Abstract
Alpha satellite DNA is a repetitive sequence known to be a major DNA component of centromeres in primates (order Primates). New World monkeys form one major taxon (parvorder Platyrrhini) of primates, and their alpha satellite DNA is known to comprise repeat units of around 340 bp. In one species (Azara's owl monkey Aotus azarae) of this taxon, we identified two types of alpha satellite DNA consisting of 185- and 344-bp repeat units that we designated as OwlAlp1 and OwlAlp2, respectively. OwlAlp2 exhibits similarity throughout its entire sequence to the alpha satellite DNA of other New World monkeys. The chromosomal locations of the two types of sequence are markedly distinct: OwlAlp1 was observed at the centromeric constrictions, whereas OwlAlp2 was found in the pericentric regions. From these results, we inferred that OwlAlp1 was derived from OwlAlp2 and rapidly replaced OwlAlp2 as the principal alpha satellite DNA on a short time scale at the speciation level. A less likely alternative explanation is also discussed.
Keywords: centromere, centromeric constriction, satellite DNA, replacement, tandem repeats
1. Introduction
Centromeres of higher eukaryotes generally contain large arrays of repetitive DNA in which the nucleotide sequences often exhibit high divergence rates between species, and even between chromosomes within species.1–3 Such high divergence rates are considered to be caused by recurrent, rapid sequence turnover in which repeat units are replaced by newly occurring sequences. If a new sequence replaces part or the entire centromere region of one chromosome, this sequence will become a chromosome-specific centromeric satellite DNA. If the sequence is transmitted to the centromere regions of other chromosomes by crossing over or other mechanisms and then maintained or amplified there, it will form a suprachromosomal family of centromeric satellite DNA. In humans, more than five suprachromosomal families of alpha satellite DNA are present in the genome,4 where alpha satellite DNA is a repetitive sequence known to be a major DNA component of centromeres of primates (order Primates). Sequences belonging to a suprachromosomal family may further expand to all chromosomes of the host organism, and the time scale on which such turnover events occur can be considered to depend on many factors, including the mutation rate of the satellite DNA, natural selection for or against new sequences, mechanisms of transmission to other chromosomes, and the population size of the host organism. Replacement by a new sequence may occur not as a satellite sequence covering the entire centromere region, but as a sequence occupying a specific region of the centromere. Examples of such sequences that are specific to centromeric constriction regions are expected to be of greater importance to centromere function because the centromeric constriction region is thought to play a central role in kinetochore formation and subsequent chromosome migration to the poles.5,6 There are examples of satellite DNA that are specific to the centromeric constriction region and that are contained in all chromosomes of the host organisms.7,8 To our knowledge, however, there is no example of such a sequence that is specific to the centromeric constriction region and that has undergone a rapid expansion to all chromosomes at the speciation level. In the present study, we identified a sequence of alpha satellite DNA for which there is evidence for such a rapid expansion.
The host species in which we identified the novel sequence of alpha satellite DNA was Azara's owl monkey (Aotus azarae). This species is a member of the New World monkey taxon (parvorder Platyrrhini). Alpha satellite DNA is known to be present in the centromere regions of all primate species so far examined, and its most basic structure consists of repeats of fundamental units of around 170 bp in length in a head-to-tail fashion.9–11 Detailed sequence comparisons of alpha satellite DNA between species have revealed that major primate taxons often have taxon-specific sequences or forms of repeat units. In New World monkeys, it is known that the repeat units are around 340 bp in length, and this is thought to be due to an event that occurred in their common ancestor: a set of two fundamental repeat units, with a sequence difference between the units, took the role of the basic repeat unit.12–15 In the present study, we found that Azara's owl monkey carries, in addition to the ‘standard’ alpha satellite DNA, another type of alpha satellite DNA. Results of chromosomal hybridization experiments were surprising in that the additional type of alpha satellite DNA resides in centromeric constrictions, whereas the ‘standard’ type was found to be located in the pericentric regions. We propose, as a plausible explanation of these puzzling results, that the ‘non-standard’ alpha satellite DNA replaced the ‘standard’ alpha satellite DNA in the centromeric constriction regions of all chromosomes after this species diverged from other extant species. Such a rapid replacement of the centromeric constriction regions by a newly occurring sequence has not, to our knowledge, been reported for alpha satellite DNA or other centromeric satellite DNAs of eukaryote species.
2. Materials and methods
2.1. Animal for collection of cells and DNA
We used a specimen of Azara's owl monkey bred at the authors' institution (Primate Research Institute of Kyoto University; KUPRI). The following is a description of this animal: individual identification number, A34; sex, female; date of birth, 8 July 1987; place of birth, KUPRI; confirmation of species, determination of the karyotype; parents, adults collected in Bolivia and legally imported to Japan in 1977. We took a tiny piece of epithelial tissue from the outer ear when the animal was 17 years and 8 months old and amplified cells on culture dishes for 7 days. The cell culture medium was AmnioMAX-II Complete Medium (Life Technologies Corp.). We collected the cultured cells and used them for extraction of genomic DNA and fluorescent in situ hybridization (FISH) analysis. All experiments were conducted according to the Guidelines for Care and Use of Nonhuman Primates (Version 3; June 2010) of KUPRI.
2.2. Experiments involving DNA manipulations
The cloning method we employed was the genomic hybridization technique that has been described in detail in our previous work.16–18 Briefly, a genomic library was prepared, individual clones were amplified in 96-well plates, transferred to membranes, hybridized with labelled genomic DNA, and clones responsible for strong signals were selected. The vector for the library was the fosmid pCC1FOS, and insert DNAs were 40- to 44-kb genomic DNA fragments produced by mechanical shearing and subsequent recovery from an electrophoresis gel. Other experiments, including subcloning, Southern hybridization analysis of genomic DNA, preparation of chromosome spreads, and FISH analysis of chromosomes, were conducted as described previously.16–18 The methods of analyses of nucleotide sequence data are explained in each case.
3. Results and discussion
3.1. Structure of owl monkey alpha satellite DNA
Owl monkeys (species of genus Aotus) are known among primates to exhibit relatively high frequencies of chromosomal fissions and fusions.19–21 The initial purpose of this study was to explore the possibility that repetitive sequences, such as transposons and satellite DNA, are causes of frequent chromosomal rearrangements. For this purpose, we collected clones of repetitive sequences present in the genome of Azara's owl monkey by the genomic hybridization method that is capable of collecting repetitive sequences irrespective of their nucleotide sequence. Among several different repetitive sequences identified, two were found to exhibit similarities to alpha satellite DNAs. The two repetitive sequences were first found by sequencing the terminal regions of fosmid clones 14 and 22. The two sequences were tandem repeats comprising 185- and 344-bp repeat units with occasional length variation. They were designated as OwlAlp1 and OwlAlp2, respectively. Figure 1 shows results of dot blot comparisons between these sequences and with the common marmoset alpha satellite DNA.
Figure 1.
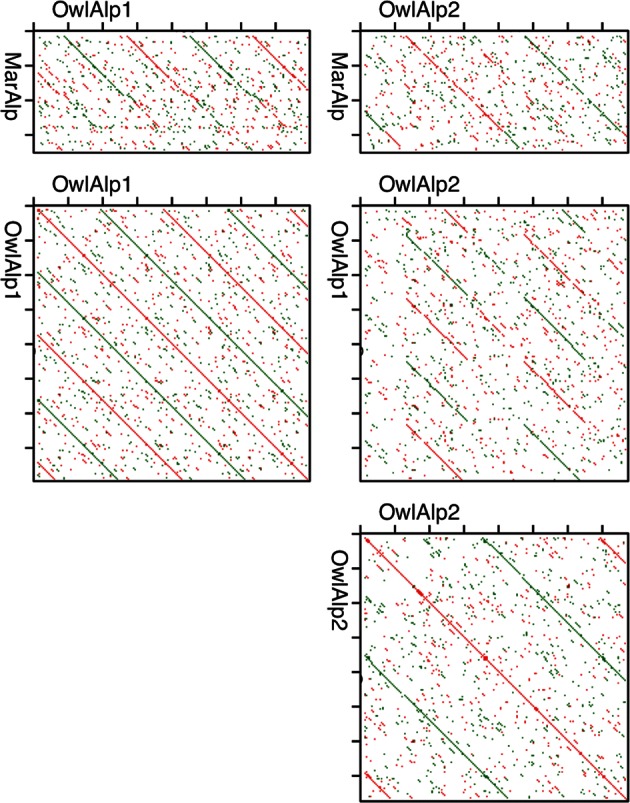
Dot blot comparison of sequences. The criterion for matching was a 70% match over a window of 10 nucleotides. OwlAlp1 and OwlAlp2 indicate here sequence reads of one terminal region of clones 14 and 22, respectively, that were the first fosmid clones found to carry the respective sequences. MarAlp is the 342-bp consensus sequence of the common marmoset alpha satellite DNA (shown in Supplementary Fig. S1). This consensus sequence was drawn by the authors from sequence data of DDBJ files FJ867326 to FJ867339 (BAC end sequences), by the same method as that for the consensus sequences of the owl monkey alpha satellite DNA (explained in the legend to Supplementary Fig. S1).
We collected additional OwlAlp1- and OwlAlp2-carrying fosmid clones by screening the owl monkey genomic library, using the insert portions of clones 14 and 22, respectively, as probes. We then sequenced the terminal regions of the clones obtained (the sequence data were deposited in DDBJ; accession numbers AB761986-AB761996 and AB761997-AB762011 for the OwlAlp1- and OwlAlp2-carrying clones, respectively) and generated consensus sequences of the repeat units identified in these sequence data. The consensus sequences are shown in Supplementary Fig. S1 together with alignments of the respective repeat units.
The length of the consensus sequence of OwlAlp2 (344 bp) is close to the length of the consensus sequence of alpha satellite DNA of the common marmoset (342 bp) and that of the other New World monkeys so far examined.7–9 In addition, as shown in Fig. 1, OwlAlp2 shows similarity along its entire sequence to the common marmoset alpha satellite DNA. The sequence similarity (gaps were counted as mismatching nucleotides) was 79, 72, 78 and 78% against the consensus sequence for the common marmoset (the MarAlp sequence described in the legend to Fig. 1), the tufted capuchin (DDBJ L07926), the pygmy marmoset (L07928), and the red-bellied titi (L07930), respectively. OwlAlp2 can, therefore, be regarded as representative of the ‘standard’ type of New World monkey alpha satellite DNA.
The entire region of the consensus sequence of OwlAlp1 was included in that of OwlAlp2 (see Fig. 1 for dot blot comparison, and Supplementary Fig. S2 for sequence alignment), with a nucleotide identity of up to 86% and with few gaps. In addition, as explained below, OwlAlp1 was found to be located in the centromeres. Based on these results, we also regard OwlAlp1 as an alpha satellite DNA.
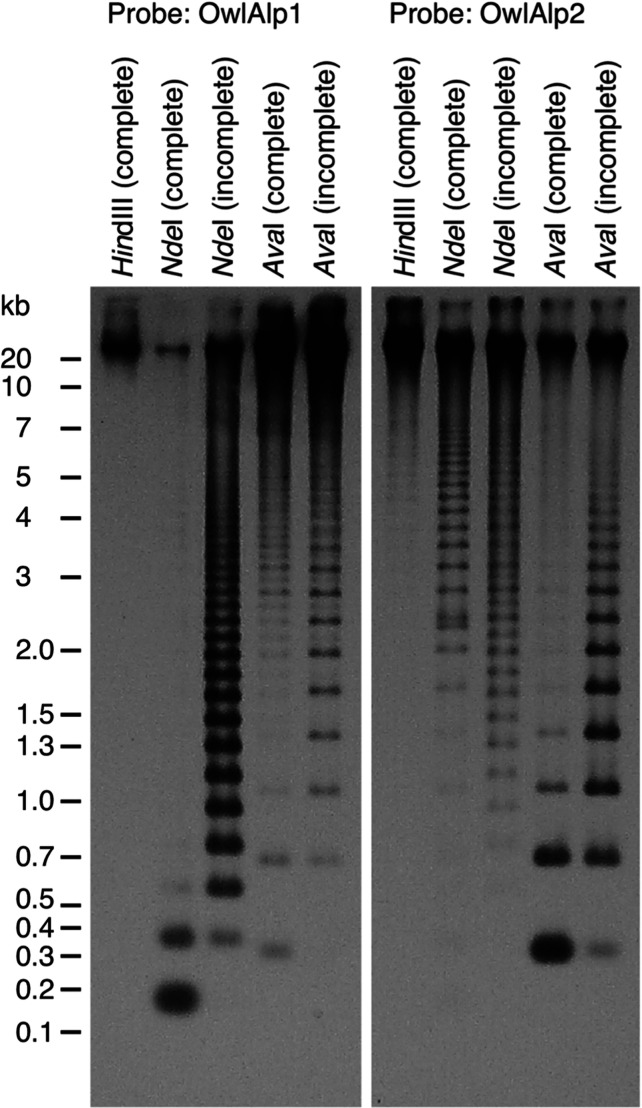
3.2. Repeat structure revealed by genomic Southern hybridization
The consensus sequence of OwlAlp1 carried a recognition site for restriction enzyme NdeI (CATATG), whereas that of OwlAlp2 did not. A site for AvaI (C(T/C)CG(A/G)G) was present in the consensus sequence of OwlAlp2, but not in that of OwlAlp1. Using these enzymes, we obtained further evidence for the presence of the two repetitive sequences in the owl monkey genome (Fig. 2). Genomic DNA was digested either completely or incompletely using these enzymes, electrophoresed on an agarose gel, and then hybridized separately with OwlAlp1 and OwlAlp2. For the incomplete digests, clear ladder patterns of the expected lengths were observed, namely, multiples of 185 and 344 bp for the OwlAlp1 and OwlAlp2 probes, respectively. Both probes gave intense signals under conditions, where detection of a single-copy sequence could not be expected (amount of genomic DNA ∼100 ng, time of exposure to X-ray film ∼30 s), suggesting that both OwlAlp1 and OwlAlp2 are present in large numbers in the owl monkey genome. Another point to be noted is that strong signals were observed, in addition to the ladder patterns, in the regions in excess of 20 kb. A plausible explanation for this result is that OwlAlp2 is the source of the upper-region signals in the lane for the NdeI digestion and OwlAlp1 is the source in the lane for the AvaI digestion.
Figure 2.
Genomic Southern blot experiments to confirm the tandem repeat structures. Genomic DNA of the owl monkey (100 ng per lane) was digested with the restriction enzymes indicated above the lanes. For complete and incomplete digestion, an excess amount (10 units) and a smaller amount (0.4 units) were used, respectively. The DNAs were electrophoresed on a 1.0% agarose gel and transferred to a nylon membrane. The membrane was then cut, and the left and right halves were hybridized with the probes for OwlAlp1 and OwlAlp2, respectively. The probes were the insert portions of fosmid clones 14 and 22, respectively. The hybridization conditions were the same as those in our previous work18 and considered to be of a medium stringency. The sizes of the marker DNA fragments are indicated along the left margin.
3.3. Chromosomal locations revealed by FISH analysis
We next examined the chromosomal locations of OwlAlp1 and OwlAlp2 by FISH analysis of metaphase chromosome spreads (Fig. 3). OwlAlp1 (green signals) was found at the centromeric constrictions of all 50 chromosomes. OwlAlp2 (red signals) was found in 46 chromosomes. The OwlAlp2 signals were located in pericentric regions. No overlap (denoted by a yellow signal) of OwlAlp1 and OwlAlp2 was observed in any of the 46 chromosomes, where both sequences occurred. Of the 46 chromosomes, 2 displayed red signals on both sides of the green signal and 40 exhibited red signals on one side only. The other four chromosomes showed a clear red signal on one side and faint, signal-like colouring on the other side that could be a noise. The merged photograph in Fig. 3 is shown with magnification in Supplementary Fig. S3, with arrowheads indicating the categories of the chromosomes. We examined the hybridization patterns of more than 10 complete metaphase spreads and confirmed identical patterns in all these spreads.
Figure 3.
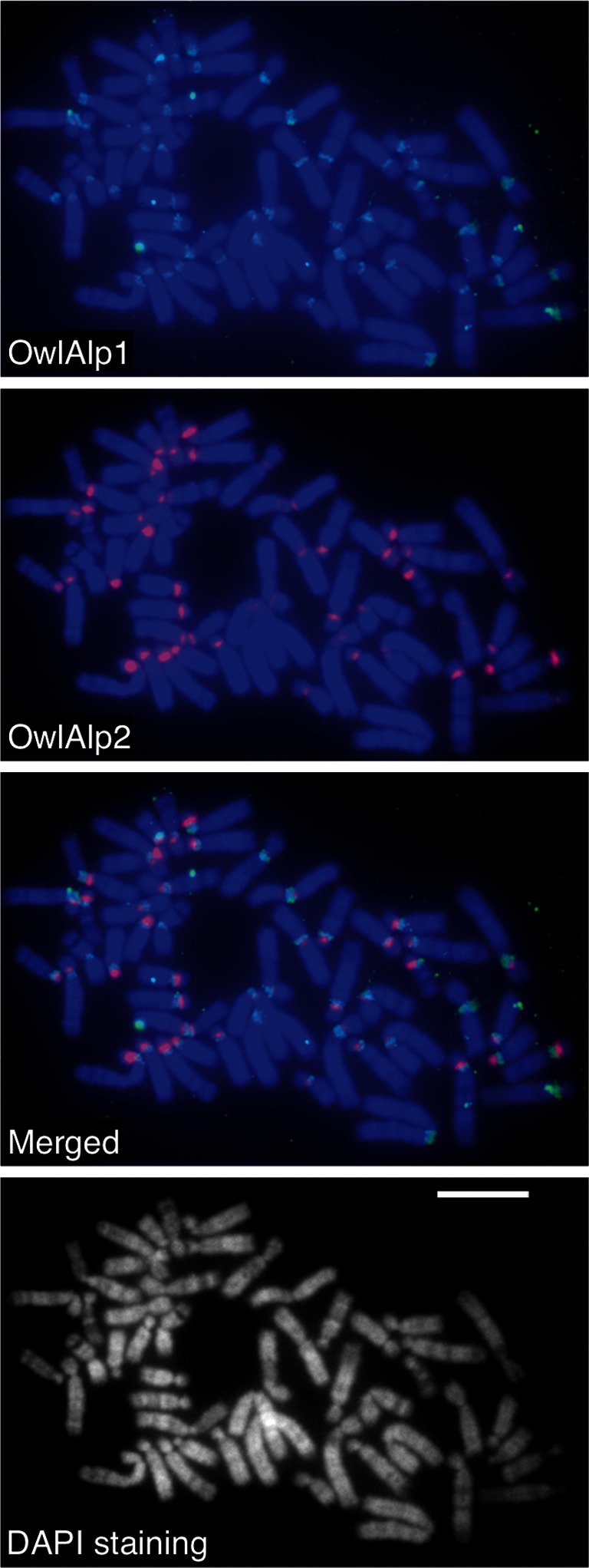
FISH analysis of OwlAlp1 and OwlAlp2 for chromosomal locations. The whole fosmid DNAs of clones 14 and 22 were labelled and hybridized with the owl monkey chromosomes. The methods and conditions were the same as those in our previous work,16 including the labelling reagents for the green (OwlAlp1) and red (OwlAlp2) colours. The bar represents 10 μm.
3.4. Confirmation of results
To exclude the possibility of an accidental mix-up of the two probes, we repeated the process consisting of the fosmid DNA preparation, sequencing of these DNAs, and FISH analysis using these newly prepared probes. We obtained the same results as those described above (data not shown).
3.5. Significance for evolution of centromeric satellite DNA
We summarize our findings as follows: (i) the owl monkey carried two types of alpha satellite DNA: OwlAlp1, consisting of 185-bp repeat units and OwlAlp2 of 344-bp repeat units, (ii) both sequences were present in large numbers in the owl monkey genome, (iii) OwlAlp2 exhibited 70–80% similarity throughout its entire sequence with alpha satellite DNA of other New World monkeys, (iv) OwlAlp1 corresponded to one partial block of OwlAlp2, with 86% sequence similarity, (v) OwlAlp1 was located at the centromeric constrictions of all chromosomes, and (vi) OwlAlp2 was found in most, but not all chromosomes and was located in the pericentric regions.
Based on Southern blot analyses and sequencing of alpha satellite DNA of numerous New World monkey species, it has been suggested that their repeat units are around 340 bp.14,15 The New World monkey species examined in these surveys included marmosets (Callimico goeldii and Cebuella pygmaea), tamarins (Leontopithecus rosalia and Saguinus labiatus), squirrel monkeys (Saimiri boliviensis), spider monkeys (Ateles fuscieps), owl monkeys (Aotus trivirgatus), titis (Callicebus moloch), woolly monkeys (Lagothrix lagotricha), capuchins (Cebus apella), and howler monkeys (Alouatta caraya). To our knowledge, there is no report of a New World monkey having two or more major types of alpha satellite DNA. Recently, an extensive analysis of trace archive sequence data of the common marmoset has reinforced the view that alpha satellite DNA of New World monkeys consists of repeat units of around 340 bp.13 The only exception to this known to date is the alpha satellite DNA of the black bearded saki (Chiropotes satanas) that comprises repeat units of about 540 bp. This can, however, be explained as a derivative of the ‘standard’ repeat units due to partial duplication.15 The alpha satellite DNA of the owl monkey is exceptional in a different sense, in that two types of alpha satellite DNA co-exist in the genome in large amounts. The two types are OwlAlp1 and OwlAlp2, and comparison of their consensus sequences with each other and with those of alpha satellite DNAs of other New World monkeys appears to support the hypothesis that OwlAlp2 is the ‘standard’ alpha satellite DNA. We discuss below a possible scenario leading to the current situation of the owl monkey alpha satellite DNA.
The most likely explanation for the generation mechanism of OwlAlp1 would be deletion of part of the OwlAlp2 sequence. Since its appearance, OwlAlp1 has undergone large-scale amplification, expansion to all chromosomes, and has replaced OwlAlp2 as the primary alpha satellite DNA. A possibility not to be disregarded is that this replacement may have taken place after Azara's owl monkey (A. azarae) diverged from another species within the genus Aotus. The species designated as A. trivirgatus was included in the genomic Southern blot analysis carried out by other authors,15 in lane 13 of their Fig. 1. A ladder pattern of multiples of ∼340 bp is seen in all of the four blots, but that of another unit size, such as 185 bp, is not observed. In addition, strong signals in the upper regions, as observed in our Fig. 2, are not seen in their blots. Accordingly, the animal sample used in their analysis15 would be considered to have carried only one type of alpha satellite DNA that corresponded to OwlAlp2. The species A. trivirgatus was divided into more than eight species, one of which is A. azarae (As we described in the Results and discussion section, species of genus Aotus exhibit high frequencies of chromosomal fissions and fusions. The species redefinition of A. trivirgatus reflected discovery of several new karyotypes.)22,23 Considering the difference in the results of Southern blotting between the two studies, it is likely that the previous study15 used an animal now designated as a species other than A. azarae. Only A. azarae was available for us to examine at the time of our study, and we intend to analyse other species when materials become available.
The hypothesis we propose here is that OwlAlp1 was derived from OwlAlp2 and that OwlAlp2 was replaced by OwlAlp1 as the principal alpha satellite DNA after the host species (Azara's owl monkey) diverged from other species. If this hypothesis is true, the results of the present study are the first example of rapid replacement of the centromeric constriction regions by a newly occurring sequence at the speciation level. To our knowledge, there are no similar reports for alpha satellite DNA or other centromeric satellite DNAs of eukaryote species.
3.6. An alternative scenario
The results of sequence comparisons support the hypothesis that OwlAlp1 was derived from OwlAlp2. Currently, we do not have sufficient information to exclude the logical possibility that OwlAlp2 was derived from OwlAlp1. If this assumption were true, the most likely situation would be that OwlAlp1 is a direct descendant of the monomeric sequence from which the ‘standard’ alpha satellite DNA of New World monkeys originates. It is implied in this assumption that the monomer sequence survived in the lineage leading to the Azara's owl monkey but was lost in other New World monkeys, including the owl monkey species designated as A. trivirgatus in the previous study.15 This leads to the conclusion that the most recent common ancestor of New World monkeys carried both monomer and dimer sequences. If this assumption is true, sequences similar in size to OwlAlp1 might be found in other New World monkeys, if a greater number of species are surveyed. The hypothesis on which this scenario stands is less likely but, even if true, the results of the present study suggest an unknown past event that may be key to the evolution of alpha satellite DNA in New World monkeys.
Supplementary data
Supplementary Data are available at www.dnaresearch.oxfordjournals.org.
Funding
This work was supported by Grants-in-Aid (23657165 to AK, 23470098 to AK, and 22247037 to HH) from the Japan Society for the Promotion of Science (JSPS). O.P. was supported by the Fellowship of Capacity Building for Kasetsart University on Internationalization.
Supplementary Material
Acknowledgements
We are grateful to Drs Hiroshi Masumoto, Toshio Mouri, and Masanaru Takai for helpful discussions.
Footnotes
Edited by Dr Osamu Ohara
References
- 1.Ma J., Wing R.A., Bennetzen J.L., Jackson S.A. Plant centromere organization: a dynamic structure with conserved functions. Trends Genet. 2007;23:134–9. doi: 10.1016/j.tig.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 2.Henikoff S., Ahmad K., Malik H.S. The centromere paradox: stable inheritance with rapidly evolving DNA. Science. 2001;293:1098–102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- 3.Ananiev E.V., Phillips R.L., Rines H.W. Chromosome-specific molecular organization of maize (Zea mays L.) centromeric regions. Proc. Natl. Acad. Sci. USA. 1998;95:13073–78. doi: 10.1073/pnas.95.22.13073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Willard H.F., Waye J.S. Chromosome-specific subsets of human alpha satellite DNA: analysis of sequence divergence within and between chromosomal subsets and evidence for an ancestral pentameric repeat. J. Mol. Evol. 1987;25:207–14. doi: 10.1007/BF02100014. [DOI] [PubMed] [Google Scholar]
- 5.Tadeu A.M., Ribeiro S., Johnston J., Goldberg I., Gerloff D., Earnshaw W.C. CENP-V is required for centromere organization, chromosome alignment and cytokinesis, EMBO J. 2008;27:2510–22. doi: 10.1038/emboj.2008.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Houben A., Schroeder-Reiter E., Nagaki K., et al. CENH3 interacts with the centromeric retrotransposon cereba and GC-rich satellites and locates to centromeric substructures in barley. Chromosoma. 2007;116:275–83. doi: 10.1007/s00412-007-0102-z. [DOI] [PubMed] [Google Scholar]
- 7.Kulikova O., Geurts R., Lamine M., et al. Satellite repeats in the functional centromere and pericentromeric heterochromatin of Medicago truncatula. Chromosoma. 2004;113:276–83. doi: 10.1007/s00412-004-0315-3. [DOI] [PubMed] [Google Scholar]
- 8.Lee H.R., Zhang W., Langdon T., et al. Chromatin immunoprecipitation cloning reveals rapid evolutionary patterns of centromeric DNA in Oryza species. Proc. Natl. Acad. Sci. USA. 2005;102:11793–98. doi: 10.1073/pnas.0503863102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maio J.J. DNA strand reassociation and polyribonucleotide binding in the African green monkey, Cercopithecus aethiops. J. Mol. Biol. 1971;56:579–95. doi: 10.1016/0022-2836(71)90403-7. [DOI] [PubMed] [Google Scholar]
- 10.Manuelidis L., Wu J.C. Homology between human and simian repeated DNA. Nature. 1978;276:92–4. doi: 10.1038/276092a0. [DOI] [PubMed] [Google Scholar]
- 11.Rudd M.K., Willard H.F. Analysis of the centromeric regions of the human genome assembly. Trends Genet. 2004;20:529–33. doi: 10.1016/j.tig.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 12.Alkan C., Ventura M., Archidiacono N., Rocchi M., Sahinalp S.C., Eichler E.E. Organization and evolution of primate centromeric DNA from whole-genome shotgun sequence data. PLoS Comput. Biol. 2007;3:1807–18. doi: 10.1371/journal.pcbi.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cellamare A., Catacchio C.R., Alkan C., et al. New insights into centromere organization and evolution from the white-cheeked gibbon and marmoset. Mol. Biol. Evol. 2009;26:1889–900. doi: 10.1093/molbev/msp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fanning T.G., Seuánez H.N., Forman L. Satellite DNA sequences in the New World primate Cebus apella Platyrrhini, Primates. Chromosoma. 1993;102:306–11. doi: 10.1007/BF00661273. [DOI] [PubMed] [Google Scholar]
- 15.Alves G., Seuanez H.N., Fanning T. Alpha satellite DNA in neotropical primates (Platyrrhini) Chromosoma. 1994;103:262–7. doi: 10.1007/BF00352250. [DOI] [PubMed] [Google Scholar]
- 16.Koga A., Hirai Y., Hara T., Hirai H. Repetitive sequences originating from the centromere constitute large-scale heterochromatin in the telomere region in the siamang, a small ape. Heredity. 2012;109:180–7. doi: 10.1038/hdy.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hara T., Hirai Y., Jahan I., Hirai H., Koga A. Tandem repeat sequences evolutionarily related to SVA-type retrotransposons are expanded in the centromere region of the western hoolock gibbon, a small ape. J. Hum. Genet. 2012;57:760–5. doi: 10.1038/jhg.2012.107. [DOI] [PubMed] [Google Scholar]
- 18.Koga A., Shimada A., Kuroki T., et al. The Tol1 transposable element of the medaka fish moves in human and mouse cells. J. Hum. Genet. 2007;52:628–35. doi: 10.1007/s10038-007-0161-2. [DOI] [PubMed] [Google Scholar]
- 19.Ma N.S., Jones T.C., Miller A.C., Morgan L.M., Adams E.A. Chromosome polymorphism and banding patterns in the owl monkey (Aotus) Lab. Anim. Sci. 1976;26:1022–36. [PubMed] [Google Scholar]
- 20.Torres O.M., Enciso S., Ruiz F., Silva E., Yunis I. Chromosome diversity of the genus Aotus from Colombia. Am. J. Primatol. 1998;44:255–75. doi: 10.1002/(SICI)1098-2345(1998)44:4<255::AID-AJP2>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 21.Ruiz-Herrera A., García F., Aguilera M., Garcia M., Ponsà Fontanals M. Comparative chromosome painting in Aotus reveals a highly derived evolution. Am. J. Primatol. 2005;65:73–85. doi: 10.1002/ajp.20098. [DOI] [PubMed] [Google Scholar]
- 22.Hershkovitz P. Two new species of night monkeys, genus Aotus (Cebidae Platyrryni): a preliminary report of Aotus taxonomy. Am. J. Primatol. 1983;4:209–43. doi: 10.1002/ajp.1350040302. [DOI] [PubMed] [Google Scholar]
- 23.Fleagle J.G. Primate Adaptation and Evolution. 2nd edition. New York: Academic Press; 1999. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.