Abstract
Nonnucleoside reverse transcriptase inhibitors (NNRTIs) play a central role in the treatment of AIDS, but their mechanisms of action are incompletely understood. The interaction of the NNRTI nevirapine (NVP) with HIV-1 reverse transcriptase (RT) is characterized by a preference for the open conformation of the fingers/thumb subdomains, and a reported variation of three orders of magnitude between the binding affinity of NVP for RT in the presence or absence of primer/template DNA. To investigate the relationship between conformation and ligand binding, we evaluated the use of methionine NMR probes positioned near the tip of the fingers or thumb subdomains. Such probes would be expected to be sensitive to changes in the local environment depending on the fractions of open and closed RT. Comparisons of the NMR spectra of three conservative mutations, I63M, L74M, and L289M, indicated that M63 showed the greatest shift sensitivity to the addition of NVP. The exchange kinetics of the M63 resonance are fast on the chemical shift timescale, but become slow in the presence of NVP due to the slow binding of RT with the inhibitor. The simplest model consistent with this behavior involves a rapid open/closed equilibrium coupled with a slow interaction of the inhibitor with the open conformation. Studies of RT in the presence of both NVP and MgATP indicate a strong negative cooperativity. Binding of MgATP reduces the fraction of RT bound to NVP, as indicated by the intensity of the NVP-perturbed M230 resonance, and enhances the dissociation rate constant of the NVP, resulting in an increase of the open/closed interconversion rate, so that the M63 resonance moves into the fast/intermediate-exchange regime. Protein-mediated interactions appear to explain most of the affinity variation of NVP for RT.
Introduction
HIV reverse transcriptase (RT), the enzyme responsible for converting viral genomic RNA into proviral double-stranded DNA, is a primary target for drug intervention. The enzyme exists as a p66/p51 heterodimer, with the catalytic sites for both the polymerase and ribonuclease H activities located in the p66 subunit. The p51 subunit includes the polymerase domain in an alternative, inactive fold, as well as a short, inactive fragment of the ribonuclease H domain. The polymerase activity of RT can be inhibited by both nucleoside and nonnucleoside RT inhibitors (NNRTIs); however, the mechanisms of action of the NNRTIs in particular are incompletely understood. Several different inhibitory mechanisms have been discussed in the literature (1–6) and involve positional distortions of catalytic residues and/or the primer grip hairpin that positions the primer terminus of the substrate (1,7), and reduced mobility of the thumb subdomain. NNRTIs can also reduce the processivity of the polymerase by interfering with the ability of the fingers/thumb to clamp onto the DNA (8,9). Apo RT exhibits a preference for a conformation in which the fingers/thumb subdomains of the polymerase adopt a closed conformation, whereas enzyme activity requires separation of these two subdomains to accommodate the substrate (Fig. 1). Each of the clinically important NNRTIs—nevirapine (NVP), efavirenz, delavirdine, etravirine, and rilpivirine—has been demonstrated to bind to a hyperextended conformation in which the thumb is positioned farther from the fingers subdomain than is the case in the RT-primer/template (RT-P/T) complex (10–15), although a recently described class of NNRTIs binds to RT with the fingers/thumb in a closed orientation (16). In general, the NNRTI-binding regions of RT-P/T and RT-NNRTI structures do not superimpose well, and thus the nature of the quaternary complexes, whose existence is supported by kinetic data, has been unclear. A recent crystallographic characterization of a ternary RT-P/T-NVP complex by Das et al. (5) indicated a dominant effect of NVP on the NNRTI-binding pocket, such that the position of the primer terminus is significantly altered relative to the RT-P/T complex. However, the authors were unable to capture a quaternary RT-P/T-NVP-dNTP complex.
Figure 1.
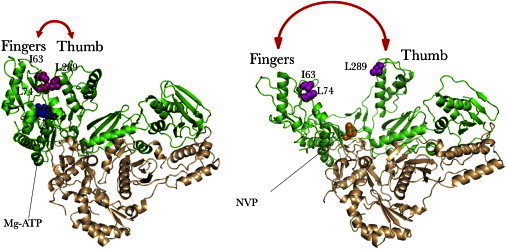
Ribbon diagrams of the RT-ATP complex (PDB code: 2IAJ) with fingers/thumb in a closed position (left panel) and the RT-NVP complex (PDB code: 1VRT) with fingers/thumb in an open orientation (right panel). Green, p66 subunit; beige, p51 subunit; magenta spheres, side chains of mutated residues I63, L74, and L289; blue spheres, MgATP ligand; orange spheres, NVP ligand.
Given the clinical significance of NVP, the reported RT-binding data are surprisingly variable. A Kd value of ∼0.02 μM has been reported for dissociation of NVP from the ternary RT-P/T-NVP complex (3), whereas a Kd value of ∼30 μM has been reported for dissociation of the RT-NVP binary complex (17), implying a >103-fold, substrate-dependent binding affinity. In sharp contrast to these results, Maga et al. (18) reported an NVP-RT dissociation constant of 0.4 μM that is essentially independent of the presence of P/T or P/T-dNTP. As noted above, crystal structure data for RT⋅NNRTI complexes show an open conformation of the fingers/thumb subdomains. Because the fingers/thumb orientation of the apo enzyme adopts a predominantly closed conformation (19), and the RT-P/T conformation has an open conformation, a P/T-induced increase in NVP affinity could be related to this conformational feature. In addition to the effect of P/T, a considerably smaller antagonistic effect of Mg2+ on NNRTI binding was reported by Spence et al. (3). Although the dNTP and NNRTI binding sites do not overlap, they are adjacent and the binding interactions induce structural changes in the methionine-containing β-hairpins that contain the active-site YMDD motif and the primer grip (20,21). Further, the crystal structure of the RT-MnATP complex reveals a closed conformation (20), whereas the structure of the RT-NVP complex adopts an open conformation (10). These structural data also support the possibility of protein-mediated interactions between dNTP and NVP binding.
The high internal mobility/low order parameter of methionine, coupled with its low copy number and three equivalent methyl protons, makes it useful as a probe for investigating the conformational behavior of high-molecular-weight proteins such as HIV RT (molecular mass = 117 kDa) (22–26). In this study, we adopted a strategy of identifying positions that were likely to provide useful information on the relative orientations of the fingers/thumb subdomain. Three such positions at the tips of the fingers and thumb subdomains that could be conservatively mutated to methionine were evaluated. Sensitivity to the conformational change was evaluated by investigating the shift sensitivity to NVP. Because the NVP-binding site is remote from each of the mutated positions, and the mutated residues were all solvent exposed, any shift perturbations that resulted from NVP binding were interpreted as a consequence of changes in the local environment of the methyl groups due to a variation in the relative domain orientation. The I63M mutation was found to be most useful for this purpose. This mutant was then used to study the interactions of RT with NVP and MgATP, and with a previously studied thumb-derived peptide that was reported to exhibit high affinity and preferential binding to the open RT conformation (27).
Materials and Methods
Labeled RT, mutations, and interacting peptide
HIV-1 RT labeled with [methyl-13C]methionine in the p66 subunit was expressed in Escherichia coli BL21 (DE3) codon plus RIPL cells as described previously (21). The p66 and p51 subunits were expressed in parallel so that independent labeling patterns could be introduced, and the cell pellets were combined during lysis. Both subunits contained the C280S mutation for enhanced resistance to oxidation, and the p66 subunit contained an additional M357K mutation to eliminate the intense M357 resonance that obscures other methionine residues (21). M357 appears to play no significant role in the activity of the enzyme and in viral isolates is often replaced nonconservatively with other residues (28,29). The C280S51,66 and M357K66 mutations are present in all of the constructs used in this study and are not explicitly noted.
Additional mutations of I63, L74, and L289 to methionine were also introduced as indicated. All mutations were introduced by using the QuikChange XL site-directed mutagenesis kit (Agilent, Santa Clara, CA), and transformed into BL21(DE)3 codon plus RIL. The purified [methyl-13C]methionine RT constructs were exchanged into NMR buffer (20 mM Tris-HCl-d11 in D2O, pD (uncorrected pH meter reading + 0.42) = 7.42, 150 mM KCl, 4mM MgCl2, 0.02% NaN3) by repeated centrifugation. NMR studies were performed at 25°C unless otherwise specified.
A 17-residue peptide derived from the thumb subdomain of RT, P1L (acetyl-G285TKALTEVIPLTEEAEL301), was obtained from Pi Proteomics (Huntsville, AL) and used without further purification. This sequence is similar to the peptide P1 studied by Agopian et al. (27) except that it contains the naturally occurring Leu-301 rather than the mutated L301C substitution present in peptide P1.
NMR methods
1H-13C heteronuclear single quantum coherence (HSQC) spectra were obtained using the gChsqc experiment in Biopack (30,31) and performed on a Varian Unity Inova 500 MHz NMR spectrometer equipped with a 5 mm Varian (500 MHz) 1H{13C,15N} triple-resonance cryogenically cooled probe with actively shielded z-gradients. The acquisition parameters for all experiments were as follows: 64 transients, 64 ms acquisition time, 1 s relaxation delay with 1024 complex points, and spectral width of 14 ppm. In the indirect dimension, 128 complex points were acquired with a spectral width of 10 ppm and the 13C offset was set to 17 ppm. All NMR data were processed using NMRPipe (32) and analyzed by NMRViewJ (33).
Measurements of the conformational exchange dynamics of the Met63 resonance were assessed with a Carr-Purcell-Meiboom-Gill (CPMG) experiment at 18.8 T (34). The CPMG data were fit with various exchange equations and tested for statistical significance as described by Tollinger et al. (35).
Data analysis
The apparent equilibrium constant for NVP was determined by fitting the observed resonance intensities to the relation
| (1) |
using the nonlinear least-squares fitting routine in Mathematica (Wolfram Research, Champaign, IL). The parameter c1, with 0 < c1 < 1, was introduced to allow for the possibility of high-affinity binding to a small amount of the open conformation of apo RT.
The lineshape of Met-184 in the 1H dimension exhibited characteristics of a two-site, intermediate-exchange process. The lineshape was analyzed using the two-site exchange formalism of Rogers and Woodbrey (36) with a program written in Mathematica.
Results
To create an RT variant containing a methionine residue that would be useful for analyzing the open/closed orientation of the fingers/thumb subdomains, we identified three candidate residues for mutation to methionine: I63, L74, and L289. The residues had to meet the following criteria: 1), aliphatic side chains so that substitution with methionine would introduce minimal structural perturbation; 2), substantial surface exposure of the side chain so that the corresponding methyl resonance would be sensitive to changes in local environment related to domain interactions; and 3), location near the tips of the fingers or thumb subdomain in positions such that the local environment would depend on the relative fingers/thumb orientation (Fig. 1). 1H-13C HSQC spectra of the [methyl-13C]methioinine66RT(I63M66), RT(L74M66), and RT(L289M66) mutants were obtained in the presence and absence of saturating NVP, and only the first of the listed mutations resulted in well-resolved resonances (Fig. 2 and Fig. S1 in the Supporting Material). Other resonance perturbations resulting from the addition of NVP followed the pattern described previously in the absence of the additional mutations (21). In particular, the M184 resonance undergoes a downfield 1H shift of ∼0.1 ppm, whereas M230 appears at δ1H,13C = (1.81, 15.97) ppm in the presence of NVP. These perturbations are consistent with the proximity of the NVP-binding site to the YMDD-containing hairpin and the primer grip hairpin that contain these two methionine residues (21).
Figure 2.
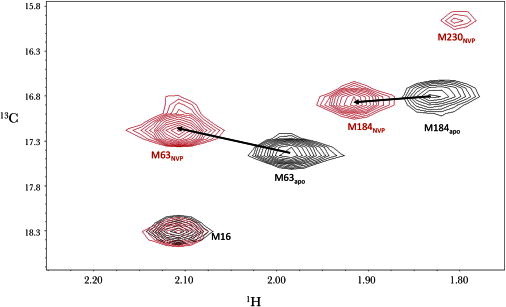
1H-13C HSQC spectra of methionine-labeled RT. (a) Superimposed 1H-13C HSQC spectra of 50 μM [13CH3-Met]66RT(I63M) in the absence (black) and presence (red) of 200 μM NVP. The sample was prepared in NMR buffer (20 mM Tris-HCl-d11 in D2O, pD = 7.42, 150 mM KCl, 4 mM MgCl2, 0.02% NaN3). Spectra were measured at 25°C.
In contrast to the NVP-binding-induced perturbations of the M184 and M230 resonances, which correspond to residues located near the NVP-binding site, I63 is located ∼30 Å away from bound NVP, and its shift is presumably dominated by the change of its local environment that results from the increased proximity to the thumb subdomain.
One significant difference relative to our previous study (21) is the absence of the M230 resonance for apo RT, which previously was thought to be obscured by the large M357 resonances. An analysis of molecular models indicated that in the closed form of RT, M230 has a very low solvent exposure and should produce a very broad, probably unobservable resonance, whereas in the open conformation, M230 has a considerably higher solvent exposure and should produce an observable resonance (21). The weakness of this resonance presumably results from the high fraction of apo RT in the closed conformation (see below), and may also result from exchange broadening between open and closed conformations. Regardless, the intensity of this resonance is more readily assessed in the M357K mutant studied here.
Titration of RT with NVP
To interpret the results of the NMR study, it is first necessary to determine how the timescale of the open ↔ closed transition compares with the chemical shift of the I63M reporter group, i.e., whether the conformational exchange is fast, slow, or intermediate on the NMR timescale. A previous electron paramagnetic resonance (EPR) study of spin-labeled RT determined that RT behaves as a bistable system, spending most of its time in conformations with the fingers/thumb orientation in either the open or closed position (19). Thermodynamic parameters relating the two states, ΔH = 34.5 ± 3 kJ/mol and ΔS = 134 ± 10 J/mol-°K, were obtained from temperature-dependent studies. Based on these parameters, the enthalpically favored open conformation predominates at low temperature, and the entropically favored closed conformation predominates at higher temperature. The fractional open and closed populations are then described by the following relations:
| (2) |
Using the ΔH and ΔS values of (19), the fractional population of the open species decreases with increasing temperature, corresponding to ∼10% at 25°C.
We obtained 1H-13C HSQC spectra of 180 μM [13CH3-Met]66 RT(I63M66) to determine whether we could observe a separate M63 resonance due to the open conformation similar to that observed in the presence of NVP (Spectrum S2 in the Supporting Material). No separate resonance corresponding to the open conformation was observed. Since the signal/noise ratio at the higher RT concentration should have been sufficient to allow observation of a resonance with ∼10% the intensity of the larger peak, this result indicates that the interconversion barrier for the open/closed orientation is low enough to ensure that the equilibrium is fast on the chemical shift timescale (τ ≪ .01 s).
The labeled RT was subsequently titrated with NVP (Fig. 3). Although the open/closed exchange is fast, the NVP binding is slow, and thus separate resonances for both M184 and M63 are observed for the predominantly closed (unbound) and predominantly open (bound) forms of RT. In addition to structural evidence, the shift values δ(1H,13C) = (2.1,17.1) ppm for M63NVP are close to those expected for an unstructured residue (37), consistent with exposure to the solvent. As illustrated in Fig. 1, Ile63 is expected to have a largely solvent-exposed local environment in the open conformation. Based on this behavior and the available structural data, the simplest model consistent with the NMR data is given by Eq. 3:
| (3) |
In particular, since the RT⋅NVP complex is observed with RT in the open conformation, the NVP apparently binds primarily with the rare open conformer, rather than with the predominant closed conformer. The data for M184 or M63 can be fit to determine an apparent NVP-RT dissociation constant. A fit of the higher-sensitivity M63 data yield an apparent KD = 4.0 μM (Fig. 3 h). We note that although there was no observable peak at the shift of the M63 resonance corresponding to the open conformation in the absence of NVP, the data fit yielded an intercept of .08, consistent with the expectation of ∼10% in the open form according to Eq. 2. The value of 4.0 μM is nearly an order of magnitude tighter than the binding constant obtained by capillary electrophoresis at 37°C (17), but at least part of this discrepancy results from the higher temperature of that measurement, which favors the open conformer (viz. Eq. 2).
Figure 3.
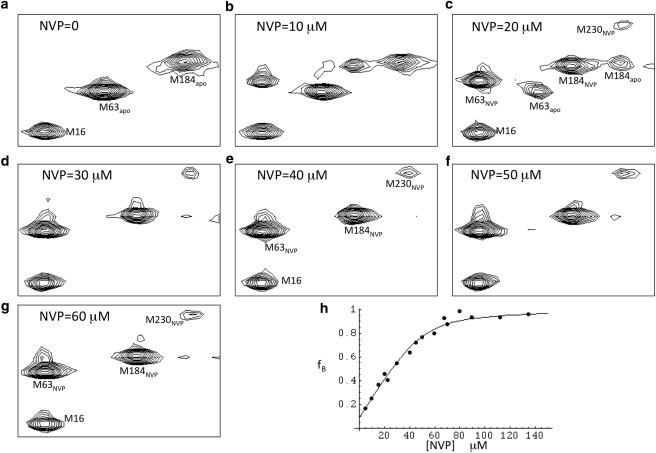
Titration of labeled RT with NVP. (a) 1H-13C HSQC spectra of 50 μM [13CH3-Met]66 RT(I63M) as a function of added NVP. (a–g) NVP concentrations in spectra are 0, 10, 20, 30, 40, 50, and 60 μM, respectively. (h) Fit of all NVP titration data. Sample buffer and temperature as described in Fig. 2.
The apparent dissociation constant obtained using the approach outlined above can be related to an intrinsic dissociation constant for RT in the open conformation using the formalism described by Vallée-Bélisle et al. (38). If it is assumed that NVP interacts exclusively with the open conformation, we can relate the two binding constants according to
| (4) |
where KDapp is the apparent dissociation constant observed for the conformational mixture, KDint is the intrinsic dissociation constant that describes binding to the open conformation, KS corresponds to the open/closed equilibrium constant (i.e., KS = [open]/[closed]), and fopen is the fraction of RT in the open conformation in the absence of NVP. Using Eq. 2 for fopen at 25°C gives KDint = 0.1 KDapp or 0.4 μM. This binding is approximately an order of magnitude weaker than the dissociation constants obtained by Spence et al. (3), which were measured in the presence of primer/template. It thus appears that the effect of the P/T is only partially explained by an increase in the open/closed ratio. This result is not surprising since both the P/T substrate and NVP interact strongly with the primer grip hairpin.
Dynamic characterization by CPMG
Based on the conclusion of fast exchange between the open and closed conformations, we subsequently attempted to characterize the open/closed RT ratio from the temperature dependence of the M63 shift (Fig. S3). However, the total 1H shift is rather small, <0.2 ppm, and the change in the fraction of open species over the available temperature range is only ∼15%. Thus, the total predicted 1H shift change is only ∼.03 ppm, which limits the feasibility of this type of analysis. Fast dynamic events involving equilibria with low-concentration states, such as those that appear to characterize the open/closed interconversion in apo RT, can often be characterized with the use of CPMG studies (see Materials and Methods). At 18.8 T and 25°C, there was a sufficient signal/noise ratio to observe a typical fast-exchange relaxation dispersion curve for M63 (Fig. 4 a). Fitting of the data revealed an exchange lifetime of ∼1.0 ms. Assuming that the chemical shift difference between the open and closed states is approximated by Δ13C = 0.23 ppm (the measured shift difference between the apo and NVP-bound M63 resonances (Fig. 1)), the CPMG analysis gives a 12% population for the open state. This is reasonably consistent with the value of 10% obtained using Eq. 2 and the ΔH and ΔS values given by Kensch et al. (19). Fixing both Δν and the fraction of the open conformation allows a Monte Carlo estimation of the error in τ of 0.8 ms (35). In contrast, a CPMG study of M63 in the RT-NVP complex showed no evidence of exchange on this timescale (Fig. 4 b), consistent with a rate that is limited by the slow dissociation of NVP from RT (Eq. 3). Attempts to include the initial, 50 Hz point yielded physically unreasonable parameters.
Figure 4.
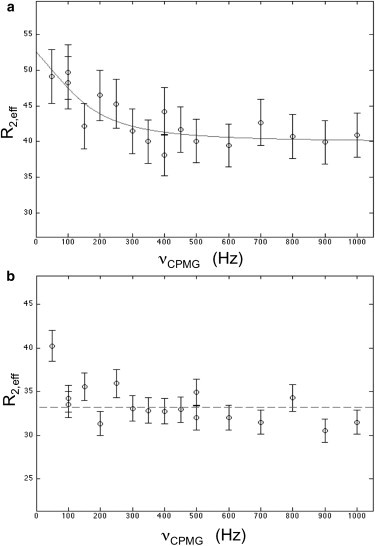
CPMG analysis of the Met63 resonance in [13CH3-Met]RT(I63M). Measurements were made on (a) 50 μM [13CH3-Met]RT(I63M) in the NMR buffer at 25°C or (b) 50 μM [13CH3-Met]RT(I63M) plus 200 μM NVP and lacking the Mg2+ in the buffer. The first data point cannot be meaningfully analyzed within the context of the other parameter restraints. Other than the Mg2+ concentration, the sample buffer is the same as described in Fig. 2. All measurements were made at 18.8 T.
Effect of MgATP
MnATP has been shown to interact with the nucleotide-binding site of RT and to induce a conformation of the active-site YMDD loop that superimposes with the conformation observed in the RT⋅P/T⋅dNTP ternary complex (20,39). Hence, although the nucleotide conformations differ, particularly regarding the position of the base, their effects on enzyme conformation are apparently similar. We investigated RT-mediated interactions between MgATP and NVP by adding millimolar concentrations of MgATP to a sample containing 50 μM [methyl-13C]methionine66 RT(I63M) plus 200 μM NVP. Surprisingly, we found that MgATP largely reversed the shift perturbations produced by the NVP (Fig. 5 a). Thus, both the M184 and M63 resonances shift back toward their positions in the apo enzyme. The shifted M230 resonance, with δ13C ∼16 ppm characteristic of the RT⋅NVP complex, also experiences an additional shift perturbation in the presence of the MgATP, as well as a reduction in intensity (Fig. 5 b). The methyl resonance from uncomplexed NVP can also be observed at δ1H,13C = (2.38,19.6) ppm in Fig. 5 b (see also Spectrum S4 in the Supporting Material). Because the methyl group is unlabeled, the resonance corresponding to bound NVP, which is expected to be shifted significantly and in slow exchange, is not observed. In contrast to the slow-exchange behavior apparent in Fig. 3, the presence of both MgATP and NVP leads to a fast/intermediate-exchange behavior for M63, so that the shift position rather than the intensity ratio between the alternate conformations is seen to vary. This result indicates that the interconversion rate between the open and closed forms of RT in the presence of both ligands is increased relative to the rate in the presence of NVP only. Thus, the binding of MgATP enhances the dissociation rate constant of NVP.
Figure 5.
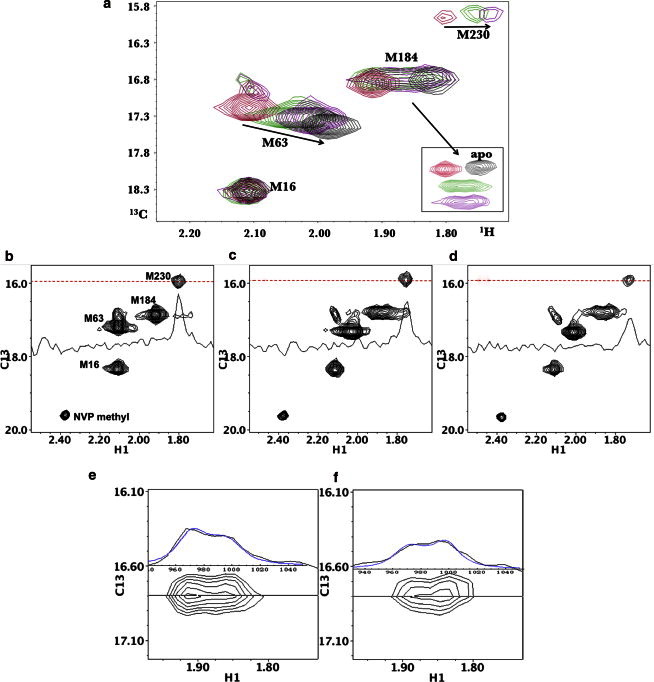
Effect of MgATP on NVP-RT(I63M). (a) 1H-13C HSQC spectra of [13CH3-Met]66 RT(I63M66) in the presence of 4 mM MgCl2 (black), after addition of 0.2 mM NVP (red), after addition of 4 mM MgATP (green), and after a second addition of 4 mM MgATP (8 mM MgATP total concentration; purple). Inset at lower right shows the M184 resonances of the same series of samples with additional 13C offsets for clarity. The arrows indicate the direction of the M63 and M230 shift perturbations with increasing MgATP. The sample was prepared in 20 mM Tris-HCl-d11 in D2O, pD 7.42, and the spectra were obtained at 25°C. (b–d) Individual spectra obtained in the presence of (b) 0.2 mM NVP, (c) 0.2 mM NVP + 4 mM MgATP, and (d) 0.2 mM NVP + 8 mM MgATP are shown along with 1D slices through δ13C = 15.97 ppm, illustrating the intensity changes of the M230 resonance as the MgATP is added. (e and f) shows the 1H-13C HSQC spectrum of M184 for the sample containing 0.2 mM NVP +4 mM MgATP (in e) or 8 mM MgATP (in f), as well as the 1D 1H slice through δ13C = 16.8 ppm. The 1D 1H slices are superimposed with simulations (blue) determined using a two-site exchange formalism with T2A = T2B = 0.02 s, Δν = 0.063 ppm × 500 Hz/ppm, τB = 0.03 s, and pB = 0.46 (e) or 0.53 (f).
Using a fast-exchange formalism to analyze the M63 1H shift behavior, the fraction of RT in the open conformation can be calculated based on the M63 shift according to
| (5) |
where the value of δopen corresponds to the shift observed in the presence of NVP, and the value for the closed shift was determined on the basis of Eq. 1, according to which the shift observed for apo RT at 25°C corresponds to 90% in the closed conformation. Based on the observed shift values, we obtain fopen = 43% and 32% at 4 mM and 8 mM MgATP, respectively.
As noted above, addition of MgATP reduces the intensity and perturbs the chemical shift of the M230 resonance. Peak volume measurements indicate that the fraction of bound NVP is reduced to 51% and 35% in the presence of 4 and 8 mM MgATP, respectively. These values are similar to the fopen values obtained above from examination of the M63 shift. It thus appears that the addition of MgATP reduces the binding affinity of RT for NVP enough to ensure that a substantial fraction of the RT has no bound NVP, despite a total NVP concentration of 200 μM, and that in the absence of bound NVP, the RT adopts a primarily closed conformation. However, in addition to this effect, there is also substantial formation of an open, ternary RT⋅NVP⋅MgATP complex present in solution, as indicated by the behavior of the M230 resonance. Since it appears that a ternary RT⋅NVP⋅MgATP complex can form, a general equilibrium scheme (Scheme 1) for the interactions of MgATP and NVP with RT can be written as:
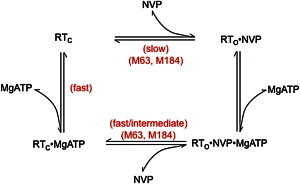
In the above scheme, the subscripts indicate that, based on our evaluations of the M63 shift behavior, the uncomplexed RT and RT⋅MgATP is predominantly closed but can adopt either conformation, whereas the RT⋅NVP and RT⋅NVP⋅MgATP complexes are predominantly open. The fast/slow/intermediate rates are defined by the shift differences of the RT resonances that report on the formation/dissociation of these complexes. Of course, although in the crystal the binary RT⋅MnATP complex RT adopts a closed conformation (PDB code: 2IAJ), MgATP must also bind to the open form of RT in the presence of primer/template to form the catalytic ternary complex.
The lineshape observed for the M184 resonance in the presence of both MgATP and NVP exhibits intermediate-exchange behavior (Fig. 5). The 1H spectrum of M184 is characteristic of an intermediate-exchange process. Based on the concentrations of NVP and MgATP present in the sample and the shift behavior of the M63 resonance, the intermediate exchange most likely corresponds to the equilibrium between the RTO⋅NVP⋅MgATP and RTC⋅MgATP complexes at the bottom in the above scheme. At the NVP concentration used in the study, the fraction of uncomplexed RT is expected to be quite small. The lineshape can be reasonably described with a two-site exchange formalism (36), with parameters T2A,B = .02 s. This provides a reasonable description of the observed linewidths in the absence of added MgATP, Δν = .063 ppm, corresponding to the M184 shifts of the RT⋅MgATP and RT⋅NVP⋅MgATP complexes, the lifetime of the ternary complex, τNVP⋅MgATP = 30 ms, and the fractions of ternary complex pNVP⋅MgATP = 0.46 and 0.53 at the two MgATP concentrations (Fig. 5, e and f). Alternatively, we obtained a significantly poorer fit if we tried to constrain the two resonance positions to the M184 shifts observed for the apo and NVP complex of RT, for which Δν = .09. This suggests that the apo enzyme does not contribute significantly to the observed exchange behavior.
Effect of the thumb-derived peptide P1L
To investigate the effects of peptides derived from the RT sequence as possible dimerization inhibitors, Divita and co-workers (27) evaluated the peptide P1, derived from residues 285–301, with an additional L301C substitution. They reported that P1 interacted only with the RT heterodimer with an apparent Kd = 7.5 μM, and exhibited a fivefold preference for the open fingers/thumb conformation. We evaluated the effect of P1L (lacking the L301C) substitution on [methyl-13C]methionine66RT(I63M). Our expectation was that at a saturating concentration of P1L, we should observe M63 resonances at both the open and closed M63 resonances with an intensity ratio of ∼5:1. Alternatively, if the peptide exchange is rapid on the NMR timescale, the M63 resonance should exhibit an intermediate shift proportionate to the fractions of open and closed species present. Addition of 0.8 mM P1L to apo-RT produced a very small shift of the M184 resonance but did not affect M16 or I63M (Fig. 6). The concentration used in the NMR study is 100-fold above the Kd value of 7.5 μM reported for P1 (27).
Figure 6.
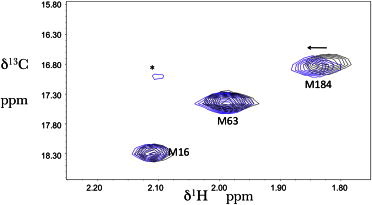
Effect of P1L on the 1H-13C HSQC spectrum of RT(I63M). 1H-13C HSQC spectra of 50 μM [13CH3-Met]66RT(I63M66) in the absence (black) and presence (blue) of 0.8 mM P1L. The resonance marked with an asterisk is unassigned but is sufficiently near the random coil values to suggest the presence of a small amount of unfolded protein. The arrow indicates the direction of the shift perturbation resulting from the P1L addition. The sample was prepared in NMR buffer (20 mM Tris-HCl-d11 in D2O, pD = 7.42, 150 mM KCl, 4 mM MgCl2 0.02% NaN3) and measured at 25°C.
Discussion
HIV RT is a conformationally complex molecule, and characterization of its behavior is fundamental to understanding mechanisms of catalysis and inhibition (1,4,40,41). Crystallographic analyses have demonstrated that the structure of the NNRTI-binding site and the orientation of the fingers/thumb subdomains are highly variable. Information on the fingers/thumb conformation in solution has been derived from EPR studies (19) and small-angle x-ray scattering analysis (42). In this study, we evaluated the introduction of a methionine residue to probe the fingers/thumb conformation of HIV-1 RT. Only one of the mutations evaluated, I63M, exhibited sufficient shift sensitivity to serve as a useful conformational indicator. Using the methionine probe-labeled RT, we were able to evaluate the effects of various ligands on the fingers/thumb conformation.
Based on a comparison with predictions of the fractions of open and closed conformers derived from the reported thermodynamic parameters (19), the NMR analysis indicates that interconversion of the open/closed forms is occurring on a fast timescale. The I63M methyl 1H shift difference of ∼0.2 ppm between open and closed conformers corresponds to an interconversion rate ≫ s−1. This conclusion is consistent with the results of molecular-dynamics simulations (14,43), which show large motions of the thumb subdomain on a submillisecond timescale. In contrast to the rapid dynamics observed for the apo enzyme, a slow conformational exchange between open and closed forms of RT is observed in the presence of the NNRTI NVP. This result is clearly indicative of the slow timescale of NVP binding and dissociation; therefore, based on the above criterion, the dissociation of the RT⋅NVP complex has a rate constant ≪ 200 s−1. Although the dissociation rate constants determined by Spence et al. (3) and Maga et al. (44) were obtained under somewhat different conditions, they are consistent with this conclusion.
Our analysis of the closed conformation in five crystallographic structures (1DLO, 1QE1, 1HMV, 2IAJ, and 3DLK) demonstrates proximity of the fingers and thumb subdomains, but also indicates substantial variability. This may indicate the absence of strong fingers/thumb interface interactions, or result from crystal-packing interactions, from ligands present in the crystal (e.g., 2IAJ) or from the mutations that are present (e.g., F160S in structure 3DLK). The small shifts we observed for each of the three substituted methionine residues at positions 63, 74, and 289 suggest that residues at the tips of the fingers and thumb subdomains are in sufficient proximity to experience small chemical shift perturbations, but do not form a very close and stable interface.
The studies presented here provide direct evidence for RT-mediated interactions between the nucleotide- and NNRTI-binding sites. Addition of MgATP to the RT⋅NVP complex altered the behavior of the M63 resonance so that the position rather than the intensities of M63 resonances corresponding to the open and closed conformations was altered. This result indicates that in the presence of MgATP, the dissociation of bound NVP no longer falls into the slow-exchange limit, as monitored by M63, but is increased sufficiently to move the M63 exchange behavior into the fast/intermediate regime. This more-rapid dissociation rate of NVP is also accompanied by a reduced level of NVP binding, as indicated by the reduced intensity of the NVP-perturbed M230 resonance. Thus, MgATP significantly interferes with NVP binding to the apo enzyme.
The observed antagonism of MgATP and NVP binding is consistent with studies demonstrating that Mg2+ reduces the affinity of RT for nonnucleoside inhibitors, although the effect reported for NVP was rather small (3). Antagonism between divalent ion and NNRTI binding was also deduced by Das et al. (20) on the basis of structural studies. They found that although manganese, ATP, and an NNRTI (HBY097) were present in the crystallization buffers, only the RT⋅MnATP and RT⋅NNRTI binary complexes crystallized; no ternary complexes were obtained. They further noted that in the MnATP complex, the active-site YMDD loop interacts directly with the MnATP ligand and is displaced, adopting an extended conformation that differs from its conformation in the NNRTI complex. The NNRTI restricts the conformational change of the YMDD loop that is needed to attain the metal-binding conformation. Although the ATP conformation in the MnATP RT complex is in poor agreement with the dNTP conformation observed in ternary complexes (e.g., 1RTD (45)), the divalent ions and the active-site carboxyl groups (D110, D185, and D186) superimpose reasonably well in the RT⋅MnATP complex and the RT⋅P/T⋅MgTTP structures (Fig. S5). Thus, the protein-mediated antagonism between MgATP and NVP likely results from conformational incompatibilities similar to those present in the active RT conformation. We note as well that the MgATP concentrations used in this study reasonably approximate estimates of intracellular concentrations (46,47). Alternatively, the structures of RT-NVP (10) and RT-MnATP (20) do not superpose well in the region of the NNRTI-binding site, consistent with the observed protein-mediated antagonism (Fig. S6).
Recent molecular-dynamics simulations demonstrated that the NNRTI efavirenz is capable of binding to RT in a conformation in which the fingers/thumb subdomains adopt a closed orientation (43). The correlation between the fraction of open RT deduced from the M63 shift and the intensity of the shifted M230 resonance supports the conclusion that NVP binding is strongly correlated with the open conformation. However, these results clearly cannot exclude the possibility that a small fraction of the RT⋅NVP complex adopts a closed conformation.
Finally, the NMR studies indicate that peptide P1L, which was previously reported to exhibit both tight binding and a fivefold preference for the open conformation, did not significantly alter the conformation of apo RT at concentration 100-fold greater than the estimated Kd as judged by the spectrum of the methionine-labeled enzyme. Thus, in contrast to NVP, P1L is not able to influence the open/closed conformation of RT.
This study demonstrates the utility of methionine as a minimally perturbing, NMR-sensitive probe for conformational studies of RT and presumably other macromolecules. Labeling of RT with [13CδH3-Ile] reveals that I63 is positioned in a highly congested region of the spectrum (unpublished results) and hence cannot readily provide the information obtained with the methionine-labeled enzyme. This result is not too surprising considering that, as discussed above, we selected a solvent-exposed residue that would likely provide a useful probe for interaction with the thumb domain. It is likely that the use of solvent-exposed probe residues will be inherently limited due to the absence of significant shift perturbations and consequent spectral congestion. For this reason, the use of an alternative residue with a lower copy number, such as methionine, becomes particularly attractive (22–26,48).
Acknowledgments
The authors wish to thank Dr. Tom Kirby and Scott Gabel for thoughtful comments on the manuscript, and the Lewis Kay group for providing the CPMG pulse sequence.
This research was supported by the Intramural National Institutes of Health (NIH) Research Program of the National Institute of Environmental Health Sciences (NIEHS; grant Z01-ES050147 to R.E.L.). E.F.D. is supported by the NIEHS, NIH, under Delivery Order HHSN273200700046U. NVP was obtained from the NIH AIDS Research and Reference Reagent Program.
Supporting Material
References
- 1.Esnouf R., Ren J.S., Stuart D. Mechanism of inhibition of HIV-1 reverse transcriptase by non-nucleoside inhibitors. Nat. Struct. Biol. 1995;2:303–308. doi: 10.1038/nsb0495-303. [DOI] [PubMed] [Google Scholar]
- 2.Rittinger K., Divita G., Goody R.S. Human immunodeficiency virus reverse transcriptase substrate-induced conformational changes and the mechanism of inhibition by nonnucleoside inhibitors. Proc. Natl. Acad. Sci. USA. 1995;92:8046–8049. doi: 10.1073/pnas.92.17.8046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spence R.A., Kati W.M., Johnson K.A. Mechanism of inhibition of HIV-1 reverse transcriptase by nonnucleoside inhibitors. Science. 1995;267:988–993. doi: 10.1126/science.7532321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sluis-Cremer N., Temiz N.A., Bahar I. Conformational changes in HIV-1 reverse transcriptase induced by nonnucleoside reverse transcriptase inhibitor binding. Curr. HIV Res. 2004;2:323–332. doi: 10.2174/1570162043351093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Das K., Martinez S.E., Arnold E. HIV-1 reverse transcriptase complex with DNA and nevirapine reveals non-nucleoside inhibition mechanism. Nat. Struct. Mol. Biol. 2012;19:253–259. doi: 10.1038/nsmb.2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xia Q., Radzio J., Sluis-Cremer N. Probing nonnucleoside inhibitor-induced active-site distortion in HIV-1 reverse transcriptase by transient kinetic analyses. Protein Sci. 2007;16:1728–1737. doi: 10.1110/ps.072829007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacobo-Molina A., Ding J.P., Clark P. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 A resolution shows bent DNA. Proc. Natl. Acad. Sci. USA. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quan Y.D., Liang C., Wainberg M.A. Enhanced impairment of chain elongation by inhibitors of HIV reverse transcriptase in cell-free reactions yielding longer DNA products. Nucleic Acids Res. 1998;26:5692–5698. doi: 10.1093/nar/26.24.5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu S.X., Abbondanzieri E.A., Zhuang X. Slide into action: dynamic shuttling of HIV reverse transcriptase on nucleic acid substrates. Science. 2008;322:1092–1097. doi: 10.1126/science.1163108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ren J.S., Esnouf R., Stammers D. High resolution structures of HIV-1 RT from four RT-inhibitor complexes. Nat. Struct. Biol. 1995;2:293–302. doi: 10.1038/nsb0495-293. [DOI] [PubMed] [Google Scholar]
- 11.Ren J., Milton J., Stammers D.K. Structural basis for the resilience of efavirenz (DMP-266) to drug resistance mutations in HIV-1 reverse transcriptase. Structure. 2000;8:1089–1094. doi: 10.1016/s0969-2126(00)00513-x. [DOI] [PubMed] [Google Scholar]
- 12.Das K., Bauman J.D., Arnold E. High-resolution structures of HIV-1 reverse transcriptase/TMC278 complexes: strategic flexibility explains potency against resistance mutations. Proc. Natl. Acad. Sci. USA. 2008;105:1466–1471. doi: 10.1073/pnas.0711209105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esnouf R.M., Ren J.S., Stuart D.I. Unique features in the structure of the complex between HIV-1 reverse transcriptase and the bis(heteroaryl)piperazine (BHAP) U-90152 explain resistance mutations for this nonnucleoside inhibitor. Proc. Natl. Acad. Sci. USA. 1997;94:3984–3989. doi: 10.1073/pnas.94.8.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ivetac A., McCammon J.A. Elucidating the inhibition mechanism of HIV-1 non-nucleoside reverse transcriptase inhibitors through multicopy molecular dynamics simulations. J. Mol. Biol. 2009;388:644–658. doi: 10.1016/j.jmb.2009.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lansdon E.B., Brendza K.M., Liu X. Crystal structures of HIV-1 reverse transcriptase with etravirine (TMC125) and rilpivirine (TMC278): implications for drug design. J. Med. Chem. 2010;53:4295–4299. doi: 10.1021/jm1002233. [DOI] [PubMed] [Google Scholar]
- 16.Freisz S., Bec G., Ennifar E. Crystal structure of HIV-1 reverse transcriptase bound to a non-nucleoside inhibitor with a novel mechanism of action. Angew. Chem. Int. Ed. Engl. 2010;49:1805–1808. doi: 10.1002/anie.200905651. [DOI] [PubMed] [Google Scholar]
- 17.Chen W.J., Li W.J., Liu J. Study on the interaction between HIV reverse transcriptase and its non-nucleoside inhibitor nevirapine by capillary electrophoresis. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2010;878:1714–1717. doi: 10.1016/j.jchromb.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 18.Maga G., Ubiali D., Spadari S. Selective interaction of the human immunodeficiency virus type 1 reverse transcriptase nonnucleoside inhibitor efavirenz and its thio-substituted analog with different enzyme-substrate complexes. Antimicrob. Agents Chemother. 2000;44:1186–1194. doi: 10.1128/aac.44.5.1186-1194.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kensch O., Restle T., Steinhoff H.J. Temperature-dependent equilibrium between the open and closed conformation of the p66 subunit of HIV-1 reverse transcriptase revealed by site-directed spin labelling. J. Mol. Biol. 2000;301:1029–1039. doi: 10.1006/jmbi.2000.3998. [DOI] [PubMed] [Google Scholar]
- 20.Das K., Sarafianos S.G., Arnold E. Crystal structures of clinically relevant Lys103Asn/Tyr181Cys double mutant HIV-1 reverse transcriptase in complexes with ATP and non-nucleoside inhibitor HBY 097. J. Mol. Biol. 2007;365:77–89. doi: 10.1016/j.jmb.2006.08.097. [DOI] [PubMed] [Google Scholar]
- 21.Zheng X.H., Mueller G.A., London R.E. Solution characterization of [methyl-(13)C]methionine HIV-1 reverse transcriptase by NMR spectroscopy. Antiviral Res. 2009;84:205–214. doi: 10.1016/j.antiviral.2009.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skrynnikov N.R., Mulder F.A.A., Kay L.E. Probing slow timescale dynamics at methyl-containing side chains in proteins by relaxation dispersion NMR measurements: application to methionine residues in a cavity mutant of T4 lysozyme. J. Am. Chem. Soc. 2001;123:4556–4566. doi: 10.1021/ja004179p. [DOI] [PubMed] [Google Scholar]
- 23.DellaVecchia M.J., Merritt W.K., London R.E. NMR analysis of [methyl-13C]methionine UvrB from Bacillus caldotenax reveals UvrB-domain 4 heterodimer formation in solution. J. Mol. Biol. 2007;373:282–295. doi: 10.1016/j.jmb.2007.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Religa T.L., Sprangers R., Kay L.E. Dynamic regulation of archaeal proteasome gate opening as studied by TROSY NMR. Science. 2010;328:98–102. doi: 10.1126/science.1184991. [DOI] [PubMed] [Google Scholar]
- 25.Kofuku Y., Ueda T., Shimada I. Efficacy of the β2-adrenergic receptor is determined by conformational equilibrium in the transmembrane region. Nat. Commun. 2012;3:1045. doi: 10.1038/ncomms2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Butterfoss G.L., DeRose E.F., London R.E. Conformational dependence of 13C shielding and coupling constants for methionine methyl groups. J. Biomol. NMR. 2010;48:31–47. doi: 10.1007/s10858-010-9436-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agopian A., Gros E., Divita G. A new generation of peptide-based inhibitors targeting HIV-1 reverse transcriptase conformational flexibility. J. Biol. Chem. 2009;284:254–264. doi: 10.1074/jbc.M802199200. [DOI] [PubMed] [Google Scholar]
- 28.Kuiken C., Foley B., Korber B. Los Alamos National Laboratory Theoretical Biology and Biophysics; Los Alamos, NM: 2012. HIV Sequence Compendium 2012. LA-UR-12-24653. 332–338. [Google Scholar]
- 29.Santos A.F., Lengruber, Soares M.A. Conservation patterns of HIV-1 RT connection and RNase H domains: identification of new mutations in NRTI-treated patients. PLoS One. 2008;3:e1781. doi: 10.1371/journal.pone.0001781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.John B.K., Plant D., Hurd R.E. Improved proton-detected heteronuclear correlation using gradient-enhanced Z and Zz Filters. J. Magn. Reson. A. 1993;101:113–117. [Google Scholar]
- 31.Kay L.E., Keifer P., Saarinen T. Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J. Am. Chem. Soc. 1992;114:10663–10665. [Google Scholar]
- 32.Delaglio F., Grzesiek S., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 33.Johnson B.A., Blevins R.A. NMR View: a computer program for the visualization and analysis of NMR data. J. Biomol. NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 34.Korzhnev D.M., Kloiber K., Kay L.E. Probing slow dynamics in high molecular weight proteins by methyl-TROSY NMR spectroscopy: application to a 723-residue enzyme. J. Am. Chem. Soc. 2004;126:3964–3973. doi: 10.1021/ja039587i. [DOI] [PubMed] [Google Scholar]
- 35.Tollinger M., Skrynnikov N.R., Kay L.E. Slow dynamics in folded and unfolded states of an SH3 domain. J. Am. Chem. Soc. 2001;123:11341–11352. doi: 10.1021/ja011300z. [DOI] [PubMed] [Google Scholar]
- 36.Rogers M.T., Woodbrey J.C. Proton magnetic resonance study of hindered internal rotation in some substituted N,N-dimethylamides. J. Phys. Chem. 1962;66:540–546. [Google Scholar]
- 37.Wishart D.S., Bigam C.G., Sykes B.D. 1H, 13C and 15N random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest-neighbor effects. J. Biomol. NMR. 1995;5:67–81. doi: 10.1007/BF00227471. [DOI] [PubMed] [Google Scholar]
- 38.Vallée-Bélisle A., Ricci F., Plaxco K.W. Thermodynamic basis for the optimization of binding-induced biomolecular switches and structure-switching biosensors. Proc. Natl. Acad. Sci. USA. 2009;106:13802–13807. doi: 10.1073/pnas.0904005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lansdon E.B., Samuel D., Swaminathan S. Visualizing the molecular interactions of a nucleotide analog, GS-9148, with HIV-1 reverse transcriptase-DNA complex. J. Mol. Biol. 2010;397:967–978. doi: 10.1016/j.jmb.2010.02.019. [DOI] [PubMed] [Google Scholar]
- 40.Seckler J.M., Barkley M.D., Wintrode P.L. Allosteric suppression of HIV-1 reverse transcriptase structural dynamics upon inhibitor binding. Biophys. J. 2011;100:144–153. doi: 10.1016/j.bpj.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Das K., Lewi P.J., Arnold E. Crystallography and the design of anti-AIDS drugs: conformational flexibility and positional adaptability are important in the design of non-nucleoside HIV-1 reverse transcriptase inhibitors. Prog. Biophys. Mol. Biol. 2005;88:209–231. doi: 10.1016/j.pbiomolbio.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 42.Zheng X.H., Mueller G.A., London R.E. Homodimerization of the p51 subunit of HIV-1 reverse transcriptase. Biochemistry. 2010;49:2821–2833. doi: 10.1021/bi902116z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wright D.W., Sadiq S.K., Coveney P.V. Thumbs down for HIV: domain level rearrangements do occur in the NNRTI-bound HIV-1 reverse transcriptase. J. Am. Chem. Soc. 2012;134:12885–12888. doi: 10.1021/ja301565k. [DOI] [PubMed] [Google Scholar]
- 44.Maga G., Amacker M., Spadari S. Resistance to nevirapine of HIV-1 reverse transcriptase mutants: loss of stabilizing interactions and thermodynamic or steric barriers are induced by different single amino acid substitutions. J. Mol. Biol. 1997;274:738–747. doi: 10.1006/jmbi.1997.1427. [DOI] [PubMed] [Google Scholar]
- 45.Huang H., Chopra R., Harrison S.C. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 46.Beis I., Newsholme E.A. The contents of adenine nucleotides, phosphagens and some glycolytic intermediates in resting muscles from vertebrates and invertebrates. Biochem. J. 1975;152:23–32. doi: 10.1042/bj1520023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Traut T.W. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 48.Kloiber K., Fischer M., Konrat R. Generation and relaxation of high rank coherences in AX3 systems in a selectively methionine labelled SH2 domain. J. Biomol. NMR. 2007;38:125–131. doi: 10.1007/s10858-007-9153-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


