Abstract
Expanded CAG repeat tracts are the cause of at least a dozen neurodegenerative disorders. In humans, long CAG repeats tend to expand during transmissions from parent to offspring, leading to an earlier age of disease onset and more severe symptoms in subsequent generations. Here, we show that the maintenance DNA methyltransferase Dnmt1, which preserves the patterns of CpG methylation, plays a key role in CAG repeat instability in human cells and in the male and female mouse germlines. SiRNA knockdown of Dnmt1 in human cells destabilized CAG triplet repeats, and Dnmt1 deficiency in mice promoted intergenerational expansion of CAG repeats at the murine spinocerebellar ataxia type 1 (Sca1) locus. Importantly, Dnmt1+/− SCA1 mice, unlike their Dnmt1+/+ SCA1 counterparts, closely reproduced the intergenerational instability patterns observed in human SCA1 patients. In addition, we found aberrant DNA and histone methylation at sites within the CpG island that abuts the expanded repeat tract in Dnmt1-deficient mice. These studies suggest that local chromatin structure may play a role in triplet repeat instability. These results are consistent with normal epigenetic changes during germline development contributing to intergenerational instability of CAG repeats in mice and in humans.
INTRODUCTION
CAG repeat expansion is the underlying cause of at least 12 neurodegenerative diseases (1, 2). In humans, repeat tracts tend to expand from one generation to the next, causing more severe phenotypes in the progeny, a phenomenon called anticipation (2). The extent of expansion bias in parent-to-offspring transmissions often depends on the sex of the transmitting parent. For example, in patients with spinocerebellar ataxia type 1 (SCA1), transmissions through the paternal germline are sharply biased toward expansions, while maternal transmissions are only slightly biased toward expansions (3–5). The mechanistic bases for these parent-of-origin differences are not known. The processes of replication, recombination, DNA repair, and transcription—acting separately or in combination—have been proposed to contribute to these differences, perhaps in conjunction with key events in male and female germline development (2). Here we explore the potential contribution of the epigenetic effects of CpG methylation on the stability of CAG repeat tracts during intergenerational transmissions.
Extensive repeat tract instability has been documented in both early embryogenesis and germline development (2). These two periods of repeat tract instability coincide with the two major cycles of epigenetic reprogramming that occur during mammalian development. Immediately after fertilization, CpG methylation is largely removed from the genome and then restored in a tissue-appropriate manner (6). A second wave of demethylation occurs during germline development, when CpG methylation is again removed and then re-established (6). The similar timing of CpG methylation changes and repeat tract instability raises the possibility that these two processes may be mechanistically linked (2, 7).
A connection between DNA methylation and repeat instability has been noted for the expanded CGG repeats in the 5' end of the FMR1 gene, which are responsible for Fragile X syndrome (8, 9). When an FMR1 CGG repeat reaches about 200 units in humans, it usually becomes methylated. In somatic tissues, methylated CGG repeat tracts at the FMR1 locus were found to be much more stable than unmethylated ones of comparable length (10, 11). Similarly, CGG expansions in the germline were observed only in male patients that carried unmethylated repeats (12). In contrast to humans, CGG repeat tracts introduced into the mouse FMR1 locus do not become methylated, even when they exceed 200 repeats (13, 14). This unmethylated locus displays an ongoing instability in intergenerational transmissions (13, 14). Finally, when vectors carrying CGG repeats were introduced into primate cells, or into bacteria, methylated repeats proved to be more stable that unmethylated ones (15).
Although these observations indicate that CpG methylation influences CGG repeat stability, they provide little mechanistic insight into how methylation might prevent such tract-length changes. For the human and mouse studies, there is a strong correlation between repeat methylation and FMR1 transcription: unmethylated repeats are transcribed; methylated repeats are not (16). Recent studies with CAG repeats in humans cells (17, 18) and in Drosophila (19) indicate that transcription through repeat tracts can trigger repeat instability, presumably by promoting formation of secondary structures in the repeat tracts that are then resolved by various DNA repair processes (17–19). Thus, a plausible hypothesis is that CpG methylation of CGG repeats stabilizes them by preventing transcription through the repeat tract.
It is less obvious what effect DNA methylation might have on CAG repeats, which are themselves not targets for DNA methylation since they contain no CG dinucleotides. Nevertheless, two observations suggest that there is also a connection between DNA methylation and CAG repeat stability. First, in bacteria, CAG repeats in plasmids that were methylated at CpG sites were mildly stabilized relative to repeats in unmethylated plasmids (15). Second, treatment of mammalian cells with 5-aza-2′-deoxycytosine (5-aza-CdR), which leads to passive depletion of DNA methylation (20) and destruction of Dnmt1 (21), or with hydralazine, which induces demethylation by inhibiting expression of Dnmt1 (22, 23), both dramatically increased CAG repeat instability (7). Again, neither of these studies provides a mechanistic link between DNA methylation and repeat stability; however, they reinforce the idea that epigenetic changes to the DNA (or to the chromatin) contribute to the stability of CAG repeats.
Studies in humans and mice have demonstrated that epigenetic changes do occur in the vicinity of long CAG repeat tracts. The most extensive studies have been carried out for the DMPK gene, in which expanded CTG repeats in the 3′ untranslated region cause myotonic dystrophy (DM1). Whereas normal individuals have a DNase hypersensitive site immediately downstream of the repeat tract, in DM1 patients with several thousand repeats, the hypersensitive site is missing (24). Absence of this hypersensitive site correlates with a change in transcription at both the DMPK gene and the downstream SIX5 gene (25–29). Aberrant transcription of the SIX5 gene apparently arises due to spreading of DNA methylation into its promoter region (29). The CTG expansion and flanking sequences in DM1 patients are also associated with changes in histone modifications—deacetylation of histone H3 and hypermethylation of histone H3 at lysine 9—changes that are indicative of heterochromatin (30). In transgenic mice carrying tandem arrays of a reporter gene linked to an adjacent 192 CTG repeat, the reporter gene is consistently silenced (31). Taken together, these results suggest that long CTG repeats can trigger the formation and spreading of heterochromatin, which causes aberrant transcription and ultimately contributes to the disease phenotype (32, 33). These kinds of epigenetic changes have not been demonstrated for the shorter repeats that characterize many CAG repeat diseases, which are typically less than 100 repeats in length.
Collectively, these observations at the FMR1 and DMPK loci suggest that repeat instability may be linked to the epigenetic changes that occur to DNA (and to the overlying chromatin), perhaps via associated changes in transcription through these genes. In this study, we investigate these potential connections, using a mouse model for spinocerebellar ataxia type 1 (SCA1), which carries 143 CAG repeats at the endogenous mouse Sca1 locus (34). We sought to perturb DNA methylation patterns in these mice by making them heterozygous for a null allele of Dnmt1, the gene for the major maintenance DNA methyltransferase (35). By comparing Dnmt1+/− SCA1 mice to Dnmt1+/+ SCA1 mice, we address four questions. Does Dnmt1 deficiency affect the stability of CAG repeats during intergenerational transmission? Do epigenetic changes to the DNA, or to the chromatin, occur in the vicinity of the repeat? Is transcription through the repeat altered? Is the effect of Dnmt1 deficiency the same in somatic tissues as it is in the germline?
RESULTS
Effects of Dnmt1 knockdown on CAG repeat tract stability in human cells
Initially, we used a well-characterized selection assay for CAG contractions in human cells (17, 18) to test whether Dnmt1 depletion could alter the instability of long CAG repeat tracts (Fig. S1). These modified HT1080 cells carry an integrated copy of a human HPRT minigene, whose function is blocked by a (CAG)95 repeat tract inserted into the intron. Contraction of this long repeat tract to less than 39 units restores the function of the HPRT gene and allows the cells to survive in selective medium (17). Treatment with either of two Dnmt1 siRNAs, each of which reduced Dnmt1 expression by about 50%, doubled the frequency of large CAG contractions (P<0.001) (Fig. S1). These results suggested that a reduced level of Dnmt1 might increase CAG repeat instability in vivo.
Dnmt1 deficiency promotes repeat expansion during germline transmission in mice
We investigated the effect of Dnmt1 CAG repeat instability in vivo by assaying germline transmission in mice. We took advantage of a null allele of Dnmt1, which was constructed to lack the conserved catalytic residues essential for methyltransferase activity, but which expresses no stable transcript or detectable protein (35). Because Dnmt1 homozygous null mice die early during embryogenesis (35), we examined repeat instability in heterozygous Dnmt1+/− mice, which do not differ from wild type mice in appearance or lifespan (36).
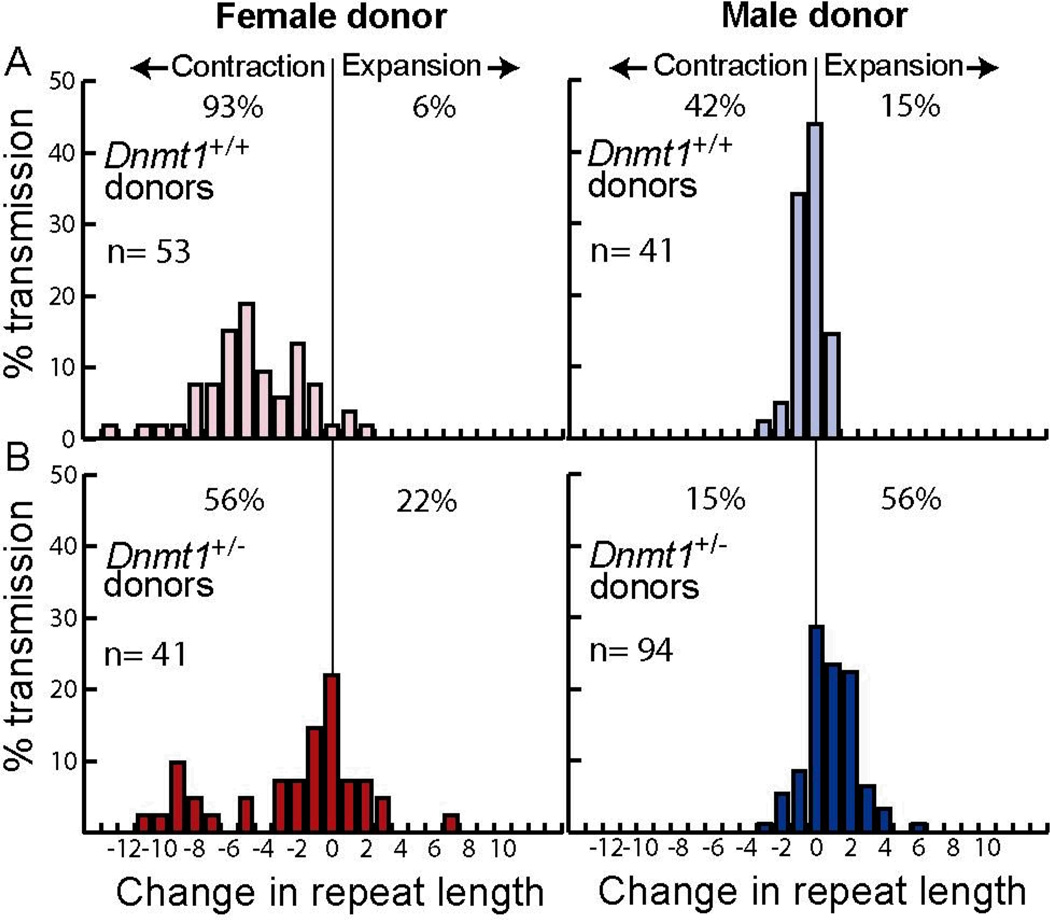
We crossed wild type mice to Dnmt1+/− SCA1 and Dnmt1+/+ SCA1 mice, which carry a long CAG tract in one allele at the Sca1 locus (34). We then compared the length of the repeat tract in each SCA1 donor parent to that of its progeny at weaning (Fig. 1). These data were gathered over a single generation. Our data with Dnmt1+/+ SCA1 mice corroborate those of a previous study (37): both paternal and maternal transmissions showed a bias toward contractions, with more extensive contractions generated in crosses with female donors (Fig. 1A). In addition, the frequency of stable transmissions—those with no change in repeat tract length—was higher among the progeny of male donors than it was among the progeny of female donors, as previously observed (37). This agreement with published results suggests that strain background does not contribute to the significant differences observed between Dnmt1+/+ SCA1 mice and their Dnmt1+/− SCA1 littermates.
Figure 1.
Intergenerational changes in CAG repeat tract lengths in SCA1 progeny mice. Percent transmission is the number of alleles with a given change in repeat length divided by the total number of alleles (n) multiplied by 100%. Change in repeat length is defined as the number of repeats in a progeny mouse minus the number of repeats in its donor parent. A) Dnmt1+/+ SCA1 mouse donors. B) Dnmt1+/− SCA1 mouse donors. The progeny from the male and female jDnmt1+/− SCA1 donors includes Dnmt1+/+ SCA1 and Dnmt1+/− SCA1 mice.
In sharp contrast to the results with Dnmt1+/+ SCA1 mice, transmissions from Dnmt1+/− SCA1 mice yielded substantially increased proportions of expansions in both paternal and maternal transmissions (15% to 56% in males, P <0.0001, and 6% to 22% in females, P = 0.0009), with an overall bias toward expansions during paternal transmissions (Fig. 1). Furthermore, the maximum expansion length increased from +1 to +6 in males and from +2 to +7 in females, which is similar to the range of CAG expansions seen in human SCA1 patients (Fig. S2). Dnmt1 deficiency decreased the percentage of stable transmissions among the progeny of male donors (from 43% to 29%), but increased their frequency during maternal transmission (2% to 22%). We conclude that Dnmt1 deficiency alters the stability of CAG repeat tracts during intergenerational transmission and increases the bias toward expansions in both male and female donors.
Age dependence of repeat expansions during germline transmission in Dnmt1+/− SCA1 mice
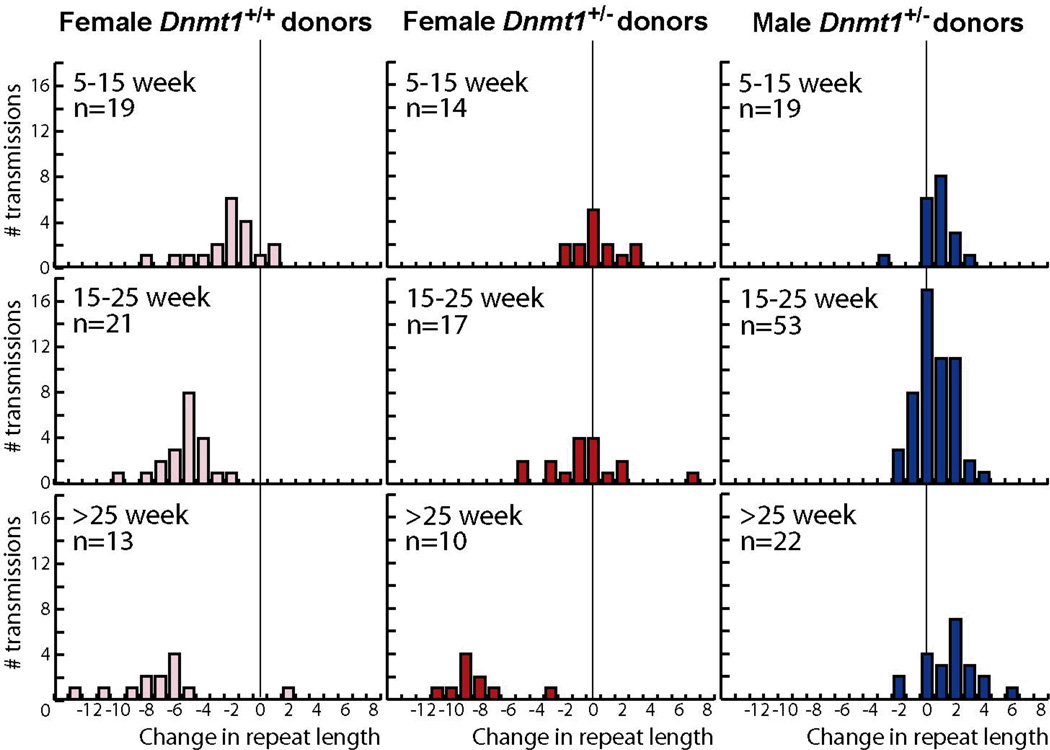
Previous characterizations of the SCA1 knock-in mouse model and of a transgenic SCA1 mouse model carrying a shorter repeat tract demonstrated that larger CAG repeat contractions occurred with advancing age of the mother (37, 38). To examine this issue for germline transmission from Dnmt1+/− SCA1 mothers, we re-plotted the data in Figure 1 after grouping the donor mice by age (Fig. 2). For female Dnmt1+/+ SCA1 donors, the dominant process is repeat contraction, with the average contraction size increasing from −2.8 to −5.3 to −7.7 with age, consistent with previous studies (37, 38). For female Dnmt1+/− SCA1 donors, the situation is more complicated. There is a similar trend in contractions, with the average contraction size increasing from −1.5 to −2.4 to −8.3 with age. Expansions, however, are observed only in the young and middle-aged donors. Thus, in females the effect of Dnmt1 deficiency on repeat expansion is apparent at early times, but is eventually overwhelmed by whatever age-dependent process generates contractions in oocytes. The confounding effects of contractions in the female germline make it difficult to determine whether expansions in Dnmt1+/− SCA1 female donors are age dependent. As a result, the distributions of repeat lengths in the progeny from young and middle-aged Dnmt1+/− SCA1 female donors are significantly different from those of Dnmt1+/+ SCA1 female donors (P = 0.001 and P = 0.0001, respectively), but the distributions in >25-week old Dnmt1+/+ SCA1 and Dnmt1+/− SCA1 female donors are indistinguishable (P = 0.1).
Figure 2.
Age association of track-length changes in progeny of Dnmt1+/+ SCA1 and Dnmt1+/− SAC1 donor mice. The number of transmissions taken from figure 1A&B were plotted as a function of age of the donor parent at conception. Top row: 5–15 week-old mouse donors. Middle row: 15 week and 1 day to 25 week-old donors. Bottom row: donors older than 25 weeks.
Previous studies in SCA1 mice did not detect age-dependence repeat instability in paternal germline transmissions (37, 39). In our experiments, as well, the distribution of repeat lengths in the progeny of Dnmt1+/+ SCA1 fathers is too narrow to reliably discern an age-dependent trend, if it exists. For male Dnmt1+/− SCA1 donors, however, we observe a gradual shift in the average size of expansions from 1.4 in young donors, to 1.7 in the middle-aged donors, to 2.5 in older donors, with a corresponding increase in maximum expansion size from 3 to 4 to 6 repeats (Fig. 2). The distribution of repeat lengths at >25 weeks is marginally different from that at 5–15 weeks (P = 0.04), consistent with a small age-dependent effect on tract-length changes in the male germline of Dnmt1+/− SCA1 mice.
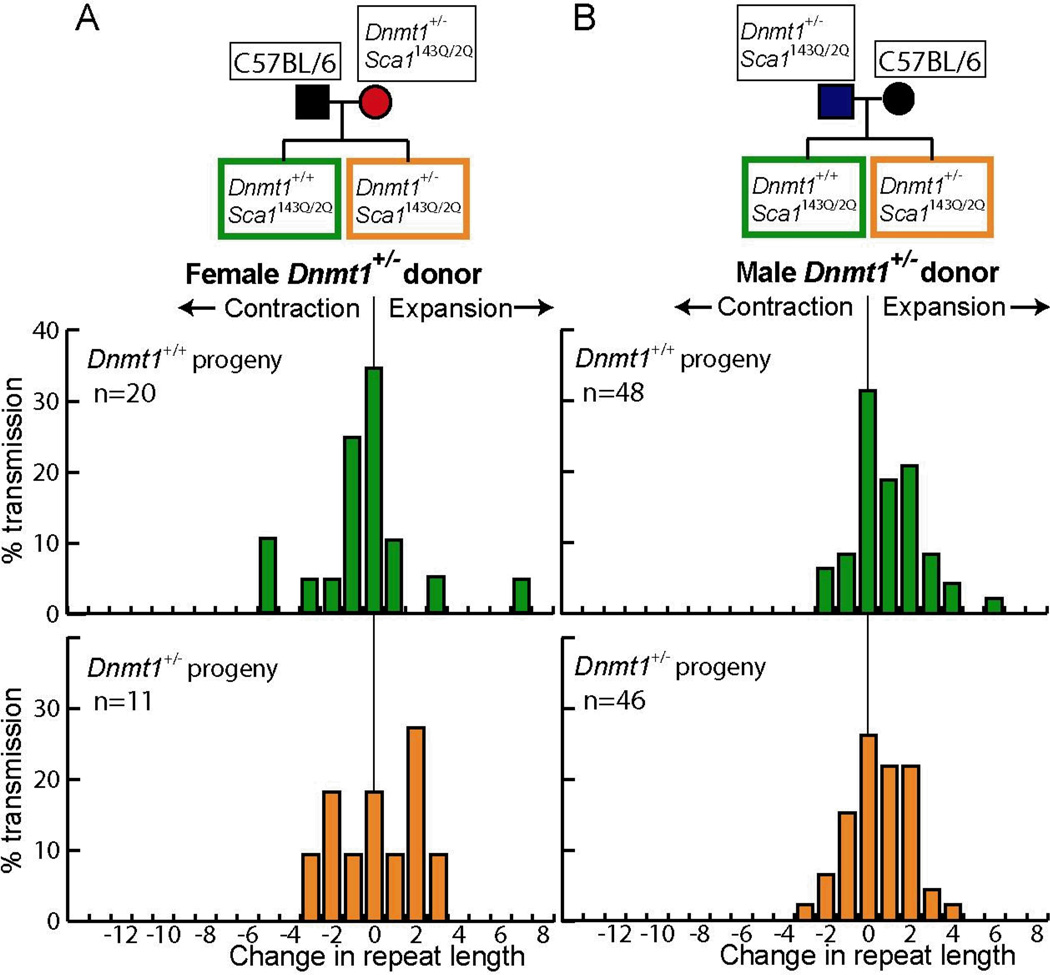
Expansions in Dnmt1+/− SCA1 donor mice depend only the genotypes of the parental mice
A strong dependence of expansions on parental age during male and female transmissions would have suggested that the changes occur in the donors themselves rather than in the first few divisions of the embryo, which is the other identified time for CAG repeat instability (2). To address these alternatives more directly, we analyzed the data in Figure 1B for effects due to the progeny genotypes. If the track-length expansions arose because of the Dnmt1 deficiency early during embryogenesis, they would be expected to be dependent on the genotype—Dnmt1+/− SCA1 or Dnmt1+/+ SCA1—of the progeny embryo. By contrast, if expansions arose due to Dnmt1 deficiency during germline development or maturation in the parents, they would be expected to depend solely on the parental genotype. Analysis of the distribution of tract-length changes in Dnmt1+/− SCA1 and Dnmt1+/+ SCA1 progeny mice (Fig. 3) showed that there was no significant difference for male donors (P = 0.8). For female donors, we analyzed progeny that arose from donors up to 25 weeks in age, where expansions were present among the progeny (Fig. 2). The progeny from older mice (>25 wks) were specifically excluded from this analysis to avoid potential bias because they display only contractions, which are known to arise in the germline (38). Once again, there was no significant difference between Dnmt1+/− SCA1 and Dnmt1+/+ SCA1 progeny mice (P = 0.5). These data indicate that the observed expansions arose in the donor mice, rather than in the progeny embryos.
Figure 3.
Progeny genotype does not alter CAG tract-length changes in Dnmt1+/− SCA1 mouse donors. The breeding schemes are shown at the top. The Dnmt1+/+ SCA1 and Dnmt1+/− SCA1 progeny are labelled in green and orange, respectively. A) Progeny from female Dnmt1+/− SCA1 donors. Only the progeny from donors that were 25 weeks of age or younger are shown. B) Progeny from male Dnmt1+/− SCA1 donors.
As one gauge of the reproducibility of these measurements, we selected Dnmt1+/− SCA1 progeny from the first-generation experiments above, bred them to wild type mice, and analyzed the repeat lengths in their progeny. Progeny of these second-generation donors replicated the data for Dnmt1+/− SCA1 mice shown in Figures 1 and 2: a similar bias toward expansion in both male and female donors, and fewer stable transmissions in males and more stable transmissions in females (data not shown).
CpG methylation in testes and ovaries from Dnmt1+/+ SCA1 and Dnmt1+/− SCA1 mice
The normal appearance and lifespan (36) of Dnmt1+/− SCA1 mice suggest that major changes in CpG methylation are unlikely. Subtle differences, however, have been detected by the coat-color readout of the methylation-sensitive Aiapy allele (40). We analyzed DNA from testes and ovaries of Dnmt1+/+ SCA1 and Dnmt1+/− SCA1 mice for differences in global CpG methylation, differences adjacent to the repeat tract, and nonstandard CpA and CpT DNA methylation within the repeat tract itself.
We used methylation-sensitive restriction enzymes and quantitative bisulfite sequencing of repetitive intracisternal A-type particles to assess DNA methylation genome-wide (41) (Fig. S3). We found that testes are hypomethylated relative to ovaries, but we detected no genome-wide differences in CpG methylation between Dnmt1+/+ SCA1 and Dnmt1+/− SCA1 gonads (Fig. S3A & B). Moreover, digestion of genomic DNA from testes by methylation sensitive enzymes did not uncover differences in genome-wide DNA methylation (Fig. S3C). Similarly, we found no differences in nonstandard cytosine methylation in CAG repeat DNA from testes and ovaries. Analysis of at least 350 CAG repeats from gonads of each genotype revealed about 1% apparent cytosine methylation, which was indistinguishable from the background levels found in control reactions with unmethylated Sca1 repeat DNA that was generated by PCR and cloning into a plasmid. Thus, altered methylation of the Sca1 CAG repeat itself does not account for the increased frequency of expansions found in Dnmt1+/− SCA1 mice.
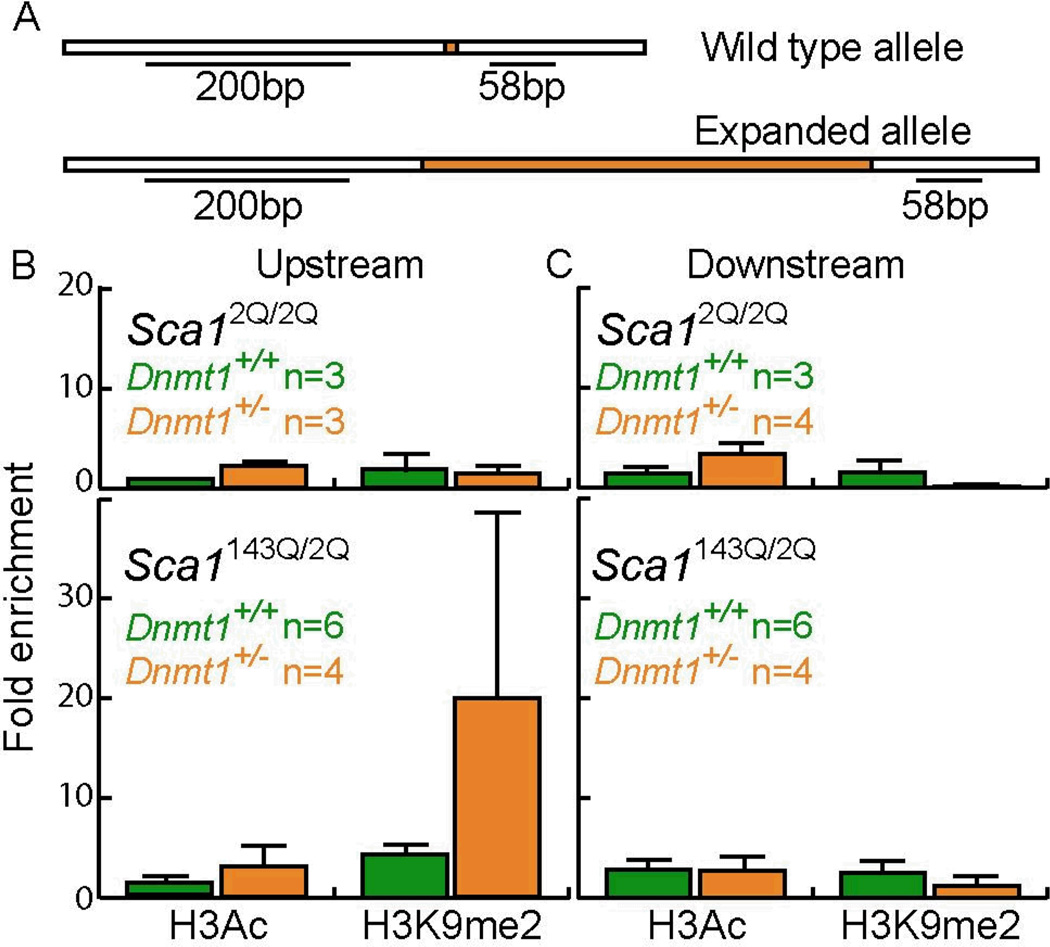
To assess DNA methylation adjacent to the repeat tract on the expanded Sca1 allele, we selectively analyzed CpG sites 17 and 20 bp upstream of the expanded CAG tract and a CpG site 32 bp downstream (Fig. 4A). No differences were apparent at the downstream site; however, at the upstream sites we found small, but significant, differences between Dnmt1+/− SCA1 mice and Dnmt1+/+ SCA1 mice in both testes (P = 0.03 and 0.02 for sites 1 and 2, respectively) and ovaries (P = 0.04 and 0.06 for sites 1 and 2, respectively) (Fig. 4B & C). Thus, Dnmt1 deficiency has opposite effects on local Sca1 DNA methylation in testes and ovaries, with elevated levels in testes and reduced levels in ovaries (P = 0.002) (Fig. 4). These observations on DNA methylation echo the opposite effects of Dnmt1 deficiency on the percentage of stable germline transmission in Dnmt1+/− SCA1 mice, which decreased in male donors, but increased in female donors (Fig. 1). We find that high local DNA methylation correlates with low frequencies of stable transmissions (P = 0.01). This correlation implies that local DNA methylation may contribute to the propensity of a repeat tract to become unstable.
Figure 4.
DNA methylation analysis of the Sca1 repeat region. A) Map of the CpG sites for the expanded allele at the Sca1 locus. The two upstream CpG sites are 17 bp and 20 bp in front of the CAG repeat tract, the third site is 32 bp downstream of the repeat. B) Representative bisulfite sequencing gel of the expanded allele of the Sca1 locus. The ovary and the testis are from Dnmt1+/− SCA1 mice. Arrows indicate the CpG sites analyzed. C) Box plot of DNA methylation for each CpG site shown in A in ovaries and testes from Dnmt1+/+ SCA1 (green) and Dnmt1+/− SCA1 (orange) mice. Boxes represent the middle 50% of all data points. The median is shown as a horizontal bar. Vertical lines represent the 95th percentile.
Histone modification at the Sca1 locus in testes from Dnmt1+/+SCA1 and Dnmt1+/− SCA1 mice
One of the most striking features of the DNA methylation data at the expanded Sca1 allele is the very broad range of methylation levels in both testes and ovaries of Dnmt1+/− SCA1 mice as compared to Dnmt1+/+ SCA1 mice. This extreme variability between mice (but not in replicate tests of the same mouse) is reminiscent of position-effect variegation, a process caused by stochastic spreading of heterochromatin to adjacent chromatin domains. Indeed, transgene arrays containing trinucleotide repeats have been shown to induce such variegation (31). As indicators of chromatin alteration, we examined specific histone modifications in the region immediately upstream and downstream of the CAG repeat at the Sca1 locus. We chose to examine acetylated histone H3 (H3Ac) and dimethylated histone H3 at lysine 9 (H3K9me2) because Dnmt1 is known to interact with the histone deacetylases HDAC1 and HDAC2 (42, 43), and with the histone methyltransferase G9a, which adds two methyl groups to H3K9 (44).
We immunoprecipitated chromatin-bound H3Ac and H3K9me2 from testes of 40-week old Dnmt1+/+ ScaI143Q/2Q and Dnmt1+/− Sca1143Q/2Q mice (SCA1 mice) and from 40-week old Dnmt1+/+ ScaI2Q/2Q and Dnmt1+/− Sca12Q/2Q mice (“wildtype” mice). We then quantified the precipitated DNA on either side of the CAG repeat (Fig. 5). We found that chromatin immunoprecipitation (ChIP) by H3Ac-specific antibodies did not enrich for DNA sequences around the site of the repeat tract, relative to the sequences precipitated by nonspecific antibodies, for any of the 33 mice that were tested. By contrast, H3K9me2-specific ChIP identified 1 mouse out of the 33 mice tested, in which DNA sequences were dramatically enriched relative to ChIP by the nonspecific antibody. This Dnmt1+/− SCA1 mouse was enriched 68 fold for upstream sequences; it yielded high enrichment values in three independent immunoprecipitations. Although these results do not rise to the level of statistical significance, they are consistent with the idea that the repeat tract itself may trigger stochastic formation of heterochromatin. These results, along with similar findings of DNA methylation changes, raise the possibility that changes in DNA methylation and altered chromatin structure in the vicinity of the repeat tract may alter repeat tract stability.
Figure 5.
Chromatin immunoprecipitation of the Sca1 repeat region. A) Map of the Sca1 locus around the repeat tract showing the regions analyzed by ChIP. H3Ac and H3K9me2 levels upstream (B) and downstream (C) of the Sca1 repeat tracts in 40-week old testes. Error bars represent one standard error.
Effects of Dnmt1 deficiency on transcription at the Sca1 locus in testes
We showed previously that increased transcription through a CAG tract in human cells destabilizes the repeats (17, 18), and transcription has recently been linked to repeat instability in the Drosophila germline (19). Given the inverse correlation between promoter methylation levels and gene expression (45), Dnmt1 deficiency could increase transcription through the CAG tract in the Sca1 gene, thereby destabilizing the repeats. To test this possibility, we used quantitative RT-PCR to measure the levels of Sca1 mRNA in testes from Dnmt1+/− SCA1 and Dnmt1+/+ SCA1 mice. Testicular levels of Sca1 transcripts were not affected by Dnmt1 genotype (Fig. S4), implying that transcription-altering changes in DNA methylation in the Sca1 promoter region are not the source of the observed increase in expansion frequencies. Consistent with this conclusion, we observed no difference in H3Ac and H3K9me2 levels in the Sca1 promoter region (~300kb upstream of the repeat tract) in Dnmt1+/+ SCA1 and Dnmt1+/− SCA1 mice (Fig. S5).
Evidence against NHEJ as a source of instability
Previously, we used a mammalian cell culture assay to determine whether long CAG repeat tracts (98 and 183 repeats) stimulated either homologous recombination or gene rearrangements (46). We found that repeat tracts in this size range did not stimulate homologous recombination above spontaneous levels, but they did increase gene rearrangements more than 50-fold—presumably via nonhomologous end joining (NHEJ) of double-strand breaks. Some of the rearrangement junctions had deletions that extended beyond the repeat tract, whereas others had extra sequences inserted at the rejoined ends, as is typically found at 10–20% of end-joining junctions (see, for example (47, 48)).
If most of the CAG repeat instability at the Sca1 locus resulted from breaks that were repaired by NHEJ, we would expect to find similar kinds of deletions and insertions at the repeat tract. We examined all the sequences gathered to analyze intergenerational triplet repeat instability, both donors and progeny, for a total of 109 Dnmt1+/+ SCA1 and 153 Dnmt1+/− SCA1 mice. In no instance did we find a deletion that extended outside the repeat tract or an insertion of extra nucleotides within the repeat tract. In addition, there were no mutations that altered either the sequence of the repeat or of any nucleotides in the 60 bp upstream of the repeat. The absence of these telltale signatures suggests that NHEJ is not responsible for the observed instability.
Dnmt1 deficiency does not substantially affect somatic instability of SCA1 repeats
Next, we asked whether Dnmt1-deficiency increases expansions specifically in the germline, or also increases expansions in somatic tissues. We analyzed DNA methylation and repeat length variations in cerebellum, liver, and striatum, which display low, moderate, and high levels of instability, respectively (37). In contrast to what we observed for IAPs in oocytes and testes, where we saw no effect of Dnmt1 deficiency, we found that DNA methylation of IAPs was slightly lower in all three tissues in Dnmt1+/− SCA1 mice compared to Dnmt1+/+ SCA1 mice (P = 0.002). We found that DNA methylation was also slightly lower for the two CpG sites upstream of the repeat tract at the Sca1 locus (P = 0.02) (Figure S6 and Figure S7), similar to our observations in oocytes.
To determine the effects of Dnmt1 deficiency on the SCA1 repeat, we carried out small-pool PCR analyses of repeat tract lengths in cerebellum, liver, and striatum in Dnmt1+/− SCA1 mice and Dnmt1+/+ SCA1 mice (Figure S8). These results, which are summarized in Table 1, indicate that repeat expansions are not enhanced in these three tissues in Dnmt1+/− SCA1 mice relative to those from Dnmt1+/+ SCA1 mice. It should be noted that the expansions we observe in liver and striatum are generally larger that those we observe in the germline, but the distributions of lengths are comparable in Dnmt1+/− and Dnmt1+/+ SCA1 mice. These results indicate that the deficiency of Dnmt1 in these somatic tissues is not sufficient to affect expansion frequencies. Thus, it appears that the effects of Dnmt1 deficiency on CAG repeat tract stability are specific for the germline.
Table 1.
Somatic instability of CAG repeats in Dnmt1+/+ SCA1 and Dnmt1+/− SCA1 mice
| Tissue | Agea (weeks) |
Dnmt1 genotype |
Mice tested |
Contractions (%) |
Stableb (%) |
Expansions (%) |
Allelesc (total) |
P-valued |
|---|---|---|---|---|---|---|---|---|
| Cerebellum | 6 | +/+ | 2 | 0 | 100 | 0 | 145 | 1 |
| 6 | +/− | 2 | 0 | 100 | 0 | 203 | ||
| 40 | +/+ | 2 | 1.1 | 89.8 | 9.1 | 614 | 0.001 | |
| 40 | +/− | 2 | 1.7 | 95.4 | 2.9 | 346 | ||
| Liver | 6 | +/+ | 2 | 12.5 | 86.1 | 1.4 | 313 | 0.11 |
| 6 | +/− | 2 | 4.9 | 93.3 | 1.8 | 238 | ||
| 40 | +/+ | 2 | 4 | 32.2 | 63.8 | 348 | 0.09 | |
| 40 | +/− | 2 | 2.9 | 40.8 | 56.3 | 128 | ||
| Striatum | 6 | +/+ | 2 | 4.2 | 95.4 | 0.4 | 262 | 0.34 |
| 6 | +/− | 2 | 3 | 95.7 | 1.3 | 368 | ||
| 24 | +/+ | 2 | 2.8 | 58.5 | 38.7 | 145 | 0.09 | |
| 24 | +/− | 2 | 8.1 | 48.7 | 43.2 | 99 |
Ages of mice to be analyzed in depth were chosen based on a preliminary screen to find times when there was very little variation (6 weeks) and times when the variation was pronounced, but not excessive.
Stable alleles are defined as being within 5 CAG repeats from the size of the repeat tract in tail at weaning. We note that most of our intergenerational instability was within the resolution that we could reliably achieve in our SP-PCR (± 5 CAGs).
Total alleles is the average number of alleles per reaction (as determined by the Poisson distribution) multiplied the number of reactions set up for a given sample.
P-value was determined by comparing Dnmt1+/+ SCA1 mice and Dnmt1+/− SCA1 mice of the same tissue and at the same age, using a Chi-squared test with 2 degrees of freedom.
DISCUSSION
Here we have investigated the potential links between epigenetics and CAG repeat stability at the Sca1 locus in mice. This study stems from three observations. First, the two identified periods of enhanced repeat instability during intergenerational transmission—during early embryogenesis and in germline development—overlap with the two major cycles of epigenetic reprogramming, when DNA methylation patterns are erased and re-established (6). Second, observations at the FMR1 and DMPK loci in humans and mice show that long repeat tracts are associated with changes in DNA methylation (for CGG repeats), histone modification, chromatin structure, and transcription (8–11, 16, 25–30, 49, 50). Third, treatments that lead to genome-wide demethylation dramatically stimulate CAG repeat instability in a cell-culture assay (7).
After confirming that siRNA knockdown of Dnmt1 in human cells altered CAG repeat stability in a cell culture assay, we sought to perturb DNA methylation in mice by making them heterozygous for a null allele of Dnmt1. We showed that Dnmt1 deficiency leads to a dramatic increase in intergenerational expansions at the Sca1 locus in both male and female mice; however, it causes no apparent increase in expansions in liver, cerebellum, and striatum. These results suggest that Dnmt1 deficiency specifically affects expansion in the germline. The dependence of the expansions on the genotype of the donating parent (and not on that of the progeny) in both males and females indicates that the critical events leading to expansions in Dnmt1-deficient mice occur during germline development. The enhanced instability of CAG repeats that is caused by genetically altering Dnmt1 levels is consistent with the notion that normal epigenetic changes during germline development may play a role in the intergenerational instability of repeat tracts at the Sca1 locus—and potentially at other repeat loci—in mice and human.
Progeny from both male and female Dnmt1+/− SCA1 donors display a clear increase in expansions—relative to progeny from Dnmt1+/+ SCA1 donors—at the earliest donor ages (5–15 weeks). Subsequently, the progeny from male donors show a small increase in expansions with donor age, while the progeny from female donors ultimately show only contractions at late donor age (>25 weeks). These data suggest that two distinct processes may operate during germline development. An expansion-biased process appears to function early on in the germlines of both male and female Dnmt1+/− SCA1 donors. It will be crucial to know the timing of this process more precisely, but the present data are consistent with the idea that it overlaps the period of hypomethylation in normal germline development. The second process operates later on and differs significantly in the male and female germlines. In male donors, a slight expansion bias continues with age. In female donors a contraction-biased process predominates at later times in both Dnmt1+/+ SCA1 and Dnmt1+/− SCA1 mice. This contraction-biased process is thought to operate in arrested oocytes and has been attributed to oocyte-specific DNA repair (38).
Because long repeat tracts at both the FMR1 and DMPK loci are associated with epigenetic changes, we examined the region around the CAG repeat tracts at the Sca1 locus to determine whether any changes in DNA methylation, or in the overlying chromatin, were present. To try to capture data that is relevant to germline transmission, we analyzed testes and ovaries. We found that the methylation changes in these tissues were in opposite directions in Dnmt1+/− SCA1 mice relative to Dnmt1+/+ SCA1 mice, with elevated levels in testes and reduced levels in ovaries. Although we had expected Dnmt1 deficiency to lead to reduced DNA methylation, as we observed in oocytes and somatic tissues, the paradoxical local increase in testes has precedent in other systems. For instance, cancer cells with decreased Dnmt1 levels are generally hypomethylated genome-wide, but local hypermethylation is common (51). In the region just upstream of the repeat tract in testes, we also found increased levels of histone H3K9me2, which is indicative of heterochromatin. These results suggest that Dnmt1 deficiency can induce epigenetic changes at the CAG repeat tract at the Sca1 locus, similar to those found at the FMR1 and DMPK loci in humans.
At the Sca1 locus the changes in DNA methylation and in histone modification were found only at sites within the CpG island that abuts the upstream side of the CAG repeat tract. This CpG island is conserved in humans and mice. A strong association between unstable repeats and nearby CpG islands has been noted for several triplet-repeat diseases (52, 53), but no mechanistic connection has been established. At the Sca1 locus, as well, we do not know whether the epigenetic changes cause the germline expansions we observe, or are merely associated with them. These epigenetic changes in the adjacent CpG island cannot be the entire story, however, because similar changes in somatic tissues are not associated with increased repeat expansions. Thus, if these epigenetic changes are causative, they must interact with germline specific factors to bring about repeat expansions. Additional experiments in SCA1 mice in which specific CpG sites have been eliminated, or the island itself removed, will be required to distinguish cause from association.
Although we have shown that Dnmt1 deficiency leads to expansions in both the male and female germline, and we have identified local epigenetic changes, we have not identified the mechanistic link between Dnmt1 deficiency and repeat expansion. Our study eliminates several potential sources of triplet repeat instability caused by Dnmt1 deficiency, thereby providing useful mechanistic information. First, we found no DNA methylation within the CAG repeat tract itself in DNA isolated from testes or ovaries, showing that CAG repeat tracts do not serve as a nidus for abnormal CpA methylation in either Dnmt1+/+ SCA1 mice or in Dnmt1+/− SCA1 mice. Second, we showed that neither histone acetylation at the Sca1 promoter nor transcription through the Sca1 locus is altered in testes from Dnmt1+/− SCA1 mice relative to those from Dnmt1+/+ SCA1 mice. These results argue against the possibility that increased transcription through the Sca1 locus causes repeat expansion during germline transmission in Dnmt1+/− SCA1 mice, an attractive hypothesis based on the link between transcription and repeat instability established recently in human cells and Drosophila germline (17–19). It is important to note that our studies with whole testes would have missed a dependence on transcription if the critical events occurred in a small proportion of the population. Third, while we cannot rule out the possibility that DNA methylation changes alter transcription of other genes, which then act on the repeat tract in trans, we analyzed mRNA levels of several genes that have been identified to participate in triplet repeat instability: Xpa (18), Xpg (17), Msh2 (18, 54–56), and Fen-1 (57). We found no differences in the abundance of their mRNA in testes from Dnmt1+/+ SCA1 mice and Dnmt1+/− SCA1 mice (data not shown). Thus, it is unlikely that changes in the levels of those particular genes are the cause of the increase in expansion in Dnmt1+/− SCA1 mice. Fourth, analysis of the repeat tracts transmitted through the germline showed none of the signature features of NHEJ, suggesting that changes in the lengths of the repeat tracts did not arise by nearby DSBs that were rejoined by the typical mechanism of NHEJ (47, 48, 58). Finally, we have recently found that 5-aza-CdR-induced genome-wide demethylation leads to instability by a mechanism that is independent of homologous recombination (7, 46, 59)
In addition, although we have demonstrated subtle changes in DNA methylation and local chromatin structure, it is possible that Dnmt1-deficiency has its primary effect on repeat stability via a mechanism that is unrelated to its enzymatic function. Dnmt1 is a component of many different complexes involved in various aspects of DNA metabolism, including chromatin structure (42–44, 60–63), DNA replication (64, 65), DNA damage response (66–68), and mismatch repair (69–72). Each of these processes has been shown to impinge on the stability of repeat tracts (2). If the deficiency of Dnmt1 in Dnmt1+/− SCA1 mice compromised the function of any of these complexes, repeat tracts might be destabilized.
One important feature of Dnmt1-deficient SCA1 mice is that they closely resemble the intergenerational instability pattern seen in SCA1 patients, which has been difficult to demonstrate for most models of repeat diseases (73). The knock-in SCA1 mouse model used in our studies is no exception. As described initially (37) and confirmed here, transmission in both male and female Dnmt1+/+ SCA1 mice is strongly biased toward contraction, so that the distributions of tract-length changes do not match those of human patients (P < 0.0001 for males and females). By contrast, the distributions in Dnmt1-deficient mice are much more similar to the patient distributions (P = 0.08 for males, and P = 0.6 for female donors less than 25 weeks old) (Fig. S2). Thus, Dnmt1+/− SCA1 mice provide the most accurate model to date for studying intergenerational repeat instability in polyglutamine diseases. Why a deficiency of Dnmt1 should bring mouse intergenerational transmission data more in line with that for humans is not yet clear. The answer to that question will likely provide insights into the human disease.
Finally, the high variability in the adjacent epigenetic marks indicates that the expanded repeat tract at the Sca1 locus creates a metastable epiallele. Typically, metastable epialleles are susceptible to environmental influences such as diet (74). If the stability of the CAG repeat tract at the Sca1 locus proves to be determined by these epigenetic changes, it may ultimately prove possible to control repeat stability through diet, a possibility that extends not only to SCA1 disease, but also potentially to other repeat-associated neurodegenerative diseases.
MATERIALS AND METHODS
Mice
Mice were handled in accordance with an approved protocol from the Animal Research Committee of Baylor College of Medicine. C57BL/6 mice were purchased from Harlan Sprague Dawley. The Dnmt1 mice were in a pure 129/Sv background (35). The null allele used, Dnmt1c, has a deletion of the catalytic domain of Dnmt1, but expresses no stable transcript or detectable protein (35). We refer to this allele as Dnmt1− for simplicity. Males carrying the knock-in allele of Sca1 with the expanded repeat tract (which we refer to as SCA1 mice) had been backcrossed to C57BL/6 females for 8 to 10 generations (34) before we bred them to Dnmt1+/− mice. The litter sizes were normal, the genetic ratios of Dnmt1+/− SCA1 progeny were as expected, and the Dnmt1 deficiency did not affect body weight of the Dnmt1+/− SCA1 mice. The male and female Dnmt1+/− SCA1 and Dnmt1+/+ SCA1 progeny from these crosses were then crossed to C57BL/6 mice to measure intergenerational repeat stability. Thus, the data reported in Figures 1, 2, and 3 were gathered over a single generation. The repeat tract lengths in the parents and offspring were determined by sequencing tail DNA at weaning. The repeat tract lengths in Dnmt1+/+ SCA1 male donors (n=5) ranged from 142 to 145; in Dnmt1+/+ SCA1 female donors (n=10), from 142 to 150; in Dnmt1+/− SCA1 male donors (n=11), from 141 to 146; and in Dnmt1+/− SCA1 female donors (n=7), from 142 to 147. We did not observe any correlation in these ranges between the repeat-tract length in the donor and the extent of the intergenerational instability.
PCR primers and sequencing
The Dnmt1 locus was genotyped as described (75): the pair of PCR primers specific for the Dnmt1+ allele amplifies an exon deleted in the Dnmt1− allele; the primer pair specific for the Dnmt1− allele amplifies sequences unique to the null allele. Genotyping primers were oVIN-24F (5'-aac atg ggc agt ctg agc cag) and oVIN-24R (5'-agc cct gct gag gtg ctg ctg) for the Sca1 locus. To determine repeat size at weaning, we amplified the CAG repeat with oVIN-106F (5'-cgt gta ccc tcc tcc tca gt) and oVIN-24R and sequenced the PCR products directly using oVIN-95F (5'-ggc cac cac tcc atc aca gc). Sequencing was performed by the Baylor College of Medicine Sequencing Core. To determine whether this sequencing approach was reliable, we independently amplified and sequenced 11 samples 4 to 5 times each (Table S1). Thirty-eight of the 46 runs (83%) were in agreement. The remaining 8 runs (17%) gave a repeat number that was either one CAG repeat higher or lower than the most commonly observed tract length. This low frequency of potential one-repeat errors does not significantly alter the data presented in this study, which were derived primarily from single sequencing runs.
Bisulfite Sequencing
To determine whether Dnmt1+/− SCA1 mice showed decreased levels of DNA methylation, we isolated DNA from ovaries and testes of 6, 12, 24, and 40 week-old Dnmt1+/− SCA1 and Dnmt1+/+ SCA1 mice. We could not obtain enough DNA from gametes for our methylation analysis at the expanded Sca1 locus and thus we opted for gonadal DNA. Methylation patterns in testis and sperm are identical (76). The differences in DNA methylation levels between wild type and heterozygous testes were not due to an altered number or morphology of somatic cells in the Dnmt1+/− SCA1 testes (data not shown). Bisulfite modification and PCR amplification were performed as described (77). The amplicons were sequenced using 33P-labelled ddNTPs (GE Healthcare) and the products were separated by electrophoresis on a polyacrylamide denaturing gel. Intensities of bands at methylation sites were quantified in a blinded fashion by phosphoimaging (77). CpG methylation was calculated as: % methylation = 100 x [Cvolume/(Cvolume+Tvolume)]. For IAP sequences, bisulfite-treated DNA was amplified using primers IAPLTRF2 and IAPLTRR1 (77). Methylation on the expanded allele at the Sca1 locus was analyzed as follows: before the bisulfite treatment, 10 µg of genomic DNA was digested with BanII (NEB), which cuts only the wild type allele, at a position 22 bp upstream of the two CAGs. PCR of the intact, expanded allele was then performed using oVIN-147F (5′- ttt ttt tta gtt gat ttt ttt att agg taa) and oVIN-61R (5′- tat aaa aaa cta aaa ata taa ata tac taa ttc tac), which prime 160 bp upstream and 57 bp downstream of the repeat tract, respectively. The amplification products were sequenced using oVIN-57F (5′- ttt att ttg ttg gtt aat atg gg), which primes 57 bp upstream of the CAG tract. We used oVIN-151F (5′- tag tag tat ttt agt agg gtt gta gga tta gtt aat t), which primes immediately adjacent to the third CpG site, to sequence it.
Chromatin immunoprecipitation
Chromatin immunoprecipitation experiments were performed as described (78), using rabbit polyclonal antibody against H3Ac (Upstate) or H3K9me2 (Upstate). We used rabbit IgG (Sigma-Aldrich) for nonspecific immunoprecipitations. We performed quantitative PCR on the immunoprecipitated DNA, using oVIN-106F (5′-cgt gta ccc tcc tcc tca gt) and oVIN-186R (5′-ctg cag ctg gga gcg) for the region upstream of the CAG repeat tract, and oVIN-200F (5′- cac cca gca gaa gta ca) and oVIN-200R (5′- gat ggg tgg ggg aga tgt) for the downstream region. For each mouse analyzed, we made two or three independent chromatin preparations. Each chromatin preparation was immunoprecipitated using nonspecific antibodies and antibodies specific for either H3Ac or H3K9me2. The DNA in each precipitation was measured by quantitative PCR using primers upstream and downstream of the repeat. Amplification values were normalized for the amount of DNA in the original chromatin preparation, which was also measured by quantitative PCR. Enrichment ratios were calculated by dividing the quantity of normalized DNA for the specific antibody by that for the nonspecific antibody. The values obtained from each chromatin preparation were averaged for each mouse, and the average of these means was calculated for all mice of the same genotype.
Small-pool PCR
DNA isolated from different mouse tissues was digested with BanII to fragment the DNA. DNA was quantified by spectrophotometry and then small-pool PCR (SP-PCR) reactions were set up at DNA concentrations of 100 pg, 50 pg, 25 pg, and 12.5 pg per reaction. An initial set of seven SP-PCRs were run to determine the optimum dilution to use for each tissue sample, so that the reactions yielded approximately 2–3 amplifiable genomes per reaction. The PCR primers were oVIN-25F (5′- gtc acc agt gca gta gcc tca g), which primes 111 bp upstream of the repeat, and oVIN-25R (5′- atg tac tgg ttc tgc tgg gtg), which primes 51 bp downstream of the repeat. We used ChromaTaq (Denville Scientific) in accordance to the supplier’s instructions, with the addition of DMSO to a final concentration of 5% in 10 µl reactions. The PCR program was 94°C for 2 min for initial denaturation, followed by 30 cycles of 94°C for 30 sec, 60°C for 30 sec, and 72°C for 1 min. The PCR products were separated by electrophoresis on a 2% agarose TAE gel at 80 V for 16 hours, transferred to a nylon membrane, and hybridized with a (CAG)10 probe that had been labelled at the 5′ end with γ-32P-ATP and polynucleotide kinase (Invitrogen). The resulting blots were analyzed by phosphorimaging. Tail DNA from the same mouse as the tissue sample was included on either side of each set of reactions. Lines bracketing the top and bottom of the tail-DNA bands were drawn, and sample bands were judged against those lines. Sample bands that were more that ¾ of the way above or below the lines were classified as expansions or contractions, respectively. Reconstruction experiments using repeats of defined length indicated that according to this criterion repeats that were more than 5 CAG units longer than the tail DNA would be classified as expansions, and those that were more than 5 CAG units shorter would be classified as contractions. The total number of alleles examined was calculated by multiplying the total number of individual samples analyzed by the average number of alleles per sample, which was estimated from the Poisson distribution by counting the number of samples that had 0 alleles.
Statistics
Distributions of tract-length changes were compared using the non-parametric Mann-Whitney test, which does not assume a normal distribution. Data on site-specific CpG methylation at IAPs and at the Sca1 locus were analyzed by repeated-measures analysis of variance (SAS PROC MIXED), which acknowledges the intercorrelation among multiple CpG sites within each individual. SP-PCR data was analyzed using a Chi-squared test with 2 degrees of freedom.
ACKNOWLEDGEMENTS
We thank Drs. Huda Zoghbi (Baylor College of Medicine) and Rudolf Jaenisch (Whitehead Institute, MIT) for gifts of mice. We thank Dr. Adam Kuspa for providing us with repeat-free lab space for our SP-PCR analyses. We acknowledge Huda Zoghbi, Fisun Hamaratoğlu, Megan N. Hersh, Waleed Nasser, and the members of the Wilson lab for helpful discussions and critical reading of the manuscript. Technical help was provided by Pooja Shivraj (2nd generation instability), Sharyl L. Fyffe (ChIP analysis) Jared Gilliam (RNA isolation) and Yi-Nan Lin and Martin M. Matzuk (testis histology and analysis). This work was funded by NSERC PGS-D scholarship (V.D.), USDA CRIS 6250-51000-049 (R.A.W.) and NIH grant GM38219 (J.H.W.).
REFERENCES
- 1.Gatchel JR, Zoghbi HY. Diseases of unstable repeat expansion: mechanisms and common principles. Nat. Rev. Genet. 2005;6:743–755. doi: 10.1038/nrg1691. [DOI] [PubMed] [Google Scholar]
- 2.Pearson CE, Edamura KN, Cleary JD. Repeat instability: mechanisms of dynamic mutations. Nat. Rev. Genet. 2005;6:729–742. doi: 10.1038/nrg1689. [DOI] [PubMed] [Google Scholar]
- 3.Chung MY, Ranum LP, Duvick LA, Servadio A, Zoghbi HY, Orr HT. Evidence for a mechanism predisposing to intergenerational CAG repeat instability in spinocerebellar ataxia type I. Nat. Genet. 1993;5:254–258. doi: 10.1038/ng1193-254. [DOI] [PubMed] [Google Scholar]
- 4.Matilla T, Volpini V, Genis D, Rosell J, Corral J, Davalos A, Molins A, Estivill X. Presymptomatic analysis of spinocerebellar ataxia type 1 (SCA1) via the expansion of the SCA1 CAG-repeat in a large pedigree displaying anticipation and parental male bias. Hum. Mol. Genet. 1993;2:2123–2128. doi: 10.1093/hmg/2.12.2123. [DOI] [PubMed] [Google Scholar]
- 5.Goldfarb LG, Vasconcelos O, Platonov FA, Lunkes A, Kipnis V, Kononova S, Chabrashvili T, Vladimirtsev VA, Alexeev VP, Gajdusek DC. Unstable triplet repeat and phenotypic variability of spinocerebellar ataxia type 1. Ann. Neurol. 1996;39:500–506. doi: 10.1002/ana.410390412. [DOI] [PubMed] [Google Scholar]
- 6.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 7.Gorbunova V, Seluanov A, Mittelman D, Wilson JH. Genome-wide demethylation destabilizes CTG.CAG trinucleotide repeats in mammalian cells. Hum. Mol. Genet. 2004;13:2979–2989. doi: 10.1093/hmg/ddh317. [DOI] [PubMed] [Google Scholar]
- 8.Oberle I, Rousseau F, Heitz D, Kretz C, Devys D, Hanauer A, Boue J, Bertheas MF, Mandel JL. Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science. 1991;252:1097–1102. doi: 10.1126/science.252.5009.1097. [DOI] [PubMed] [Google Scholar]
- 9.Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DP, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang FP, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- 10.Taylor AK, Tassone F, Dyer PN, Hersch SM, Harris JB, Greenough WT, Hagerman RJ. Tissue heterogeneity of the FMR1 mutation in a high-functioning male with fragile X syndrome. Am. J. Med. Genet. 1999;84:233–239. [PubMed] [Google Scholar]
- 11.Wohrle D, Salat U, Glaser D, Mucke J, Meisel-Stosiek M, Schindler D, Vogel W, Steinbach P. Unusual mutations in high functioning fragile X males: apparent instability of expanded unmethylated CGG repeats. J. Med. Genet. 1998;35:103–111. doi: 10.1136/jmg.35.2.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reyniers E, Vits L, De Boulle K, Van Roy B, Van Velzen D, de Graaff E, Verkerk AJ, Jorens HZ, Darby JK, Oostra B, et al. The full mutation in the FMR-1 gene of male fragile X patients is absent in their sperm. Nat. Genet. 1993;4:143–146. doi: 10.1038/ng0693-143. [DOI] [PubMed] [Google Scholar]
- 13.Brouwer JR, Mientjes EJ, Bakker CE, Nieuwenhuizen IM, Severijnen LA, Van der Linde HC, Nelson DL, Oostra BA, Willemsen R. Elevated Fmr1 mRNA levels and reduced protein expression in a mouse model with an unmethylated Fragile X full mutation. Exp. Cell Res. 2007;313:244–253. doi: 10.1016/j.yexcr.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Entezam A, Biacsi R, Orrison B, Saha T, Hoffman GE, Grabczyk E, Nussbaum RL, Usdin K. Regional FMRP deficits and large repeat expansions into the full mutation range in a new Fragile X premutation mouse model. Gene. 2007;395:125–134. doi: 10.1016/j.gene.2007.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nichol K, Pearson CE. CpG methylation modifies the genetic stability of cloned repeat sequences. Genome Res. 2002;12:1246–1256. doi: 10.1101/gr.74502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sutcliffe JS, Nelson DL, Zhang F, Pieretti M, Caskey CT, Saxe D, Warren ST. DNA methylation represses FMR-1 transcription in fragile X syndrome. Hum. Mol. Genet. 1992;1:397–400. doi: 10.1093/hmg/1.6.397. [DOI] [PubMed] [Google Scholar]
- 17.Lin Y, Wilson JH. Transcription-induced CAG repeat contraction in human cells is mediated in part by transcription-coupled nucleotide excision repair. Mol. Cell. Biol. 2007;27:6209–6217. doi: 10.1128/MCB.00739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin Y, Dion V, Wilson JH. Transcription promotes contraction of CAG repeat tracts in human cells. Nat. Struct. Mol. Biol. 2006;13:179–180. doi: 10.1038/nsmb1042. [DOI] [PubMed] [Google Scholar]
- 19.Jung J, Bonini N. CREB-binding protein modulates repeat instability in a Drosophila model for polyQ disease. Science. 2007;315:1857–1859. doi: 10.1126/science.1139517. [DOI] [PubMed] [Google Scholar]
- 20.Haaf T. The effects of 5-azacytidine and 5-azadeoxycytidine on chromosome structure and function: implications for methylation-associated cellular processes. Pharmacol. Ther. 1995;65:19–46. doi: 10.1016/0163-7258(94)00053-6. [DOI] [PubMed] [Google Scholar]
- 21.Ghoshal K, Datta J, Majumder S, Bai S, Kutay H, Motiwala T, Jacob ST. 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Mol. Cell. Biol. 2005;25:4727–4741. doi: 10.1128/MCB.25.11.4727-4741.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Deng C, Lu Q, Zhang Z, Rao T, Attwood J, Yung R, Richardson B. Hydralazine may induce autoimmunity by inhibiting extracellular signal-regulated kinase pathway signaling. Arthritis Rheum. 2003;48:746–756. doi: 10.1002/art.10833. [DOI] [PubMed] [Google Scholar]
- 23.Segura-Pacheco B, Trejo-Becerril C, Perez-Cardenas E, Taja-Chayeb L, Mariscal I, Chavez A, Acuna C, Salazar AM, Lizano M, Duenas-Gonzalez A. Reactivation of tumor suppressor genes by the cardiovascular drugs hydralazine and procainamide and their potential use in cancer therapy. Clin. Cancer Res. 2003;9:1596–1603. [PubMed] [Google Scholar]
- 24.Otten AD, Tapscott SJ. Triplet repeat expansion in myotonic dystrophy alters the adjacent chromatin structure. Proc. Natl. Acad. Sci. U. S. A. 1995;92:5465–5469. doi: 10.1073/pnas.92.12.5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klesert TR, Otten AD, Bird TD, Tapscott SJ. Trinucleotide repeat expansion at the myotonic dystrophy locus reduces expression of DMAHP. Nat. Genet. 1997;16:402–406. doi: 10.1038/ng0897-402. [DOI] [PubMed] [Google Scholar]
- 26.Thornton CA, Wymer JP, Simmons Z, McClain C, Moxley RT., 3rd Expansion of the myotonic dystrophy CTG repeat reduces expression of the flanking DMAHP gene. Nat. Genet. 1997;16:407–409. doi: 10.1038/ng0897-407. [DOI] [PubMed] [Google Scholar]
- 27.Eriksson M, Ansved T, Edstrom L, Anvret M, Carey N. Simultaneous analysis of expression of the three myotonic dystrophy locus genes in adult skeletal muscle samples: the CTG expansion correlates inversely with DMPK and 59 expression levels, but not DMAHP levels. Hum. Mol. Genet. 1999;8:1053–1060. doi: 10.1093/hmg/8.6.1053. [DOI] [PubMed] [Google Scholar]
- 28.Frisch R, Singleton KR, Moses PA, Gonzalez IL, Carango P, Marks HG, Funanage VL. Effect of triplet repeat expansion on chromatin structure and expression of DMPK and neighboring genes, SIX5 and DMWD, in myotonic dystrophy. Mol. Genet. Metab. 2001;74:281–291. doi: 10.1006/mgme.2001.3229. [DOI] [PubMed] [Google Scholar]
- 29.Filippova GN, Thienes CP, Penn BH, Cho DH, Hu YJ, Moore JM, Klesert TR, Lobanenkov VV, Tapscott SJ. CTCF-binding sites flank CTG/CAG repeats and form a methylation-sensitive insulator at the DM1 locus. Nat. Genet. 2001;28:335–343. doi: 10.1038/ng570. [DOI] [PubMed] [Google Scholar]
- 30.Cho DH, Thienes CP, Mahoney SE, Analau E, Filippova GN, Tapscott SJ. Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol. Cell. 2005;20:483–489. doi: 10.1016/j.molcel.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 31.Saveliev A, Everett C, Sharpe T, Webster Z, Festenstein R. DNA triplet repeats mediate heterochromatin-protein-1-sensitive variegated gene silencing. Nature. 2003;422:909–913. doi: 10.1038/nature01596. [DOI] [PubMed] [Google Scholar]
- 32.Klesert TR, Cho DH, Clark JI, Maylie J, Adelman J, Snider L, Yuen EC, Soriano P, Tapscott SJ. Mice deficient in Six5 develop cataracts: implications for myotonic dystrophy. Nat. Genet. 2000;25:105–109. doi: 10.1038/75490. [DOI] [PubMed] [Google Scholar]
- 33.Jansen G, Groenen PJ, Bachner D, Jap PH, Coerwinkel M, Oerlemans F, van den Broek W, Gohlsch B, Pette D, Plomp JJ, et al. Abnormal myotonic dystrophy protein kinase levels produce only mild myopathy in mice. Nat. Genet. 1996;13:316–324. doi: 10.1038/ng0796-316. [DOI] [PubMed] [Google Scholar]
- 34.Watase K, Weeber EJ, Xu B, Antalffy B, Yuva-Paylor L, Hashimoto K, Kano M, Atkinson R, Sun Y, Armstrong DL, et al. A long CAG repeat in the mouse Sca1 locus replicates SCA1 features and reveals the impact of protein solubility on selective neurodegeneration. Neuron. 2002;34:905–919. doi: 10.1016/s0896-6273(02)00733-x. [DOI] [PubMed] [Google Scholar]
- 35.Lei H, Oh SP, Okano M, Juttermann R, Goss KA, Jaenisch R, Li E. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122:3195–3205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
- 36.Ray D, Wu A, Wilkinson JE, Murphy HS, Lu Q, Kluve-Beckerman B, Liepnieks JJ, Benson M, Yung R, Richardson B. Aging in heterozygous Dnmt1-deficient mice: effects on survival, the DNA methylation genes, and the development of amyloidosis. J. Gerontol. A. Biol. Sci. Med. Sci. 2006;61:115–124. doi: 10.1093/gerona/61.2.115. [DOI] [PubMed] [Google Scholar]
- 37.Watase K, Venken KJ, Sun Y, Orr HT, Zoghbi HY. Regional differences of somatic CAG repeat instability do not account for selective neuronal vulnerability in a knock-in mouse model of SCA1. Hum. Mol. Genet. 2003;12:2789–2795. doi: 10.1093/hmg/ddg300. [DOI] [PubMed] [Google Scholar]
- 38.Kaytor MD, Burright EN, Duvick LA, Zoghbi HY, Orr HT. Increased trinucleotide repeat instability with advanced maternal age. Hum. Mol. Genet. 1997;6:2135–2139. doi: 10.1093/hmg/6.12.2135. [DOI] [PubMed] [Google Scholar]
- 39.Burright EN, Clark HB, Servadio A, Matilla T, Feddersen RM, Yunis WS, Duvick LA, Zoghbi HY, Orr HT. SCA1 transgenic mice: a model for neurodegeneration caused by an expanded CAG trinucleotide repeat. Cell. 1995;82:937–948. doi: 10.1016/0092-8674(95)90273-2. [DOI] [PubMed] [Google Scholar]
- 40.Gaudet F, Rideout WM, Meissner A, 3rd, Dausman J, Leonhardt H, Jaenisch R. Dnmt1 expression in pre- and postimplantation embryogenesis and the maintenance of IAP silencing. Mol. Cell. Biol. 2004;24:1640–1648. doi: 10.1128/MCB.24.4.1640-1648.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32:e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rountree MR, Bachman KE, Baylin SB. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat. Genet. 2000;25:269–277. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- 43.Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet. 2000;25:338–342. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- 44.Esteve PO, Chin HG, Smallwood A, Feehery GR, Gangisetty O, Karpf AR, Carey MF, Pradhan S. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev. 2006;20:3089–3103. doi: 10.1101/gad.1463706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 46.Meservy JL, Sargent RG, Iyer RR, Chan F, McKenzie GJ, Wells RD, Wilson JH. Long CTG tracts from the myotonic dystrophy gene induce deletions and rearrangements during recombination at the APRT locus in CHO cells. Mol. Cell. Biol. 2003;23:3152–3162. doi: 10.1128/MCB.23.9.3152-3162.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sandor Z, Calicchio ML, Sargent RG, Roth DB, Wilson JH. Distinct requirements for Ku in N nucleotide addition at V(D)J- and non-V(D)J-generated double-strand breaks. Nucleic Acids Res. 2004;32:1866–1873. doi: 10.1093/nar/gkh502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin Y, Waldman AS. Capture of DNA sequences at double-strand breaks in mammalian chromosomes. Genetics. 2001;158:1665–1674. doi: 10.1093/genetics/158.4.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coffee B, Zhang F, Warren ST, Reines D. Acetylated histones are associated with FMR1 in normal but not fragile X-syndrome cells. Nat. Genet. 1999;22:98–101. doi: 10.1038/8807. [DOI] [PubMed] [Google Scholar]
- 50.Coffee B, Zhang F, Ceman S, Warren ST, Reines D. Histone modifications depict an aberrantly heterochromatinized FMR1 gene in fragile x syndrome. Am. J. Hum. Genet. 2002;71:923–932. doi: 10.1086/342931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.El-Osta A. DNMT cooperativity--the developing links between methylation, chromatin structure and cancer. Bioessays. 2003;25:1071–1084. doi: 10.1002/bies.10345. [DOI] [PubMed] [Google Scholar]
- 52.Gourdon G, Dessen P, Lia AS, Junien C, Hofmann-Radvanyi H. Intriguing association between disease associated unstable trinucleotide repeat and CpG island. Ann. Genet. 1997;40:73–77. [PubMed] [Google Scholar]
- 53.Brock GJ, Anderson NH, Monckton DG. Cis-acting modifiers of expanded CAG/CTG triplet repeat expandability: associations with flanking GC content and proximity to CpG islands. Hum. Mol. Genet. 1999;8:1061–1067. doi: 10.1093/hmg/8.6.1061. [DOI] [PubMed] [Google Scholar]
- 54.Manley K, Shirley TL, Flaherty L, Messer A. Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nat. Genet. 1999;23:471–473. doi: 10.1038/70598. [DOI] [PubMed] [Google Scholar]
- 55.Savouret C, Garcia-Cordier C, Megret J, te Riele H, Junien C, Gourdon G. MSH2-dependent germinal CTG repeat expansions are produced continuously in spermatogonia from DM1 transgenic mice. Mol. Cell. Biol. 2004;24:629–637. doi: 10.1128/MCB.24.2.629-637.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Savouret C, Brisson E, Essers J, Kanaar R, Pastink A, te Riele H, Junien C, Gourdon G. CTG repeat instability and size variation timing in DNA repair-deficient mice. EMBO J. 2003;22:2264–2273. doi: 10.1093/emboj/cdg202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Freudenreich CH, Kantrow SM, Zakian VA. Expansion and length-dependent fragility of CTG repeats in yeast. Science. 1998;279:853–856. doi: 10.1126/science.279.5352.853. [DOI] [PubMed] [Google Scholar]
- 58.Ma Y, Lu H, Schwarz K, Lieber MR. Repair of Double-Strand DNA Breaks by the Human Non-Homologous End Joining Pathway. Cell Cycle. 2005;4:1193–1200. doi: 10.4161/cc.4.9.1977. [DOI] [PubMed] [Google Scholar]
- 59.Dion V, Lin Y, Price BA, Fyffe SL, Seluanov A, Gorbunova V, Wilson JH. Genome-wide demethylation promotes trinucleotide repeat instability independently of homologous recombination. DNA Repair. 2007 doi: 10.1016/j.dnarep.2007.11.002. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Macaluso M, Cinti C, Russo G, Russo A, Giordano A. pRb2/p130-E2F4/5-HDAC1-SUV39H1-p300 and pRb2/p130-E2F4/5-HDAC1-SUV39H1-DNMT1 multimolecular complexes mediate the transcription of estrogen receptor-alpha in breast cancer. Oncogene. 2003;22:3511–3517. doi: 10.1038/sj.onc.1206578. [DOI] [PubMed] [Google Scholar]
- 61.Fuks F, Hurd PJ, Deplus R, Kouzarides T. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 2003;31:2305–2312. doi: 10.1093/nar/gkg332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smallwood A, Esteve PO, Pradhan S, Carey M. Functional cooperation between HP1 and DNMT1 mediates gene silencing. Genes Dev. 2007;21:1169–1178. doi: 10.1101/gad.1536807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kimura H, Shiota K. Methyl-CpG-binding protein, MeCP2, is a target molecule for maintenance DNA methyltransferase, Dnmt1. J. Biol. Chem. 2003;278:4806–4812. doi: 10.1074/jbc.M209923200. [DOI] [PubMed] [Google Scholar]
- 64.Schermelleh L, Haemmer A, Spada F, Rosing N, Meilinger D, Rothbauer U, Cardoso MC, Leonhardt H. Dynamics of Dnmt1 interaction with the replication machinery and its role in postreplicative maintenance of DNA methylation. Nucleic Acids Res. 2007;35:4301–4312. doi: 10.1093/nar/gkm432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chuang LS, Ian HI, Koh TW, Ng HH, Xu G, Li BF. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science. 1997;277:1996–2000. doi: 10.1126/science.277.5334.1996. [DOI] [PubMed] [Google Scholar]
- 66.Reale A, Matteis GD, Galleazzi G, Zampieri M, Caiafa P. Modulation of DNMT1 activity by ADP-ribose polymers. Oncogene. 2005;24:13–19. doi: 10.1038/sj.onc.1208005. [DOI] [PubMed] [Google Scholar]
- 67.Chen T, Hevi S, Gay F, Tsujimoto N, He T, Zhang B, Ueda Y, Li E. Complete inactivation of DNMT1 leads to mitotic catastrophe in human cancer cells. Nat. Genet. 2007;39:391–396. doi: 10.1038/ng1982. [DOI] [PubMed] [Google Scholar]
- 68.Mortusewicz O, Schermelleh L, Walter J, Cardoso MC, Leonhardt H. Recruitment of DNA methyltransferase I to DNA repair sites. Proc. Natl. Acad. Sci. U. S. A. 2005;102:8905–8909. doi: 10.1073/pnas.0501034102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Trinh BN, Long TI, Nickel AE, Shibata D, Laird PW. DNA methyltransferase deficiency modifies cancer susceptibility in mice lacking DNA mismatch repair. Mol. Cell. Biol. 2002;22:2906–2917. doi: 10.1128/MCB.22.9.2906-2917.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang KY, James Shen CK. DNA methyltransferase Dnmt1 and mismatch repair. Oncogene. 2004;23:7898–7902. doi: 10.1038/sj.onc.1208111. [DOI] [PubMed] [Google Scholar]
- 71.Kim M, Trinh BN, Long TI, Oghamian S, Laird PW. Dnmt1 deficiency leads to enhanced microsatellite instability in mouse embryonic stem cells. Nucleic Acids Res. 2004;32:5742–5749. doi: 10.1093/nar/gkh912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guo G, Wang W, Bradley A. Mismatch repair genes identified using genetic screens in Blm-deficient embryonic stem cells. Nature. 2004;429:891–895. doi: 10.1038/nature02653. [DOI] [PubMed] [Google Scholar]
- 73.Gomes-Pereira M, Foiry L, Gourdon G. Transgenic mouse models of unstable trinucleotide repeats: toward an understanding of disease-associated repeat size mutation. In: Wells RD, Ashizawa T, editors. Genetic instabilities and neurological diseases. second edition ed. Academic Press; 2006. pp. 563–586. [Google Scholar]
- 74.Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol. Cell. Biol. 2003;23:5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Trasler J, Deng L, Melnyk S, Pogribny I, Hiou-Tim F, Sibani S, Oakes C, Li E, James SJ, Rozen R. Impact of Dnmt1 deficiency, with and without low folate diets, on tumor numbers and DNA methylation in Min mice. Carcinogenesis. 2003;24:39–45. doi: 10.1093/carcin/24.1.39. [DOI] [PubMed] [Google Scholar]
- 76.Oakes CC, La Salle S, Smiraglia DJ, Robaire B, Trasler JM. A unique configuration of genome-wide DNA methylation patterns in the testis. Proc. Natl. Acad. Sci. U. S. A. 2007;104:228–233. doi: 10.1073/pnas.0607521104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Waterland RA, Lin JR, Smith CA, Jirtle RL. Post-weaning diet affects genomic imprinting at the insulin-like growth factor 2 (Igf2) locus. Hum. Mol. Genet. 2006;15:705–716. doi: 10.1093/hmg/ddi484. [DOI] [PubMed] [Google Scholar]
- 78.McGill BE, Bundle SF, Yaylaoglu MB, Carson JP, Thaller C, Zoghbi HY. Enhanced anxiety and stress-induced corticosterone release are associated with increased Crh expression in a mouse model of Rett syndrome. Proc. Natl. Acad. Sci. U. S. A. 2006;103:18267–18272. doi: 10.1073/pnas.0608702103. [DOI] [PMC free article] [PubMed] [Google Scholar]