Abstract
Probiotics are live microorganisms that potentially confer beneficial outcomes to host by modulating gut microbiota in the intestine. The aim of this study was to comprehensively investigate effects of probiotics on human intestinal microbiota using 454 pyrosequencing of bacterial 16S ribosomal RNA genes with an improved quantitative accuracy for evaluation of the bacterial composition. We obtained 158 faecal samples from 18 healthy adult Japanese who were subjected to intervention with 6 commercially available probiotics containing either Bifidobacterium or Lactobacillus strains. We then analysed and compared bacterial composition of the faecal samples collected before, during, and after probiotic intervention by Operational taxonomic units (OTUs) and UniFrac distances. The results showed no significant changes in the overall structure of gut microbiota in the samples with and without probiotic administration regardless of groups and types of the probiotics used. We noticed that 32 OTUs (2.7% of all analysed OTUs) assigned to the indigenous species showed a significant increase or decrease of ≥10-fold or a quantity difference in >150 reads on probiotic administration. Such OTUs were found to be individual specific and tend to be unevenly distributed in the subjects. These data, thus, suggest robustness of the gut microbiota composition in healthy adults on probiotic administration.
Keywords: probiotics, gut microbiota, 16S ribosomal RNA gene, pyrosequencing
1. Introduction
Probiotics are defined as live bacterial strains conferring various benefits to the consumer by modulating the intestinal ecosystem, thereby potentially promoting host health and improving host disease risk.1–11 Various probiotic strains have been industrially developed and marketed as a variety of products and applications such as fermented foods and supplements, including yogurt12–15 Most probiotics taxonomically belong to two genera, Bifidobacterium and Lactobacillus, that originate from various environments, including the human intestine, and both species are generally regarded as safe.16–18
The interaction between administrated probiotics and indigenous microbiota is one of the most attractive and important research areas, particularly because gut microbiota have been shown to be profoundly associated with various host physiology states, including disease, diet, and age through the shift of bacterial composition, as well as metabolic and nutritional processes.19–23 The ability of probiotics to survive through the intestine and to modulate gut microbiota is a critical factor in determining their potential for health-related outcomes.
There have been a large number of probiotic intervention studies to assess the impact of probiotics on gut microbiota in healthy adults,24–34 infants, and children,35,36 and in clinical trials on patients with a variety of diseases.37,38 Most probiotic intervention studies were carried out by comparison between probiotic-treated groups and placebo controls and examined only one or two samples from periods before and during intervention or post-intervention for each subject. These experimental designs make evaluation of results obscure from a statistical viewpoint due to the high inter-individual variability of gut microbiota.4 In addition, most of the analyses focussed on the composition of specific bacterial species or groups by conventional methods such as culturing, quantitative polymerase chain reaction (qPCR), fluorescence in situ hybridization, denaturing gradient gel electrophoresis, or terminal-restriction fragment length polymorphism based on the bacterial 16S ribosomal RNA gene (16S). These conventional methodologies may also overlook subtle changes in bacterial community structure and change of species other than targeted species. Thus, the effect of probiotic administration on the overall structure of gut microbiota is largely unknown.
Recently, a high-throughput sequencing-based analysis has been conducted for gut microbiota fed with a probiotic yogurt that provided new insights into probiotics research by utilizing a large-scale dataset.39 However, much more data are required to understand the impact of probiotics on gut microbiota. Recent advances in sequencing technology have enabled us to elucidate complex bacterial communities, including human gut microbiota.40,41 Particularly, 454 pyrosequencing of bacterial 16S gene tags coupled with bioinformatics provides a high-throughput and cost-effective approach for the comprehensive analysis of bacterial communities at the species level.42–48
In this study, we developed an analysis pipeline for bacterial communities based on barcoded 454 pyrosequencing of 16S gene tags using modified PCR primers that improved the quantitative accuracy of inferred species composition in human gut microbiota. Using this pipeline, we analysed faecal samples longitudinally collected from individuals with and without probiotic administration to evaluate the effect of probiotics on gut microbiota with respect to species richness and diversity. The results revealed the robustness and stability of gut microbiota of healthy adults in response to probiotic administration.
2. Materials and methods
2.1. Subjects, faecal sample collection, and probiotic intervention
Eighteen healthy volunteers (age: 22 ± 3.16 yrs, 6 male, 12 female) were recruited through Azabu University, Kanagawa, Japan (Supplementary Table S1). All subjects were informed of the purpose of this study. This study was approved by the ethical committee of Azabu University, and written consent was obtained from all subjects. No subjects were treated with antibiotics during faecal sample collection. The subjects were divided into six groups (three subjects per group), and each group consumed six different commercially available probiotics supplied from Yakult Honsha Co., Ltd, Kagome Co., Ltd, Morinaga Milk Industry Co., Ltd, Takanashi Milk Products Co., Ltd, Meiji Co., Ltd, and Danone Japan Co., Ltd, respectively (Supplementary Table S1). The number of each bacterial strain contained in the probiotic products was estimated as the genome equivalent by qPCR of 16S ribosomal RNA genes using 27Fmod-338R, followed by pyrosequencing of the 16S amplicons (see below). The genome equivalent per gram or millilitre and the total genome equivalent of each bacterial strain in one probiotic product are summarized in Supplementary Table S1. Three subjects in each group consumed the same probiotics daily for 8 weeks according to the schedule of sampling and probiotic intervention (Supplementary Fig. S1). Faecal samples from 4 weeks before (S00) and 8 weeks during probiotic intervention (S01–S04), and 8 weeks after cessation of probiotic intervention (S05–S08), were collected every 2 weeks from each subject. In total, we collected 158 faecal samples from the 18 subjects because we could not collect 1 sample each from 4 of the subjects.
2.2. Recovery of bacteria from faecal samples
Freshly collected faeces (1.0 g) were suspended in 20% glycerol (Wako Pure Chemical Industries, Ltd) and phosphate buffered saline solution (Life Technologies Japan, Ltd, Tokyo, Japan), frozen in liquid nitrogen, and stored at −80°C until ready for use. Bacterial pellets were prepared from frozen faecal samples as described previously.49
2.3. DNA isolation from bacteria
Faecal DNA was isolated and purified according to the literature, with minor modifications.49 The bacterial pellet was suspended and incubated with 15 mg/ml lysozyme (Sigma-Aldrich Co., LCC) at 37°C for 1 h in TE10. Purified achromopeptidase (Wako Pure Chemical Industries, Ltd) was added at a final concentration of 2000 units/ml and then incubated at 37°C for 30 min. The suspension was treated with 1% (wt/vol) sodium dodecyl sulphate and 1 mg/ml proteinase K (Merck Japan) and incubated at 55°C for 1 h. The lysate was treated with phenol/chloroform/isoamyl alcohol (Life Technologies Japan, Ltd). DNA was precipitated by adding ethanol and pelleted by centrifugation at 3,300 g at 4°C for 15 min. The DNA pellet was rinsed with 75% ethanol, dried, and dissolved in 10 mM Tris-HCl/1 mM EDTA (TE). DNA samples were purified by treating with 1 mg/ml RNase A (Wako Pure Chemical Industries, Ltd) at 37°C for 30 min and precipitated by adding equal volumes of 20% polyethylene glycol solution (PEG6000-2.5M NaCl). DNA was pelleted by centrifugation at 8,060 g at 4°C, rinsed with 75% ethanol, and dissolved in TE.
2.4. 454 barcoded pyrosequencing of 16S rRNA gene
The V1–V2 region of the 16S rRNA gene was amplified using forward primer (5′-CCATCTCATCCCTGCGTGTCTCCGACTCAGNNNNNNNNNNagrgtttgatymtggctcag-3′) containing the 454 primer A, a unique 10-bp barcode sequence for each sample (indicated in N), and 27Fmod (5′-agrgtttgatymtggctcag) in which the third base A in the original primer 27F was changed to R, and reverse primer (5′-CCTATCCCCTGTGTGCCTTGGCAGTCTCAGtgctgcctcccgtaggagt-3′) containing the 454 primer B and reverse primer 338R (5′-tgctgcctcccgtaggagt). PCR was performed in 1 × Ex Taq PCR buffer (50 µl), deoxynucleoside triphosphate (2.5 mM), Ex Taq polymerase (Takara Bio, Inc., Shiga), each primer (10 μM), and 40 ng of extracted DNA under conditions of 2 min at 96°C, 20 cycles of 96°C for 30 s, 55°C for 45 s, and 72°C for 1 min, and a final extension of 72°C for 10 min on a 9700 PCR system (Life Technologies Japan, Ltd, Tokyo, Japan). PCR products of approximately 370 bp were confirmed by agarose gel electrophoresis, purified by AMPure XP magnetic purification beads (Beckman Coulter, Inc., Brea, CA, USA), and quantified using the Quant-iT PicoGreen dsDNA Assay Kit (Life Technologies Japan, Ltd, Tokyo, Japan). Mixed samples were prepared by pooling approximately equal amounts of PCR amplicons from each sample and subjected to 454 GS FLX Titanium or 454 GS JUNIOR (Roche Applied Science) sequencing according to the manufacturer's instructions.
2.5. Analysis pipeline for 454 barcoded pyrosequencing of 16S PCR amplicons
We developed an analysis pipeline for 454 barcoded pyrosequencing of PCR amplicons of the V1-2 region amplified by 27Fmod-338R primers. First, 16S reads were assigned to each sample based on the barcode sequence information. Second, 16S reads that did not have PCR primer sequences at both sequence termini and those with an average quality value < 25 were filtered out. Third, 16S reads containing possible chimaeric sequences that had BLAST match lengths of < 90% with reference sequences in the database were removed. Reads removed in these processes accounted for about 35% of all reads, most of which represented reads lacking PCR primer sequences (Supplementary Table S2). Finally, filter-passed reads were obtained for further analysis by trimming off both primer sequences.
All 3000 filter-passed reads of the 16S V1-2 sequences obtained from each subject were deposited in DDBJ/GenBank/EMBL with accession numbers DRA000869–DRA000886.
2.6. Assessment of the quantitative accuracy of 16S data using artificial bacterial communities
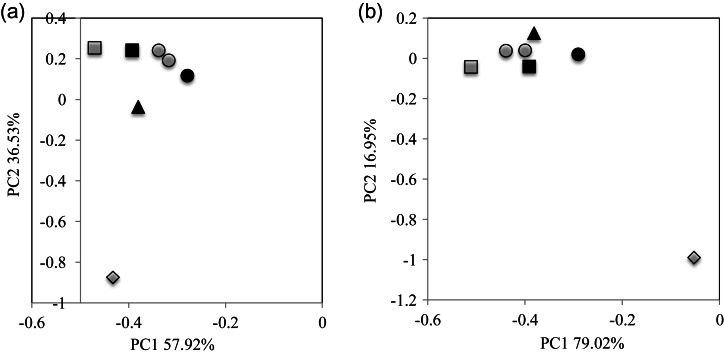
Two artificial bacterial communities (designated ‘mock01’ and ‘mock02’) were constructed by mixing genomic DNA from 10 and 11 different human gut-associated bacterial strains with an appropriate ratio, respectively (Supplementary Table S3). Genome sequences of these microbes were completely sequenced and are publicly available. From these communities, we amplified the V1-2 region by PCR using 27F-338R and 27Fmod-338R primers, the V5-6 region by 787F-1061R primers, and the V1-9 region by 27F-1492R primers. V1-2 and V5-6 amplicons were subjected to 454 pyrosequencing, and V1-9 amplicons were cloned in Escherichia coli, and 3000 clones were sequenced by the Sanger method, and the products were analysed with the ABI3730xl (Life Technologies Japan, Ltd, Tokyo). We also performed duplicate qPCR experiments targeting a specific genomic region of the bacterial strains in the two mock communities. All filter-passed 16S de novo sequences and qPCR data were then analysed by principal component analysis (PCA) to compare and assess the quantitative accuracy (Fig. 1).
Figure 1.
Assessment of the quantitative accuracy of the analysis of the bacterial composition of two mock communities by various methods. PCA analysis of the data was obtained from various methods using mock01 (a) and mock02 (b). Closed circle: expected, open circle: duplicate qPCR, closed square: pyrosequencing of 16S V1-2 region using 27Fmod, open square: pyrosequencing of 16S V1-2 region using 27F, closed triangle: pyrosequencing of 16S V5-6 region, open diamond: Sanger sequencing of nearly full-length 16S clone.
The error rate of the filter-passed sequences using 27Fmod-338R primers obtained from the two mock communities was estimated by aligning the 16S V1-2 de novo sequences with the reference 16S sequences in the two mock communities (Supplementary Table S4).
2.7. Data analysis
2.7.1. Database
Two databases were constructed for the analysis of 16S sequences. One is the 16S rRNA gene sequence database constructed by collecting 16S sequences of ≥1200 bp of bacterial isolates from the Ribosomal Database Project v. 10.27. Another database is the reference genome database constructed by collecting genome sequences from the NCBI FTP site (ftp://ftp.ncbi.nih.gov/genbank/, Dec 2011) that includes 1482 complete and 605 draft bacterial genomes.
2.7.2. Operational taxonomic unit (OTU) and UniFrac distance analysis
We used 3000 filter-passed reads of 16S sequences for operational taxonomic unit (OTU) and UniFrac distance analysis for each sample. For OTU analysis, clustering of 16S reads was done using a 96% pairwise-identity cutoff with the UCLUST program (www.drive5.com). Representative sequences for each OTU were assigned to bacterial species by BLAST search with a 96% pairwise-identity cutoff against the two databases mentioned above. UniFrac distance analysis was used to determine the dissimilarity (distance) between two communities based on the fraction of branch length shared between two communities within a phylogenetic tree constructed from 16S sequence datasets.44
2.7.3. Other
Estimation of OTU numbers by extrapolation (Chao1 and ACE) was calculated with the vegan package (v2.0-5) for R (v2.15.2).
3. Results and discussion
3.1. Quantitative accuracy of 16S data produced by 454 pyrosequencing
Pyrosequencing of PCR amplicons of bacterial 16S short variable regions is the most popular and a high-throughput approach to infer and characterize the species composition in bacterial communities.42,45,46,48 The 454 pyrosequencing platform, which can produce over 400 bases per read, is also superior to shorter read-length sequencers with respect to sequence accuracy for single-end sequencing.50,51 However, this PCR-based method has a problem particularly in quantification of the composition of the genus Bifidobacterium, a dominant species in human gut microbiota because the 16S sequence of Bifidobacterium has a few base mismatches with the commonly used PCR primer 27F (or 8F), underestimating this genus in the community.52–55 To improve this, we modified primer 27F to 27Fmod by changing the third base G to R (G or A) in 27F-YM53 that perfectly matched with the annealing site of the Bifidobacterium 16S gene (see Materials and methods).
To assess the 16S data using 27Fmod, we compared various 16S sequence and qPCR data obtained from two mock communities (Supplementary Table S3) that are useful to evaluate the quantitative accuracy of 16S-based data and the sequencing error rate.56,57 Quantitative accuracy of the overall bacterial composition was evaluated by comparing the similarity of each data to the expected (‘Expected’) using PCA (Fig. 1). From the PCA data, Euclidean distance was calculated for evaluation of the similarity of each data with the ‘Expected’. The results revealed that the order of their similarities with the ‘Expected’ was the qPCR data ≥ the V1-2 data using 27Fmod > the V5-6 data > the V1-2 data using 27F >> the data of Sanger sequencing-based full-length V1-9, indicating that the use of 27Fmod greatly improved the quantitative accuracy for evaluation of the overall bacterial composition (Supplementary Table S5). This improvement was largely dependent on the improved estimation of the Bifidobacterium content by the use of 27Fmod. The average relative Bifidobacterium content in the two mock communities estimated from the data of V1-2 using 27F was only 1.5% of the ‘Expected’ (100%), whereas the use of 27Fmod increased the relative content to 61% that was also better than that estimated from the data of V5-6 and Sanger full-length analyses (Supplementary Fig. S2). Because qPCR can be used only when genomes of all bacteria in a given community are known, or only for a limited number of specific known species, we concluded that 454 pyrosequencing of the V1-2 region using 27Fmod-338R provided more quantitatively accurate data for bacterial composition in human gut microbiota than that using the conventional 27F primer.
We estimated the average error rate of filter-passed V1-2 data using 27Fmod-338R by aligning the V1-2 and reference 16S sequences of bacterial strains used in the two mock communities. The error rate was estimated to be 0.58 and 0.66% for mock01 and mock02 by local alignment, respectively (Supplementary Table S4). These error rates are similar to the previously published data,43,45,50 but lower than in another study.58 The latter may be due to differences in the examined alignment length and between local and global alignments. Errors in 454 pyrosequencing data can be the primary cause for overestimation of the OTU number that is an issue which needs to be improved for accurate estimation of species richness in bacterial communities.59,60 We compared OTU numbers generated from clustering of various qualities of 16S reads with a 96% and a 97% pair-wise identity cutoff. For this comparison, we made and used three datasets: only primer check-passed reads having the highest error rates, filter-passed reads, and selected filter-passed reads having the lowest error rates. The results indicated that a 96% cutoff clustering of error-rich reads and a 97% cutoff clustering of filter-passed reads gave the worse results than a 96% cutoff clustering of filter-passed and selected filter-passed reads (Supplementary Fig. S3). A 97% cutoff was defined for clustering of highly accurate Sanger full-length 16S sequences.61 Therefore, in clustering of pyrosequencing data having higher error rate than Sanger data, the use of a cutoff identity lower than 97% and a lower number of reads are reasonable to reduce overestimation of the OTU number. A 96% cutoff clustering of filter-passed reads gave similar OTU numbers up to 30–50 reads to those of filter-passed reads having the lowest error rates. These read numbers are approximately three to five times the number of input strains. After several trials testing the mock communities, we decided to use 3000–5000 reads per sample for clustering with a 96% cutoff for the analysis of human gut microbiota. Indeed, OTU numbers using a 96% cutoff clustering of 3000 reads decreased about 15% when compared with those using a 97% cutoff clustering.
3.2. Species richness and diversity in human faecal microbiota with probiotic intervention
We randomly selected 3000 reads of 16S V1-2 sequences from all filter-passed reads for each sample (Supplementary Table S2) and used 474 000 reads in total from 158 faecal DNA samples of 18 subjects for the analysis of species richness and composition in human gut microbiota. Clustering of all reads with a 96% pairwise-identity cutoff gave a total of 2758 OTUs. After removing the minority OTUs having <0.1% abundance in any samples, 1175 OTUs having ≥0.1% abundance in at least 1 sample, accounting for 99.1% of all 16S reads, were used for further analysis.
3.2.1. Detection of administrated probiotic strains in faecal samples
We investigated whether administrated strains contained in the probiotic products can be detected in faecal DNA. We sequenced the 16S V1-2 region of all bacterial strains contained in probiotic products used in this study. The BLAST search to the databases indicated that except for the Bifidobacterium longum strain used in Group III, the 16S sequences of all strains in the probiotic products significantly differed from those of the indigenous species phylogenetically closest to the probiotic strains. The 16S sequence of the B. longum strain used in Group III was almost identical to that of an indigenous Bifidobacterium species, so that we used a distinguishable additive Lactococcus lactis strain in this product for the detection of administrated bacteria in Group III samples. The 16S sequences of these probiotic strains were included in the databases constructed in this study, and the 16S reads assigned to administrated strains had the average similarity between 99.4 and 99.9% identities with the reference sequences (data not shown). The 16S reads assigned to probiotic or additive strains were detected in samples (S01–S04) during probiotic intervention [designated ‘Pro(+)’] at various frequencies, but almost none were detected in samples (S00 and S05–S08) without probiotic administration [designated ‘Pro(−)’] (Supplementary Table S6). The administrated probiotic strains were shown to be more frequently detected in samples during the intervention than in the pre- and post-intervention periods using different detection methods such as culturing, targeted PCR, and hybridization.24,26–28,30,32,33,62 In the present study, two probiotic Lactobacillus and one additive Lactococcus strains were detected in post-intervention samples in three subjects with a minimum count, respectively. The similarity of three 16S sequences was 99.4, 99.7, and 100% identity with those of administrated Lactobacillus and Lactococcus strains, indicating that these are administrated strains. The survival of some probiotic strains in the post-intervention period was also reported previously.28,30 Our data suggested that some probiotic strains seem to be able to persistently colonize the intestine and their survivability may be related to metabolic activity in the intestine.63,64 Probiotic Bifidobacterium strains were not detected in any Pro(−) samples. However, we found two distinct 16S sequences both assigned to Bifidobacterium animalis in two subjects APr37 and APr39. One showed a high similarity of >98% identity with the 16S sequence of the administrated B. animalis and was detected with high frequency only in the Pro(+) samples, whereas another showed a low similarity of 96.5–97.4% identity (a mean of 97.2%) with low frequency in both the Pro(−) and Pro(+) samples. These data suggest the presence of unknown indigenous species phylogenetically close to, but distinct from, probiotic B. animalis in human gut microbiota. The total number of bacteria contained in each probiotic product was varied between 109 and 1010, showing no large difference in quantity among them (Supplementary Table S1). No clear correlation was also observed between the number of bacteria in the products and the frequency in detection of the administrated strains in the Pro(+) samples. From these observations, the frequency of administrated bacteria detected in faeces may not be largely affected by their amounts in the products. Therefore, detection of Lactobacillus brevis and Lactobacillus delbrueck at relatively low level in faeces cannot be simply explained by the difference in a dose, but could be considered the association with several factors such as their survivability in the intestine, diet, or physiological conditions of subjects.
3.2.2. Change of species richness in samples with and without probiotics
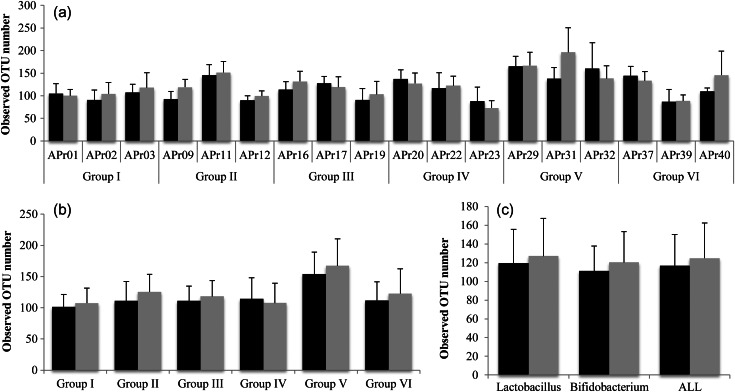
We analysed species richness (OTU number) in the Pro(+) and Pro(−) samples. Supplementary Figure S4 shows the change in OTU numbers for every sample in each subject, indicating that OTU numbers vary dramatically for every sample. Most of the variation can be attributed to single OTUs representing the minority species. We averaged the OTU numbers of the Pro(−) and Pro(+) samples and compared them for subject, group, type of probotics (Lactobacillus and Bifidobacterium), and all combined samples, respectively (Fig. 2). The average OTU numbers in 6 out of 18 subjects were decreased in the range of the ratio of 0.83–0.95 in the Pro(+) samples when compared with the Pro(−) samples, whereas those in other 12 subjects were increased in the range of the ratio of 1.01–1.43. For group, only Group IV showed a decrease in the average OTU number in the Pro(+) samples with the ratio of 0.94. For type of probiotics and all samples, the average OTU numbers in the Pro(+) samples were slightly more abundant (approximately 1.07-fold) than those in the Pro(−) samples, but no statistical significance was observed in any dataset. The increase in OTU number in the Pro(+) samples was largely due to the minority species (Supplementary Fig. S4), whereas the abundance of the majority species (OTUs containing ≥10 reads) was almost constant over time. We performed the same analysis using different sets of 3000 reads for each subject. The analysis reproducibly showed the similar pattern and the degree of the change in OTU numbers to which the minority species is largely attributed (data not shown). These data indicate that administration of probiotics tends to increase species richness in faecal microbiota that may be beneficial for the consumer because the species richness in faecal microbiota of subjects afflicted with disease such as inflammatory bowel disease is significantly reduced when compared with that of healthy subjects.65
Figure 2.
Change in OTU number in faecal microbiota with and without probiotic administration. (a) Individual, (b) group, (c) type of probiotics. Black bar indicates Pro(−) samples. Grey bar indicates Pro(+) samples. The error bars represent standard deviation.
3.2.3. Change of species composition in samples with and without probiotics
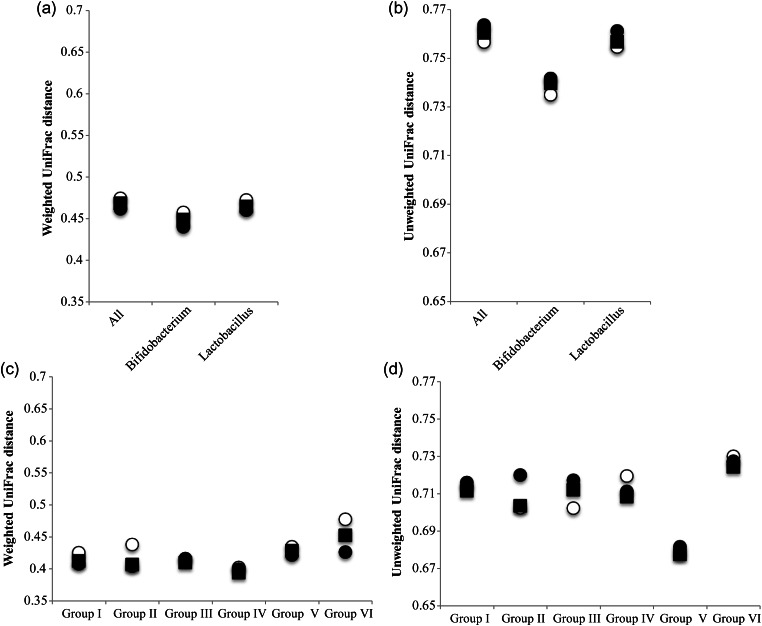
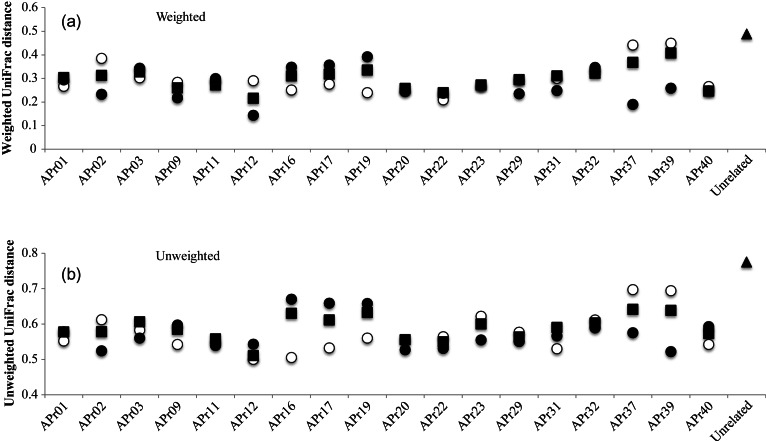
We obtained the average weighted and unweighted UniFrac distances within Pro(−), within Pro(+), and between Pro(−) and Pro(+) samples for every group, probiotic types, and all subjects, respectively (Fig. 3). High UniFrac distance implies high variability of microbiota structure within and between samples. If the difference between any pair of the three distances is statistically significant, it can be considered that probiotic administration significantly affected the overall microbiota composition. We found the largest difference between weighted UniFac distances of the Pro(+) and Pro(−) samples in Group VI. However, statistical evaluation of this difference by the Student's t-test showed no significance (P-value > 0.05) for 781 out of 1000 times (Supplementary Table S7). These data imply high stability of gut microbiota to probiotic administration for all subjects examined. We also analysed UniFrac distances of intra-subject gut microbiota (Fig. 4). Although 5 subjects (APr02, 12, 16, 37, and 39) showed a significant difference in the UniFrac distances between Pro(−) and Pro(+) samples, the results showed that both weighted and unweighted distances between Pro(−) and Pro(+) of all intra-subjects were significantly lower than the average distance of the 18 unrelated subjects. The Welch's t-test for these differences showed statistical significance (Supplementary Table S8). We also performed the UniFrac distance analysis using different 16S datasets of 5000 reads for group, type of probiotics, all subjects, and intra-subject. The results similarly showed no statistical significance in differences between any pair of the 3 UniFrac distances and the significantly lower UniFrac distance of each intra-subject than that of the 18 unrelated subjects (data not shown). Thus, these data suggested that the perturbation of microbiota elicited by probiotics in an intra-subject did not overcome the inter-subject variations of gut microbiota, supporting high intra-specificity and stability of gut microbiota.66,67 This robustness of gut microbiota of adults is in contrast with the profound effect of antibiotic administration on adult gut microbiota68 and the observed response of gut microbiota of infants fed with probiotics, in which the infant gut microbiota composition was considerably affected by probiotics.36 A short-term dietary intervention study showed that in controlled feeding of the same diet to subjects over 10 days, a marked change was observed within 1 day after the intervention initiation.69 In the present study, no significant difference was observed between samples before (S00) and first samples (S01) after the intervention initiation (data not shown). It would be valuable to analyse faecal samples collected within a few days after administration of probiotics for evaluation of the short-term effect of probiotics.
Figure 3.
Average UniFrac distance within Pro (−) and Pro(+) and between Pro(−) and Pro(+) for each group, type of probitotics, and all subjects. Average UniFrac distance between any pair of the three distances for type of probiotics and all subjects (a and b), and each group (c and d). Open circle, closed circle, and closed square indicate average UniFrac distance within Pro (−), within Pro (+), and between Pro(−) and Pro(+) samples, respectively.
Figure 4.
Average UniFrac distance within Pro(−) and Pro(+) and between Pro(−) and Pro(+) for each subject. Open circles, closed circles, and closed squares indicate average UniFrac distance within Pro(−), within Pro(+), and between Pro(−) and Pro(+) samples, respectively. Closed triangles indicate average UniFrac distance between samples (S00) of 18 unrelated individuals.
4. Identification of bacterial species showing significant increase or decrease by probiotic administration
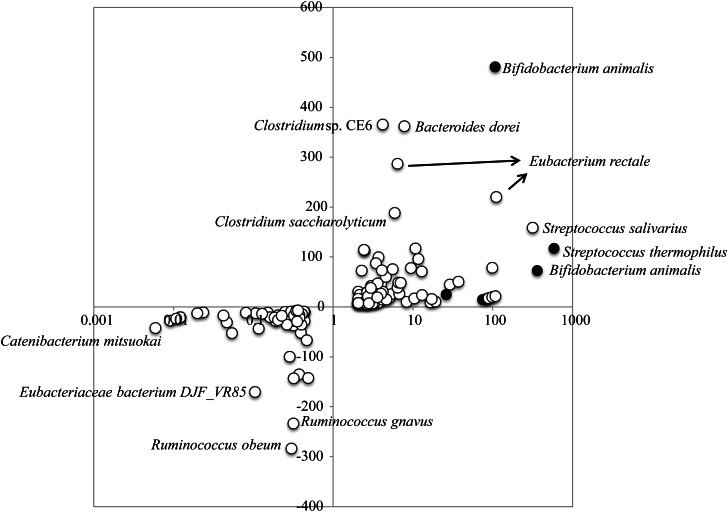
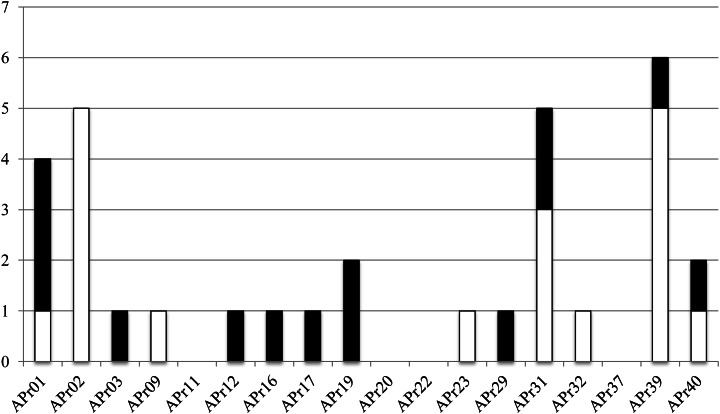
Although our results suggested that administration of probiotics had almost no effect on the overall structure of gut microbiota, it is possible to identify bacterial species largely responding to the administrated probiotics at the OTU/species level. We surveyed OTUs showing an increase or a decrease between the Pro(+) and Pro(−) samples by comparing the number of 16S reads for each OTU. We first enumerated the OTUs showing ≥2-fold change between the Pro(−) and Pro(+) samples for each subject, and the quantity difference was also obtained by subtracting the 16S read number of the Pro(+) samples from that of the Pro(−) samples. This is because OTUs showing a high quantity difference, but less fold change may also have substantial influence on gut microbiota composition. We found several OTUs significantly changed by probiotic administration, including OTUs assigned to both the indigenous and administrated strains (Fig. 5). We listed 88 OTUs (7.5% of all analysed 1175 OTUs) showing significant change of ≥3-fold, among which 30 OTUs changed by ≥10-fold (Supplementary Fig. S5). We excluded 6 OTUs assigned to the administrated strains from the 30 OTUs and obtained 24 OTUs assigned to the indigenous species, including OTU00072 assigned to Streptococcus salivarius that showed significant change in 2 subjects (Supplementary Table S9). We also found seven OTUs showing significant difference in quantity between both samples (Supplementary Table S10). Of the combined 32 OTUs (2.7%), 18 were increased and 14 were decreased by probiotic administration. Many of the OTUs showing a significant increase were assigned to minority species in the Pro(−) samples, but some increased up to nearly 7% in abundance (e.g. OTU00372 assigned to Eubacterium rectale). On the other hand, the OTUs showing a significant decrease were almost undetected in the Pro(+) samples. Phylum-level species assignment showed that species belonging to the phylum Firmicutes were most largely affected by both probiotics, and all species belonging to the phylum Bacteroidetes were affected only by Lactobacillus probiotics (Table 1). The 32 OTUs were assigned to 27 indigenous species, among which 4 species (Clostridium clostridioforme, Eubacterium eligens, E. rectale, and Faecalibacterium prausnitzii) were assigned by 8 different OTUs and 1 species (S. salivarius) was assigned by the 2 same OTUs as described above. All these species except for S. salivarius were found to show significant change only in one subject, indicating that response of the indigenous species to probiotics is highly individual specific (Supplementary Fig. S6). Two different OTUs (OTU02677 and OTU02748) assigned to F. prausnitzii, of which the reduction is known to be correlated with inflammatory bowel disease,70 were found to both decrease and increase in the same subject (APr40) by probiotic administration, suggesting that these two phylogenetically close species may have the diversity of response to probiotic action. We also examined distribution of the 32 OTUs in the subjects. The results revealed that 4 subjects (APr11, 20, 22, and 37) did not have such OTUs and 8 subjects had only 1 OTU, whereas 4 subjects (APr01, 02, 31, and 39) had more than 4 OTUs showing significant change (Fig. 6), suggesting their uneven distribution in the 18 subjects. These data imply existence of the sensitive and less sensitive responders to probiotic action and if so, it would be interesting to investigate the relation between gut microbiota type and its response to probiotics.
Figure 5.
OTUs showing ≥2-fold change and their difference in quantity between the Pro(−) and Pro(+) samples. The x-axis represents the scale of fold change between the Pro(+) and Pro(−) samples. The y-axis represents the difference (number of reads) in quantity between the Pro(+) and Pro(−) samples. Closed and open circles indicate the administrated probiotic and indigenous species, respectively.
Table 1.
Phylum-level species assignment of OTUs showing significant fold change or quantity difference by administration of probiotics
| Type of probiotics | Change | aNumber of varied OTUs | Fold change (≥10-fold) |
Number of varied OTUs | Difference (≥150 reads) |
||||
|---|---|---|---|---|---|---|---|---|---|
| Firmicutes | Actinobacteria | Bacteroidetes | Unclassified bacterium | Firmicutes | Bacteroidetes | ||||
| Lactobacillus | Increase | 9 | 7 | 0 | 1 | 1 | 3 | 2 | 1 |
| Decrease | 7 | 3 | 1 | 3 | 0 | 1 | 1 | 0 | |
| Total | 16 | 10 | 1 | 4 | 1 | 4 | 3 | 1 | |
| Bifidobacterium | Increase | 5 | 5 | 0 | 0 | 0 | 1 | 1 | 0 |
| Decrease | 4 | 4 | 0 | 0 | 0 | 2 | 2 | 0 | |
| Total | 9 | 9 | 0 | 0 | 0 | 3 | 3 | 0 | |
| All | Increase | 14 | 12 | 0 | 1 | 1 | 4 | 3 | 1 |
| Decrease | 11 | 7 | 1 | 3 | 0 | 3 | 3 | 0 | |
| Total | 25 | 19 | 1 | 4 | 1 | 7 | 6 | 1 | |
aAdministrated probiotic strains were excluded, and only OTUs with a P-value < 0.05 are shown.
Figure 6.
Distribution of 32 OTUs showing a significant change in 18 subjects. The y-axis indicates the number of OTUs showing significant change between the Pro(−) and Pro(+) samples in each subject (see Supplementary Tables S9 and S10). Open and closed bars indicate increased and decreased OTUs, respectively.
In summary, we analysed changes of the gut microbiota composition of healthy adults fed with probiotics using the 454 pyrosequencing platform with the improved quantitative accuracy for evaluation of the overall bacterial composition. The present study using large datasets enabled us to more comprehensively and precisely evaluate the effect of probiotics on gut microbiota than the previous probiotic intervention researches in which the analysis exclusively focussed on only several limited bacterial species using conventional methods. Our data further support the high inter-subject variability and the high intra-subject stability that is the current common view for the feature of adult gut microbiota. A recent study of gut microbiota in twins demonstrated that probiotics had almost no effect on the community structure, but affected the gene expression of microbiota.39 To more deeply understand the potential function of probiotics, the analysis of bacterial and host cell's transcriptome and intestinal metabolome is required.
Supplementary data
Supplementary Data are available at www.dnaresearch.oxfordjournals.org.
Funding
This work was supported in part by the global COE project of ‘Genome Information Big Bang’ from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan (to M.H. and K.O.), a research project grant from Azabu University to H.M. and by a grant from the Core Research for Evolutional Science and Technology (CREST) program of the Japan Science and Technology Agency (JST) to K.O.
Supplementary Material
Acknowledgements
We thank Dr Todd D. Taylor for critical reading of the manuscript, and K. Furuya, C. Shindo, H. Inaba, E. Iioka, Y. Takayama, E. Ohmori, M. Kiuchi, Y. Hattori (The University of Tokyo), and A. Nakano (Azabu University) for technical support.
Footnotes
Edited by Dr Katsumi Isono
References
- 1.Preidis G.A., Versalovic J. Targeting the human microbiome with antibiotics, probiotics, and prebiotics: gastroenterology enters the metagenomics era. Gastroenterology. 2009;136:2015–31. doi: 10.1053/j.gastro.2009.01.072. doi:10.1053/j.gastro.2009.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patel R.M., Lin P.W. Developmental biology of gut-probiotic interaction. Gut Microbes. 2010;1:186–95. doi: 10.4161/gmic.1.3.12484. doi:10.4161/gmic.1.3.12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerritsen J., Smidt H., Rijkers G.T., de Vos W.M. Intestinal microbiota in human health and disease: the impact of probiotics. Genes Nutr. 2011;6:209–40. doi: 10.1007/s12263-011-0229-7. doi:10.1007/s12263-011-0229-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanders M.E., Heimbach J.T., Pot B., et al. Health claims substantiation for probiotic and prebiotic products. Gut Microbes. 2011;2:127–33. doi: 10.4161/gmic.2.3.16174. doi:10.4161/gmic.2.3.16174. [DOI] [PubMed] [Google Scholar]
- 5.Aureli P., Capurso L., Castellazzi A.M., et al. Probiotics and health: an evidence-based review. Pharmacol. Res. 2011;63:366–76. doi: 10.1016/j.phrs.2011.02.006. doi:10.1016/j.phrs.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Rauch M., Lynch S.V. The potential for probiotic manipulation of the gastrointestinal microbiome. Curr. Opin. Biotechnol. 2012;23:192–201. doi: 10.1016/j.copbio.2011.11.004. doi:10.1016/j.copbio.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 7.Fujimura K.E., Slusher N.A., Cabana M.D., Lynch S.V. Role of the gut microbiota in defining human health. Expert Rev. Anti Infect. Ther. 2010;8:435–54. doi: 10.1586/eri.10.14. doi:10.1586/eri.10.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deshpande G.C., Rao S.C., Keil A.D., Patole S.K. Evidence-based guidelines for use of probiotics in preterm neonates. BMC Med. 2011;9:92. doi: 10.1186/1741-7015-9-92. doi:10.1186/1741-7015-9-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bron P.A., van Baarlen P., Kleerebezem M. Emerging molecular insights into the interaction between probiotics and the host intestinal mucosa. Nat. Rev. Microbiol. 2011;10:66–78. doi: 10.1038/nrmicro2690. [DOI] [PubMed] [Google Scholar]
- 10.Thomas D.W., Greer F.R. American Academy of Pediatrics Committee on Nutrition; American Academy of Pediatrics Section on Gastroenterology, Hepatology, and Nutrition. Probiotics and prebiotics in pediatrics. Pediatrics. 2010;126:1217–31. doi: 10.1542/peds.2010-2548. doi:10.1542/peds.2010-2548. [DOI] [PubMed] [Google Scholar]
- 11.Indrio F., Neu J.N. The intestinal microbiome of infants and the use of probiotics. Curr. Opin. Pediatr. 2011;23:145–50. doi: 10.1097/MOP.0b013e3283444ccb. doi:10.1097/MOP.0b013e3283444ccb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saxelin M., Tynkkynen S., Mattila-Sandholm T., de Vos W.M. Probiotic and other functional microbes: from markets to mechanisms. Curr. Opin. Biotechnol. 2005;16:204–11. doi: 10.1016/j.copbio.2005.02.003. doi:10.1016/j.copbio.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 13.Nagpal R., Kumar A., Kumar M., Behare P.V., Jain S., Yadav H. Probiotics, their health benefits and applications for developing healthier foods: a review. FEMS Microbiol. Lett. 2012;334:1–15. doi: 10.1111/j.1574-6968.2012.02593.x. doi:10.1111/j.1574-6968.2012.02593.x. [DOI] [PubMed] [Google Scholar]
- 14.Bisanz J.E., Reid G. Unraveling how probiotic yogurt works. Sci. Transl. Med. 2011;3:106ps41. doi: 10.1126/scitranslmed.3003291. [DOI] [PubMed] [Google Scholar]
- 15.Bron P.A., Kleerebezem M. Engineering lactic acid bacteria for increased industrial functionality. Bioeng. Bugs. 2011;2:80–7. doi: 10.4161/bbug.2.2.13910. doi:10.4161/bbug.2.2.13910. [DOI] [PubMed] [Google Scholar]
- 16.Kleerebezem M., Vaughan E.E. Probiotic and gut lactobacilli and bifidobacteria: molecular approaches to study diversity and activity. Annu. Rev. Microbiol. 2009;63:269–90. doi: 10.1146/annurev.micro.091208.073341. doi:10.1146/annurev.micro.091208.073341. [DOI] [PubMed] [Google Scholar]
- 17.Ventura M., O'Flaherty S., Claesson M.J., et al. Genome-scale analyses of health-promoting bacteria: probiogenomics. Nat. Rev. Microbiol. 2009;7:61–71. doi: 10.1038/nrmicro2047. doi:10.1038/nrmicro2047. [DOI] [PubMed] [Google Scholar]
- 18.Snydman D.R. The safety of probiotics. Clin. Infect. Dis. 2008;46:S104–11. doi: 10.1086/523331. doi:10.1086/523331. [DOI] [PubMed] [Google Scholar]
- 19.Lozupone C.A., Stombaugh J.I., Gordon J.I., Jansson J.K., Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–30. doi: 10.1038/nature11550. doi:10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clemente J.C., Ursell L.K., Parfrey L.W., Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148:1258–70. doi: 10.1016/j.cell.2012.01.035. doi:10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicholson J.K., Holmes E., Kinross J., et al. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–7. doi: 10.1126/science.1223813. doi:10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 22.Walter J., Ley R. The human gut microbiome: ecology and recent evolutionary changes. Annu. Rev. Microbiol. 2011;65:411–29. doi: 10.1146/annurev-micro-090110-102830. doi:10.1146/annurev-micro-090110-102830. [DOI] [PubMed] [Google Scholar]
- 23.Hooper L.V., Littman D.R., Macpherson A.J. Interactions between the microbiota and the immune system. Science. 2012;336:1268–73. doi: 10.1126/science.1223490. doi:10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tannock G.W., Munro K., Harmsen H.J., Welling G.W., Smart J., Gopal P.K. Analysis of the fecal microflora of human subjects consuming a probiotic product containing Lactobacillus rhamnosus DR20. Appl. Environ. Microbiol. 2000;66:2578–88. doi: 10.1128/aem.66.6.2578-2588.2000. doi:10.1128/AEM.66.6.2578-2588.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.García-Albiach R., Pozuelo de Felipe M.J., Angulo S., et al. Molecular analysis of yogurt containing Lactobacillus delbrueckii subsp. bulgaricus and Streptococcus thermophilus in human intestinal microbiota. Am. J. Clin. Nutr. 2008;87:91–6. doi: 10.1093/ajcn/87.1.91. [DOI] [PubMed] [Google Scholar]
- 26.Alvaro E., Andrieux C., Rochet V., et al. Composition and metabolism of the intestinal microbiota in consumers and non-consumers of yogurt. Br. J. Nutr. 2007;97:126–33. doi: 10.1017/S0007114507243065. doi:10.1017/S0007114507243065. [DOI] [PubMed] [Google Scholar]
- 27.Rochet V., Rigottier-Gois L., Levenez F., et al. Modulation of Lactobacillus casei in ileal and fecal samples from healthy volunteers after consumption of a fermented milk containing Lactobacillus casei DN-114 001Rif. Can. J. Microbiol. 2008;54:660–7. doi: 10.1139/w08-050. doi:10.1139/W08-050. [DOI] [PubMed] [Google Scholar]
- 28.Rochet V., Rigottier-Gois L., Ledaire A., et al. Survival of Bifidobacterium animalis DN-173 010 in the faecal microbiota after administration in lyophilized form or in fermented product – a randomised study in healthy adults. J. Mol. Microbiol. Biotechnol. 2008;14:128–36. doi: 10.1159/000106092. doi:10.1159/000106092. [DOI] [PubMed] [Google Scholar]
- 29.Ouwehand A.C., Bergsma N., Parhiala R., et al. Bifidobacterium microbiota and parameters of immune function in elderly subjects. FEMS Immunol. Med. Microbiol. 2008;53:18–25. doi: 10.1111/j.1574-695X.2008.00392.x. doi:10.1111/j.1574-695X.2008.00392.x. [DOI] [PubMed] [Google Scholar]
- 30.Firmesse O., Mogenet A., Bresson J.L., Corthier G., Furet J.P. Lactobacillus rhamnosus R11 consumed in a food supplement survived human digestive transit without modifying microbiota equilibrium as assessed by real-time polymerase chain reaction. J. Mol. Microbiol. Biotechnol. 2008;14:90–9. doi: 10.1159/000106087. doi:10.1159/000106087. [DOI] [PubMed] [Google Scholar]
- 31.Lahtinen S.J., Tammela L., Korpela J., et al. Probiotics modulate the Bifidobacterium microbiota of elderly nursing home residents. Age (Dordr) 2009;31:59–66. doi: 10.1007/s11357-008-9081-0. doi:10.1007/s11357-008-9081-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Savard P., Lamarche B., Paradis M.E., Thiboutot H., Laurin É., Roy D. Impact of Bifidobacterium animalis subsp. lactis BB-12 and, Lactobacillus acidophilus LA-5-containing yoghurt, on fecal bacterial counts of healthy adults. Int. J. Food Microbiol. 2011;149:50–7. doi: 10.1016/j.ijfoodmicro.2010.12.026. doi:10.1016/j.ijfoodmicro.2010.12.026. [DOI] [PubMed] [Google Scholar]
- 33.Yamano T., Iino H., Takada M., Blum S., Rochat F., Fukushima Y. Improvement of the human intestinal flora by ingestion of the probiotic strain Lactobacillus johnsonii La1. Br. J. Nutr. 2006;95:303–12. doi: 10.1079/bjn20051507. doi:10.1079/BJN20051507. [DOI] [PubMed] [Google Scholar]
- 34.Engelbrektson A.L., Korzenik J.R., Sanders M.E. Analysis of treatment effects on the microbial ecology of the human intestine. FEMS Microbiol. Ecol. 2006;57:239–50. doi: 10.1111/j.1574-6941.2006.00112.x. doi:10.1111/j.1574-6941.2006.00112.x. [DOI] [PubMed] [Google Scholar]
- 35.Marzotto M., Maffeis C., Paternoster T., et al. Lactobacillus paracasei A survives gastrointestinal passage and affects the fecal microbiota of healthy infants. Res. Microbiol. 2006;157:857–66. doi: 10.1016/j.resmic.2006.06.007. doi:10.1016/j.resmic.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 36.Cox M.J., Huang Y.J., Fujimura K.E., et al. Lactobacillus casei abundance is associated with profound shifts in the infant gut microbiome. PLoS One. 2010;5:e8745. doi: 10.1371/journal.pone.0008745. doi:10.1371/journal.pone.0008745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Culligan E.P., Hill C., Sleator R.D. Probiotics and gastrointestinal disease: successes, problems and future prospects. Gut Pathog. 2009;1:19. doi: 10.1186/1757-4749-1-19. doi:10.1186/1757-4749-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gareau M.G., Sherman P.M., Walker W.A. Probiotics and the gut microbiota in intestinal health and disease. Nat. Rev. Gastroenterol. Hepatol. 2010;7:503–14. doi: 10.1038/nrgastro.2010.117. doi:10.1038/nrgastro.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McNulty N.P., Yatsunenko T., Hsiao A., et al. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci. Transl. Med. 2011;3:106ra106. doi: 10.1126/scitranslmed.3002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Metzker M.L. Sequencing technologies—the next generation. Nat. Rev. Genet. 2010;11:31–46. doi: 10.1038/nrg2626. doi:10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 41.Qin J., Li R., Raes J., et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. doi:10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huse S.M., Dethlefsen L., Huber J.A., Mark WD., Relman D.A., Sogin M.L. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 2008;4:e1000255. doi: 10.1371/journal.pgen.1000255. doi:10.1371/journal.pgen.1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huse S.M., Huber J.A., Morrison H.G., Sogin M.L., Welch D.M. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 2007;8:R143. doi: 10.1186/gb-2007-8-7-r143. doi:10.1186/gb-2007-8-7-r143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hamady M., Lozupone C., Knight R. Fast Unifrac: facilitating high-throughput phylogenetic analysis of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 2010;4:17–27. doi: 10.1038/ismej.2009.97. doi:10.1038/ismej.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Droege M., Hill B. The Genome Sequencer FLX System–longer reads, more applications, straight forward bioinformatics and more complete data sets. J. Biotechnol. 2008;136:3–10. doi: 10.1016/j.jbiotec.2008.03.021. doi:10.1016/j.jbiotec.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 46.Andersson A.F., Lindberg M., Jakobsson H., Bäckhed F., Nyrén P., Engstrand L. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One. 2008;3:e2836. doi: 10.1371/journal.pone.0002836. doi:10.1371/journal.pone.0002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuczynski J., Lauber C.L., Walters W.A., et al. Experimental and analytical tools for studying the human microbiome. Nat. Rev. Genet. 2011;13:47–58. doi: 10.1038/nrg3129. doi:10.1038/nrg3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hamady M., Walker J.J., Harris J.K., Gold N.J., Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat. Methods. 2008;5:235–7. doi: 10.1038/nmeth.1184. doi:10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morita H., Kuwahara T., Ohshima K., et al. An improved isolation method for metagenomic analysis of the microbial flora of the human intestine. Microbes Environ. 2007;22:214–22. doi:10.1264/jsme2.22.214. [Google Scholar]
- 50.Claesson M.J., Wang Q., O'Sullivan O., et al. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res. 2010;38:e200. doi: 10.1093/nar/gkq873. doi:10.1093/nar/gkq873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou H.W., Li D.F., Tam N.F., et al. BIPES, a cost-effective high-throughput method for assessing microbial diversity. ISME J. 2011;5:741–9. doi: 10.1038/ismej.2010.160. doi:10.1038/ismej.2010.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hattori M., Taylor T.D. The human intestinal microbiome: a new frontier of human biology. DNA Res. 2009;16:1–12. doi: 10.1093/dnares/dsn033. doi:10.1093/dnares/dsn033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frank J.A., Reich C.I., Sharma S., Weisbaum J.S., Wilson B.A., Olsen G.J. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl. Environ. Microbiol. 2008;74:2461–70. doi: 10.1128/AEM.02272-07. doi:10.1128/AEM.02272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hill J.E., Fernando W.M., Zello G.A., Tyler R.T., Dahl W.J., Van Kessel A.G. Improvement of the representation of bifidobacteria in fecal microbiota metagenomic libraries by application of the cpn60 universal primer cocktail. Appl. Environ. Microbiol. 2010;76:4550–2. doi: 10.1128/AEM.01510-09. doi:10.1128/AEM.01510-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Palmer C. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:1556–73. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haas B.J., Gevers D., Earl A.M., et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21:494–504. doi: 10.1101/gr.112730.110. doi:10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schloss P.D., Gevers D., Westcott S.L. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One. 2011;6:e27310. doi: 10.1371/journal.pone.0027310. doi:10.1371/journal.pone.0027310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gilles A., Meglécz E., Pech N., Ferreira S., Malausa T., Martin J.F. Accuracy and quality assessment of 454 GS-FLX Titanium pyrosequencing. BMC Genomics. 2011;12:245. doi: 10.1186/1471-2164-12-245. doi:10.1186/1471-2164-12-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Quince C., Lanzén A., Curtis T.P., et al. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat. Methods. 2009;6:639–41. doi: 10.1038/nmeth.1361. doi:10.1038/nmeth.1361. [DOI] [PubMed] [Google Scholar]
- 60.Diaz P.I., Dupuy A.K., Abusleme L., et al. Using high throughput sequencing to explore the biodiversity in oral bacterial communities. Mol. Oral Microbiol. 2012;27:182–201. doi: 10.1111/j.2041-1014.2012.00642.x. doi:10.1111/j.2041-1014.2012.00642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schloss P.D., Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 2005;71:1501–6. doi: 10.1128/AEM.71.3.1501-1506.2005. doi:10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.del Campo R., Bravo D., Cantón R., et al. Scarce evidence of yogurt lactic acid bacteria in human feces after daily yogurt consumption by healthy volunteers. Appl. Environ. Microbiol. 2005;71:547–9. doi: 10.1128/AEM.71.1.547-549.2005. doi:10.1128/AEM.71.1.547-549.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oozeer R., Leplingard A., Mater D.D., et al. Survival of Lactobacillus casei in the human digestive tract after consumption of fermented milk. Appl. Environ. Microbiol. 2006;72:5615–7. doi: 10.1128/AEM.00722-06. doi:10.1128/AEM.00722-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marco M.L., de Vries M.C., Wels M., et al. Convergence in probiotic Lactobacillus gut-adaptive responses in humans and mice. ISME J. 2010;4:1481–4. doi: 10.1038/ismej.2010.61. doi:10.1038/ismej.2010.61. [DOI] [PubMed] [Google Scholar]
- 65.Manichanh C., Rigottier-Gois L., Bonnaud E., et al. Reduced diversity of fecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut. 2006;55:205–11. doi: 10.1136/gut.2005.073817. doi:10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Turnbaugh P.J., Hamady M.H., Yatsuneko T., et al. A core gut microbiome in obese and lean twins. Nature. 2009;475:480–4. doi: 10.1038/nature07540. doi:10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kurokawa K., Itoh T., Kuwahara T., et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007;14:169–81. doi: 10.1093/dnares/dsm018. doi:10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jernberg C., Löfmark S., Edlund C., Jansson J.K. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J. 2007;1:56–66. doi: 10.1038/ismej.2007.3. doi:10.1038/ismej.2007.3. [DOI] [PubMed] [Google Scholar]
- 69.Wu G.D., Chen J., Hoffmann C., et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–8. doi: 10.1126/science.1208344. doi:10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sokol H., Seksik P., Furet J.P., et al. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm. Bowel Dis. 2009;15:1183–9. doi: 10.1002/ibd.20903. doi:10.1002/ibd.20903. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.