SUMMARY
AAA+ unfoldases denature and translocate polypeptides into associated peptidases. We report direct observations of mechanical, force-induced protein unfolding by the ClpX unfoldase from E. coli, alone, and in complex with the ClpP peptidase. ClpX hydrolyzes ATP to generate mechanical force and translocate polypeptides through its central pore. Threading is interrupted by pauses that are found to be off the main translocation pathway. ClpX’s translocation velocity is force dependent, reaching a maximum of 80 aa/s near-zero force and vanishing at around 20 pN. ClpX takes 1, 2, or 3 nm steps, suggesting a fundamental step-size of 1 nm and a certain degree of intersubunit coordination. When ClpX encounters a folded protein, it either overcomes this mechanical barrier or slips on the polypeptide before making another unfolding attempt. Binding of ClpP decreases the slip probability and enhances the unfolding efficiency of ClpX. Under the action of ClpXP, GFP unravels cooperatively via a transient intermediate.
INTRODUCTION
ATP-dependent proteases of the AAA+ (ATPases associated with various cellular activities) superfamily power the degradation of abnormal, denatured, or otherwise damaged polypeptides, as well as the removal of short-lived regulatory proteins (King et al., 1996). The hydrolytic active sites of these proteases are sequestered in the internal chamber of a barrel-shaped peptidase complex, preventing the diffusion and nonspecific degradation of folded or even large unfolded polypeptides (Wang et al., 1997). To facilitate specific protein degradation, the peptidases pair with energy-dependent hexameric AAA+ unfoldases that recognize appropriately tagged protein substrates and utilize the energy from ATP hydrolysis to unfold and translocate the polypeptide into the associated peptidase chamber for degradation (Baker and Sauer, 2006). It has been suggested previously that AAA+ unfoldases may exert mechanical force to unravel the tertiary and secondary structures of protein substrates. Even though there have been initial single-molecule fluorescence studies of ClpX (Shin et al., 2009), direct evidence for the generation of force and a detailed characterization of the mechanochemistry of these molecular machines are still lacking. Here we investigate the motor properties of ClpX, a homohexameric AAA+ ATPase from Escherichia coli that recognizes proteins with a C-terminal ssrA tag and uses cycles of ATP hydrolysis to unfold and translocate the substrates into its associated peptidase, ClpP (Gottesman et al., 1998). We use a single-molecule optical tweezers-based assay to demonstrate that ClpX generates mechanical force to unfold its substrates. We characterize the dynamics of ClpX as it encounters a folded substrate such as GFP and translocates unfolded polypeptides through its central processing pore. Furthermore, we investigate the effects of ClpP on the translocation activity and unfolding efficiency of ClpX. These studies thus provide important new insight into the general operating principles used by energy-dependent proteases to unfold and degrade protein substrates inside the cell.
RESULTS
ClpX Unfolding Trajectories
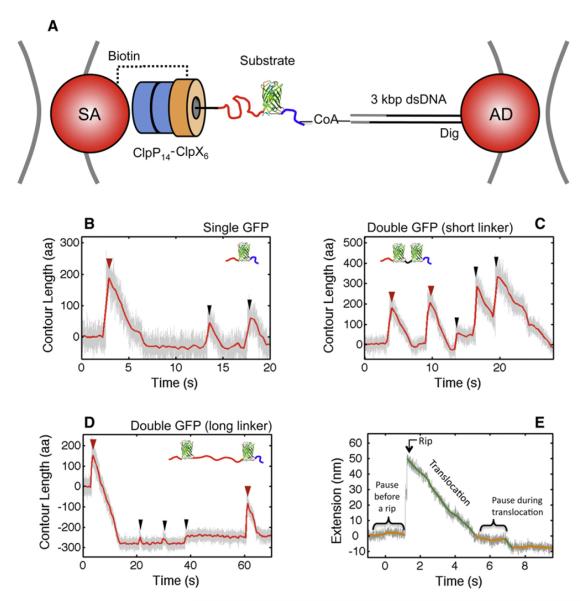
A dual-trap optical tweezers geometry was used to monitor real-time trajectories of individual ClpX hexamers or ClpXP complexes as they unfold and translocate GFP-titin fusion substrates. In these substrates, either one GFP molecule or two GFP moieties in tandem separated by either a short ora long unstructured linker were C-terminally fused to a permanently unfolded ssrA-tagged titin I27 module (Ticm) and attached by their N termini to a dsDNA handle (Figure 1, see Experimental Procedures). The DNA-tethered substrates and a single-chain variant of ClpX were immobilized on different polystyrene beads coated with anti-digoxigenin antibody and streptavidin, respectively. ClpX was allowed to bind and engage the ssrA-tagged substrate by bringing the two beads into close proximity in the presence of saturating ATP and an ATP regeneration system (Figure 1A).
Figure 1. Unfolding and Translocation of GFP-Titin Fusion Proteins by ClpX and ClpXP.
(A) The geometry of our single-molecule assay in dual-trap optical tweezers: ClpX(P) complexes were immobilized on a streptavidin polystyrene bead (SA) via a biotin tag on ClpX. The GFP-titin fusion substrate is covalently linked to a 3 kbp dsDNA handle with a Dig tag that binds to an antibody-coated polystyrene bead (AD). All substrates included one or two GFP molecules (green) fused to a Ticm-ssrA moiety (red and black). The blue flexible linker corresponds to the ybbR tag.
(B–D) GFP unfolding (red arrowheads), motor slips (black arrowheads) and translocation trajectories for three different substrates obtained at ~7 pN. (B) Single GFP-titin substrate. (C) Double GFP substrate with a short linker (10 aa) between the GFP molecules. (D) Double GFP substrate with a long flexible linker of 200 aa corresponding to two Ticm domains (red) between the GFP molecules.
(E) Basic features of our single-molecule trajectories. The rip corresponding to a GFP unfolding is preceded by a pause (orange). After GFP is unfolded, ClpX translocates the unfolded polypeptide chain (green) with occasional pauses (orange). In all cases the raw data were filtered and decimated to 1000 Hz (in gray) or 2.5 Hz (in red). For the protocol describing the attachment of the dsDNA handle to the fusion substrates, see Figure S3. In the presence of ATP-γ-S, none of the unfolding or translocation events described in (B)–(E) were observed (Figure S5).
After successful substrate engagement, we monitored ClpX unfolding and translocation activities by measuring the changes in extension between the two beads in passive mode (i.e., the trap positions are fixed and the force load on the motor is allowed to vary with motor activity). Traces for the three different substrates displayed sudden extension gains (rips) followed by a slower decrease in extension, together resembling a sawtooth-like pattern (Figures 1B–1D). We assigned rips either to ClpX-induced unfolding events of GFP or to temporary disengagement of the motor along the polypeptide backbone track (Figure 1, red and black arrowheads, respectively). The continuous decrease in extension after a rip was identified as the translocation of the unfolded polypeptide chain through the ClpX pore. As seen in Figures 1B–1E, translocation is interspersed by pauses of various lengths. In >95% of all trajectories, translocation events were not observed before the first rip. We therefore conclude that the motor had already translocated the unfolded Ticm-ssrA segment by the time we started recording ClpX unfolding activity. The changes in extension (in nm) associated with rips and translocation can be expressed in terms of numbers of amino acids (aa) using the worm-like chain (WLC) force-extension formula (Bustamante et al., 1994) (see Experimental Procedures).
ClpX Translocation and Force Dependence
The characteristics of the translocation process of ClpX are fundamentally distinct from those of other molecular motors that have been studied under external force (Bustamante et al., 2011; Yildiz and Selvin, 2005). For molecular motors moving along stiff tracks such as dsDNA or microtubules, the track persistence length (P) is orders of magnitude larger than the step-size of the motor (for example, PdsDNA = ~50 nm compared to motor step-sizes of just 0.34 and 0.85 nm for RNA polymerase and ϕ29 packaging motor, respectively [Abbondanzieri et al., 2005; Moffitt et al., 2009]; and Pmicrotubule = ~1 mm compared to a step-size of 8 nm for kinesin [Svoboda et al., 1993]). We observe ClpX(P) taking 1 nm steps (see below), which are larger than the persistence length of the polypeptide track (Ppeptide = 0.65 nm). As a result, on a lengthscale comparable to the step-size, ClpX(P) translocates a flexible, irregular polymer, whereas motors such as kinesin or dsDNA translocases move along much more rigid, periodic tracks.
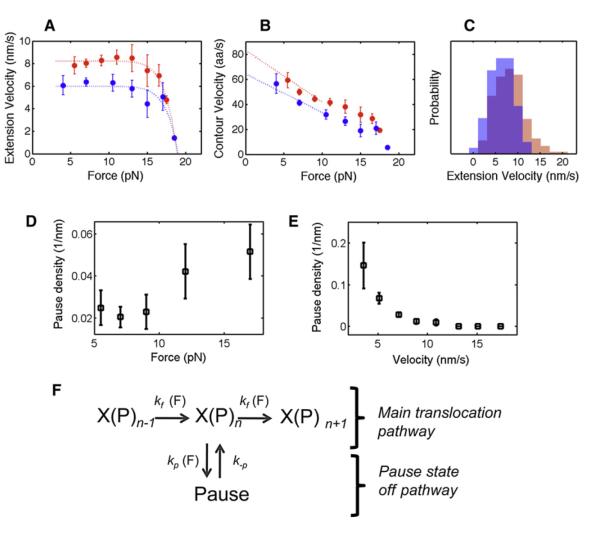
The highly elastic nature of unfolded polypeptides and the unique characteristics of this AAA+ unfoldase compelled us to define two distinct quantities: contour velocity (in aa/s) and extension velocity (in nm/s). Extension velocity refers to the geometric length (in nm) of the translocated polypeptide chain, reflecting the size of the motor step, whereas contour velocity measures how many amino acids pass through the ClpX pore per unit time. For example, 1 nm/s of extension velocity corresponds to ~8 aa/s of contour velocity at 4 pN external force and only ~4 aa/s at 13 pN. To illustrate this point, imagine ClpX as an ant walking on a rubber band stretched under force. At a low force the ant will traverse the entire length of the rubber band faster than at a high force, even though the ant maintains a constant step-size and stepping rate in both cases. The product of the step-size and the stepping rate in the ant analogy corresponds to the extension velocity (nm/s) of ClpX, whereas the amount of rubber traveled by the ant corresponds to the contour velocity. Although contour velocity (aa/s) has been widely used in bulk studies (zero-external force), the extension velocity (nm/s) is necessary to properly characterize polypeptide translocation under external forces. Because all of our single-molecule experiments were performed under external force, most of our discussions refer to the extension velocity (nm/s). Our analysis yielded a pause-free extension velocity of 8.2 ± 0.3 nm/s (mean ± standard error of the mean [SEM]) for ClpX at opposing forces between 5–13 pN (Figure 2A, red symbols), whereas the extension velocity for the ClpXP complex in the same force range was 6.0 ± 0.3 nm/s (mean ± SEM, Figure 2A, blue symbols). This 27% decrease in velocity can be explained by the repression in ATP hydrolysis activity of ClpX when bound to ClpP (Table S1 available online). We used the contour velocity (aa/s) to estimate near-zero force velocities of ~80 aa/s and ~60 aa/s for ClpX and ClpXP, respectively (Figure 2B). The latter value is in excellent agreement with bulk estimates for ClpXP translocation (Martin et al., 2008b).
Figure 2. Motor Properties and Mechanochemical Cycle of ClpX(P).
(A) Extension translocation velocity (mean ± SEM in nm/s) as a function of external force for ClpX (red symbols) and ClpXP (blue symbols). The dotted curves represent phenomenological fits to the data.
(B) Contour length translocation velocity (mean ± SEM in aa/s) as a function of external force for ClpX (red symbols) and ClpXP (blue symbols). The dotted lines extrapolate velocity to zero-external force.
(C) Histograms of extension velocity for ClpX (red) and ClpXP (blue) at forces below 13 pN, where velocity is force independent.
(D) The pause density (or frequency) during ClpX(P) translocation (mean ± SEM) as a function of force.
(E) Correlation plot between ClpX(P) pause density (mean ± SEM) versus translocation velocity, when velocity is force independent (<13 pN).
(F) The proposed minimal mechanochemical cycle for ClpX(P) during translocation, including associated force-dependent rates. X(P)n-1, X(P)n, and X(P)n+1 refer to consecutive steps of ClpX(P) during translocation. The translocation rate (kf) and pause entry (kp), reflected by the pause-free velocity and pause density, respectively, are force dependent whereas the exit from a pause (k-p) is independent of force. For additional information on pause desnsity and duration, see Table S2.
Interestingly, a rather broad distribution of the mean pause-free translocation velocities for ClpX and ClpXP was seen regardless of the force range (Figure 2C), revealing an intrinsic heterogeneity in the activity of individual ClpX hexamers. Although similar heterogeneity has been described previously for other molecular motor enzymes (Neuman et al., 2003), it is possible that this dispersion in part reflects the chemical and physical heterogeneity of the unfolded polypeptide track.
ClpXP: Allosteric or Force-Generating Enzyme
A long-standing question about AAA+ unfoldases is whether these enzymes in fact exert mechanical force or just use an allosteric binding mechanism to unfold their substrates. To investigate the unfolding mechanism used by ClpX, we monitored the response of the motor to an opposing force while translocating a substrate. The force versus velocity plot shows that ClpX is capable of working against and therefore generating mechanical force (Figure 2A, red symbols). This plot also reveals that the ClpX translocation velocity is nearly constant up to ~13 pN, implying that within this force range and at saturating ATP concentrations, chemical steps (hydrolysis or product release) are rate limiting. At opposing external forces exceeding 13 pN, the pause-free velocity decreases monotonically, indicating that conformational changes driving translocation become rate limiting. We were able to place a lower bound of ~20 pN for the stall force of ClpX (Figure 2A). ClpXP’s stall force and force-velocity dependence is very similar to that of ClpX (Figure 2A, blue symbols). It has been proposed that protein unraveling by a AAA+ unfoldase results from the enzyme’s attempts to translocate the folded structure through its narrow central pore. Thus, the stall force for translocating a resisting polypeptide gives a measure of the maximum mechanical force that ClpX can apply to unfold a substrate.
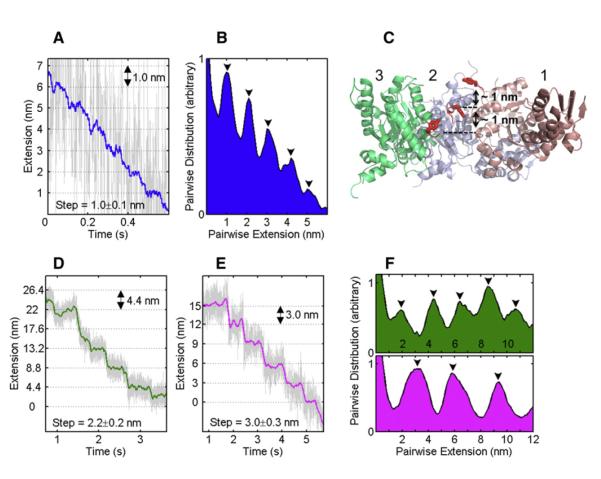
At high opposing loads, it was possible to observe well-defined translocation steps for ClpXP (Figure 3). The pairwise distribution analysis of these traces revealed a remarkable stepping periodicity of 1, 2, and 3 nm (Figures 3B and 3F). The observed 1 nm step is in good agreement with high-resolution structural data of ClpX (Figure 3C), which show distinct conformations of subunits in different nucleotide states and indicating potential transitions and loop movements of ~1 nm per ClpX subunit that might lead to substrate translocation (Glynn et al., 2009). It is assumed that conformational changes of the pore 1 loops with their highly conserved Tyr residues (Figure 3C, red loops) together with rigid-body movements of subunits in the ClpX hexamer propel the substrate through the central pore (Martin et al., 2008a). Based on mutational studies, it was suggested that subunits contribute additively to ClpX activity and that ATP hydrolysis in one subunit at a time drives the conformational changes for substrate translocation (Martin et al., 2005). Our measured extension velocities of 6 and 8 nm/s, along with the smallest observed step-size of 1 nm, predict hydrolysis rates of 360 and 480 ATP•min−1 for ClpXP and ClpX, respectively (assuming 1 ATP consumed per step). These values are in good agreement with our bulk measurements of the ATPase activities during translocation of permanently unstructured substrates (Table S1 and Figure S4). Our observation of distinct 2 and 3 nm steps (Figures 3D–3F) suggests a coordinated, near-simultaneous stepping of two or three motor subunits that cannot be resolved in our measurements.
Figure 3. ClpXP Translocation Stepping Displays Coordination between Subunits.
(A) Fragment of a ClpXP translocation region at ~10 pN displaying stepping periodicity of 1.0 nm. Raw data filtered and decimated to 500 Hz are shown in gray, and the raw data boxcar-filtered to 20 Hz are shown in blue.
(B) The pairwise distance distribution for the trace in (A) shows a 1 nm periodicity (black arrows).
(C) Side view of the nucleotide-bound ClpX hexamer (Glynn et al., 2009) with three subunits removed to allow visualization of the pore 1 loops (red) with the critical Tyr153 (stick representation) shows distinct staggering relative to the pore axis. The loop of the nucleotide-free subunit 1 is close to the top of the pore, whereas the loops of the nucleotide-bound subunits 2 and 3 are in an intermediate and bottom position, respectively. The distances between Tyr153 in each of these staggered positions are ~1 nm.
(D and E) Fragment of a ClpXP translocation region at ~14 pN displaying a stepping periodicity of 2.2 and 3.0 nm, respectively.
(F) The pairwise distance distribution for the traces in (D) and (E) shows the 2.2 and 3.0 nm periodicity (upper and lower panels, respectively).
For data comparing the ATP hydrolysis rates of ClpX and ClpXP, see Table S1 and Figure S4.
Power Stroke versus Brownian Ratchet Model
Two general models of motor operation have been proposed for AAA+ molecular machines. In the Brownian ratchet model, the motor uses ATP binding/hydrolysis or product release to rectify its Brownian motion, cross the energy barrier, and move in one direction (Astumian, 1997). In the power-stroke model, the motor uses the energy of ATP binding/hydrolysis or product release to directly drive the motion. Given a lower bound of ~20 pN for the stall force and a step-size of 1 nm, the work performed by a single ClpX subunit near stall is ΔWsubunit = 20 pN•nm = 5 kBT per hydrolyzed ATP. Similar calculations for E. coli RNA polymerase (Brownian ratchet) and ϕ29 ATPase (power-stroke motor) yield near-stall work values of ~2 kBT and ~10 kBT, respectively (Moffitt et al., 2009; Neuman et al., 2003; Smith et al., 2001). Because the maximum work done by a ClpX subunit is >5 kBT, we favor the power-stroke model over the Brownian ratchet for ClpX. Moreover, we estimated that the free energy of hydrolyzing one ATP molecule in our buffer is ~55 pN•nm. Therefore, the maximum thermodynamic efficiency of ClpX per step is 20 pN•1 nm/55 pN•nm = ~35%, within the efficiency range of other power-stroke motors (Smith et al., 2001). Future experiments, in which the concentration of ATP and hydrolysis products are changed systematically under varying external force, will be required to identify the force-generating step of the mechanochemical cycle and will help confirm a power-stroke mechanism for ClpX.
Mechanochemistry and Minimal Kinetic Cycle of ClpX
To better understand the mechanochemistry of ClpX, we analyzed the motor’s pause density (number of pauses per amino acid translocated) and pause duration during translocation of the unfolded polypeptide in two opposing force regimes, 5–12 pN and 12–20 pN. We found that higher force loads increased the probability of ClpX entering a pause state by a factor of two from 0.025 ± 0.005 nm−1 to 0.045 ± 0.008 nm−1 (p = 0.008, Table S2). On the other hand, the pause duration (which reflects the probability to exit a pause state) was not affected by high opposing force load (p = 0.99, Table S2). The dissimilar effect of the external force on pause entry versus exit can be explained if the transition state, x‡, is located very close to the pause state (Tinoco and Bustamante, 2002). For a displacement (Δx) between the active and pause states, this means that Δx – x‡ ≈0. Calculation of x‡ based on the pause density distribution (Figure 2D) allowed us to estimate Δx ≈x‡ = 1.7 ± 0.1 Å (mean ± standard deviation [SD], Table S2). Analysis of pause durations shows that they are distributed according to a single exponential (k = 2.3 ± 0.6 s−1, R2 = 0.99), indicating that exiting from the pause state involves a single kinetic event.
In order to establish whether or not pauses are states off the main translocation pathway of ClpX, we analyzed the natural fluctuations of the pause-free velocity in a force range that does not affect the rate of motor translocation and calculated the correlation between the translocation rate and the probability of entering a pause (pause density in units of 1/nm). Between 5–12 pN, the translocation rate and pause density were negatively correlated (R2 = −0.5, Table S2). Thus, we observe an increase in the number of pauses as the pause-free velocity decreases (Figure 2E), indicating that pausing and translocation compete kinetically. This type of kinetic competition is expected when pauses are states off the main translocation pathway. These results also explain the increase in pause density for forces between 12–20 pN, as the force-induced reduction of the motor velocity increases the pause entry probability. The analysis of pause density, pause duration, and their force dependence was statistically indistinguishable between ClpX and the ClpXP complex (Table S2). Based on these results, we propose the kinetic cycle shown in Figure 2F for the translocation of unfolded polypeptide by ClpXP, as well as the effects of mechanical forces on the various steps.
ClpX Unfolds GFP via a Well-Defined Intermediate
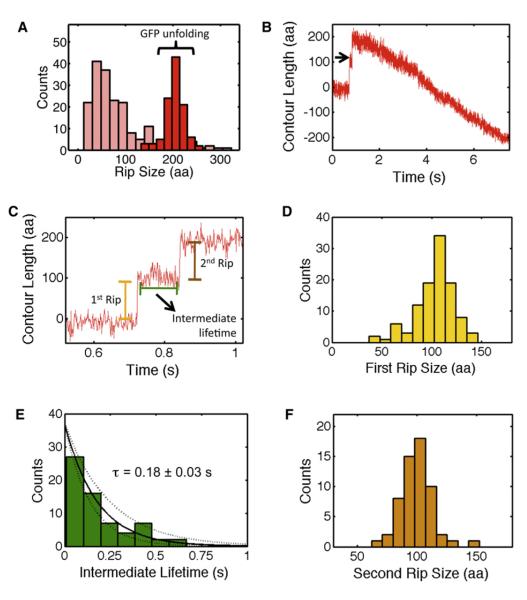
Another fundamental question about AAA+ unfoldases is whether denaturation of single-domain substrates is primarily determined by the protein’s energy landscape and occurs in a single cooperative unfolding transition or proceeds through several unfolding events depending on the presence of distinct mechanical barriers along the unfolding trajectory. To address this question, we analyzed the extensions of all rips for the three different GFP-titin substrates (Figure 1). Unfolding trajectories of the fusion construct with a single GFP molecule showed mainly two or three rips, each one followed by translocation of unfolded polypeptide chain (Figure 1B). On average, we observed twice as many rips throughout the unfolding trajectory for substrates with two GFP molecules (Figures 1C and 1D). From all these trajectories, we clearly recognized identical rip extensions that indicate a common unfolding signature of GFP (Figure 1, red arrowheads). In fact, a histogram of the rip extensions (in number of amino acids) obtained from all GFP substrates revealed a higher probability for a transition centered at 207 ± 2 aa (mean ± SEM,Figure 4A, dark red bars). In addition to this 207 aa rip, we observed rips whose extensions are less regular and vary between 20 and 300 aa, with the highest probability at 37 ± 6 aa (mean ± SEM, Figure 4A, pink bars). We interpret these irregular extensions as slipping events of ClpX backward along an already unfolded polypeptide (black arrowheads, Figure 1).
Figure 4. Unraveling of GFP by ClpX(P) Shows a Well-Defined Structural Transition and Reveals a Single-Unfolding Intermediate.
(A) The distribution of rip sizes (in aa) for all observed GFP unfolding events (red, n = 107) and slips (light red, n = 250) in ClpX and ClpXP traces.
(B) ClpX trace containing the unfolding of the first GFP in the double GFP construct with the long linker (Figure 1D). The unfolding event is followed by the translocation of the unfolded GFP and the long linker. Raw data filtered and decimated to 800 Hz.
(C) A detailed view of the short-lived GFP unfolding intermediate (black arrow in panel B).
(D) Histogram of the first rip size during GFP unfolding.
(E) The distribution of GFP unfolding intermediate lifetimes is well described by a single exponential. Dashed lines represent 95% confidence interval of the fit.
(F) Histogram of the second rip size during GFP unfolding.
For the rip size distribution before the long translocation marker (panel B), see Figure S1, and for side-by-side comparison of ClpX and ClpXP unfolding of GFP, see Figure S2.
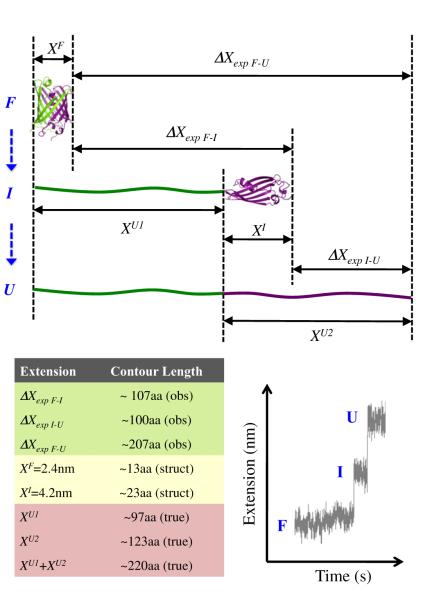
Given the extension of the folded GFP molecule (XF), and the experimentally observed extension change upon GFP unfolding (ΔXexp F-U), the true extension of the unfolded GFP (XU) can be calculated from ΔXexp F-U = XU – XF (Figure 5, upper portion). Based on the crystal structure (Ormö et al., 1996; Yang et al., 1996), the extension of folded GFP in our experimental geometry is 2.4 nm, equivalent to 13 ± 1 aa in the force range of 6–10 pN. Thus, the ΔXexp F-U = 207 aa rip corresponds to the unfolding of 220 ± 3 aa (mean ± SEM). This result is in excellent agreement with the number of amino acids that show well-defined secondary structures in GFP (residues 5 to 227, Figure 7A). To corroborate the assignment of GFP unfolding events in our trajectories, we used the specific extension signature of the construct with two Ticm domains inserted between two GFP molecules. This long stretch of unfolded polypeptide (~200 aa) served as an independent internal marker to identify the unfolding event of the first C-terminal GFP before the long translocation (Figure 1D, red arrowheads). When we analyzed the size distribution of those rips immediately preceding the translocation of the long unfolded polypeptide, we observed a peak at 207 aa (Figure S1), thus corroborating our previous structural assignment.
Figure 5. A Schematic Representation of GFP Unfolding via a Short-Lived Intermediate.
(F), (I), and (U) denote the folded, intermediate, and unfolded conformations of GFP. ΔX quantities correspond to experimentally observed extension changes (obs). XF and XI are the dimensions of the folded and intermediate states of GFP, estimated from structural data (struct). XU1 and XU2 are the true extensions (true) corresponding to the unfolding of the first and second portions of GFP (shown as flexible chains in green and purple, respectively). The true extension of the entire unfolded GFP molecule is XU1 + XU2. The table provides reference values estimated for 6–10 pN. Bottom right panel displays a characteristic “rip-transition-rip” during GFP unfolding.
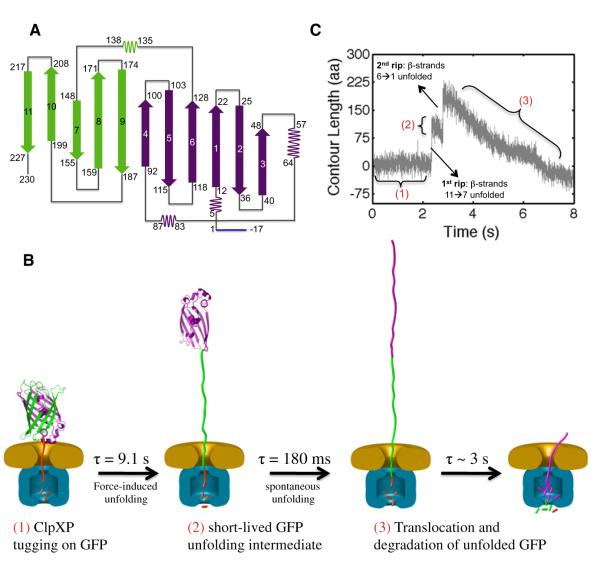
Figure 7. Model for the Mechanical Unfolding of GFP by ClpX(P).
(A) A two-dimensional representation of the folded GFP topology: β strands 11→7 are shown in green, and β strands 6→1 in purple. The ybbR tag is in blue.
(B) A schematic diagram of GFP (green/purple) unfolding by a ClpX hexamer (orange) in complex with ClpP (blue). The left panel shows the entire complex with an intact GFP molecule before unfolding. The center-left panel shows the unfolding intermediate (in purple). The center-right panel shows the fully unfolded GFP after the spontaneous unfolding of the intermediate. The right panel shows ClpXP toward the end of unfolded GFP translocation. The GFP structure has been enlarged by ~53 for display purposes.
(C) Assignment of the stages of GFP unfolding and translocation by ClpXP (in C-to-N direction) to a typical single-molecule trajectory. After several unfolding attempts (1), ClpXP unravels β strands 11→7 of GFP, generating a short-lived intermediate (2). After the spontaneous unfolding of the remaining GFP structure (β strands 6→1), ClpXP translocates the unfolded polypeptide through its central pore into ClpP (3). Note that ClpXP commenced translocation while the GFP unfolding intermediate was still present (2). The number labeling (1–3) scheme is the same as in panel B.
Close inspection of the GFP unfolding events showed that the 207 aa rip consisted of two steps, separated by a transient intermediate with a lifetime of ~180 ms (Figure 4B, black arrow). In fact, this “rip-transition-rip” signature (Figure 4C) was present in >70% of all events of the 207 aa peak. The presence of a well-defined transition indicates that the ClpX-induced mechanical unfolding of GFP from the C terminus proceeds via a short-lived intermediate state. Furthermore, a plot of the size distribution of each rip segment revealed that the first rip is 107 ± 2 aa, whereas the second portion consists of 100 ± 2 aa (mean ± SEM, Figures 4D and 4F, respectively).
In order to estimate the secondary structures involved in such an intermediate, we mapped the first and second segments of the 207 aa rip onto the tertiary structure of GFP. Given the dimension of fully native GFP (XF), the dimension of the folded portion of the unfolding intermediate (XI), and the experimentally observed extension change upon the unfolding of the first, C-terminal GFP segment (ΔXexp F-I), we can compute the true extension (XU1) corresponding to the first rip using the following relation: ΔXexp F-I = XU1 – (XF – XI) (Figure 5). Then XU1 can easily be converted into amino acids via the WLC formalism. Because the dimensions of the remaining folded structure (i.e., the unfolding intermediate) are not known, we defined the lower and upper bounds to XI as 2.4 and 4.2 nm, which correspond to the short and long axes of the folded GFP. In the force range of 6–10 pN, these numbers translate into 13 and 23 aa, respectively. Therefore, the first rip of 107 aa corresponds to the unfolding of a C-terminal GFP segment anywhere between 97 and 107 residues, leaving between 120 and 130 N-terminal residues still folded (see Figure 7A).
Based on the topology map of GFP, an unfolding intermediate with the N-terminal residues 1–120 still folded would require part of b strand 6 to be unstructured. We do not favor this intermediate boundary because unfolding of β strands is highly cooperative, as observed in previous mechanical unfolding experiments (Marszalek et al., 1999). In contrast, residue 130 is located within a long loop, near the end of β strand 6. Thus, we surmise that ClpX unfolds 97 aa from the C terminus corresponding to β strands 7–11, generating a GFP unfolding intermediate with ~130 N-terminal residues still structured (β strands 1–6).
In order to corroborate the structural elements assigned to the unfolding intermediate at the N terminus of GFP, we analyzed the second segment of the 207 aa rip (Figure 4C). As described above, we estimated the dimensions of the folded portion of the GFP intermediate (XI) to be ~4.2 nm or 23 aa at 6–10 pN. Calling XU2 the true extension corresponding to the second rip (the size of the unfolding intermediate when unfolded), we can calculate the observed change in extension upon unfolding of the intermediate ΔXexp I-U using the following expression: ΔXexp I-U = XU2 – XI (Figure 5). Thus, XU2 = 100 aa + 23 aa = 123 aa. Unfolding 123 amino acids starting at residue 130 would end at position 7 (as counted from the N terminus), which is in excellent agreement with structural data for GFP that show Glu5 to be the first residue involved in secondary structures (Figure 7A). Analysis of the structural transitions of GFP when unfolded by the ClpXP protease is essentially indistinguishable from that of ClpX alone (Figure S2).
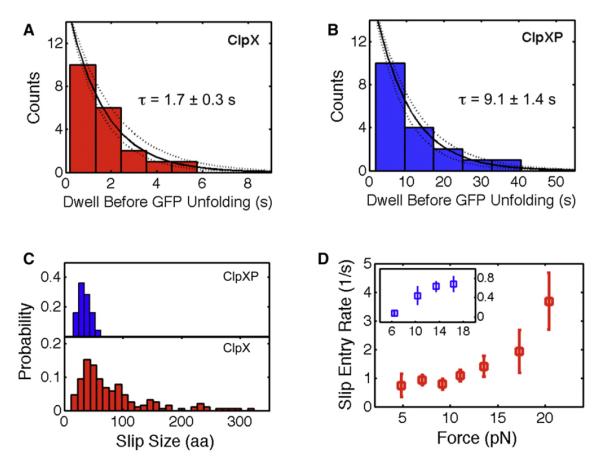
In addition to providing a structural assignment for the unfolding of GFP, we also sought to investigate the dynamic components during GFP unraveling by ClpX. The distributions of the dwell times preceding the first and second rip segments are well described by a single exponential (Figure 6A and Figure 4E, respectively). However, the time constant for the first rip is nine times longer than for the second one (1.7 ± 0.3 s versus 0.18 ± 0.03 s). We interpret the time constant of the first rip dwell as the average time required by ClpX to destabilize and unravel the fully folded GFP molecule. In contrast, the second rip time constant corresponds to the time required for the spontaneous unfolding of the 130 N-terminal residues of GFP. During the 180 ms lifetime of this N-terminal intermediate, ClpX would be able to translocate only about 15 residues of the chain unraveled in the first step of GFP unfolding. Therefore, by the time the N-terminal portion of GFP unfolds, ClpX is still several nm away and unlikely to play a major role in this second stage of GFP unfolding.
Figure 6. ClpXP Is a More Robust Motor with Higher Unfolding Efficiency and Lower Slipping Frequency than ClpX Alone.
(A and B) The dwell time before each ClpX- or ClpXP-mediated unfolding event is exponentially distributed. Dashed lines represent 95% confidence interval of the fit.
(C) Slip size histograms for ClpX (red) and ClpXP (blue).
(D) The slip entry rate (mean ± SEM) for ClpX (red) and ClpXP (blue). Slip entry rate is the inverse of the average waiting time before a slip occurs.
Interestingly, for the ClpXP complex, the time constant for the first stage of GFP unfolding was about five times longer compared to that for ClpX alone (9.1 ± 1.4 s versus 1.7 ± 0.3 s,Figures 6B and 6A, respectively). The former value is similar to the previously reported time constant τ = 5.6 s for the loss of GFP fluorescence in single-turnover degradation (Martin et al., 2008b). These degradation experiments had also suggested that ClpX initiates GFP unfolding by extracting the C-terminal β strand 11 and trapping it through at least four subsequent translocation steps. This requirement for rapid translocation leads to a strictly nonlinear dependence of GFP unfolding on the rate of ClpX ATP hydrolysis. In fact, reducing the ATP hydrolysis rate by 30% was found to decrease GFP degradation ~3-fold (Martin et al., 2008b). Therefore, our observed 5-fold difference in unfolding rate between ClpX and ClpXP is consistent with the ~30% lower translocation velocity of ClpXP at all forces observed in our experiments (Figure 2A), which is, in turn, a consequence of the reduced ATP hydrolysis rate and pulling frequency of ClpX when bound to ClpP.
ClpP Enhances the Unfolding Activity of ClpX
Besides rips corresponding to successful GFP unfolding, the trajectories for all fusion substrates showed frequent slippage events, in which ClpX apparently failed to unfold GFP, disengaged the substrate, and moved backward along the polypeptide track (Figure 1, black arrowheads). Such behavior is consistent with previous degradation studies that have shown that hard-to-unfold substrates with a short ssrA tag are frequently released and rebound by ClpXP before successful unfolding (Kenniston et al., 2005). We can imagine two possible mechanisms by which ClpX can resume tugging at the substrate after a failed unfolding attempt. Either it can remain engaged to the substrate, making an immediate new unfolding attempt, or it can completely disengage the polypeptide and diffuse backward for a short time before re-engaging the substrate. The first scenario is beyond the spatiotemporal resolution of the present experiments. The second scenario should manifest itself as rips of several amino acids due to the applied opposing load. We therefore sought to investigate in greater detail the motor slips observed during GFP unfolding.
The probability distribution of slip extensions for ClpX peaked at ~30–40 aa but also displayed longer slips well beyond 50 aa (Figure 6C, lower panel). Because ClpX alone cannot hydrolyze polypeptides, it can slip backward up to the entire length of the already translocated polypeptide chain. Remarkably, the observed slip distribution for the ClpXP complex also peaked between 30–40 aa; however, it lacked longer slips (Figure 6C, upper panel). This absence of longer slips for ClpXP is expected, as the ClpP peptidase constantly trims the polypeptide chain inside its proteolytic cavity down to ~38 aa, a length sufficient to span the distance between the ClpX-pore entry and the ClpP active sites (Martin et al., 2008b). Slips of ClpXP longer than 38 aa result, therefore, in tether rupture. The fact that the distribution of slip sizes peak between 30–40 aa for both ClpX and ClpXP indicate that the time ClpX takes to re-engage the substrate is the same with or without ClpP. The size of slips depends on the product of the substrate re-engagement time by the motor and the speed at which the polypeptide is dragged out of the pore. Thus, the presence of an external force in our experiments greatly amplifies the sizes of the slips observed here relative to those that occur in the cell and in bulk assays (Martin et al., 2008a).
Importantly, we found that ClpXP complexes are much less prone to slipping compared to ClpX hexamers alone: for ClpX, 70% of the rips observed correspond to slips and the rest to unfolding events, whereas for ClpXP, this number is only 27%. Thus, binding of ClpP dramatically decreases the slip frequency of ClpX, potentially due to additional contacts between the polypeptide and the extended processing pore of ClpXP. These additional interactions may prevent substrate release after an unsuccessful unfolding attempt.
We also analyzed the slip entry rate as a function of force for ClpX alone and in complex with ClpP. The slip entry rate is defined as the inverse of the average waiting time before a slip occurs in front of a mechanical barrier. We found that the slip entry rate for ClpXP is 5–10 times lower than that of ClpX at all forces (Figure 6D). For instance, when using the double GFP + long linker substrate, ClpXP was able to process both GFP molecules in 45% of all traces, whereas ClpX alone was successful in only 10%–15% of the cases. These numbers are in good agreement with the probabilities of successful unfolding versus slips determined above. For ClpX, the probability of unfolding both GFP molecules within a single trace is 0.27•0.27 (~10%), whereas for ClpXP it is 0.70•0.70 (~50%). Because in the cell there is no opposing force on the substrate, we surmise that the probabilities described above would correspond to a lower bound for the successful unfolding and translocation of this tandem substrate.
DISCUSSION
Our studies provide direct experimental evidence that ClpX is able to generate mechanical force to induce protein denaturation. Moreover, the force dependence of the motor velocity favors a power-stroke mechanism for ClpX. Although the stall force of the motor is just 20 pN, this force should be sufficient to unfold most cellular proteins, given the low loading rates at which AAA+ unfoldases may operate in the cell. The loading rate is a measure of the speed at which the force is applied to the protein substrate. Because protein unfolding always involves the stochastic thermal crossing of an energy barrier, pulling at lower loading rates gives the substrate more time to unfold at low forces. The most likely force at which a protein unfolds scales as the log of the loading rate (Bustamante et al., 2004). We estimate that the loading rate at which ClpX pulls the folded substrate in our experimental conditions is ~0.15 pN/s (Extended Experimental Procedures) and is five orders of magnitude smaller than the loading rate in mechanical protein unfolding experiments using atomic force microscopy (AFM) (Mickler et al., 2007; Perez-Jimenez et al., 2006). Given that typical protein unfolding forces in AFM experiments are between 50–150 pN, we expect that the corresponding unfolding forces should be at least five times smaller under the loading rates applied by ClpXP. Although the motor is hence strong enough to unfold most proteins mechanically, it is possible that ClpX also uses allosteric contacts to distort the local structure of its substrates and thereby weaken their thermodynamic stabilities.
The single-molecule trajectories obtained here allow us to address two additional important questions about the motor properties of ClpX, namely, the mechanisms involved in motor slow-down and motor stall under force. First, why does the motor pause-free velocity decrease at forces above 13 pN? One possible explanation is that the external force slows down the mechanochemical cycle of the motor and decreases the frequency of pulling events. Alternatively, high forces could cause the translocation loops of individual subunits of ClpX to slip on the substrate, resulting in futile translocation attempts that fail to move the substrate through the central pore. We favor the former explanation because in >98% of all traces we do not observe any small rips that may be indicative of motor slippage during translocation. In fact, even if we could not resolve these rips, their presence should manifest itself as an increased “noise” in velocity at high force, which we do not observe.
The second question concerns the mechanism of motor stalling. Is the maximum force generated by the motor equivalent to its thermodynamic stall force (Bustamante et al., 2001) or an “operational” stall force at which the motor is rendered unable to translocate on its track (due, for example, to mechanical unfolding of the motor itself, slippage on the track, etc.)? At forces around 20 pN, near the stall, a large fraction of the traces (>80%) are interrupted by tether ruptures that appear to be caused by backsliding of the entire, already translocated polypeptide out of the ClpX(P) motor pore. In support of this interpretation we observed that the motor slip entry rate increases significantly from ~1 s−1 at forces <13 pN to ~3.5 s−1 at forces near the stall (Figure 6D, p = 0.006, Kolmogorov-Smirnov test).
Thus, it seems likely that the reduction of the motor’s ATPase rate and frequency of pulling events at higher opposing loads increases the chance of complete loss of grip and disengagement of the substrate, causing an operational stall.
ClpX(P) maintains a constant extension velocity (nm/s) up to forces of 13 pN (Figure 2A). At low forces ClpX translocates ~8 aa per 1 nm step compared to only ~4 aa/step at 13 pN. Because the number of residues translocated per step changes as a function of external force, the 1 nm step-size of ClpX must be dictated by the relevant conformational change of the motor during the power stroke rather than any spatial periodic features of the substrate. Moreover, because of the chemical heterogeneity of the track, it is likely that nonspecific steric contacts between ClpX pore loops and the substrate are more important than specific chemical interactions when threading an unfolded polypeptide through the central pore. These nonspecific steric contacts might be key to ClpX’s ability to translocate its irregular and diverse polypeptide substrates in either C-to-N or N-to-C direction (Barkow et al., 2009; Martin et al., 2008a).
Our analysis of the ClpX-induced unfolding of GFP in the C-to-N terminal direction revealed a well-defined, short-lived intermediate (Figure 7). This intermediate, comprising the N-terminal 130 residues that form β strands 1 thru 6, is in good agreement with previous mechanical unfolding experiments of GFP (Bertz et al., 2008; Perez-Jimenez et al., 2006). Importantly, ClpX and ClpXP lead to the formation of the same transient intermediate (Figure 4E and Figure S2H), indicating that the unfolding mechanism is determined largely by the energy landscape of the substrate and the presence of cooperative folding units that unravel after ClpX or ClpXP disrupt critical interactions at the C terminus.
AAA+ unfoldases target structurally and functionally diverse proteins in all cells. Moreover, their client proteins are found not only in a folded, soluble conformation, but also in hyperstable misfolded or aggregated states (Horwich et al., 1999). These molecular machines must therefore utilize efficient mechanisms to unravel proteins with a wide range of thermodynamic stabilities, topologies, and sequence characteristics. The present study shows that ClpX(P) is able to generate and apply mechanical forces sufficient to unfold most target proteins.
ClpX processes substrates in a linear fashion, applying force and overcoming only the local mechanical barriers encountered along the unfolding trajectory of the protein. As a result, it is the linear profile of these barriers, as defined by the pulling end and the topology of the substrate, but not the global protein stability that determines the kinetics of substrate processing. Because the chance of slipping on the polypeptide track increases significantly near the stall force, ClpX will work on a hard-to-unfold substrate much longer, repeatedly tugging and slipping until an unfolding attempt is successful. Mechanical unfolding ultimately involves the thermally induced crossing of an energy barrier. By maintaining a constant tugging on a hard-to-unfold substrate, ClpX decreases the magnitude of this barrier while increasing the chance that sooner or later a thermal fluctuation within the protein substrate will allow its crossing. When facing a high mechanical barrier, the motor will thus require more time and consume larger amounts of ATP before such spontaneous crossing occurs. In this way, the motor is able to process proteins with a wide range of thermodynamic and mechanical stabilities.
As the ClpX motor shares its basic design and operating principles with other AAA+ unfoldases, including the prokaryotic ClpA, ClpB, HslU, FtsH, or Lon and the eukaryotic 26S protea-some, it is conceivable that all these enzymes utilize very similar mechanisms to generate mechanical force and disrupt the secondary, tertiary, and quarternary structures of their protein substrates. However, it remains to be determined how differences in the rate of ATP hydrolysis, the length of the central processing channel, or the heterohexameric versus homohexameric architecture of the AAA+ unfoldases affect pulling forces, translocation velocities, and the frequency of pausing or slipping on hard-to-unfold substrates.
EXPERIMENTAL PROCEDURES
Protein Modifications and dsDNA-Handle Attachment to Protein Substrates
Single-chain ClpX hexamers and GFP-titin I27 fusion proteins were expressed and purified as described previously (Martin et al., 2005, 2008b). In all fusion substrates, the titin I27 domains were permanently unfolded (Ticm) by carboxymethylation (Martin et al., 2008b). Single-chain ClpX hexamers included an avi tag that was biotinylated using purified BirA in vitro (Chen et al., 2005). We covalently attached a 3 kbp dsDNA handle to the N termini of the GFP-Ticm fusion proteins by utilizing the ybbR tag/Sfp system (Yin et al., 2005). A detailed protocol for the dsDNA-handle attachment to the protein substrates is described in Extended Experimental Procedures (Figures S3A and S3B).
Single-Molecule Sample Preparation
All single-molecule unfolding trajectories were obtained in ClpX-100 buffer (25 mM HEPES-KCl, pH 7.4, 20 mM MgCl2, 100 mM KCl, and 1 mM EDTA), 5 mM ATP and ATP regeneration system (16 mM creatine phosphate and 32 μg/ml of creatine phosphokinase) (Kenniston et al., 2005). Before an experiment, the buffer was passed through a 0.22 μm pore filter and degassed thoroughly. In the presence of ATP-γ-S, tethers between ClpX and the DNA-tethered substrate were obtained but no rips or continuous decrease in extension were observed. Details of the control experiments with ATP-γ-S are described in Extended Experimental Procedures (Figure S5). Tethers in the absence of ATP or ATP-γ-S were not observed. Experiments conducted with ClpP contained 500 nM in all chambers to ensure formation of the ClpXP complex (Kd = 90 nM) (Joshi et al., 2004).
Single-Molecule Data Collection
Data was collected in a dual-trap instrument with differential detection (Moffitt et al., 2006). Raw single-molecule data were acquired at 2000 Hz. Sudden extension changes and translocation (in nm) were converted into polypeptide contour length (in amino acids) in a two-step procedure. We first removed the extension contribution of the 3 kbp dsDNA handle using the WLC formalism (Extended Experimental Procedures). We then calculated the unfolded polypeptide contour length using the WLC model and Ppeptide = 0.65 nm (Cecconi et al., 2005).
Single-Molecule Data Analysis
During data acquisition, we monitored the dsDNA handle + polypeptide extension. For our 3 kbp dsDNA handle, the expected extension is between 930–1000 nm in the force range of our experiments (4–20 pN, respectively). Any tethers with extensions out of the expected range were discarded. During data analysis we had an additional internal control—the characteristic signature of GFP unfolding by ClpX(P): a double rip with a short-lived intermediate in between (Figure 4C). If the two rips did not add up to the expected ~200 aa change in contour length, we discarded the trace. A total of 101 ClpX traces and 62 ClpXP traces passed our screening and were used in further analysis. We analyzed three distinct events (Figure 1E): the time before a sudden extension gain occurs, the length of the sudden change in extension (rip) caused by substrate unfolding or motor slippage, and the continuous decrease in length due to translocation of the unfolded polypeptide (translocation velocity). In the analysis of pause durations before a sudden rip, we only included pauses after the first rip is observed; that is, pauses preceding rip events # 2, 3, and so on within a single trajectory. This is due to the uncertainty in the time at which a tether between ClpX and the substrate was formed and the time before the first rip was observed. In order to locate the occurrence of sudden rips and their extensions, we analyzed our traces using the Student’s t test (Figure S5). For velocity calculation, data were filtered and decimated to 2.5 Hz, and any pauses longer than 1 s were removed. Velocity data for a given force range were computed for each molecule. Velocity values corresponding to different molecules were used to compute the mean and standard error. The calculated velocity was essentially the same for filter bandwidths ranging from 1 Hz to 10 Hz.
Supplementary Material
ACKNOWLEDGMENTS
We thank Lacramioara Bintu, Craig Hetherington, and Phillip Elms for helpful discussions. M.S. acknowledges support from the NSF Graduate Research Fellowship. C.M.K. acknowledges support from the QB3 Institute (Distinguished Postdoctoral Fellowship) and the NIH K99 Award (5K99GM086516). This research was supported in part by the Searle Scholars Program (A.M.), the NIH grant R01-GM0325543, the Lawrence Berkeley National Laboratory, and the Howard Hughes Medical Institute (C.B.).
Footnotes
SUPPLEMENTAL INFORMATION Supplemental Information includes Extended Experimental Procedures, five figures, and two tables and can be found with this article online at doi: 10.1016/j.cell.2011.04.010.
REFERENCES
- Abbondanzieri EA, Greenleaf WJ, Shaevitz JW, Landick R, Block SM. Direct observation of base-pair stepping by RNA polymerase. Nature. 2005;438:460–465. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astumian RD. Thermodynamics and kinetics of a Brownian motor. Science. 1997;276:917–922. doi: 10.1126/science.276.5314.917. [DOI] [PubMed] [Google Scholar]
- Baker TA, Sauer RT. ATP-dependent proteases of bacteria: recognition logic and operating principles. Trends Biochem. Sci. 2006;31:647–653. doi: 10.1016/j.tibs.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkow SR, Levchenko I, Baker TA, Sauer RT. Polypeptide translocation by the AAA+ ClpXP protease machine. Chem. Biol. 2009;16:605–612. doi: 10.1016/j.chembiol.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertz M, Kunfermann A, Rief M. Navigating the folding energy landscape of green fluorescent protein. Angew. Chem. Int. Ed. Engl. 2008;47:8192–8195. doi: 10.1002/anie.200802987. [DOI] [PubMed] [Google Scholar]
- Bustamante C, Marko JF, Siggia ED, Smith S. Entropic elasticity of lambda-phage DNA. Science. 1994;265:1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- Bustamante C, Keller D, Oster G. The physics of molecular motors. Acc. Chem. Res. 2001;34:412–420. doi: 10.1021/ar0001719. [DOI] [PubMed] [Google Scholar]
- Bustamante C, Chemla YR, Forde NR, Izhaky D. Mechanical processes in biochemistry. Annu. Rev. Biochem. 2004;73:705–748. doi: 10.1146/annurev.biochem.72.121801.161542. [DOI] [PubMed] [Google Scholar]
- Bustamante C, Cheng W, Meija YX. Revisiting the central dogma one molecule at a time. Cell. 2011;144:480–497. doi: 10.1016/j.cell.2011.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecconi C, Shank EA, Bustamante C, Marqusee S. Direct observation of the three-state folding of a single protein molecule. Science. 2005;309:2057–2060. doi: 10.1126/science.1116702. [DOI] [PubMed] [Google Scholar]
- Chen I, Howarth M, Lin W, Ting AY. Site-specific labeling of cell surface proteins with biophysical probes using biotin ligase. Nat. Methods. 2005;2:99–104. doi: 10.1038/nmeth735. [DOI] [PubMed] [Google Scholar]
- Glynn SE, Martin A, Nager AR, Baker TA, Sauer RT. Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine. Cell. 2009;139:744–756. doi: 10.1016/j.cell.2009.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S, Roche E, Zhou Y, Sauer RT. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwich AL, Weber-Ban EU, Finley D. Chaperone rings in protein folding and degradation. Proc. Natl. Acad. Sci. USA. 1999;96:11033–11040. doi: 10.1073/pnas.96.20.11033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi SA, Hersch GL, Baker TA, Sauer RT. Communication between ClpX and ClpP during substrate processing and degradation. Nat. Struct. Mol. Biol. 2004;11:404–411. doi: 10.1038/nsmb752. [DOI] [PubMed] [Google Scholar]
- Kenniston JA, Baker TA, Sauer RT. Partitioning between unfolding and release of native domains during ClpXP degradation determines substrate selectivity and partial processing. Proc. Natl. Acad. Sci. USA. 2005;102:1390–1395. doi: 10.1073/pnas.0409634102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RW, Deshaies RJ, Peters JM, Kirschner MW. How proteolysis drives the cell cycle. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- Marszalek PE, Lu H, Li H, Carrion-Vazquez M, Oberhauser AF, Schulten K, Fernandez JM. Mechanical unfolding intermediates in titin modules. Nature. 1999;402:100–103. doi: 10.1038/47083. [DOI] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Rebuilt AAA + motors reveal operating principles for ATP-fuelled machines. Nature. 2005;437:1115–1120. doi: 10.1038/nature04031. [DOI] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Pore loops of the AAA+ ClpX machine grip substrates to drive translocation and unfolding. Nat. Struct. Mol. Biol. 2008a;15:1147–1151. doi: 10.1038/nsmb.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Protein unfolding by a AAA+ protease is dependent on ATP-hydrolysis rates and substrate energy landscapes. Nat. Struct. Mol. Biol. 2008b;15:139–145. doi: 10.1038/nsmb.1380. [DOI] [PubMed] [Google Scholar]
- Mickler M, Dima RI, Dietz H, Hyeon C, Thirumalai D, Rief M. Revealing the bifurcation in the unfolding pathways of GFP by using single-molecule experiments and simulations. Proc. Natl. Acad. Sci. USA. 2007;104:20268–20273. doi: 10.1073/pnas.0705458104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt JR, Chemla YR, Izhaky D, Bustamante C. Differential detection of dual traps improves the spatial resolution of optical tweezers. Proc. Natl. Acad. Sci. USA. 2006;103:9006–9011. doi: 10.1073/pnas.0603342103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt JR, Chemla YR, Aathavan K, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Intersubunit coordination in a homomeric ring ATPase. Nature. 2009;457:446–450. doi: 10.1038/nature07637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman KC, Abbondanzieri EA, Landick R, Gelles J, Block SM. Ubiquitous transcriptional pausing is independent of RNA polymerase backtracking. Cell. 2003;115:437–447. doi: 10.1016/s0092-8674(03)00845-6. [DOI] [PubMed] [Google Scholar]
- Ormö M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- Perez-Jimenez R, Garcia-Manyes S, Ainavarapu SR, Fernandez JM. Mechanical unfolding pathways of the enhanced yellow fluorescent protein revealed by single molecule force spectroscopy. J. Biol. Chem. 2006;281:40010–40014. doi: 10.1074/jbc.M609890200. [DOI] [PubMed] [Google Scholar]
- Shin Y, Davis JH, Brau RR, Martin A, Kenniston JA, Baker TA, Sauer RT, Lang MJ. Single-molecule denaturation and degradation of proteins by the AAA+ ClpXP protease. Proc. Natl. Acad. Sci. USA. 2009;106:19340–19345. doi: 10.1073/pnas.0910484106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. The bacteriophage straight phi29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- Svoboda K, Schmidt CF, Schnapp BJ, Block SM. Direct observation of kinesin stepping by optical trapping interferometry. Nature. 1993;365:721–727. doi: 10.1038/365721a0. [DOI] [PubMed] [Google Scholar]
- Tinoco I, Jr., Bustamante C. The effect of force on thermodynamics and kinetics of single molecule reactions. Biophys. Chem. 2002;101-102:513–533. doi: 10.1016/s0301-4622(02)00177-1. [DOI] [PubMed] [Google Scholar]
- Wang J, Hartling JA, Flanagan JM. The structure of ClpP at 2.3 A resolution suggests a model for ATP-dependent proteolysis. Cell. 1997;91:447–456. doi: 10.1016/s0092-8674(00)80431-6. [DOI] [PubMed] [Google Scholar]
- Yang F, Moss LG, Phillips GN., Jr. The molecular structure of green fluorescent protein. Nat. Biotechnol. 1996;14:1246–1251. doi: 10.1038/nbt1096-1246. [DOI] [PubMed] [Google Scholar]
- Yildiz A, Selvin PR. Kinesin: walking, crawling or sliding along? Trends Cell Biol. 2005;15:112–120. doi: 10.1016/j.tcb.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Yin J, Straight PD, McLoughlin SM, Zhou Z, Lin AJ, Golan DE, Kelleher NL, Kolter R, Walsh CT. Genetically encoded short peptide tag for versatile protein labeling by Sfp phosphopantetheinyl transferase. Proc. Natl. Acad. Sci. USA. 2005;102:15815–15820. doi: 10.1073/pnas.0507705102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.