S rRNA gene sequence similarities.
3.3. Biochemical Assay
Biochemical analysis of the genus Bacillus gave positive results for Indole, Citrate, Oxidase, and Urease and positive results for Methyl red Voges-Proskaeur, Catalase, and Gelatinase tests. Genus Methylobacterium gave positive results for Citrate, Catalase, and Urease and negative results for Indole, Methyl Red, Voges-Proskaeur, Oxidase, and Gelatinase. Genus Cupriavidus gave positive results for Citrate, Oxidase, Catalase, and Urease and negative results for Indole, Methyl red, Voges-Proskaeur, and Gelatinase. Delftia gave positive results for Citrate and Urease and negative ones for Indole, Methyl red, Voges-Proskaeur, Oxidase, Catalase, and Gelatinase. Genus Acinetobacter gave positive results for Citrate and Gelatinas and negative results for Indole, Methyl red, Voges-Proskaeur, Oxidase, Catalase, and Urease (A. ursingii gave positive results even for Citrate and Oxidase and Catalase). Genus Aeromonas (Ar. aquariorum gave positive results for all except Catalase and Gelatinase, whereas Ar. hydrophila, Ar. Ichthiosmia, and Ar. punctata gave positive results for all tests except Methyl red). Genus Citrobacter gave positive results for Methyl red, Oxidase, Catalase, and Urease and negative results for Indole, VP, Citrate, and Gelatinase. Genus Pantoea gave positive results for VP, Citrate, and Catalase and negative results for Indole, Methyl red, Oxidase, Urease, and Gelatinase. Genus Pseudomonas gave positive results for Citrate, Oxidase, Catalase, and Gelatinase and negative ones for Indole, Methyl red, VP, and Urease. Kocuria rhizophila gave positive results for Oxidase and Catalase and negative ones for the remaining tests. Exiguobacterium indicum gave positive results for Methyl red, Citrate, and Catalase and negative ones for Indole,VP, Oxidase, Urease, and Gelatinase. Lysinibacillus fusiformis gave positive results for Oxidase and Gelatinase and negative results for the remaining tests (Table 3).
Table 3.
Biochemical tests for the strains obtained.
| Sl no. | Strain no. | Nearest phylogenetic neighbor | Biochemical tests | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ind | MR | VP | Cit | Oxi | Cat | Ure | Gel | |||
| 1 | D24 | Methylobacterium chloromethanicum CM4T CP001298 | − | − | − | + | − | + | + | − |
| 2 | AL1 | Methylobacterium zatmanii DSM 5688T AB175647 | − | − | − | + | − | − | + | − |
| 3 | AP1 | Cupriavidus pauculus LMG 3413T AF085226 | − | − | − | + | + | + | + | − |
| 4 | AL3 | Delftia lacustris DSM 21246T EU888308 | − | − | − | + | − | − | + | − |
| 5 | JD2 | Acinetobacter calcoaceticus DSM 30006T X81661 | − | − | − | + | − | − | + | + |
| 6 | KL1 | Acinetobacter calcoaceticus DSM 30006T X81661 | − | − | − | + | − | − | − | + |
| 7 | KL5 | Acinetobacter calcoaceticus DSM 30006T X81661 | − | − | − | + | − | − | − | + |
| 8 | MB1 | Acinetobacter calcoaceticus DSM 30006T X81661 | − | − | − | + | − | − | − | + |
| 9 | MK4 | Acinetobacter calcoaceticus DSM 30006T X81661 | − | − | − | + | − | − | − | + |
| 10 | NR4 | Acinetobacter calcoaceticus DSM 30006T X81661 | − | − | − | + | − | − | − | + |
| 11 | GD5 | Acinetobacter johnsonii DSM 6963T X81663 | − | − | − | − | − | − | − | − |
| 12 | NG1 | Acinetobacter johnsonii DSM 6963T X81663 | − | − | − | − | − | − | − | − |
| 13 | AP2 | Acinetobacter junii LMG998T AM410704 | − | − | − | − | − | − | − | − |
| 14 | AP4 | Acinetobacter junii LMG998T AM410704 | − | − | − | − | − | − | − | − |
| 15 | GD2 | Acinetobacter junii LMG998T AM410704 | − | − | − | − | − | − | − | − |
| 16 | JD4 | Acinetobacter junii LMG998T AM410704 | − | − | − | − | − | − | − | − |
| 17 | KL3 | Acinetobacter junii LMG998T AM410704 | − | − | − | − | − | − | − | − |
| 18 | KL4 | Acinetobacter junii LMG998T AM410704 | − | − | − | − | − | − | − | − |
| 19 | MB3 | Acinetobacter junii LMG998T AM410704 | − | − | − | − | − | − | − | − |
| 20 | R1 | Acinetobacter junii LMG998T AM410704 | − | − | − | − | − | − | − | − |
| 21 | WN1 | Acinetobacter junii LMG998T AM410704 | − | − | − | − | − | − | − | − |
| 22 | WN3 | Acinetobacter junii LMG998T AM410704 | − | − | − | − | − | − | − | − |
| 23 | WN4 | Acinetobacter junii LMG998T AM410704 | − | − | − | − | − | − | − | − |
| 24 | WN5 | Acinetobacter junii LMG998T AM410704 | − | − | − | − | − | − | − | − |
| 25 | MB4 | Acinetobacter ursingii LUH 3792T AJ275038 | − | − | − | + | + | + | − | − |
| 26 | JD1 | Aeromonas aquariorum MDC47T EU085557 | + | + | + | + | + | − | + | − |
| 27 | R31 | Aeromonas aquariorum MDC47T EU085557 | + | + | + | + | + | − | + | − |
| 28 | R30 | Aeromonas aquariorum MDC47T EU085557 | + | + | + | + | + | − | + | − |
| 29 | GD4 | Aeromonas hydrophila subsp. hydrophila ATCC 7966T CP000462 | + | − | + | + | + | + | + | + |
| 30 | JD3 | Aeromonas ichthiosmia DSM 6393T X71120 | + | − | + | + | + | + | + | + |
| 31 | MK2 | Aeromonas ichthiosmia DSM 6393T X71120 | + | − | + | + | + | + | + | + |
| 32 | R26 | Aeromonas punctata subsp. punctate NCIMB 13016T X60408 | + | − | + | + | + | + | + | + |
| 33 | NG2 | Citrobacter freundii DSM 30039T AJ233408 | − | + | − | − | + | + | + | − |
| 34 | R21 | Pantoea anthophila LMG2558T EF688010 | − | − | + | + | − | + | − | − |
| 35 | R22 | Pantoea anthophila LMG2558T EF688010 | − | − | + | + | − | + | − | − |
| 36 | NR2 | Pseudomonas aeruginosa LMG 1242T Z76651 | − | − | − | + | + | + | − | + |
| 37 | R6 | Pseudomonas alcaligenes LMG 1224T Z76653 | − | − | − | + | + | + | − | + |
| 38 | R7 | Pseudomonas alcaligenes LMG 1224T Z76653 | − | − | − | + | + | + | − | + |
| 39 | R23 | Pseudomonas alcaligenes LMG 1224T Z76653 | − | − | − | + | + | + | − | + |
| 40 | D22 | Pseudomonas otitidis MCC10330T AY953147 | − | − | − | + | + | + | − | + |
| 41 | GD6 | Pseudomonas otitidis MCC10330T AY953147 | − | − | − | + | + | + | − | + |
| 42 | MB2 | Pseudomonas otitidis MCC10330T AY953147 | − | − | − | + | + | + | − | + |
| 43 | MK3 | Pseudomonas otitidis MCC10330T AY953147 | − | − | − | + | + | + | − | + |
| 44 | R8 | Kocuria rhizophila DSM 11926T Y16264 | − | − | − | − | + | + | − | − |
| 45 | D12 | Bacillus anthracis ATCC 14578T AB190217 | − | + | + | − | − | + | − | + |
| 46 | KL2 | Bacillus aryabhattai B8W22T EF114313 | − | + | + | − | − | + | − | + |
| 47 | NR3 | Bacillus aryabhattai B8W22T EF114313 | − | + | + | − | − | + | − | + |
| 48 | R5 | Bacillus cereus ATCC 14579T AE016877 | − | + | + | − | − | + | − | + |
| 49 | JU1 | Bacillus infantis SMC4352-1T AY904032 | − | + | + | − | − | + | − | + |
| 50 | JU3 | Bacillus licheniformis ATCC 14580 CP000002 | − | + | + | − | − | + | − | + |
| 51 | JU4 | Bacillus niabensis 4T19T AY998119 | − | + | + | − | − | + | − | + |
| 52 | AL4 | Bacillus stratosphericus 41KF2aT AJ831841 | − | + | + | − | − | + | − | + |
| 53 | R2 | Bacillus stratosphericus 41KF2aT AJ831841 | − | + | + | − | − | + | − | + |
| 54 | D25 | Exiguobacterium indicum HHS31 T AJ846291 | − | + | − | + | − | + | − | − |
| 55 | JU2 | Lysinibacillus fusiformis NBRC 15717T AB271743 | − | − | − | − | + | − | − | + |
Ind: Indole; MR: Methyl red; VP: Voges Proskeur; Cit: Citrate; Oxi: Oxidase; Cat: Catalase; Ure: Urease; and Gel: Gelatinase.
Coliform is the name of a test adopted in 1914 by the Public Health Service for the Enterobacteriaceae family. It is the commonly used bacterial indicator of sanitary quality of foods and water. Coliform bacteria live in soil or vegetation and in the gastrointestinal tract of animals. Coliform bacteria enter water supplies from the direct disposal of waste into streams or lakes, or from runoff, from wooded areas, pastures, feedlots, septic tanks, and sewage plants into streams or groundwater. Coliform bacteria are not a single type of bacteria, but a group of bacteria that includes many strains, such as E. coli. They are ubiquitous in nature, and many types are harmless. Coliform bacteria belong to the genera Citrobacter, Enterobacter, Escherichia, Hafnia, Klebsiella, Pantoea, and Serratia. Out of 55 isolates only 3 isolates belonged to coliform group which are NG2 which is phylogenetically close to Citrobacter freundii and R21 and R22 which are phylogenetically close to Pantoea anthophila. This shows that the water samples NG and R collected from Nagarkurnool and Mahabubnagar-2 might be contaminated with fecal pollution and a consequent hazard of contracting a disease through pathogenic organisms. Nonetheless, the disease-causing organisms (pathogens) mostly transmitted via drinking water are predominantly of fecal origin. A total of 2 and 12 strains were isolated from the samples NG and R which include 3 coliform bacteria, and out of 14 strains, 7 were likely to be pathogenic which include NG1 which is phylogenetically close to Acinetobacter johnsonii, R1 which is phylogenetically close to Acinetobacter junii, NG2 which is phylogenetically close to Citrobacter freundii, R6, 7, and 23 which are phylogenetically close to Pseudomonas alcaligenes, and R5 which is phylogenetically close to Bacillus cereus.
In the present study, species that belonged to the genera Aeromonas, Methylobacterium, Bacillus, and Pseudomonas were isolated which is supported by the different studies where several novel species which belong to the genera Aeromonas [11], Methylobacterium [12], Bacillus [13], and Pseudomonas [14] were reported from drinking water systems. Aeromonas spp. are ubiquitous bacteria found in diverse aquatic environments worldwide such as bottled water, chlorinated water, well water, and heavily polluted waters. Aeromonas hydrophila survives easily in waters polluted by feces and seems resistant to various disinfectants, insecticides, and chemicals.
Recent studies on drinking water in rural areas in Northern Rajasthan, India, [4] and in drinking water distribution system of a hospital in Hungary, [5] showed the presence of several bacteria in the drinking water which included Acinetobacter lwoffii, Alcaligenes faecalis, Bacillus cereus, Corynebacterium tuberculostearicum, Enterobacter aerogenes, Escherichia coli, Escherichia albertii, Klebsiella sp., Micrococcus luteus, Proteus vulgaris, Pseudomonas aeruginosa, Staphylococcus aureus, and Streptococcus lactis. In the present study, species belonging to the genus Acinetobacter, Bacillus cereus, and Pseudomonas aeruginosa which are opportunistic pathogens were isolated from 9 drinking water samples such as AP, GD, JD, KL, MB, NG, NR, R, and WN out of 13.
Of the 27 groups, only few (GD4, R5, AP1, and NR2) of the phylogenetic neighbors had been isolated earlier from the drinking water sources including Aeromonas hydrophila [4], Bacillus cereus [4], Cupriavidus pauculus [15], and Pseudomonas aeruginosa [4, 5]. More than half (52.7%, 29 strains) of the phylogenetic neighbors which belonged to 12 groups were reported to be pathogenic and were isolated from clinical specimens, and other 15 groups were reported to be isolated from several distinct habitats. Most of the drinking water sources studied for bacterial diversity include bacteria which are phylogenetically close to pathogenic bacteria except one sample AL from which 3 strains AL1, AL3, and AL4 were isolated which belong to the phylogenetic neighbors Methylobacterium zatmanii, Delftia lacustris, and Bacillus stratosphericus, respectively. The isolates which are likely to be pathogenic include strain AP1 which is phylogenetically close to Cupriavidus pauculus which causes human infections sporadically and were isolated earlier from a variety of human clinical sources and also from drinking water sources [15] (Table 4); strains GD5 and NG1 which are phylogenetically close to Acinetobacter johnsonii which causes vascular catheter-related bloodstream infection were isolated from clinical specimens and milk [16] (Table 4); strains AP2, AP4, GD2, JD4, KL3, KL4, MB3, R1, WN1, WN3, WN4, and WN5 which are phylogenetically close to Acinetobacter junii which causes septicemia, meningitis, peritonitis, and so forth were isolated from human clinical specimens like urine [17] (Table 4); strain MB4 which is phylogenetically close to Acinetobacter ursingii which causes bacteremia was isolated from blood [18] (Table 4); strain GD4 which is phylogenetically close to Aeromonas hydrophila subsp. Hydrophila which causes acute diarrheal disease was isolated from humans, animals, fish, and fresh water [11, 19] (Table 4); strain NG2 which is phylogenetically close to Citrobacter freundii which causes opportunistic infections was isolated from canal water [20] (Table 4); strain NR2 which is phylogenetically close to Pseudomonas aeruginosa which causes several infections in immunocompromised individuals was isolated from wide variety habitats including clinical samples and also from drinking water samples [4, 5] (Table 4); strains R6, R7, and R23 which are phylogenetically close to Pseudomonas alcaligenes which causes endocarditis occasionally were isolated from soil, water, and also from clinical samples [21] (Table 4); strains D22, GD6, MB2, and MK3 which are phylogenetically close to Pseudomonas otitidis which causes inflammation of the ear were isolated from the ear [22] (Table 4); strain D12 which is phylogenetically close to Bacillus anthracis which causes anthrax disease was isolated from sheep blood (Table 4); strain R5 which is phylogenetically close to Bacillus cereus which causes food poisoning was isolated from the soil and also food materials [23] (Table 4); and strain JU1 which is phylogenetically close to Bacillus infantis which causes sepsis was isolated from clinical specimens [24] (Table 4).
Table 4.
Isolation of source of the type strains associate with disease.
| Sl no. | Nearest phylogenetic neighbor | Isolation source | Associated human disease | Reference |
|---|---|---|---|---|
| 1 | Cupriavidus pauculus LMG 3413T AF085226 | Isolated from a variety of human clinical sources including blood, wounds, sputum, urine, eye, throat, and peritoneal fluid. In addition strains have been isolated from pool water, ground water, and bottled mineral water | Sporadically cause human infections | Vandamme et al., 1999 [15] |
|
| ||||
| 2 | Acinetobacter johnsonii DSM 6963T X81663 | Isolated from clinical specimens and from eviscerated chickens and may cause ropiness in milk. Isolated from duodenum | Vascular catheter-related bloodstream infection | Seifert et al., 1993 [16] |
|
| ||||
| 3 | Acinetobacter junii LMG 998T AM410704 | Isolated from human clinical specimens like urine | Septicemia, community-acquired bacterial meningitis, peritoneal dialysis-related peritonitis, and infections associated with corneal perforation | Hung et al., 2009 [17] |
|
| ||||
| 4 | Acinetobacter ursingii LUH 3792T AJ275038 | Isolated from blood of a hospitalized patient with endocarditis | Bacteremia | Loubinoux et al., 2003 [18] |
|
| ||||
| 5 | Aeromonas hydrophila subsp. hydrophila ATCC 7966T CP000462 | Isolated from humans, animals, fish, and fresh water | Acute diarrheal disease | Ljungh et al., 1977 [19] |
|
| ||||
| 6 | Citrobacter freundii DSM 30039T AJ233408 | Isolated from canal water | Opportunistic infections like neonatal meningitis | Badger et al., 1999 [20] |
|
| ||||
| 7 | Pseudomonas aeruginosa LMG 1242T Z76651 | Found in soil, water, skin flora, and most manmade environments throughout the world | Localized infection of eye, ear, skin, urinary, and respiratory. Bone, joint, gastrointestinal tract, central nervous system, and systemic infection with bacteremia. Secondary pneumonia. Endocarditis | |
|
| ||||
| 8 | Pseudomonas alcaligenes LMG 1224T Z76653 | Common soil and water inhabitant that has rarely been proven a human pathogen | Endocarditis | Valenstein et al., 1983 [21] |
|
| ||||
| 9 | Pseudomonas otitidis MCC 10330T AY953147 | Isolated from the ears of patients with acute otitis externa (inflammation of the ear) | Inflammation of the ear | Clark et al., 2006 [22] |
|
| ||||
| 10 | Bacillus anthracis ATCC 14578T AB190217 | Isolated from the blood of sheep suffering from anthrax | Cutaneous anthrax, pulmonary anthrax, and gastrointestinal anthrax | |
|
| ||||
| 11 | Bacillus cereus ATCC 14579T AE016877 | Isolated from soil and food materials | Food poisoning | Todar, 2008 [23] |
|
| ||||
| 12 | Bacillus infantis SMC 4352-1T AY904032 | Isolated from a newborn child with sepsis | Sepsis | Ko et al., 2006 [24] |
Out of 27 representative taxa are affiliated have eight representative genera in drinking water except for those affiliated with the genera Exiguobacterium, Delftia, Kocuria, and Lysinibacillus.
4. Conclusion
This study is an overview of drinking water quality in the rural and urban parts of Mahabubnagar District, India. Recent investigations on bacterial contaminations of drinking water in the hospitals suggested risks of secondary infection to the patients, staff, and also patient relatives. These observations were just a drinking water quality assessment, and no data on human health scenario in this region were recorded. Further detailed studies on the health issues of the patients and other people using this contaminated drinking water are still required to trace the impact of this water consumption on people in those hospitals. The lack of awareness about good sanitation and personal hygienic practices, among governmental rural and also urban hospitals is an important factor for poor drinking water quality in these sources.
Conflict of Interests
The authors have no conflict of interests.
Acknowledgments
The authors are grateful to Profesor Bagyanarayana, Vice-Chancellor, and Profesor K Venkata Chalam, Registrar, Palamuru University, for their encouragement and support. The authors wish to acknowledge the Department of Science and Technology (DST) for their financial assistance for the Project “Advanced Molecular Methods for rapid detection of coliforms in potable water” DST/TM/WTI/2 K10/265.
References
- 1.Suthar S. Contaminated drinking water and rural health perspectives in Rajasthan, India: an overview of recent case studies. Environmental Monitoring and Assessment. 2011;173:837–849. doi: 10.1007/s10661-010-1427-2. [DOI] [PubMed] [Google Scholar]
- 2.WHO. Emerging Issues in Water and Infectious Disease. Geneva, Switzerland: World Health Organization (WHO); 2003. [Google Scholar]
- 3.Ashbolt NJ. Microbial contamination of drinking water and disease outcomes in developing regions. Toxicology. 2004;198(1–3):229–238. doi: 10.1016/j.tox.2004.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suthar S, Singh S, Chhimpa V, et al. Puranik. The problem of safe drinking water in Northern Rajasthan, India. Proceedings of the 16th National Symposium on Environment; 2008; Hisar, India. Department of Environmental Science and Technology, Guru Jambheshwar University of Science and Technology; pp. 491–496. [Google Scholar]
- 5.Felföldi T, Heéger Z, Vargha M, Márialigeti K. Detection of potentially pathogenic bacteria in the drinking water distribution system of a hospital in Hungary. Clinical Microbiology and Infection. 2010;16:89–92. doi: 10.1111/j.1469-0691.2009.02795.x. [DOI] [PubMed] [Google Scholar]
- 6.Lane DJ. 16S/23S rRNA sequencing. In: Stackebrandt E, Good Fellow M, editors. Nucleic Acid Techniques in Bacterial Systematics. Chichester, UK: John Wiley & Sons; 1991. pp. 115–175. [Google Scholar]
- 7.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 8.Chun J, Lee JH, Jung Y, et al. EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. International Journal of Systematic and Evolutionary Microbiology. 2007;57(10):2259–2261. doi: 10.1099/ijs.0.64915-0. [DOI] [PubMed] [Google Scholar]
- 9.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;25(24):4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology. 2003;52(5):696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 11.Huys G, Kämpfer P, Albert MJ, Kühn I, Denys R, Swings J. Aeromonas hydrophila subsp. dhakensis subsp. nov., isolated from children with diarrhoea in Bangladesh, and extended description of Aeromonas hydrophila subsp. hydrophila (Chester 1901) stanier 1943 (approved lists 1980) International Journal of Systematic and Evolutionary Microbiology. 2002;52(3):705–712. doi: 10.1099/00207713-52-3-705. [DOI] [PubMed] [Google Scholar]
- 12.Lee SW, Oh HW, Lee KH, Ahn TY. Methylobacterium dankookense sp. nov., isolated from drinking water. Journal of Microbiology. 2010;47(6):716–720. doi: 10.1007/s12275-009-0126-6. [DOI] [PubMed] [Google Scholar]
- 13.Zhang T, Fan X, Hanada S, Kamagata Y, Fang HHP. Bacillus macauensis sp. nov., a long-chain bacterium isolated from a drinking water supply. International Journal of Systematic and Evolutionary Microbiology. 2006;56(2):349–353. doi: 10.1099/ijs.0.63868-0. [DOI] [PubMed] [Google Scholar]
- 14.Ivanova EP, Gorshkova NM, Sawabe T, et al. Pseudomonas extremorientalis sp. nov., isolated from a driking water reservoir. International Journal of Systematic and Evolutionary Microbiology. 2002;52(6):2113–2120. doi: 10.1099/00207713-52-6-2113. [DOI] [PubMed] [Google Scholar]
- 15.Vandamme P, Goris J, Coenye T, et al. Assignment of Centers for Disease Control group IVc-2 to the genus Ralstonia as Ralstonia paucula sp. nov. International Journal of Systematic Bacteriology. 1999;49(2):663–669. doi: 10.1099/00207713-49-2-663. [DOI] [PubMed] [Google Scholar]
- 16.Seifert H, Strate A, Schulze A, Pulverer G. Vascular catheter-related bloodstream infection due to Acinetobacter johnsonii (formerly Acinetobacter calcoaceticus var. lwoffii): report of 13 cases. Clinical Infectious Diseases. 1993;17(4):632–636. doi: 10.1093/clinids/17.4.632. [DOI] [PubMed] [Google Scholar]
- 17.Hung YT, Lee YT, Huang LJ, et al. Clinical characteristics of patients with Acinetobacterjunii infection. Journal of Microbiology, Immunology and Infection. 2009;42(1):47–53. [PubMed] [Google Scholar]
- 18.Loubinoux J, Mihaila-Amrouche L, Le Fleche A, et al. Bacteremia caused by Acinetobacter ursingii . Journal of Clinical Microbiology. 2003;41(3):1337–1338. doi: 10.1128/JCM.41.3.1337-1338.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ljungh A, Popoff M, Wadstrom T. Aeromonas hydrophila in acute diarrheal disease: detection of enterotoxin and biotyping of strains. Journal of Clinical Microbiology. 1977;6(2):96–100. doi: 10.1128/jcm.6.2.96-100.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Badger JL, Stins MF, Kim KS. Citrobacter freundii invades and replicates in human brain microvascular endothelial cells. Infection and Immunity. 1999;67(8):4208–4215. doi: 10.1128/iai.67.8.4208-4215.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valenstein P, Bardy GH, Cox CC, Zwadyk P. Pseudomonas alcaligenes endocarditis. American Journal of Clinical Pathology. 1983;79(2):245–247. doi: 10.1093/ajcp/79.2.245. [DOI] [PubMed] [Google Scholar]
- 22.Clark LL, Dajcs JJ, McLean CH, Bartell JG, Stroman DW. Pseudomonas otitidis sp. nov., isolated from patients with otic infections. International Journal of Systematic and Evolutionary Microbiology. 2006;56(4):709–714. doi: 10.1099/ijs.0.63753-0. [DOI] [PubMed] [Google Scholar]
- 23.Todar K. Todar’s Online Textbook of Bacteriology. 2008. Bacillus cereus food poisoning. http://www.textbookofbacteriology.net/B.cereus.html. [Google Scholar]
- 24.Ko KS, Oh WS, Lee MY, et al. Bacillus infantis sp. nov. and Bacillus idriensis sp. nov., isolated from a patient with neonatal sepsis. International Journal of Systematic and Evolutionary Microbiology. 2006;56(11):2541–2544. doi: 10.1099/ijs.0.64213-0. [DOI] [PubMed] [Google Scholar]


