Abstract
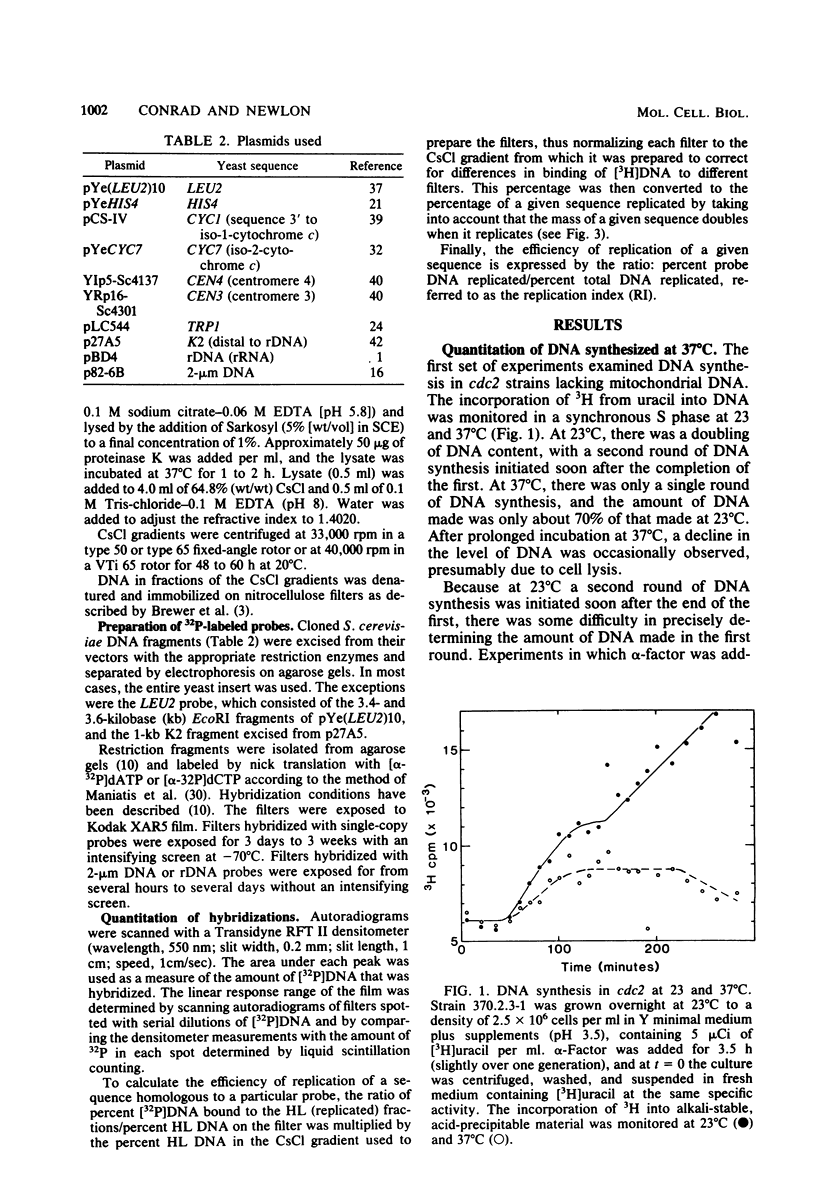
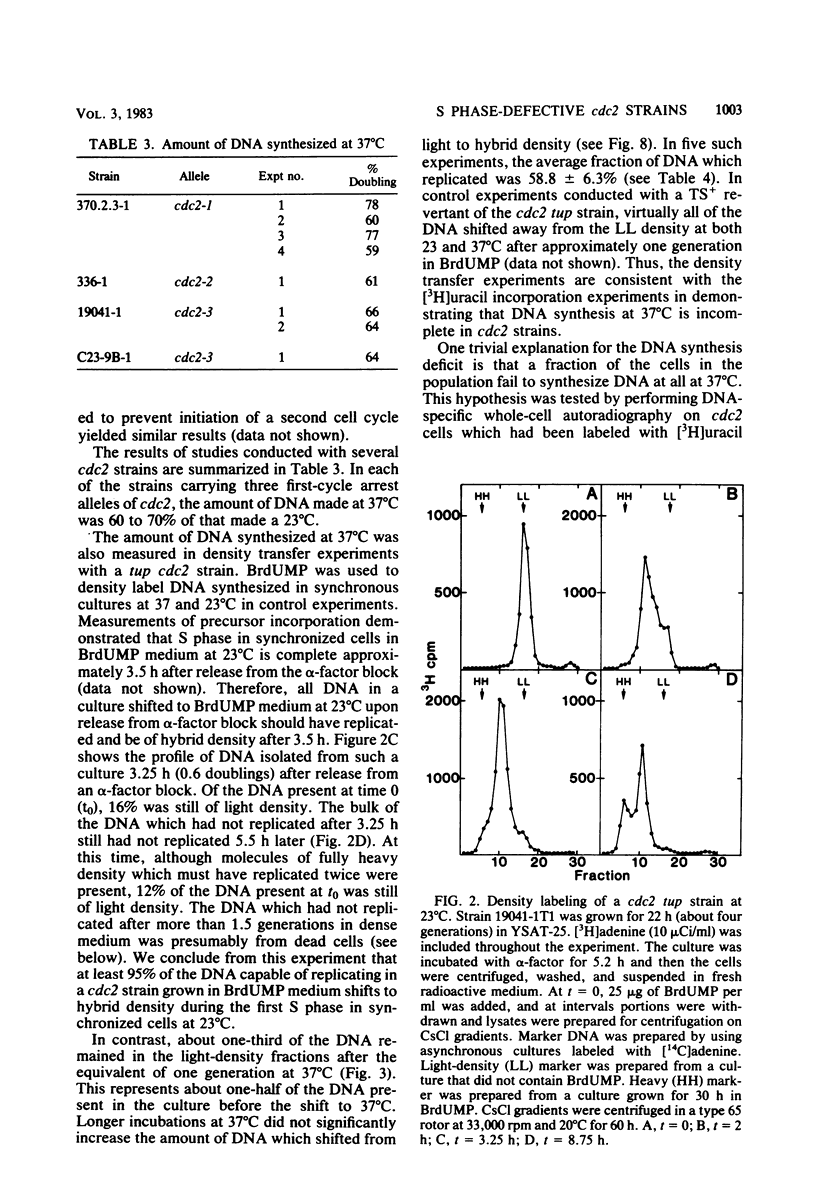
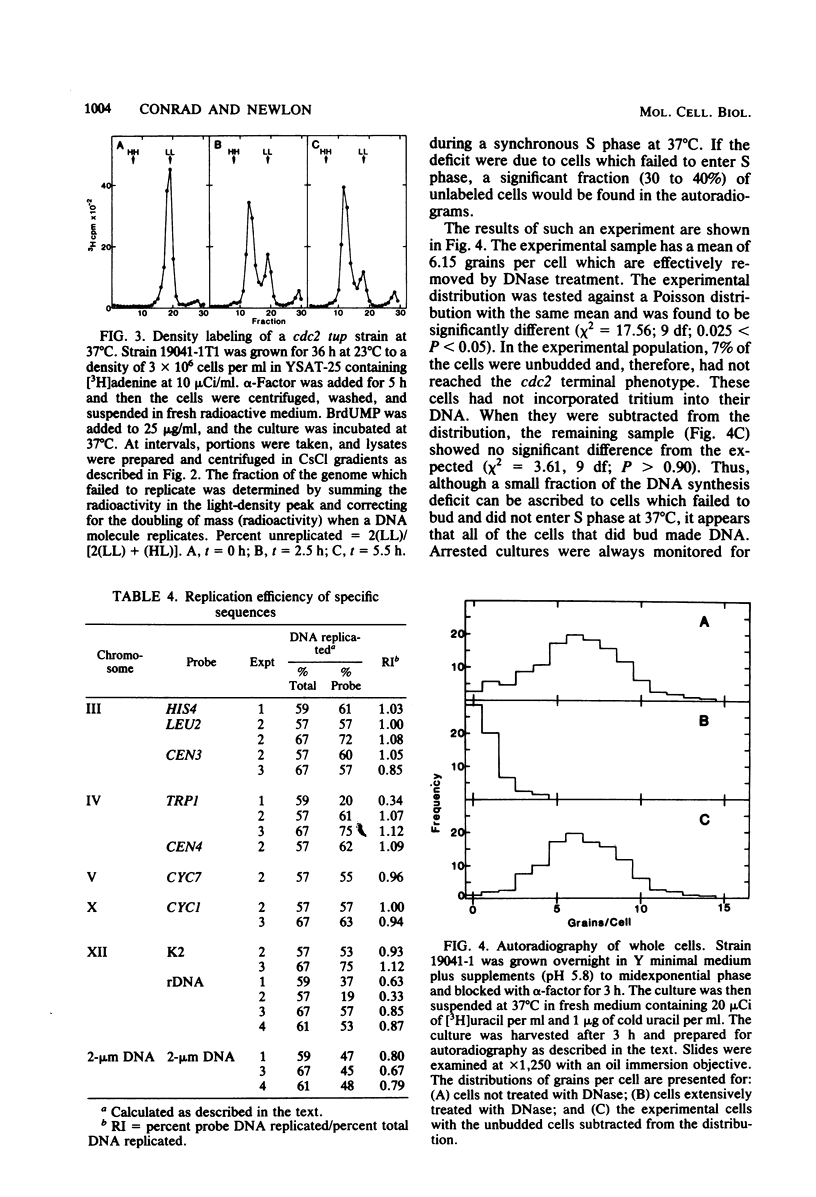
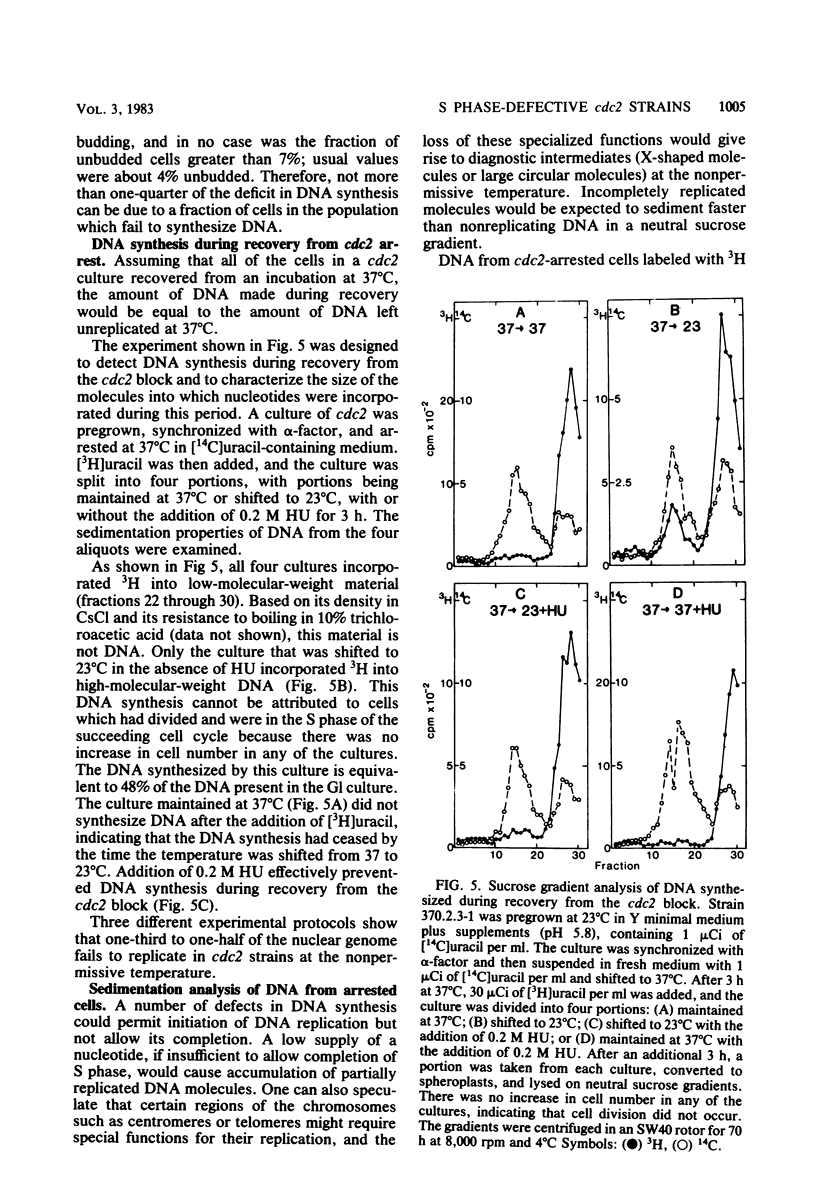
Chromosomal DNA replication was examined in temperature-sensitive mutants of Saccharomyces cerevisiae defective in a gene required for the completion of S phase at the nonpermissive temperature, 37 degrees C. Based on incorporation of radioactive precursors and density transfer experiments, strains carrying three different alleles of cdc2 failed to replicate approximately one-third of their nuclear genome at 37 degrees C. Whole-cell autoradiography experiments demonstrated that 93 to 96% of the cells synthesized DNA at 37 degrees C. Therefore, all cells failed to replicate part of their genome. DNA isolated from terminally arrested cells was of normal size as measured on neutral and alkaline sucrose gradients, suggesting that partially replicated DNA molecules do not accumulate and that DNA strands are ligated properly in cdc2 mutants. In addition, electron microscopic examination of the equivalent of more than one genome's DNA from arrested cells failed to reveal any partially replicated molecules. The sequences which failed to replicate at 37 degrees C were not highly specific; eight different cloned sequences replicated to the same extent as total DNA. The 2-microns plasmid DNA and rDNA replicated significantly less well than total DNA, but approximately one-half of these sequences replicated at 37 degrees C. These observations suggest that cdc2 mutants are defective in an aspect of initiation of DNA replication common to all chromosomes such that a random fraction of the chromosomes fail to initiate replication at 37 degrees C, but that once initiated, replication proceeds normally.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bell G. I., DeGennaro L. J., Gelfand D. H., Bishop R. J., Valenzuela P., Rutter W. J. Ribosomal RNA genes of Saccharomyces cerevisiae. I. Physical map of the repeating unit and location of the regions coding for 5 S, 5.8 S, 18 S, and 25 S ribosomal RNAs. J Biol Chem. 1977 Nov 25;252(22):8118–8125. [PubMed] [Google Scholar]
- Blumenthal A. B., Kriegstein H. J., Hogness D. S. The units of DNA replication in Drosophila melanogaster chromosomes. Cold Spring Harb Symp Quant Biol. 1974;38:205–223. doi: 10.1101/sqb.1974.038.01.024. [DOI] [PubMed] [Google Scholar]
- Brewer B. J., Zakian V. A., Fangman W. L. Replication and meiotic transmission of yeast ribosomal RNA genes. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6739–6743. doi: 10.1073/pnas.77.11.6739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B., Goetsch L. Duplication of spindle plaques and integration of the yeast cell cycle. Cold Spring Harb Symp Quant Biol. 1974;38:123–131. doi: 10.1101/sqb.1974.038.01.016. [DOI] [PubMed] [Google Scholar]
- Callan H. G. DNA replication in the chromosomes of eukaryotes. Cold Spring Harb Symp Quant Biol. 1974;38:195–203. doi: 10.1101/sqb.1974.038.01.023. [DOI] [PubMed] [Google Scholar]
- Cramer J. H., Farrelly F. W., Rownd R. H. Restriction endonuclease analysis of ribosomal DNA from Saccharomyces cerevisiae. Mol Gen Genet. 1976 Nov 17;148(3):233–241. doi: 10.1007/BF00332897. [DOI] [PubMed] [Google Scholar]
- Crossen P. E., Pathak S., Arrighi F. E. A high resolution study of the DNA replication patterns of chinese hamster chromosomes using sister chromatid differential staining technique. Chromosoma. 1975 Nov 11;52(4):339–347. doi: 10.1007/BF00364018. [DOI] [PubMed] [Google Scholar]
- Culotti J., Hartwell L. H. Genetic control of the cell division cycle in yeast. 3. Seven genes controlling nuclear division. Exp Cell Res. 1971 Aug;67(2):389–401. doi: 10.1016/0014-4827(71)90424-1. [DOI] [PubMed] [Google Scholar]
- Devenish R. J., Newlon C. S. Isolation and characterization of yeast ring chromosome III by a method applicable to other circular DNAs. Gene. 1982 Jun;18(3):277–288. doi: 10.1016/0378-1119(82)90166-4. [DOI] [PubMed] [Google Scholar]
- Duntze W., Stötzler D., Bücking-Throm E., Kalbitzer S. Purification and partial characterization of -factor, a mating-type specific inhibitor of cell reproduction from Saccharomyces cerevisiae. Eur J Biochem. 1973 Jun;35(2):357–365. doi: 10.1111/j.1432-1033.1973.tb02847.x. [DOI] [PubMed] [Google Scholar]
- Forte M. A., Fangman W. L. Naturally occurring cross-links in yeast chromosomal DNA. Cell. 1976 Jul;8(3):425–431. doi: 10.1016/0092-8674(76)90155-0. [DOI] [PubMed] [Google Scholar]
- Gimmler G. M., Schweizer E. rDNA replication in a synchronized culture of Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1972 Jan 14;46(1):143–149. doi: 10.1016/0006-291x(72)90642-0. [DOI] [PubMed] [Google Scholar]
- Goldring E. S., Grossman L. I., Krupnick D., Cryer D. R., Marmur J. The petite mutation in yeast. Loss of mitochondrial deoxyribonucleic acid during induction of petites with ethidium bromide. J Mol Biol. 1970 Sep 14;52(2):323–335. doi: 10.1016/0022-2836(70)90033-1. [DOI] [PubMed] [Google Scholar]
- Hartley J. L., Donelson J. E. Nucleotide sequence of the yeast plasmid. Nature. 1980 Aug 28;286(5776):860–865. doi: 10.1038/286860a0. [DOI] [PubMed] [Google Scholar]
- Hartwell L. H. Macromolecule synthesis in temperature-sensitive mutants of yeast. J Bacteriol. 1967 May;93(5):1662–1670. doi: 10.1128/jb.93.5.1662-1670.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H. Periodic density fluctuation during the yeast cell cycle and the selection of synchronous cultures. J Bacteriol. 1970 Dec;104(3):1280–1285. doi: 10.1128/jb.104.3.1280-1285.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H. Sequential function of gene products relative to DNA synthesis in the yeast cell cycle. J Mol Biol. 1976 Jul 15;104(4):803–817. doi: 10.1016/0022-2836(76)90183-2. [DOI] [PubMed] [Google Scholar]
- Johnston L. H., Nasmyth K. A. Saccharomyces cerevisiae cell cycle mutant cdc9 is defective in DNA ligase. Nature. 1978 Aug 31;274(5674):891–893. doi: 10.1038/274891a0. [DOI] [PubMed] [Google Scholar]
- Kassir Y., Simchen G. Meiotic recombination and DNA synthesis in a new cell cycle mutant of Saccharomyces cerevisiae. Genetics. 1978 Sep;90(1):49–68. doi: 10.1093/genetics/90.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsman A. J., Gimlich R. L., Clarke L., Chinault A. C., Carbon J. Sequence variation in dispersed repetitive sequences in Saccharomyces cerevisiae. J Mol Biol. 1981 Feb 5;145(4):619–632. doi: 10.1016/0022-2836(81)90306-5. [DOI] [PubMed] [Google Scholar]
- Klein H. L., Byers B. Stable denaturation of chromosomal DNA from Saccharomyces cerevisiae during meiosis. J Bacteriol. 1978 May;134(2):629–635. doi: 10.1128/jb.134.2.629-635.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leff J., Lam K. B. Bromodeoxyuridine 5'-monophosphate incorporation into yeast nuclear and mitochondrial deoxyribonucleic acid. J Bacteriol. 1976 Jul;127(1):354–361. doi: 10.1128/jb.127.1.354-361.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. F., Liu C. C., Alberts B. M. T4 DNA topoisomerase: a new ATP-dependent enzyme essential for initiation of T4 bacteriophage DNA replication. Nature. 1979 Oct 11;281(5731):456–461. doi: 10.1038/281456a0. [DOI] [PubMed] [Google Scholar]
- Liu L. F., Liu C. C., Alberts B. M. Type II DNA topoisomerases: enzymes that can unknot a topologically knotted DNA molecule via a reversible double-strand break. Cell. 1980 Mar;19(3):697–707. doi: 10.1016/s0092-8674(80)80046-8. [DOI] [PubMed] [Google Scholar]
- MICHELSON A. M., DONDON J., GRUNBERG-MANAGO M. The action of polynucleotide phosphorylase on 5-halogenouridine-5' pyrophosphates. Biochim Biophys Acta. 1962 Apr 2;55:529–540. doi: 10.1016/0006-3002(62)90986-1. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., Kleid D. G. Nucleotide sequence of the rightward operator of phage lambda. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1184–1188. doi: 10.1073/pnas.72.3.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery D. L., Leung D. W., Smith M., Shalit P., Faye G., Hall B. D. Isolation and sequence of the gene for iso-2-cytochrome c in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1980 Jan;77(1):541–545. doi: 10.1073/pnas.77.1.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newlon C. S., Fangman W. L. Mitochondrial DNA synthesis in cell cycle mutants of Saccharomyces cerevisiae. Cell. 1975 Aug;5(4):423–428. doi: 10.1016/0092-8674(75)90061-6. [DOI] [PubMed] [Google Scholar]
- Newlon C. S., Petes T. D., Hereford L. M., Fangman W. L. Replication of yeast chromosomal DNA. Nature. 1974 Jan 4;247(5435):32–35. doi: 10.1038/247032a0. [DOI] [PubMed] [Google Scholar]
- Petes T. D., Fangman W. L. Sedimentation properties of yeast chromosomal DNA. Proc Natl Acad Sci U S A. 1972 May;69(5):1188–1191. doi: 10.1073/pnas.69.5.1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratzkin B., Carbon J. Functional expression of cloned yeast DNA in Escherichia coli. Proc Natl Acad Sci U S A. 1977 Feb;74(2):487–491. doi: 10.1073/pnas.74.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivin C. J., Fangman W. L. Replication fork rate and origin activation during the S phase of Saccharomyces cerevisiae. J Cell Biol. 1980 Apr;85(1):108–115. doi: 10.1083/jcb.85.1.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalit P., Loughney K., Olson M. V., Hall B. D. Physical analysis of the CYC1-sup4 interval in Saccharomyces cerevisiae. Mol Cell Biol. 1981 Mar;1(3):228–236. doi: 10.1128/mcb.1.3.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcomb D. T., Thomas M., Kelly J., Selker E., Davis R. W. Eukaryotic DNA segments capable of autonomous replication in yeast. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4559–4563. doi: 10.1073/pnas.77.8.4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathern J. N., Newlon C. S., Herskowitz I., Hicks J. B. Isolation of a circular derivative of yeast chromosome III: implications for the mechanism of mating type interconversion. Cell. 1979 Oct;18(2):309–319. doi: 10.1016/0092-8674(79)90050-3. [DOI] [PubMed] [Google Scholar]
- Zamb T. J., Petes T. D. Analysis of the junction between ribosomal RNA genes and single-copy chromosomal sequences in the yeast Saccharomyces cerevisiae. Cell. 1982 Feb;28(2):355–364. doi: 10.1016/0092-8674(82)90353-1. [DOI] [PubMed] [Google Scholar]



