Abstract
A large number of really interesting new gene (RING) E3 ligases contribute to the post-translational modification of target proteins during plant responses to environmental stresses. However, the physical interactome of RING E3 ligases in rice remains largely unknown. Here, we evaluated the expression patterns of 47 Oryza sativa RING finger protein (OsRFP) genes in response to abiotic stresses via semi-quantitative reverse transcription polymerase chain reaction (RT-PCR) and in silico analysis. Subsequently, molecular dissection of nine OsRFPs was performed by the examination of their E3 ubiquitin ligase activity, subcellular localization, and physical interaction with target proteins. Most of the OsRFPs examined possessed E3 ligase activity and showed diverse subcellular localization. Yeast two-hybrid analysis was then employed to construct a physical interaction map of seven OsRFPs with their 120 interacting proteins. The results indicated that these OsRFPs required dynamic translocation and partitioning for their cellular activation. Heterogeneous overexpression of each of the OsRFP genes in Arabidopsis suggested that they have functionally diverse responses to abiotic stresses, which may have been acquired during evolution. This comprehensive study provides insights into the biological functions of OsRFPs, which may be useful in understanding how rice plants adapt to unfavourable environmental conditions.
Keywords: E3 ligase, environmental stress, functional diversity, interactome, RING finger protein
1. Introduction
Unfavourable environmental conditions, such as salinity, drought, cold, heat, and nutrient limitation, cause extensive losses to agricultural productivity worldwide. In fact, environmental conditions are responsible for reducing average yields for most major crop plants by >50%.1 Drought and soil salinity, which are caused by osmotic stresses, are two of the major stresses that adversely affect plant growth and productivity. Both of these stresses lead to cellular dehydration and reactive oxygen species production, causing a reduction in cytosolic and vacuolar volumes, which then negatively affects cellular structures and metabolism. Therefore, molecular dissection of the complex plant response to abiotic stresses, such as drought and salinity, has attracted much interest since understanding these responses may enable researchers to develop transgenic plants that have improved survival rates following exposure to stressful environmental conditions.
The covalent attachment of ubiquitin(s) to target proteins is a central mechanism in the regulation of protein degradation via the Ub-26S proteasome pathway in eukaryotes. In particular, accumulating genetic analyses support the notion that the Ub-26S proteasome pathway regulates various key aspects of growth, development, and defence mechanisms in plants.2–4 In this pathway, Ub, a highly evolutionary conserved 76-residue polypeptide, is attached to protein substrates in a multi-step reaction involving three enzymes, such as Ub-activating enzyme (E1), Ub-conjugating enzyme (E2), and Ub ligase enzyme (E3).5 In an adenosine triphosphate (ATP)-dependent process, E1 activates the Ub molecule by forming a thioester bond between the carboxy-terminal end of the Ub molecule and the catalytic cysteine (Cys) of E1. Thioester-linked Ub is then transferred to a Cys residue of an E2 enzyme. The enzyme transfers the activated Ub from an E2-Ub thioester intermediate to a lysine residue of the substrate protein, either directly or in cooperation with a specific E3 enzyme. Most eukaryotes, including plants, have only a small number of E1 enzymes and multiple E2 and E3 enzymes. It is believed that the Arabidopsis genome encodes only two E1s, 45 E2s and E2-like proteins, and more than 1200 E3 ligases.4 E3 ligases are the most numerous because they confer specificity to ubiquitination by recognizing substrate proteins and by mediating the transfer of Ub from an E2 protein to a substrate.6 Therefore, the E3 ligase is considered the major component that recognizes target substrates in the pathway. E3 ligases can be grouped into two main classes based on the presence of their functional domains; one class has a single subunit and includes ligases, such as the homologous to E6-AP C-terminus (HECT), and a RING/U-box domain. The other class forms a multi-subunit complex and includes the S-phase kinase-associated protein 1 (SKP1)-cullin1 (CUL1)-F-box (SCF) complex and the anaphase-promoting complex (APC).7,8
Cys-rich really interesting new gene (RING) proteins are characterized either by the presence of a zinc-binding motif, or by the RING finger domain. The RING domain (Cys-X2-Cys-X9-39-Cys-X3-His-X2-3-Cys/His-X2-Cys-X4-48-Cys-X2-Cys, where X can be any amino acid) is similar to the DNA-binding zinc finger domain in that Cys or His residues coordinate two zinc ions. The RING domain is believed to play a role in protein–protein interactions (PPIs).9 The well-characterized plant genomes of Arabidopsis and rice are known to harbour 426 and 425 predicted RING-type E3 ligases in high proportions, respectively.10,11 The canonical RING domains can be sub-categorized into RING-H2 and RING-HC subtypes based on the fifth coordination Cys or His residues, respectively. A number of RING E3 ligases have been reported to play important roles in response to environmental stresses, such as drought and salt conditions. For example, salt- and drought-induced ring finger 1 (SDIR1) and small RING-H2 protein RHA2a E3 ligases have been reported to enhance salt and drought stress-responsive abscisic acid (ABA) signalling.12,13 The dehydration-responsive element-binding protein 2A (DREB2A)-interacting protein 1 and 2 (DRIP1 and DRIP2) E3 ligases negatively regulate the DREB2A transcription factor, leading to the down-regulation of drought–stress response genes.14 Hot pepper RING membrane-anchor 1 homologue 1 (Rma1H1) E3 ligase positively regulates the plasma membrane aquaporin PIP2;1 under water-deficient conditions.15 In addition, AtAIRP2 (RING-HC type) has been well characterized as a positive regulator ABA-dependent drought responses.16 In contrast, Oryza sativa drought-induced seven in absentia (SINA) protein 1 (OsDIS1) E3 ligase plays a negative role in drought–stress tolerance through transcriptional regulation of stress-related genes by post-translational regulation of OsNek6 in rice.17 However, the functional relationship between rice RING E3 ligases and abiotic stresses is largely unknown.
Previously, we identified 425 genes harbouring RING domains and classified them into four groups: RING-H2, RING-HC, RING-v, and RING-C2. We then evaluated the expressional diversity and evolutionary dynamics of 369 RING finger protein (RFP) genes by using a rice public genome array dataset.11 In this study, the expression patterns of 47 O. sativa RFP (OsRFP) genes in response to abiotic stresses were analysed via semi-quantitative RT-PCR and in silico analysis. Subsequently, molecular dissection of nine OsRFPs was performed by examining their E3 Ub ligase activity, subcellular localization, and physical interaction with other proteins (interactors). We further constructed a physical interaction network with 120 interactors. In addition, heterogeneous overexpression of OsRFPs in Arabidopsis was performed in order to reveal their molecular function in response to drought and salinity stresses.
2. Materials and methods
2.1. Plant materials for RT-PCR
Rice seedlings (O. sativa L. cv. Donganbyeo) were grown on mesh supported in plastic containers with Murashige and Skoog (MS) solution in a growth chamber (16/8-h light/dark photoperiod at 25°C with 70% relative humidity) for 14 days. For abiotic stress treatments, seedlings were treated with salinity (200 mM NaCl), dehydration, cold (4°C), and heat (45°C). Leaf tissues were harvested 0, 1, 6, 12, and 24 h after stress treatment, and control leaf tissues were also sampled at the same time. The samples were frozen using liquid nitrogen and immediately stored at −80°C. Total RNAs were extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer's protocol. Semi-quantitative RT-PCR was performed as described previously.11 First-strand cDNA synthesis was performed using 1 μg total RNA and a cDNA synthesis kit (Takara-Bio, Shiga, Japan). A total of 47 gene-specific primer pairs for OsRFPs were designed using Primer-BLAST (NCBI, http://www.ncbi.nlm.nih.gov/tools/primer-blast/). Os18S-rRNA18 was used as an internal control. A list of all primer pairs are provided in Supplementary Table S1.
2.2. Subcellular localization
For subcellular localization, the coding sequence of the enhanced green fluorescent protein (EGFP) and multiple cloning sites were PCR-amplified using primers 5′-GAGATCTAGAGGATCCCCCGGGATGGTGAGCAAGGGCGAG-3′ (forward) and 5′-GAGAGAGCTCTTACTTGTACAGCTCGTC-3′ (reverse) from the pEGFP-C1 vector as a template (Clontech, Palo Alto, CA, USA). Both the PCR product and the pBIN35S vector were digested with XbaI and SacI and were ligated together. The coding regions of the full-length cDNAs of OsRFPH2-3, OsRFPH2-14, OsRFPH2-16, OsRFPH2-23, OsRFPHC-2, OsRFPHC-3, OsRFPHC-4, OsRFPHC-13, and OsRFPV-6 were amplified with the appropriate primer pairs and were then cloned into a 35S:EGFP vector (Supplementary Table S1). The recombinant vectors were verified by sequencing. The resulting plasmids were transformed into Agrobacterium tumefaciens strain GV3101 and used for agro-infiltration. For transient expression of OsRFPs in Nicotiana benthamiana leaves, the Agrobacterium strains harbouring each construct were grown in liquid yeast extract peptone (YEP) medium and resuspended in infiltration medium [10 mM 4-Morpholineethanesulfonic acid hydrate (MES), pH 5.6, 10 mM MgCl2, and 200 mM acetosyringone]. The cell cultures were injected into 4-week-old N. benthamiana leaves (final optical density at 600 nm = 0.5). Several leaves were cut and then observed under confocal microscopy at the Korea Basic Science Institute, Chuncheon Center.
2.3. In vitro ubiquitination assay
Full-length OsRFP coding sequences (OsRFPH2-3, OsRFPH2-7, OsRFPH2-14, OsRFPH2-16, OsRFPH2-23, OsRFPHC-2, OsRFPHC-3, OsRFPHC-13, and OsRFPV-6) were amplified by RT-PCR with the appropriate primer pairs (Supplementary Table S1). The PCR products were digested and cloned into a pMAL-c5X vector (New England BioLabs, Ipswich, MA, USA). Nine recombinant maltose-binding protein (MBP)-OsRFP plasmids and the MBP-empty plasmid (as a negative control) were expressed in E. coli strain BL21 (DES) pLysS (Promega, Madison, WI, USA) and purified by affinity chromatography using amylose resin (New England BioLabs). The full-length coding sequences of AtUBC10 and AtUBC11 were cloned into the pET-28a (+) vector (Novagen, Gibbstown, NJ, USA). 6X His-AtUBC10 and 6X His-AtUBC11 were expressed in E. coli strain BL21 and purified using a Ni-NTA Purification System (Invitrogen).
In vitro self-ubiquitination assays were conducted according to the methods of Hardtke et al.19 with some modifications. Briefly, each MBP-OsRFP (250 ng) was purified and mixed with 50 ng yeast E1 (Boston Biochemicals, Cambridge, MA, USA), 250 ng Arabidopsis E2s, and 10 μg bovine Ub (Sigma-Aldrich, St. Louis, MO, USA) before incubation in ubiquitination reaction buffer [50 mM Tris–HCl, pH 7.5, 10 mM MgCl2, 0.05 mM ZnCl2, 1 mM ATP, 0.2 mM dithiothreitol (DTT), 10 mM phosphocreatine, and 0.1 U creatine kinase (Sigma-Aldrich)]. The reaction was stopped by the addition of 6× SDS sample buffer followed by a 3-h incubation at 30°C. Twenty microliters of each reaction mixture was analysed via 10–14% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) depending on the molecular weight of different fusion proteins and then transferred to a nitrocellulose membrane. Immunoblot analyses were conducted using anti-Ub antibodies (Sigma-Aldrich) with secondary goat anti-rabbit IgG peroxidase antibodies (Sigma-Aldrich). Detection was carried out using the chemiluminescent substrate SuperSignal West Pico (Thermo Scientific, Waltham, MA, USA) for horse-radish peroxidase and imaged on an X-ray film (Kodak).
2.4. Yeast two-hybrid screening
A rice cDNA library in the pGADT7-AD (GAL4 activation domain) was generated from salt-treated 14-day-old seedlings. Full-length coding sequences of OsRFPH2-3, OsRFPH2-14, OsRFPH2-16, OsRFPH2-23, OsRFPHC-2, OsRFPHC-13, and OsRFPv-6 were amplified and cloned into pGBKT7-BD (GAL4 DNA-binding domain) vector as bait. A library screening for proteins that interact with OsRFPs was conducted in accordance with the recommended commercial procedures (Make Your Own ‘Mate & Plate TM’ Library System; Matchmaker Gold Yeast Two-Hybrid System; Yeastmaker Yeast Transformation System 2, Clontech). A total of 1540 yeast transformants were selected on a synthetic defined (SD) medium lacking Leu and Trp with 40 μg/ml X-α-Gal and 70 ng/ml aureobasidin A (DDO/X/A). The selected transformants were repatched or dotted on SD medium lacking His, Leu, and Trp with 40 μg/ml X-α-Gal and 70 ng/ml AbA (QDO/X/A). A total of 154 clones were identified as positive interactors with the OsRFPs and were sequenced and verified by BLASTp searches in the GenBank database (http://blast.ncbi.nlm.nih.gov/).
2.5. Arabidopsis transformation
Agrobacterium strains GV3101 containing 35S:OsRFP-EGFPs or 35S:EGFP (control) were used to transform Arabidopsis plants according to the floral-dip method.20 For selection of transgenic lines, seeds harvested from T3 transformants were placed on MS agar plates containing 50 μg/ml kanamycin.
2.6. Plant growth and treatments
Three-independent 35S:OsRFP-EGFP- and 35S:EGFP-overexpressing Arabidopsis lines were tested to observe the effects of salt and drought stresses. For seed germination assays, seeds of OsRFPH2-3-EGFP, OsRFPH2-14-EGFP, OsRFPH2-16-EGFP, OsRFPH2-23-EGFP, OsRFPHC-2-EGFP, OsRFPHC-3-EGFP, OsRFPHC-4-EGFP, OsRFPHC-13-EGFP, OsRFPV-6-EGFP, and 35S:EGFP (control) were sterilized in 70% ethanol for 5 min followed by 20% bleach for 3 min and were then rinsed three times with double-distilled water. Sterilized seeds were sown and monitored on 1/2 MS medium containing different concentrations (0, 100, or 150 mM) of NaCl and (−0.25, −0.5, or −0.7 MPa) of PEG 8000 (Sigma-Aldrich). Germination percentages were scored and calculated at 1-day interval for 7 days. PEG plates were prepared as described by Verslues et al.21 Briefly, 40 ml of 1.5% agar containing 1/2 MS medium was solidified for 16 h and then overlaid with 60 ml of a solution containing 0, 250, or 400 g/l PEG 8000, yielding final MPas of −0.25, −0.5, or −0.7, respectively. The PEG solution was allowed to stand for 24 h and was then removed from the plates. For root elongation assays, transgenic seeds were germinated on 1/2 MS medium for 2 days and transferred into medium containing 100 mM NaCl and −0.5 MPa PEG. Plant growth was then monitored and photographed after 7 days, at which point root length was analysed using Image J software.
2.7. Transcriptome analysis and in silico subcellular localization prediction
The cell intensity file (CEL) files of Affymetrix GeneChip genome arrays for four abiotic stresses [drought, salt, cold (GSE6901), and heat stress (GSE14245)] of O. sativa were downloaded from the Gene Expression Omnibus (GEO) datasets of the NCBI database (http://www.ncbi.nlm.nih.gov/geo/) and were then normalized by the robust multi-array average (RMA) method with R statistical software (http://www.r-project.org/). The similarities of expression patterns between genes under the different stresses were evaluated via Pearson's correlation coefficient (PCC). The regulatory pathways of genes were predicted by using MapMan software.22 Heat maps of the genes were visualized by the MEV v4.6 program (http://www.tm4.org/) using hierarchical clustering with default parameters. In silico subcellular localization was predicted by WoLF PSORT tools23 with default parameters, and the predicted localizations with the highest PSORT feature values were then chosen. An interaction map consisting of OsRFPs and interactors was visualized using Cytoscape software.24
3. Results
3.1. Expression patterns of 47 OsRFP genes in response to abiotic stresses
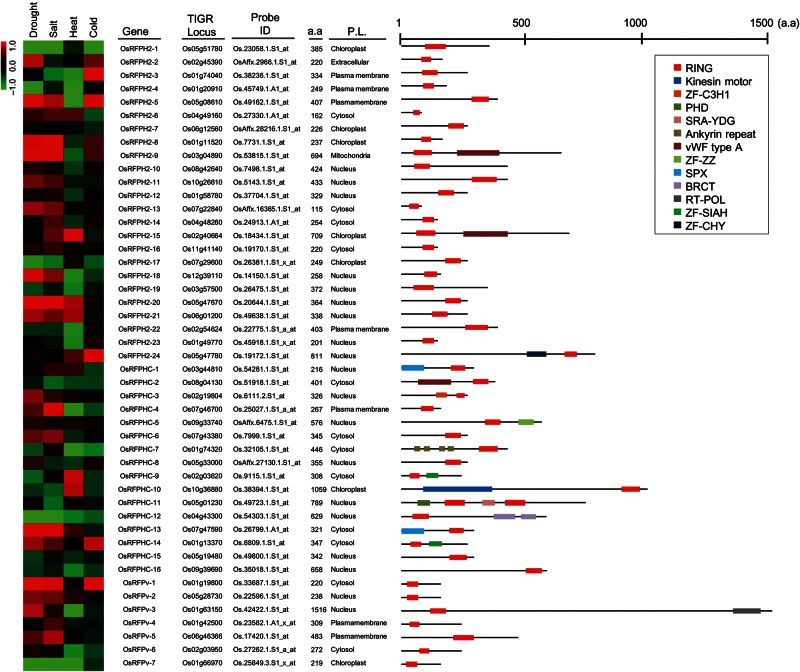
A total of 47 OsRFP genes, including 24 RING-H2, 16 RING-HC, and 7 RING-v subtypes, were randomly selected from targets with high levels of expression in specific tissues and/or stress treatments.11 In an effort to evaluate the expression patterns of 47 OsRFP genes in response to abiotic stresses, we retrieved microarray datasets for different stressors, including drought (GSE6901), salt (GSE6901), heat (GSE6901), and cold (GSE14245) in rice deposited to the GEO database (Fig. 1).25
Figure 1.
Expression patterns, general information, and gene structures of 47 RFPs in rice. Expression patterns under conditions such as drought, salt, cold (GSE6901), and heat (GSE14245) were retrieved from the GEO database (http://www.ncbi.nlm.nih.gov/geo/). Probe ID and a.a indicate the probe of Affymetrix GeneChip genome arrays and the protein length (number of amino acids), respectively. P.L. indicates the predicted subcellular localizations by WoLF PSORT Server (http://psort.hgc.jp/). The gene structures were retrieved from the Interpro database (http://www.ebi.ac.uk/interpro/). The colours of boxes represent different protein domains.
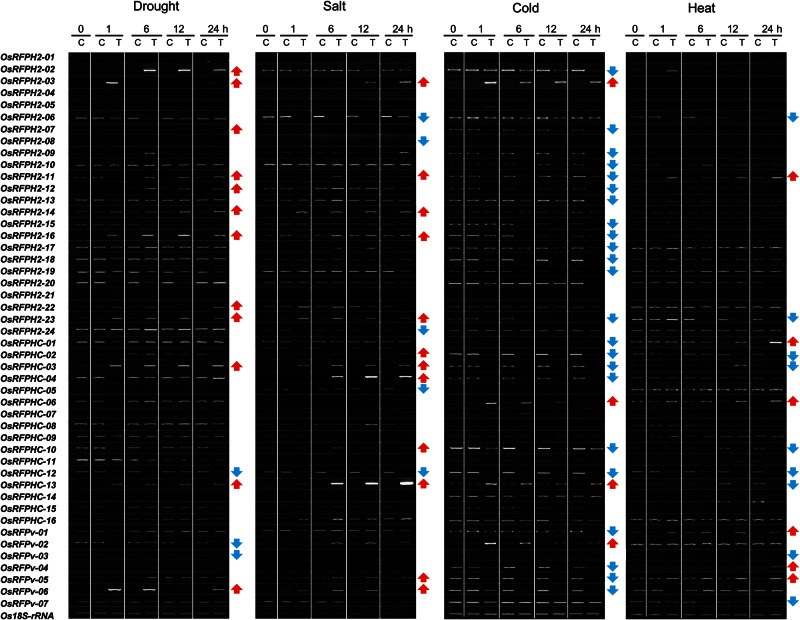
We next conducted semi-quantitative RT-PCR analysis with leaf tissues of rice seedlings (O. sativa) subjected to dehydration, high salinity (200 mM NaCl), cold (4°C), and basal heat (45°C) stress conditions for 2 days and then evaluated the transcript levels of 47 OsRFP genes (Fig. 2). At least 60% (8 of 12) of the seedlings up-regulated OsRFP genes as a result of dehydration stress. The seedlings also showed a tendency to increase OsRFP transcript levels in response to salinity. For example, eight genes, i.e. OsRFPH2-3, OsRFPH2-11, OsRFPH2-14, OsRFPH2-16, OsRFPH2-23, OsRFPHC-03, OsRFPHC-13, and OsRFPv-6, were up-regulated under drought and salinity conditions. In addition, OsRFPH2-2, OsRFPH2-7, OsRFPH2-12, and OsRFPH2-22 were up-regulated under drought, whereas OsRFPHC-2, OsRFPHC-4, OsRFPHC-10, and OsRFPv-6 were down-regulated under salt stress. One gene (OsRFPHC-12) and five genes (OsRFPH2-6, OsRFPH2-8, OsRFPH2-24, OsRFPHC-5, and OsRFPHC-12) were down-regulated under drought and salt, respectively. Under cold stress conditions, transcripts levels of four OsRFP genes increased, whereas 23 OsRFP genes were down-regulated. For heat stress conditions, six genes were up-regulated, whereas nine genes were down-regulated. These findings support the hypothesis regarding evolutional fate of duplicate genes, i.e. there is functional diversity and redundancy in OsRFP genes.
Figure 2.
Expression analysis of 47 OsRFP genes in rice plants. Expression patterns of OsRFP genes subjected to plant abiotic stresses. Fourteen-day-old rice seedlings were treated with dehydration (dehydration on two pieces of tissue papers), cold (4°C), or heat (45°C). Leaf tissues were sampled at 0, 1, 6, 12, and 24 h. Os18S-rRNA was used as an internal control. C and T indicate control and stress-treated samples, respectively. Red and blue arrows indicate up-regulation or down-regulation of indicated genes, respectively.
To better understand the subcellular distribution of OsRFPs, we performed in silico prediction using WolF PSORT.23 At least 66.0% (31 of 47) of OsRFPs were associated with the nucleus (19 OsRFPs) and the cytosol (12 OsRFPs). The other OsRFPs were predicted to localize to the chloroplast (seven OsRFPs), plasma membrane (seven OsRFPs), mitochondria (one OsRFP), and extracellular matrix (one OsRFP), as shown in Fig. 1 and Supplementary Fig. S1. To further define the structures of the 47 OsRFPs, we examined whether these OsRFPs harboured any other functional domains using the InterPro search program. A total of 32 OsRFPs did not harbour any other detectable domains, whereas 15 proteins had one or more functional domains (Fig. 1). Three of these domains harboured zinc finger domains, such ZF-C3H1, ZF-SIAH, and ZF-CHY, which have been characterized as nucleic acid-binding zinc fingers, while one was the zinc finger domain, ZF-ZZ, which is believed to mediate PPIs. Furthermore, other PPI domains were found, including ankyrin repeats, after the C_terminal domain of a breast cancer susceptibility protein (BRCT), and kinesin motor domains, which may function as the substrate-binding domains of E3 ligases.10
3.2. Subcellular localization of OsRFP–EGFP fusion proteins
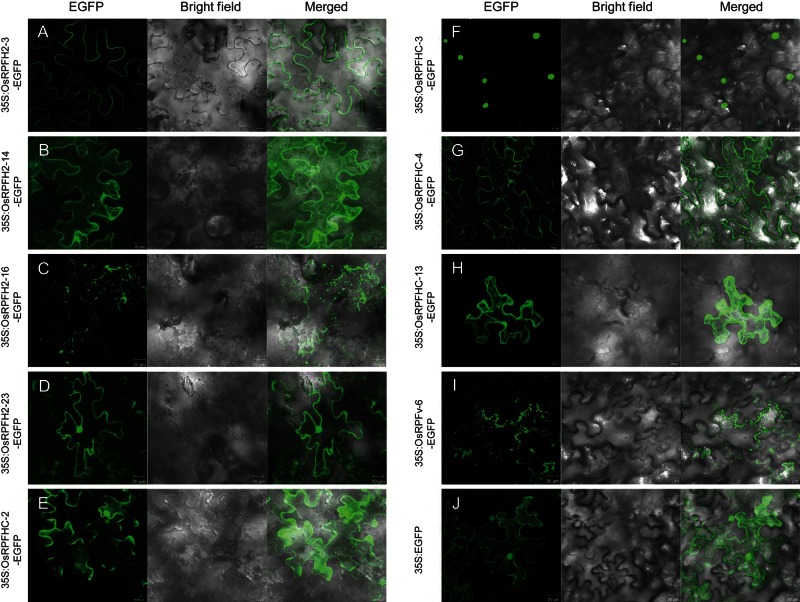
It is generally believed that the subcellular localization of a gene is important in understanding its cellular function. Therefore, we decided to further examine the subcellular localization of 10 OsRFP genes (OsRFPH2-2, OsRFPH2-3, OsRFPH2-14, OsRFPH2-16, OsRFPH2-23, OsRFPHC-2, OsRFPHC-3, OsRFPHC-4, OsRFPHC-13, and OsRFPv-6), chosen depending on their expression patterns under salt and dehydration conditions. To do this, full-length OsRFP cDNAs were cloned into 35S:EGFP and transformed into Agrobacterium strain GV3101. Each of the 10 OsRFP-EGFPs and the control (35S:EGFP) were transiently expressed in N. benthamiana leaves. As shown in Fig. 3J, transient expression of 35S:EGFP was found in both the cytosol and nucleus. Fluorescent signals of each of the fusion proteins (OsRFPH2-14-EGFP, OsRFPHC-2-EGFP, and OsRFPHC-13-EGFP) were localized to the cytoplasm. Three proteins, OsRFPH2-16-EGFP, OsRFPHC-4-EGFP, and OsRFPv-6-EGFP, appeared as small, punctuate spots with their aggregates in the cytoplasm. Interestingly, subcellular localization of OsRFPHC-4-EGFP was observed as punctate spots not only in the cytoplasm, but also in the plasma membrane. In addition, OsRFPH2-3-EGFP and OsRFPHC-3-EGFP were localized to both the plasma membrane and nucleus, respectively. OsRFPH2-23-EGFP was localized to both the plasma membrane and nucleus, respectively. However, OsRFPH2-2-EGFP did not show any fluorescence in cells (data not shown).
Figure 3.
Subcellular localization of the OsRFP–EGFP fusion proteins in tobacco leaves. Agrobacterium strains GV3101 harbouring each construct of 35S:OsRFPH2-3-EGFP (A), 35S:OsRFPH2-14-EGFP (B), 35S:OsRFPH2-16-EGFP (C), 35S:OsRFPH2-23-EGFP (D), 35S:OsRFPHC-2-EGFP (E), 35S:OsRFPHC-3-EGFP (F), 35S:OsRFPHC-4-EGFP (G), 35S:OsRFPHC-13-EGFP (H), 35S:OsRFPv-6-EGFP (I), or 35S:EGFP (J) were transiently expressed with p19 in Nicotiana leaves. Images were captured and merged by z-series optical sections after 3 days of agro-infiltration. A 35S:EGFP construct was used as a control.
3.3. Rice RING domain possess E3 Ub ligase activity
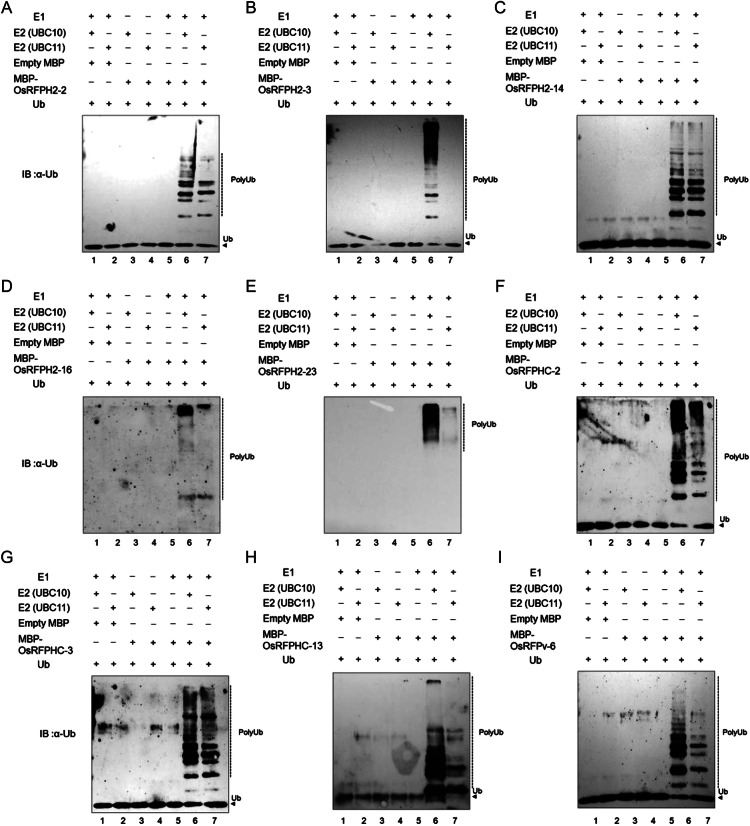
Proteins harbouring RING domains are involved in numerous cellular processes. Functions attributed to the RING domain itself include PPIs and degradation via Ub-26S proteasome. RFPs can be classified based on the presence of either a Cys or His residue in their RING domains. However, it is still an open question as to whether the modified RING domains function as E3 ligases. We therefore used in vitro ubiquitination assays to examine a total of 10 OsRFPs (OsRFPH2-2, OsRFPH2-3, OsRFPH2-14, OsRFPH2-16, OsRFPH2-23, OsRFPHC-2, OsRFPHC-3, OsRFPHC-4, OsRFPHC-13, and OsRFPv-6), chosen on the basis of their expression patterns under salt and dehydration conditions. Each of the 10 OsRFPs was fused with MBP and expressed in E. coli. However, MBP-OsRFPHC-4 was never expressed in this E. coli system, even under several modified conditions. A total of nine MBP-OsRFPs, with the exception of OsRFPHC-4, were purified by affinity chromatography using amylose resin. The purified MBP-OsRFP fusion proteins were mixed with Ub, ATP, E1 (yeast), and E2s (AtUBC10 or AtUBC11) and then incubated for 3 h. Immunoblot analysis with anti-Ub antibodies showed that a high-molecular mass poly-Ub chain was present in eight OsRFP samples (Fig. 4). However, no poly-Ub chain was observed when MBP-OsRFPH2-3 was incubated with AtUBC11 (Fig. 4B, lane 7). In addition, omission of E2s, E1, or MBP-OsRFPs from the activity assay resulted in a loss of protein ubiquitination. Thus, most of the selected OsRFPs were capable of mediating E2-dependent protein ubiquitination in vitro.
Figure 4.
In vitro E3 ligase ubiquitin ligase activity of MBP-OsRPFs. The ubiquitination reaction contained E1, atE2s (Arabidopsis UBC10 and UBC11), maltose-binding protein-tagged OsRFPH2-2 (A), OsRFPH2-3 (B), OsRFPH2-14 (C), OsRFPH2-16 (D), OsRFPH2-23 (E), OsRFPHC-2 (F), OsRFPHC-3 (G), OsRFPHC-13 (H), or OsRFPv-6 (I), Ub, and ATP. Poly-Ub chains were detected by immunoblotting with a Ub-specific antibody.
3.4. Physical interaction map of OsRFPs
A yeast two-hybrid (Y2H) screen was conducted in order to retrieve proteins that interacted with each of the 10 OsRFPs Ub E3 ligases. Of these, seven OsRFP genes (OsRFPH2-3, OsRFPH2-14, OsRFPH2-16, OsRFPH2-23, OsRFPHC-2, OsRFPHC-13, and OsRFPv-6) were screened, and their interactors were identified via sequencing analysis. Unfortunately, the transformed yeast cells expressing OsRFPHC-3 did not survive on the selection medium for more than three replications and, thus, the gene was excluded from further experiments. Positive interactors were not found for two genes, such as OsRFPH2-2 and OsRFPHC-4. A total of 229 positive clones were screened and selected depending on their α-galactosidase activity, resulting in 147 positive interactors (Supplementary Fig. S2). To understand their cellular distribution, we performed in silico prediction analysis by examining their subcellular localization (Fig. 5A and Supplementary Fig. S2).
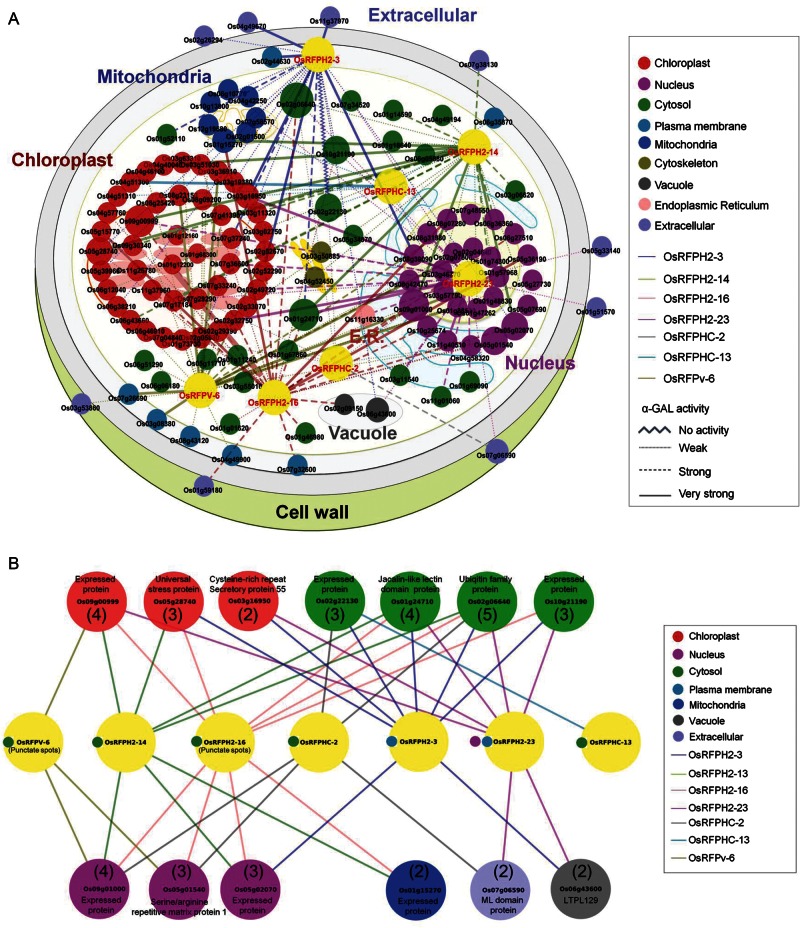
Figure 5.
The interaction network of rice OsRFPs. (A) A total of 147 interactors with drought- and/or salt-induced OsRFPH2-3, OsRFPH2-14, OsRFPH2-16, OsRFPH2-23, OsRFPHC-2, OsRFPHC-13, and OsRFPv-6 proteins detected by yeast two-hybrid screens are shown as a physical interaction network. Visualization of yeast two-hybrid screening of OsRFPs using cytoscape (http://www.cytoscape.org/). Different node colours and node line colours indicate subcellular localization and each OsRFP protein, respectively. Large nodes indicate multiple interaction proteins with OsRFPs and different forms of node lines indicate α-GAL activity. Subcellular localization and α-GAL activity of all of the interacting proteins based on prediction and experimental data are shown in Supplementary Fig. S2. (B) Proteins that exhibited multiple interactions with OsRFPs. Numbers in parentheses indicate the number of interaction with OsRFPs.
The Y2H screenings also showed that the seven OsRFPs interacted with various environmental stress-related proteins (Supplementary Fig. S2). For example, membrane-localized OsRFPH2-3 interacted with peptidyl-proly cis-trans isomerase, Peptidyl-prolyl cis-trans isomerase, FKBP-type (Os02g52290), aquaporin protein (Os02g44630), wound-induced protein (Os11g37970), Cys-rich repeat secretory protein 55 (Os03g16950), coiled-coil domain-containing protein (Os08g39090), and DUF581 domain-containing protein (Os04g49670). OsRFPH2-14 interacted with copper methylamine oxidase (Os04g40040), jacalin-like lectin domain-containing protein (Os01g24710), profilin domain-containing protein (Os06g05880), and universal stress protein (Os05g28740). The OsRFPH2-16 protein interacted with glycosylhydrolase and OsFBT7-F box and tubby domain-containing protein (Os05g36190). OsRFPH2-23 interacted with RNA recognition motif-containing protein (Os11g40510), Cys-rich repeat secretory protein 55 precursor (Os03g16950), glycosyltransferase (Os02g32750), and jaclin-like lectin domain-containing protein (Os01g24710). OsRFPHC-2 and OsRFPHC-13 interacted with myeloid differentiation protein (MD)-2-related lipid-recognition domain protein (Os07g06590) and peroxidase precursor (Os04g51300), respectively. OsRFPv-6 interacted with early flowering 3 (ELF3) protein (Os01g38530), indole-3-glycerol phosphate synthase (Os08g23150), ABC transporter (Os03g08380), Myb-like DNA-binding domain-containing protein (Os02g04640), S-adenosylmethionine-dependent methyltransferase (Os03g36910), and aquaporin protein (Os07g26690). Furthermore, we found multiple proteins that interacted with more than two OsRFPs, including Ub family protein (Os02g06640) and jacalin-like lectin domain-containing protein (Os01g24710), six expressed proteins (Os09g00999, Os02g22130, Os10g21190, Os09g01000, Os05g02070, and Os01g15270), universal stress protein (Os05g28740), and serine/arginine repetitive matrix protein 1 (Os05g01540; Fig. 5B).
An obvious difference was observed in the frequencies of subcellular localizations of these interactors. As shown in Fig. 5A and Supplementary Table S2, about 36.7% (44 of 120) of interactors of the seven OsRFPs were predicted to localize to chloroplasts. In contrast, 19.2% (23 of 120) and 20.8% (25 of 120) of the interactors were predicted to localize to both the cytosol and nucleus, respectively.
An in silico analysis of the subcellular localizations of the interactors and in vivo subcellular localizations of OsRFPs demonstrated the dynamic interactions of OsRFPs with other subcellularly localized protein partners (Fig. 5A and Supplementary Table S2). For example, even though OsRFPH2-23-EGFP was targeted to the nucleus in vivo, it showed physical interactions with chloroplast-localized proteins (i.e. Os03g16950, Os02g32750, Os02g49720, Os07g36600, and Os07g29290) as well as nuclear-targeted proteins (Os11g40510 and Os08g31980). In addition, the plasma membrane-associated protein, OsRFPH2-3, exhibited interactions with eight chloroplast-targeting proteins (i.e. Os02g52290, Os07g37240, Os03g19380, Os03g16950, Os06g38210, Os05g28740, Os05g39960, and Os02g05830) and two nuclear-targeted proteins (Os05g02070 and Os08g39090).
3.5. Comparative transcriptome analysis of OsRFPs with interactors
To further investigate the functional relationships of seven OsRFP genes and their interactors against four abiotic stresses (drought, salt, heat, and cold), their expression patterns in response to these stresses were evaluated using a public microarray dataset (Supplementary Table S3). A total of 7 OsRFP genes and 112 interactor genes were found to correspond to one or more probes, respectively, whereas no probes were found for eight interactors. Differentially expressed genes (DEGs) were classified as either up-regulated (log2-fold change >1) or down-regulated (log2-fold change <−1) via a comparison of probe signal intensities with treated plants and untreated plants. Of the 119 genes, 52 and 25 genes were either up-regulated or down-regulated under one or more stress treatments, respectively, and were considered as DEGs (Fig. 6A). A total of 25 (48%) up-regulated and 2 (8.0%) down-regulated genes commonly responded to both drought and salt, supporting the hypothesis that the genes may exhibit similar molecular functions in both stresses. In contrast, heat stress resulted in the up-regulation of 13 genes and the down-regulation 12 genes. These genes were not modulated in response to any other stresses.
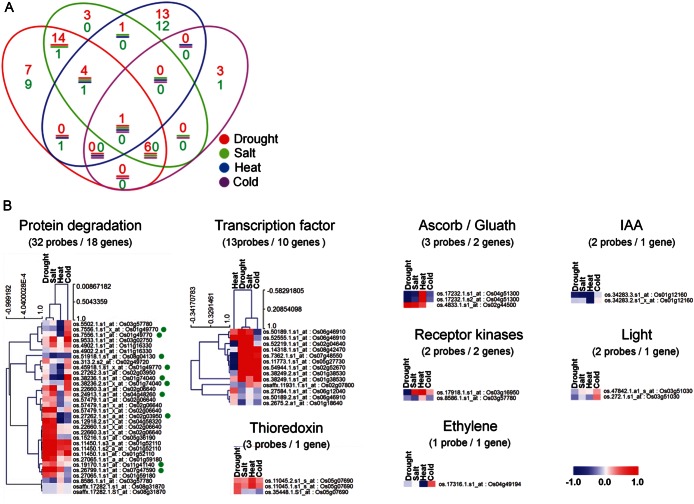
Figure 6.
Distribution of DEGs among seven OsRFPs and their interactors in the context of four abiotic stresses in rice. (A) Dendrogram of DEGs. The red and green numbers indicate up-regulation (log2 > 1) and down-regulation (log2 < −1), respectively. The circle colours represent different abiotic stresses. (B) Expression patterns of OsRFPs and their interactors in different functions detected by MapMan (http://mapman.gabipd.org). Green circles in protein degradation indicate the seven OsRFPs.
We then grouped the DEGs depending on their activities in different regulatory pathways by using the MapMan program. A total of eight pathways, i.e. protein degradation, transcription factor, thioredoxin, ascorb/gluath, receptor kinases, ethylene, indole-3-acetic acid (IAA), and light, were mapped (Fig. 6B). As expected, the genes with the highest frequency were those involved in protein degradation (18 genes). The genes involved in transcription factors had the second highest frequency (10 genes). Similarly, genes included in both pathways exhibited closer relationships in expression patterns between drought and salt stresses.
3.6. Tolerance evaluation by overexpression of each OsRFP gene in Arabidopsis grown under salt and drought conditions
The finding that the selected OsRFP genes were up-regulated in response to salt and/or dehydration treatment may provide some clues regarding the molecular functions of these genes against stresses. Therefore, to further examine their functions, we generated eight transgenic Arabidopsis overexpressing each gene (35S:RFPH2-3-EGFP, 35S:RFPH2-14-EGFP, 35S:RFPH2-16-EGFP, 35S:RFPH2-23-EGFP, 35S:RFPHC-2-EGFP, 35S:RFPHC-3-EGFP, 35S:RFPHC-13-EGFP, and 35S:RFPv-6-EGFP) and examined their tolerances against both stresses. The Agrobacterium strains GV3101 containing 35S:OsRFP-EGFPs and 35S:EGFP (control) were used to transform Arabidopsis plants. Three-independent transgenic lines (T3) were selected depending on the expression levels of the OsRFP genes by RT-PCR under normal conditions (Supplementary Fig. S3).
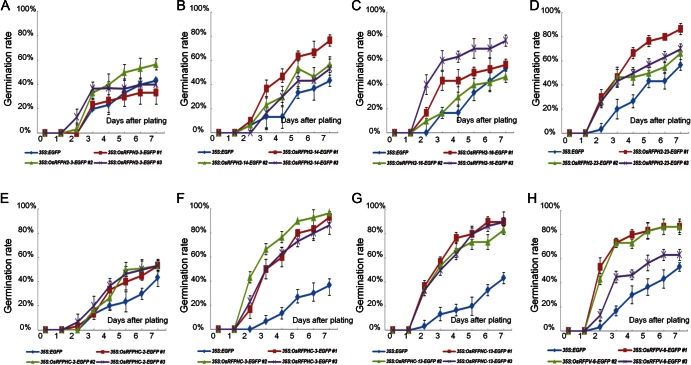
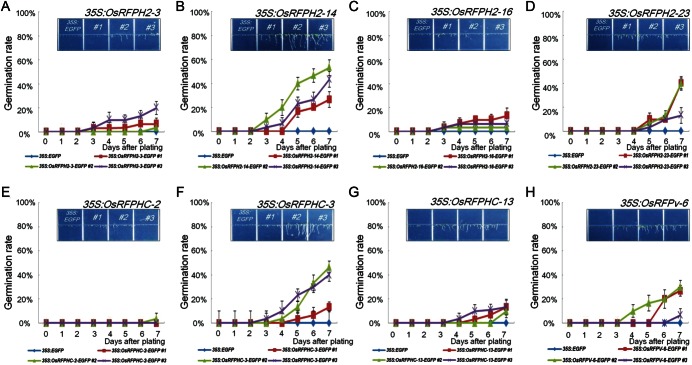
For germination rate analysis, sterilized seeds of three-independent lines of eight transgenic plants as well as 35S:EGFP were placed on 1/2 MS medium containing different concentrations of NaCl (0, 100, or 150 mM) and PEG (0 or −0.7 MPa). Germination rates of transgenic lines and control seeds were not significantly different on 1/2 MS medium without treatment (Supplementary Fig. S4). In contrast, parts of the transgenic seeds of OsRFP-EGFPs germinated faster than control seeds when seeds were plated on 1/2 MS medium supplemented with NaCl (150 mM) and PEG (−0.7 MPa; Figs 7 and 8). For example, even though about 46.6% of control seeds germinated 7 days after plating with 150 mM NaCl treatment, four transgenic lines exhibited >70% germination rations, including OsRFPH2-23 (74.4%), OsRFPHC-3 (92.2%), OsRFPHC-13 (87.7%), and OsRFPv-6 (78.8%; Fig. 7). In dehydration stress, control seeds were not germinated on PEG (−0.7 MPa) plates until 7 days after plating, but transgenic seeds of OsRFPH2-14, OsRFPH2-23, OsRFPHC-3, and OsRFPv-6 showed high germination rates with a range of 31–79% (Fig. 8).
Figure 7.
Increased germination rates of seven overexpressed OsRFP genes in Arabidopsis under salt stress conditions. Three-independent seeds from 35S:OsRFPH2-3-EGFP (A), 35S:OsRFPH2-14-EGFP (B), 35S:OsRFPH2-16-EGFP (C), 35S:OsRFPH2-23-EGFP (D), 35S:OsRFPHC-2-EGFP (E), 35S:OsRFPHC-3-EGFP (F), 35S:OsRFPHC-13-EGFP (G), and 35S:OsRFPv-6-EGFP (H) with control plants (35S:EGFP) were plated on 1/2 MS containing 150 mM NaCl. Germination percentages for each experiment were scored and calculated at 1-day interval for 7 days. Data represent means ± SDs of three-independent experiments (n = 30).
Figure 8.
An increased germination rate of overexpressed OsRFP genes in Arabidopsis under PEG stress conditions. Three-independent seeds from 35S:OsRFPH2-3-EGFP (A), 35S:OsRFPH2-14-EGFP (B), 35S:OsRFPH2-16-EGFP (C), 35S:OsRFPH2-23-EGFP (D), 35S:OsRFPHC-2-EGFP (E), 35S:OsRFPHC-3-EGFP (F), 35S:OsRFPHC-13-EGFP (G), and 35S:OsRFPv-6-EGFP (H) with control plants (35S:EGFP) were plated on 1/2 MS containing −0.7 MPa PEG. Germination percentages for each experiment were scored and calculated at 1-day intervals for 7 days. Data represent means ± SDs of three-independent experiments (n = 30).
For root growth assays, sterilized seeds of three-independent lines of eight OsRFP-EGFPs and 35S:EGFP were plated on 1/2 MS medium. Germinated seeds were then transferred onto 1/2 MS medium containing 100 mM NaCl and −0.5 MPa PEG. The root lengths of OsRFPH2-3, OsRFPH2-14, OsRFPH2-16, OsRFPH2-23, OsRFPHC-2, and OsRFPHC-13 were not significantly different from those of control plants; however, the root lengths of two transgenic lines (OsRFPHC-3 and OsRFPv-6) were significantly longer than those of control plants (Supplementary Fig. S5A). Overexpression of OsRFPHC-3 and OsRFPv-6 genes therefore improved seedling growth and root length under normal conditions. We further assessed the effects of salt (100 mM NaCl) and PEG (−0.5 MPa) treatments on the roots of transgenic seedlings. As shown in Supplementary Fig. S5B and C, most of the root lengths of transgenic lines were significantly longer than those of control plants.
Since parts of the transgenic plants exhibited tolerance to PEG treatment, we compared accumulation of hydrogen peroxide (H2O2) in transgenic and control plant leaves after dehydration treatment. To do this, each leaf was dehydrated for 90 min and then incubated with 3,3′-diaminobenzidine (DAB) staining solution. The analysis of DAB staining indicated that H2O2 accumulation was greater in all transgenic leaves than in control plants (Supplementary Fig. S6), suggesting that overexpression of OsRFPs in Arabidopsis caused dehydration-induced H2O2 accumulation. Finally, 2-week-old control and transgenic plants were exposed to drought stress for 10 days by being deprived of water. After the drought treatment, control and transgenic plants were irrigated for 5 days, and their survival rates were determined (Supplementary Fig. S7). When grown under normal conditions in soil, no significant difference in phenotypes was observed between control and transgenic plants. However, after exposure to drought stress (i.e. no irrigation for 10 days), the transgenic plants exhibited more tolerance than control plants. After watering again, the survival rates of the transgenic plants were significantly higher than those of the control plants (Supplementary Fig. S7). For example, the survival rates of OsRFPH2-3 and OsRFPHC-13 transgenic plants reached 67.6% (48 of 71 for #2) and 58.9% (43 of 73 for #1), while those in control plants only reached 4% (50 of 275) and 7.5% (6 of 79), respectively. These results implied that heterogeneous overexpression of the selected OsRFP genes in Arabidopsis can promote the germination rate, seedling growth, and survival rate under salt and drought stress conditions.
4. Discussion
Previously, we identified a gene family encoding rice RFPs and divided their members on the basis of the presence of a zinc-coordinating Cys or His residue in their RING domain.11 In addition, their expression patterns across a wide variety of tissues and environments were evaluated via in silico analysis, with a dataset of 155 GEO samples on a rice genome microarray. By using these methods, we were able to divide the 369 OsRFP genes into five groups. Interestingly, members in three of the groups showed distinct expression patterns in specific tissues and/or environments, leading us to further investigate their molecular functions. We randomly selected about half of the proteins in the three groups (47 of 102) and analysed their molecular functions to elucidate their evolutionary dynamics and fate.
The analysis of PPIs is critical to understanding the biological functions of different proteins during cellular processes. The interactions of RING E3 ligases are generally localized to the same organelles or an interactable subcellular compartment in the cell. For example, the Arabidopsis RING E3 ligase, DRIP1, which can interact with the transcription factor, DREB2A, and regulates DRIP1 protein levels via the 26S proteasome system, is localized to the nucleus.14 In addition, the endoplasmic reticulum membrane-localized Rma1H1 inhibits subcellular trafficking of ER-localized PIP2;1 to the plasma membrane.15 In this study, we demonstrated the subcellular localization of nine OsRFP-EGFPs via transient expression in tobacco cells and predicted that they had 120 interactors via in silico analysis (Figs 1–3). However, we observed inconsistencies in the subcellular colocalization of OsRFPs and their interactors. For example, OsRFPH2-14 was localized to the cytoplasm, and only 7 of its 22 interactors were associated with the same location. The plasma membrane-localized OsRFPH2-3 had only one interactor, aquaporin protein, which was also targeted to the plasma membrane. Interestingly, 44 of the 120 interactors of seven OsRFPs were localized to chloroplasts, even though there were no chloroplast-localized OsRFPs (Fig. 5A and Supplementary Table S2). Our findings therefore raise questions regarding the physical interactions of OsRFPs and their interactors in cases in which each OsRFP/interactor is localized to different compartments. An increasing body of evidence suggests that proteins can be translocated to other cellular organelles during certain physiological and environmental conditions.26,27 Additionally, a variety of studies have indicated that plant RING E3 ligases show dynamic translocation and partitioning for the activation of cellular process during specific conditions. For example, cytoplasm-localized Arabidopsis HOS1 RING E3 ligase accumulates in the nucleus in response to low temperature treatments and localizes to the cytoplasm without cold treatment.28 In addition, nuclear-localized COP1 RING E3 ligase is translocated to the cytoplasm under light signals for the activation of photomorphogenic development.29,30 Therefore, the translocation (or partitioning) of RING E3 ligases may be a general cellular mechanism through which plants adapt to the environment via post-translational modifications. Interestingly, one of the genes we studied, OsRFPHC-10, harbours a kinesin motor domain. The presence of this domain might be a reflection of the functional evolution of the members of the E3 ligase family to acquire their own mobility. Further analysis of OsRFPHC-10 is required to conclusively determine whether this is the case.
Our finding that some interactors exhibit multiple PPIs with one or more OsRFPs may support the hypothesis that there is complexity and redundancy in the post-translational regulation of substrate proteins via the 26S proteasome pathway. For example, the OsSalT (Os01g24710, jacalin-like lectin domain protein) protein appeared to interact physically with four OsRFPs (OsRFPH2-3, OsRFPH2-14, OsRFPH2-16, and OsRFPH2-23). Transcript levels and protein accumulation of OsSalT were reported to have increased significantly in response to salt and drought stresses.31–33 A possible hypothesis is that the stress-inducible protein, which is induced under a variety of environments, participates in multiple stress regulation pathways and is regulated by unique OsRFPs.
In the ubiquitination pathway, RING E3 ligases are generally known to recognize specific target proteins as substrates and directly transfer Ub from an E2 ligase to the substrate without an E3-Ub intermediate.6 The finding that five OsRFPs may exhibit physical interactions with the Ub protein is somewhat inconsistent with this notion. In contrast, a large number of Ub E3 ligases have been reported to have the ability to catalyse their own ubiquitination.34 Self-ubiquitination, which can occur in substrate-independent and substrate-dependent modes, supports the notion that RING E3 ligases can protect the substrate proteins or their own proteins from self-destruction.35,36 Thus, our results support the hypothesis that a large number of OsRFPs may have the ability for self-ubiquitination under specific conditions.
Increasing numbers of plant RING E3 ligases have been reported to regulate molecular responses to abiotic stresses, supporting the hypothesis that they play an important role in the regulation of stress-induced cellular processes. RING E3 ligases are known to positively and negatively regulate target proteins via the 26S proteasomal degradation pathway. For example, salt- and drought-induced SDIR1 are believed to be a positive regulator of ABA signalling.12 Drought-induced Rma1H1 also plays a role in responses to dehydration via inhibition of aquaporin trafficking between compartments, which lead to subsequent aquaporin degradation.15 In addition, OsDIS1 and DRIP1 E3 ligases play negative roles in drought–stress tolerance through post-translational modification of their transcription factors.14,17 The finding that overexpressing E3 ligases in most plants confer protection against salt and/or dehydration stress may be explained by their ability to regulate proteins via the 26S proteasomal degradation pathway. However, further research is required to confirm these results. The OsRFPH2-3 protein, whose overexpression in plants conferred protection against dehydration, may interact with an aquaporin protein and, thus, the inhibition of the aquaporin protein through OsRFPH2-3 E3 ligase could improve drought tolerance in transgenic plants. Similarly, the finding that OsRFPv-6 clearly confers salt and dehydration stress tolerance and interacts with aquaporin as well as ABC transporters37 may also support this hypothesis. Furthermore, Aharon et al.38 reported that overexpression of the plasma membrane aquaporin, PIP1b, in tobacco caused a hypersensitive phenotype to drought stress. The study suggested that enhanced water transport activity via plasma membrane aquaporins and ABC transporters may negatively affect plants during drought stress. Therefore, our findings regarding the dynamic responses of OsRFPs to abiotic stresses, the tolerance of their overexpression in plants, and their interactors' profiles may provide some clues regarding their molecular regulation of abiotic stress responses. Furthermore, the fact that heterogeneous overexpression of OsRFPs enhances drought and/or salt tolerance warrants further investigation, since it is a mechanism through which we can enhance monocot plants (including rice) using genetic approaches.
The finding that there was high accumulation of H2O2 during drought stress in all tested plants overexpressing E3 ligases compared with control plants may provide strong evidence regarding the relationship between H2O2 and stress tolerance in plants. H2O2 production may involve ABA-induced stomatal closure and act as an intermediate in ABA signalling.39 A host of studies have demonstrated the positive relationship between H2O2 accumulation and ABA-mediated drought–stress tolerance in overexpressing plants. For example, plants overexpressing AtAIRP2 (a cytosolic RING-HC type Arabidopsis E3 ligase) exhibited strong tolerance to drought stress, producing higher levels of H2O2 than knockout plants and wild-type plants.16 Therefore, our findings may be a good example of the positive relationship between H2O2 accumulation and stress tolerance in plants overexpressing the RING E3 ligase gene. In conclusion, the findings obtained in our comprehensive study may provide some insights into the biological functions of RING E3 ligases and may provide genetic strategies that promote plant adaptation to unfavourable environmental conditions.
Supplementary Data
Supplementary Data are available at www.dnaresearch.oxfordjournals.org.
Funding
This work was supported by grants from the BioGreen21 Program (no. PJ007043), the Next-Generation BioGreen 21 Program (Plant Molecular Breeding Center No. PJ009084), Rural Development Administration, Republic of Korea, and 2011 Research Grant from Kangwon National University (no. 120110129) to C.S.J.
Supplementary Material
Footnotes
Edited by Dr Mikio Nishimura
References
- 1.Wang W., Vinocur B., Altman A. Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta. 2003;218:1–14. doi: 10.1007/s00425-003-1105-5. doi:10.1007/s00425-003-1105-5. [DOI] [PubMed] [Google Scholar]
- 2.Hellmann H., Estelle M. Plant development: regulation by protein degradation. Science. 2002;297:793–7. doi: 10.1126/science.1072831. doi:10.1126/science.1072831. [DOI] [PubMed] [Google Scholar]
- 3.Bachmair A., Novatchkova M., Potuschak T., Eisenhaber F. Ubiquitylation in plants: a post-genomic look at a post-translational modification. Trends Plant Sci. 2001;6:463–70. doi: 10.1016/s1360-1385(01)02080-5. doi:10.1016/S1360-1385(01)02080-5. [DOI] [PubMed] [Google Scholar]
- 4.Vierstra R.D. The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci. 2003;8:135–42. doi: 10.1016/S1360-1385(03)00014-1. doi:10.1016/S1360-1385(03)00014-1. [DOI] [PubMed] [Google Scholar]
- 5.Vierstra R.D. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 2009;10:385–97. doi: 10.1038/nrm2688. doi:10.1038/nrm2688. [DOI] [PubMed] [Google Scholar]
- 6.Deshaies R.J., Joazeiro C.A. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. doi:10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 7.Smalle J., Vierstra R.D. The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 2004;55:555–90. doi: 10.1146/annurev.arplant.55.031903.141801. doi:10.1146/annurev.arplant.55.031903.141801. [DOI] [PubMed] [Google Scholar]
- 8.Moon J., Parry G., Estelle M. The ubiquitin-proteasome pathway and plant development. Plant Cell. 2004;16:3181–95. doi: 10.1105/tpc.104.161220. doi:10.1105/tpc.104.161220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borden K.L. RING domains: master builders of molecular scaffolds? J. Mol. Biol. 2000;295:1103–12. doi: 10.1006/jmbi.1999.3429. doi:10.1006/jmbi.1999.3429. [DOI] [PubMed] [Google Scholar]
- 10.Stone S.L., Hauksdottir H., Troy A., Herschleb J., Kraft E., Callis J. Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. 2005;137:13–30. doi: 10.1104/pp.104.052423. doi:10.1104/pp.104.052423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim S.D., Yim W.C., Moon J.C., Kim D.S., Lee B.M., Jang C.S. A gene family encoding RING finger proteins in rice: their expansion, expression diversity, and co-expressed genes. Plant Mol. Biol. 2010;72:369–80. doi: 10.1007/s11103-009-9576-9. doi:10.1007/s11103-009-9576-9. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y.Y., Yang C.W., Li Y., et al. SDIR1 is a RING finger E3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. Plant Cell. 2007;19:1912–29. doi: 10.1105/tpc.106.048488. doi:10.1105/tpc.106.048488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bu Q., Li H., Zhao Q., et al. The Arabidopsis RING finger E3 ligase RHA2a is a novel positive regulator of abscisic acid signaling during seed germination and early seedling development. Plant Physiol. 2009;150:463–81. doi: 10.1104/pp.109.135269. doi:10.1104/pp.109.135269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qin F., Sakuma Y., Tran L.S.P., et al. Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell. 2008;20:1693–707. doi: 10.1105/tpc.107.057380. doi:10.1105/tpc.107.057380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee H.K., Cho S.K., Son O., Xu Z.Y., Hwang I., Kim W.T. Drought stress-induced Rma1H1, a RING membrane-anchor E3 ubiquitin ligase homolog, regulates aquaporin levels via ubiquitination in transgenic arabidopsis plants. Plant Cell. 2009;21:622–41. doi: 10.1105/tpc.108.061994. doi:10.1105/tpc.108.061994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho S.K., Ryu M.Y., Seo D.H., Kang B.G., Kim W.T. The Arabidopsis RING E3 ubiquitin ligase AtAIRP2 plays combinatory roles with AtAIRP1 in abscisic acid-mediated drought stress responses. Plant Physiol. 2011;157:2240–57. doi: 10.1104/pp.111.185595. doi:10.1104/pp.111.185595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ning Y., Jantasuriyarat C., Zhao Q., et al. The SINA E3 ligase OsDIS1 negatively regulates drought response in rice. Plant Physiol. 2011;157:242–55. doi: 10.1104/pp.111.180893. doi:10.1104/pp.111.180893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim B.R., Nam H.Y., Kim S.U., Kim S.I., Chang Y.J. Normalization of reverse transcription quantitative-PCR with housekeeping genes in rice. Biotechnol. Lett. 2003;25:1869–72. doi: 10.1023/a:1026298032009. doi:10.1023/A:1026298032009. [DOI] [PubMed] [Google Scholar]
- 19.Hardtke C.S., Okamoto H., Stoop-Myer C., Deng X.W. Biochemical evidence for ubiquitin ligase activity of the Arabidopsis COP1 interacting protein 8 (CIP8) Plant J. 2002;30:385–94. doi: 10.1046/j.1365-313x.2002.01298.x. doi:10.1046/j.1365-313X.2002.01298.x. [DOI] [PubMed] [Google Scholar]
- 20.Clough S.J., Bent A.F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–43. doi: 10.1046/j.1365-313x.1998.00343.x. doi:10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 21.Verslues P.E., Agarwal M., Katiyar-Agarwal S., Zhu J., Zhu J.K. Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J. 2006;45:523–39. doi: 10.1111/j.1365-313X.2005.02593.x. doi:10.1111/j.1365-313X.2005.02593.x. [DOI] [PubMed] [Google Scholar]
- 22.Usadel B., Nagel A., Thimm O., et al. Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol. 2005;138:1195–204. doi: 10.1104/pp.105.060459. doi:10.1104/pp.105.060459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horton P., Park K.J., Obayashi T., et al. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35:W585–7. doi: 10.1093/nar/gkm259. doi:10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shannon P., Markiel A., Ozier O., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. doi:10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hwang S.G., Kim D.S., Jang C.S. Comparative analysis of evolutionary dynamics of genes encoding leucine-rich repeat receptor-like kinase between rice and Arabidopsis. Genetica. 2011;139:1023–32. doi: 10.1007/s10709-011-9604-y. doi:10.1007/s10709-011-9604-y. [DOI] [PubMed] [Google Scholar]
- 26.Shaffer K.L., Sharma A., Snapp E.L., Hegde R.S. Regulation of protein compartmentalization expands the diversity of protein function. Dev. Cell. 2005;9:545–54. doi: 10.1016/j.devcel.2005.09.001. doi:10.1016/j.devcel.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 27.Singh R., Lee M.O., Lee J.E., et al. Rice mitogen-activated protein kinase interactome analysis using the yeast two-hybrid system. Plant Physiol. 2012;160:477–87. doi: 10.1104/pp.112.200071. doi:10.1104/pp.112.200071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee H., Xiong L., Gong Z., Ishitani M., Stevenson B., Zhu J.K. The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo-cytoplasmic partitioning. Genes Dev. 2001;15:912–24. doi: 10.1101/gad.866801. doi:10.1101/gad.866801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.von Arnim A.G., Deng X.W. Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell. 1994;79:1035–45. doi: 10.1016/0092-8674(94)90034-5. doi:10.1016/0092-8674(94)90034-5. [DOI] [PubMed] [Google Scholar]
- 30.Deng L., Wang C., Spencer E., et al. Activation of the kappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–61. doi: 10.1016/s0092-8674(00)00126-4. doi:10.1016/S0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 31.Claes B., Dekeyser R., Villarroel R., et al. Characterization of a rice gene showing organ-specific expression in response to salt stress and drought. Plant Cell. 1990;2:19–27. doi: 10.1105/tpc.2.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garcia A.B., Engler J., Iyer S., Gerats T., Van Montagu M., Caplan A.B. Effects of osmoprotectants upon NaCl stress in rice. Plant Physiol. 1997;115:159–69. doi: 10.1104/pp.115.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moons A., Prinsen E., Bauw G., Van Montagu M. Antagonistic effects of abscisic acid and jasmonates on salt stress-inducible transcripts in rice roots. Plant Cell. 1997;9:2243–59. doi: 10.1105/tpc.9.12.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Bie P., Ciechanover A. Ubiquitination of E3 ligases: self-regulation of the ubiquitin system via proteolytic and non-proteolytic mechanisms. Cell Death Differ. 2011;18:1393–402. doi: 10.1038/cdd.2011.16. doi:10.1038/cdd.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petroski M.D., Deshaies R.J. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. doi:10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 36.Okamoto K., Taya Y., Nakagama H. Mdmx enhances p53 ubiquitination by altering the substrate preference of the Mdm2 ubiquitin ligase. FEBS Lett. 2009;583:2710–4. doi: 10.1016/j.febslet.2009.07.021. doi:10.1016/j.febslet.2009.07.021. [DOI] [PubMed] [Google Scholar]
- 37.Kang J., Hwang J.U., Lee M., et al. PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc. Natl Acad. Sci. USA. 2010;107:2355–60. doi: 10.1073/pnas.0909222107. doi:10.1073/pnas.0909222107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aharon R., Shahak Y., Wininger S., Bendov R., Kapulnik Y., Galili G. Overexpression of a plasma membrane aquaporin in transgenic tobacco improves plant vigor under favorable growth conditions but not under drought or salt stress. Plant Cell. 2003;15:439–47. doi: 10.1105/tpc.009225. doi:10.1105/tpc.009225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X., Zhang L., Dong F., Gao J., Galbraith D.W., Song C.P. Hydrogen peroxide is involved in abscisic acid-induced stomatal closure in Vicia faba. Plant Physiol. 2001;126:1438–48. doi: 10.1104/pp.126.4.1438. doi:10.1104/pp.126.4.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.