Knowledge of the pore-size distribution of biopolymer networks is fundamental for understanding their mechanical properties, diffusive transport, and cell migration through such networks. Molteni et al. (1) have recently proposed a novel approach to determine the average and standard deviation of the three-dimensional (3D) pore size distribution of biopolymer networks from thin, quasi two-dimensional (2D) confocal sections. Their approach is based on placing circles, or bubbles, of maximum diameter that can fit into the 2D pores and produce maximum coverage of the entire 2D image. The center location of a bubble is computed using an iterative procedure that gradually converges; this procedure is repeated many times at various starting points in the image until a sufficient number of bubbles are found. Molteni et al. (1) then compare the statistics of the size distribution of these 2D bubbles to that obtained from fitting 3D bubbles into the complete 3D structure of a biopolymer network, and show that the 3D pore-size statistics can be faithfully estimated from the analysis of a few 2D sections. This method provides users with a more intuitive and faster way of analyzing pores of random 3D biopolymer networks than previous approaches (2,3).
Here, we propose a simple and fast two-step algorithm based on readily available image processing routines to compute the centers of all bubbles in the entire image simultaneously. First, after preprocessing of the 2D optical section by segmenting the image into fiber pixels of value 1 and fluid pixels of value 0, the Euclidean distance map (EDM) is determined for the fluid space, as shown in Fig. 1 A. The EDM assigns each pixel p of the fluid phase the Euclidean distance to the nearest fiber pixel. This EDM value determines the largest radius a circle centered at p can have without overlapping any fibers. Second, the coordinates of all local maxima of the EDM are determined (Fig. 1 A, open crosses). A local maximum is defined as a pixel whose eight neighbors all have smaller values (Fig. 1 A, inset). A circle centered at a local maximum m with a radius given by EDM(m) is trapped by the surrounding fibers: It cannot be moved to any of its neighboring pixels without overlapping a fiber, because each of the neighboring pixels has a smaller distance to a fiber than the radius of the circle. Therefore, the local maxima of the EDM define the locations of the center points of all possible 2D bubbles (Fig. 1 B). This also holds true in three dimensions, when local maxima are defined with respect to their 26 neighboring pixels. To suppress artifacts due to the pixelwise calculation of the EDM, the EDM can be smoothed with a small Gaussian kernel before its local maxima are determined. Moreover, to avoid boundary effects, bubbles touching the edge of the image should be discarded.
Figure 1.
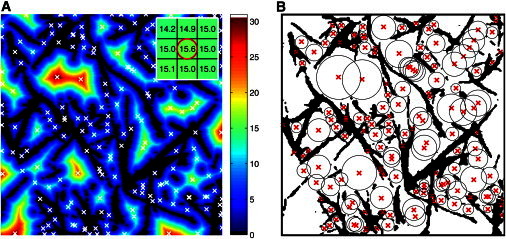
Simplified bubble analysis of the pore space of a random biopolymer network in two steps. (A) First, the Euclidean distance map (EDM) of the fluid space of the network structure is computed (shades of gray (colors online) indicate the distance of each fluid pixel to the nearest fiber pixel). (Black) Fibers. Second, the local maxima of the EDM (white crosses) determine the centers of all 2D bubbles. To avoid bubbles of similar size in close proximity, the EDM was smoothed with a 5 × 5 Gaussian kernel with a sigma of one before the local maxima were determined. (Inset) A local maximum of the EDM (red circle) is a pixel whose eight neighbors all have smaller values. (B) Resulting 2D bubbles (black circles) fit into the pore zones of the fiber structure (black). (Red crosses) Bubble centers.
The computation of the EDM as well as the determination of local maxima are readily implemented in the software MATLAB (The MathWorks, Natick, MA; Vers. 2006a or higher) through the functions bwdist and imregionalmax, and are also available in the software IMAGEJ (National Institutes of Health, Bethesda, MD) through plug-ins (4,5). Hence, this simple implementation of the bubble analysis of biopolymer networks from confocal images requires only a few lines of user code. The time to calculate all bubble centers in a typical 512 × 512 pixels image on a standard PC laptop (Intel Core-i7 2.7 GHz, 8 GB RAM, Windows 7 OS) in MATLAB 2012a is <100 ms. Furthermore, with the same MATLAB function calls, this two-step algorithm can be easily applied to 3D image stacks to determine the center locations and radii of 3D bubbles. After the 2D or 3D bubbles are found with this approach, the including/discarding procedure proposed by Molteni et al. (1) can be applied in the same manner, if desired.
We hope that this simplified implementation of the bubble analysis will help spread the acceptance and use of this method. A sample MATLAB script can be found in the Supporting Material.
Supporting Material
References
- 1.Molteni M., Magatti D., Ferri F. Fast two-dimensional bubble analysis of biopolymer filamentous networks pore size from confocal microscopy thin data stacks. Biophys. J. 2013;104:1160–1169. doi: 10.1016/j.bpj.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mickel W., Münster S., Schröder-Turk G.E. Robust pore size analysis of filamentous networks from three-dimensional confocal microscopy. Biophys. J. 2008;95:6072–6080. doi: 10.1529/biophysj.108.135939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaufman L.J., Brangwynne C.P., Weitz D.A. Glioma expansion in collagen I matrices: analyzing collagen concentration-dependent growth and motility patterns. Biophys. J. 2005;89:635–650. doi: 10.1529/biophysj.105.061994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iannuccelli E., Mompart F., Boudier T. NEMO: a tool for analyzing gene and chromosome territory distributions from 3D-FISH experiments. Bioinformatics. 2010;26:696–697. doi: 10.1093/bioinformatics/btq013. [DOI] [PubMed] [Google Scholar]
- 5.ImageJ Documentation Wiki. http://imagejdocu.tudor.lu. Accessed March 18, 2013.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


