Abstract
Context
Polymorphisms in the gene encoding the glucocorticoid receptor (GR) regulating co-chaperone FKBP5 have been shown to alter GR sensitivity and are associated with an increased risk to develop posttraumatic stress disorder (PTSD).
Objective
To investigate interactions of the FKBP5 single-nucleotide polymorphism rs9296158 and PTSD symptoms on baseline cortisol level, low-dose dexamethasone suppression, and whole-blood gene expression.
Design
Association of FKBP5 genotypes and PTSD symptoms with endocrine measures and genome-wide expression profiles.
Setting
Waiting rooms of general medical and gynecological clinics of an urban hospital at Emory University.
Participants
The 211 participants were primarily African American (90.05%) and of low socioeconomic status and had high rates of trauma and PTSD.
Main Outcome Measures
Baseline and post–dexamethasone suppression cortisol measures and gene expression levels.
Results
In our endocrine study, we found that only risk allele A carriers of rs9296158 showed GR supersensitivity with PTSD; in contrast, baseline cortisol levels were decreased in PTSD only in patients with the GG genotype. Expression of 183 transcripts was significantly correlated with PTSD symptoms after multiple testing corrections. When adding FKBP5 genotype and its interaction with PTSD symptoms, expression levels of an additional 32 genes were significantly regulated by the interaction term. Within these 32 genes, previously reported PTSD candidates were identified, including FKBP5 and the IL18 and STAT pathways. Significant overrepresentation of steroid hormone transcription factor binding sites within these 32 transcripts was observed, highlighting the fact that the earlier-described genotype and PTSD-dependent differences in GR sensitivity could drive the observed gene expression pattern. Results were validated by reverse transcriptase–polymerase chain reaction and replicated in an independent sample (N=98).
Conclusions
These data suggest that the inheritance of GR sensitivity–moderating FKBP5 polymorphisms can determine specific types of hypothalamic-pituitary-adrenal axis dysfunction within PTSD, which are also reflected in gene-expression changes of a subset of GR-responsive genes. Thus, these findings indicate that functional variants in FKBP5 are associated with biologically distinct subtypes of PTSD.
Posttraumatic stress disorder (PTSD) is a severely debilitating psychiatric condition characterized by persistent symptoms of intrusive reexperiencing, avoidance, and hyperarousal following exposure to a traumatic event. Although a lifetime trauma incidence of 40% to 90% in the general population has been reported, the overall lifetime prevalence for PTSD ranges between 7% and 12%.1,2 This suggests that individuals with a specific genetic susceptibility might have a higher risk of developing PTSD after experiencing traumatic events. Twin and family studies have provided evidence for a genetic component in the development of PTSD.3–5
While studies investigating main genetic effects associated with PTSD have yielded inconsistent findings,6–8 there is growing evidence supporting the role of gene×environment interactions in PTSD.9 For example, polymorphisms in the serotonin transporter gene as well as in the glucocorticoid receptor–regulating co-chaperone FK506 binding protein 5 (FKBP5) have been found in more than 1 cohort to have significant interactions with environmental exposure on risk for PTSD.9–11 Additionally, the syndrome of PTSD, as with many other psychiatric disorders, is clinically variable. The identification of biologically distinct subsets of patients with PTSD would provide an important tool for improving our mechanistic, diagnostic, and therapeutic approaches to PTSD. This study is predicated on the possibility that there are subtypes of PTSD that can be differentiated based on differential physiological stress reactivity that may, in part, be genetically determined.
The stress hormone system, or hypothalamus-pituitary-adrenal (HPA) axis, is likely an important mediator of such gene×environment interactions because it is activated by traumatic events and dysregulated in a number of psychiatric disorders, including PTSD.12 In fact, HPA axis dysfunction with higher glucocorticoid receptor (GR) sensitivity is described to be one of the endocrine hallmarks of PTSD.13 A number of studies have shown altered stress hormone/cortisol levels in PTSD and enhanced GR sensitivity as measured by a low-dose dexamethasone suppression test as well as ex vivo binding assays.14–17
Overall, these studies reflect a sensitization of the HPA axis in response to exposure to stressors, with changes in GR function playing a critical role in the pathophysiology of PTSD. The function of the GR is dependent on a large molecular complex, including chaperones and co-chaperones as well as coactivators and corepressors.18,19 One of the co-chaperones strongly regulating GR function is FKBP5, which is part of the mature GR heterocomplex and is known to regulate GR sensitivity.20 When bound to the steroid receptor, FKBP5 decreases its affinity for the ligand and prevents translocation to the nucleus.20,21 By being a modulator of GR sensitivity, FKBP5 is an interesting candidate gene for PTSD. In fact, 2 studies so far have reported interactions of functional polymorphisms within FKBP5 with early trauma to predict adult PTSD.10,11 In addition to these interaction effects on PTSD symptoms, we have also reported FKBP5 genotype–dependent difference in GR sensitivity in PTSD.10 Only patients with PTSD carrying the risk alleles displayed increased GR sensitivity, suggesting that endocrine changes observed with PTSD might be restricted to a genetically defined subset of patients.
In our previous report, rs9296158 showed the strongest interaction with early trauma on PTSD symptoms10 and was chosen as a tagging single-nucleotide polymorphism (SNP) for a functional haplotype that includes rs1360780 and rs3800373. Within this haplotype, risk alleles for PTSD are associated with an increased induction of FKBP5 messenger RNA by glucocorticoids, increased FKBP5 protein levels, and, as a consequence, a decrease in GR sensitivity and negative feedback regulation of the HPA axis in healthy controls.10,22,23 Genotype-dependent endocrine and messenger RNA effects in PTSD are less well studied.
We hypothesized that functional polymorphisms within FKBP5 are associated with differential GR sensitivity, which together may define biologically distinct subsets of patients with PTSD. To test this, we investigated interactions of the FKBP5 SNP rs9296158 and PTSD symptoms on baseline cortisol level, low-dose dexamethasone suppression, and whole-blood gene expression in a highly traumatized, low-income, primarily African American cohort with high rates of PTSD.24,25
METHODS
SAMPLES
The participants were a subset of a larger study investigating the contribution of genetic and environmental factors in PTSD10,25,26 and they had each experienced at least 1 traumatic event. Individuals were recruited between 2007 and 2010 at Grady Memorial Hospital, Atlanta, Georgia.24 All study procedures were approved by the institutional review boards of Emory University School of Medicine and Grady Memorial Hospital and all subjects gave written informed consent to the study.
Of the 219 participating individuals, 211 had available serum samples for baseline cortisol level only, while 115 also had cortisol measures after low-dose dexamethasone suppression. All individuals had DNA samples from whole blood or saliva and whole-blood RNA. Of 219 individuals, 10 samples were excluded from RNA analysis because of a low RNA integrity number (<5.0).27 Of 209 individuals, 111 individuals served as discovery samples and 98 served as replication samples for gene expression (eFigure 1 and eTable 1, http://www.archgenpsychiatry.com).
PSYCHOLOGICAL ASSESSMENTS
PTSD Symptom Scale
The PTSD Symptom Scale (PSS) was used as a measure of PTSD symptoms.28 The PSS frequency items were summed to obtain a continuous measure ranging from 0 to 48. The mean (SD) PSS score across all 219 individuals was 13.12 (12.33) points, with values more than 20.0 usually considered to show clinically significant PTSD symptoms. Application of the DSM-IV criteria for PTSD to the PSS frequency items was used to create a proxy variable for PTSD diagnosis status.
Clinician-Administered PTSD Scale
The Clinician-Administered PTSD Scale (CAPS) score was available for a subset of 156 of the 219 individuals.29 The mean (SD) current CAPS score in the 156 individuals was 39.95 (40.38). There was a significant correlation between PSS score and current CAPS score (Pearson r=0.657; P<.01).
Beck Depression Inventory
The Beck Depression Inventory was administered to measure depression symptoms.30 In this sample, the Beck Depression Inventory score ranged from 0 to 55 (mean [SD], 14.26 [11.63]). Values more than 18.0 are considered to show clinically relevant depressive symptoms.
TRAUMA EXPOSURE MEASUREMENTS
Trauma Events Inventory
The Trauma Events Inventory was the primary measure of non–child abuse trauma in this study.31 Total numbers of different types of non–child abuse trauma among the participants were summed. The mean (SD) score for non–child abuse trauma reported within this study sample was 3.90 (2.61).
Childhood Trauma Questionnaire
The Childhood Trauma Questionnaire was used as a continuous measure of child abuse.32 The Childhood Trauma Questionnaire score ranged from 25 to 111, with a mean (SD) of 42.20 (18.59).
Substance Abuse
Open-ended questions as well as the Kreek-McHugh-Schluger-Kellogg Scale were used to quantify self-exposure to opiates, cocaine, alcohol, and tobacco33 and to assess overall current and past substance use.
Dexamethasone Suppression Test
The subjects were characterized using a low-dose dexamethasone suppression test. After patients fasted, blood was collected between 8 and 9 AM for baseline serum cortisol measurements. The subjects received 0.5 mg of dexamethasone orally at 11 PM. Blood was collected between 8 and 9 AM the next day for serum cortisol measurement after dexamethasone stimulation. Serum cortisol concentration was measured with a commercial radioimmunoassay kit (Diagnostic Systems Laboratories, Webster, Texas).
DNA Extraction and SNP Genotyping
DNA was extracted from whole blood or saliva collected in Oragene saliva kits (DNA Genotek, Ottawa, Ontario, Canada) using the Qiagen M48 automated extraction system (Qiagen, Valencia, California). We genotyped rs9296158 using a TaqMan-based assay, as described before.10
RNA Extraction and Microarray Procedures for the Atlanta PTSD Samples
After patients fasted, whole blood was collected between 8 and 9 AM in Tempus RNA tubes (Applied Biosystems, Darmstadt, Germany) for baseline cortisol measurements. RNA was isolated using the Versagene kit (Gentra Systems, Minneapolis, Minnesota) and quantified using the NanoPhotometer (Implen, Westlake Village, California) and quality checks were performed on the Agilent Bioanalyzer (Agilent Technologies, Santa Clara, California). RNA was not globin reduced since our preliminary data showed that although globin reduction increased the sensitivity, it altered the expression of nonglobin transcripts in a nonreproducible manner (data not shown). Two hundred fifty nanograms of total RNA were reverse transcribed to complementary DNA, converted to complementary RNA, and biotin labeled using the Ambion kit (AMIL1791; Applied Biosystems). Seven hundred fifty nanograms of complementary RNA were hybridized to Illumina HT-12 version 3.0 arrays (Illumina, San Diego, California) and incubated overnight (16 hours) at 55°C. Arrays were washed, stained with Cy3-labeled streptavidin, dried, and scanned on the Illumina Bead-Scan confocal laser scanner.
Quantitative Polymerase Chain Reaction Procedures
For validation and replication of the results, complementary DNA was synthesized from 200 ng of total RNA using Superscript II Reverse Transcriptase (Invitrogen, Darmstadt, Germany). Quantitative polymerase chain reaction (qPCR) was performed using the Universal Probe Library for FKBP5, IL18R1, and TBP on the Roche LightCycler 480 (Roche Applied Science, Penzberg, Germany). Assays were designed using Probe Finder Software (Roche Applied Science) and run in duplicate according to the manufacturer’s protocol, except for a total reaction volume of 10 μL (eTable 2).
STATISTICAL ANALYSIS
Microarray Data Analysis
Raw microarray scan files were exported using the Illumina BeadStudio program and loaded into R for downstream analysis (http://www.R-project.org). The data were transformed and normalized using the variance-stabilizing normalization.34 Of 48 804 probes present on the Human HT-12 version 3.0 assays, probes were selected that fulfilled the criteria of the Illumina probe detection P value of <.01 in 5% of the individuals. A total of 15 583 probes passing the filter criteria were used for subsequent analysis. Association analysis was performed in R. General linear models were constructed using the quantitative PSS scores, rs9296158 A carrier genotypic model, and interactions between the 2 as predictors, controlling for age and sex. The significance of association was estimated by a 2-tailed P value corresponding to the t ratio based on a t reference distribution. To assess the contributions of model covariates, pairwise comparison between the full and reduced models was performed using an analysis of variance F test. The results were corrected for multiple testing using the permutation of regressor residuals test implemented in the R package glmperm (http://cran.r-project.org/web/packages/glmperm/index.html). For each transcript, 10 000 permutations were performed. The permutation of regressor residuals test permutes the residuals from the linear regression models using the shuffle-Z method for permutation.35 Repeated-measures analysis was performed in SPSS (SPSS Inc, Chicago, Illinois) using the analysis of variance design. The Huynh-Feldt method was applied to correct for potential violations of the sphericity assumption.36
The BiblioSphere data mining software from Genomatix was used for pathway analysis of transcripts.37 BiblioSphere is a data-mining solution for extracting and studying gene relationships from literature databases and genome-wide promoter analysis.38 The signal transduction association filter with the highest specificity was used where 2 genes were co-cited in a sentence containing a pathway-associated term, with at least 1 of the co-cited genes bearing a Genomatix signal transduction pathway annotation used for the pathway (http://www.genomatix.de). The JASPAR CORE database via the cREMaG interface (http://149.156.177.116/cremag/) was used to assess the distribution of steroid receptor binding sites.39
qPCR Data Analysis
Raw data from the Roche LightCycler 480 System were extracted using LinRegPCR software.40 The crossing thresholds (CT) from the technical replicates were averaged across all samples. The transcripts were normalized to housekeeping gene TBP. Normalized ΔCT values were converted into the linear form by 2−ΔCT transformation41. The transformed ΔCT values were regressed against the PSS score, rs9296158, and their interaction, while adjusting for age and sex, in R. For calculation of correlations between expression and PSS scores, negative correlations of the ΔCT were used to make the direction of effect comparable with the microarray data.
RESULTS
DESCRIPTIVE ANALYSIS OF INDIVIDUALS USED FOR ENDOCRINE ANALYSIS AND MICROARRAY ANALYSIS
Phenotypic characteristics of the 211 individuals used for endocrine analysis and 111 individuals used for microarray analysis are shown in Table 1. For comparisons, individuals were separated into groups of PTSD and non-PTSD based on DSM-IV criteria for PTSD using the responses to the PSS interview.
Table 1.
Comparison of 211 Individuals Used for Endocrine Analysis and 111 Individuals Used for Microarray Analysisa
| Endocrine Analysis (n=211)
|
Microarray Analysis (n=111)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Mean (SD)
|
P Value | Mean (SD)
|
P Value | |||||
| Non-PTSD (n=136) | PTSD (n=75) | Non-PTSD (n=70) | PTSD (n=41) | |||||
| Age, y | 43.28 (13.86) | 40.12 (12.72) | .10 | 44.24 (13.24) | 40.66 (12.72) | .17 | ||
| Sex, No. (%) | ||||||||
| M | 45 (33.1) | 22 (29.3) |
|
.34 | 21 (30.4) | 11 (27.5) |
|
.45 |
| F | 91 (66.9) | 53 (70.7) | 49 (69.6) | 30 (72.5) | ||||
| Ethnicity, No. (%) | ||||||||
| African American or black | 123 (90.4) | 67 (89.3) |
|
.80 | 64 (91.42) | 35 (85.37) |
|
.44 |
| White | 8 (5.9) | 6 (8) | 3 (4.29) | 0 | ||||
| Hispanic or Latino | 2 (1.5) | 0 | 1 (1.4) | 4 (9.75) | ||||
| Mixed | 2 (1.5) | 1 (1.3) | 1 (1.4) | 2 (4.88) | ||||
| Unknown | 1 (0.7) | 1 (1.3) | 1 (1.4) | 0 | ||||
| Trauma exposure | ||||||||
| TEI total trauma adult score | 5.34 (3.9) | 7.15 (3.58) | .001 | 3.0 (2.0) | 4.0 (2.0) | .11 | ||
| CTQ total score | 39.03 (14.27) | 49.96 (22.67) | <.001 | 39.18 (14.66) | 49.58 (19.00) | .004 | ||
| PSS total score | 5.74 (5.93) | 26.17 (9.85) | <.001 | 6.70 (6.69) | 25.17 (9.70) | <.001 | ||
| CAPS current trauma total score | 24.53 (27.54) | 68.50 (46.91) | <.001 | 25.78 (29.62) | 67.89 (44.93) | <.001 | ||
| CAPS lifetime trauma total score | 59.2 (46.13) | 106.43 (53.99) | <.001 | 64.90 (46.53) | 109.46 (51.73) | <.001 | ||
| BDI total score | 9.82 (8.8) | 22.69 (11.98) | <.001 | 10.49 (9.27) | 22.72 (11.52) | <.001 | ||
| Substance abuse current, No. (%) | ||||||||
| Yes | 3 (2.2) | 5 (6.7) |
|
.11 | 23 (32.9) | 17 (41.5) |
|
.56 |
| No | 131 (97.8) | 70 (93.3) | 47 (67.1) | 24 (58.5) | ||||
| Substance abuse past, No. (%) | ||||||||
| Yes | 41 (30.8) | 31 (41.3) |
|
.09 | 3 (4.3) | 4 (9.8) |
|
.40 |
| No | 92 (69.2) | 44 (58.7) | 67 (95.7) | 37 (90.2) | ||||
| KMSK alcohol current score | 2.6 (3.7) | 2.54 (3.5) | .93 | 2.5 (3.4) | 1.87 (3.38) | .60 | ||
| KMSK tobacco current score | 1.79 (3.3) | 3.21 (3.9) | .12 | 1.72 (3.4) | 3.18 (4.1) | .27 | ||
| KMSK cocaine current score | 0.09 (0.61) | 0.83 (2.7) | .10 | 0.22 (0.94) | 0 | .37 | ||
| KMSK marijuana current score | 1.0 (2.49) | 1.0 (2.4) | >.99 | 1.056 (2.69) | 1.067 (2.46) | .99 | ||
| Day 1 cortisol level, μg/dL | 15.37 (7.70) | 15.61 (7.59) | .51 | 14.92 (7.61) | 15.59 (7.47) | .65 | ||
| Day 2 cortisol level, μg/dL | 4.94 (6.01) | 4.35 (4.77) | .59 | 4.97 (5.50) | 5.46 (5.54) | .44 | ||
Abbreviations: BDI, Beck Depression Inventory; CAPS, Clinician-Administered PTSD Scale; CTQ, Childhood Trauma Questionnaire; KMSK, Kreek-McHugh-Schluger-Kellogg Scale; PSS, PTSD Symptom Scale; PTSD, posttraumatic stress disorder; TEI, Trauma Events Inventory.
SI conversion factor: To convert cortisol to nanomoles per liter, multiply by 27.588.
Phenotypic group comparisons between individuals in the PTSD and non-PTSD groups using the proxy PTSD variable among individuals used in the endocrine analysis (n=211) and individuals used in the microarray analysis (n=111). Group comparisons were calculated using analysis of variance, χ2 test, or Fisher exact test.
No differences in age, sex, ethnicity, and substance abuse were observed between the groups (P>.05) in both samples. There were no significant differences in adult trauma exposure (P=.11) in the microarray data set, but there was a significant difference in the endocrine data set (P=.001). Differences in child abuse were significant (P<.001 for the endocrine data set; P=.004 for the microarray data set). The severity of the PTSD symptoms measured by the PSS and current CAPS was significantly higher among individuals in the PTSD group (P<.001 for PSS score and P<.001 for CAPS score in the endocrine data set; P<.001 for PSS score and P<.001 for CAPS score in the microarray data set). Individuals with PTSD had higher Beck Depression Inventory scores than those without PTSD (P<.001 for the endocrine data set and P<.001 for the microarray data set). To determine whether the comorbidity between PTSD and depression would lead to multicollinearity, diagnostic tests for variance inflation factor and tolerance were estimated using SPSS. The variance inflation factor was 1.1 and the tolerance was 0.903, indicating no evidence for multicollinearity.
BASELINE CORTISOL LEVEL AND DEXAMETHASONE SUPPRESSION IN THE ATLANTA COHORT
Previously, we reported a possible genotype-dependent difference in the effects of PTSD on GR sensitivity (n=80),with higher GR sensitivity only in patients with PTSD carrying the risk allele.10 We now reinvestigated the effects of both PTSD symptoms and rs9296158 genotype in a second larger independent sample (n=211 for baseline cortisol level and n=115 for the dexamethasone suppression test).
Baseline Cortisol Level
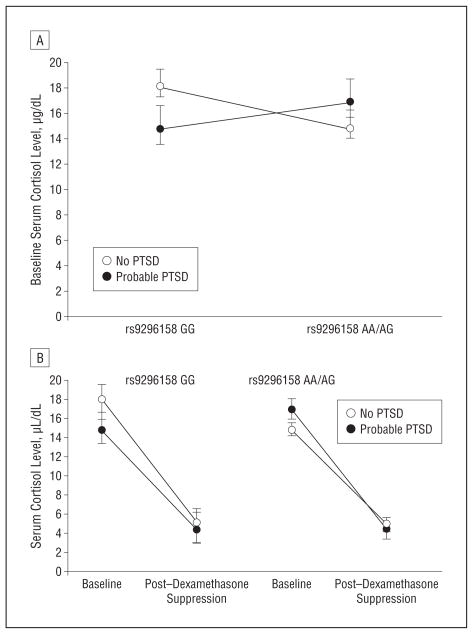
We first regressed baseline cortisol levels against the PSS score, FKBP5 SNP rs9296158 A carrier status, and their interaction term, adjusting for age and sex. Significant main effects of PSS scores (P =.003) and rs9296158 (P = .007) and significant interaction between the 2 (P=.002) on baseline cortisol levels were observed. Post-traumatic stress disorder symptom severity was negatively correlated with baseline cortisol level only in individuals carrying rs9296158 GG (Pearson correlation=−0.328 for GG and 0.157 for AA/AG, both significant at P=.01) (Figure 1A). The association remained significant after correcting for possible confounding effects of child abuse and adult trauma severity (PSS scores: P=.03; rs9296158: P=.04; and PSS score×rs9296158: P=.02). Overall, FKBP5 genotype was not associated with the level of child abuse (P=.55).
Figure 1.
Cortisol levels at baseline and after low-dose dexamethasone suppression. A, Baseline cortisol levels in individuals with and without posttraumatic stress disorder (PTSD) stratified by the rs9296158 A carrier status. In individuals carrying rs9296158 GG, PTSD symptom severity was negatively correlated with baseline cortisol level (Pearson correlation=−0.328), and in individuals carrying AA/AG, PTSD symptom severity was positively correlated with baseline cortisol level (Pearson correlation=0.157). B, Levels of serum cortisol suppression after low-dose (0.5-mg) dexamethasone suppression in individuals with and without PTSD stratified by rs9296158 A carrier status. Posttraumatic stress disorder was associated with increased dexamethasone suppression only in risk allele A carriers. The triple interaction of rs9296158 A carrier status and PTSD status on cortisol suppression was significant (P=.03). To convert cortisol to nanomoles per liter, multiply by 27.588.
Dexamethasone Suppression Test
Using repeated-measures analysis of variance, we next investigated the interaction of rs9296158 A carrier status and PTSD status on cortisol suppression. We confirmed a significant triple interaction of the genotype and PTSD on the change in cortisol levels (P=.03; F114,1=4.963).
We observed a significant PTSD status×rs9296158 genotype carrier status interaction (P=.04), but no main genotype or disease effect, on the change in cortisol levels. As observed before, PTSD was associated with increased dexamethasone suppression only in risk allele A carriers. The triple interaction of the genotype and PTSD status on cortisol suppression remained significant after adjusting for child abuse and adult trauma (P=.04) (Figure 1B).
MAIN EFFECTS OF PTSD SYMPTOM SEVERITY ON GENE EXPRESSION IN THE ATLANTA COHORT
To analyze the effects of PTSD symptom severity on gene expression, the continuous PSS scores were regressed against the filtered 15 583 expression profiles. After age and sex adjustment and multiple testing correction using the permutation of regressor residuals, 183 transcripts were significant with P values <.01, none of which had been previously reported to be associated with PTSD. Of these, 85 transcripts remained significant after adjusting for possible confounding effects of child abuse and adult trauma (eTable 3 and eFigure 2).
INTERACTION OF FKBP5 SNP rs9296158 WITH PTSD SYMPTOM SEVERITY ON GENE EXPRESSION IN THE ATLANTA COHORT
To investigate whether the observed FKBP5 genotype-dependent difference in GR sensitivity in PTSD would influence gene expression patterns, we used linear regression analyses to test the contribution of the continuous PTSD symptom severity, rs9296158 A carrier status, and their interaction to expression levels. Forty-one transcripts were significant for the genotype×PTSD symptom interaction after correcting for multiple testing using the permutation of regressor residuals test and an interaction P value of <.05 (Table 2 and eFigure 3). Nine of these 41 transcripts had already been identified using only PTSD symptoms as a predictor as described earlier, so that 32 novel transcripts were identified using this model. These 32 genes included FKBP5 and IL18R1, where either the molecule itself or the ligand had previously been reported to be regulated by PTSD.42–44
Table 2.
List of 41 Transcripts Significant for the Interaction Between PSS Score and rs9296158a
| Gene Symbol | Illuminab Probe ID | Significant With PSS Score Alone | Expression~PSS Score Pearson r
|
PSS Score×rs9296158 P Value
|
|||
|---|---|---|---|---|---|---|---|
| rs9296158 GG | rs9296158 AA/AG | ANOVA | Trauma Adjusted | Corrected | |||
| P76 | ILMN_1734184 | No | 0.47817 | −0.33669 | .00015 | .00043 | .00020 |
| TK2 | ILMN_1766814 | No | −0.76436 | −0.02777 | .00016 | .00028 | .00010 |
| CTSC | ILMN_1696347 | No | 0.61956 | −0.09383 | .00031 | .03900 | .00020 |
| ODF2 | ILMN_1730698 | No | 0.49463 | −0.14273 | .00073 | .09206 | .00150 |
| GTPBP2 | ILMN_1694475 | No | −0.59951 | 0.10253 | .00081 | .02476 | .00070 |
| LOC643452 | ILMN_1682736 | No | −0.56890 | 0.09748 | .00086 | .05438 | .00130 |
| FKBP5 | ILMN_1778444 | No | −0.49987 | 0.02563 | .00282 | .04444 | .00330 |
| BIRC5 | ILMN_2349459 | No | 0.57429 | 0.05468 | .00355 | .03030 | .00450 |
| MCM4 | ILMN_2412860 | Yes | 0.50162 | −0.03105 | .00381 | .03824 | .00340 |
| C3orf1 | ILMN_2193175 | No | −0.33791 | 0.30346 | .00396 | .07704 | .00450 |
| EZH2 | ILMN_1708105 | No | 0.50143 | −0.11772 | .00488 | .28239 | .00480 |
| ZBTB16 | ILMN_2305407 | No | −0.53328 | −0.04405 | .00673 | .02190 | .00720 |
| BHLHB2 | ILMN_1768534 | No | 0.35849 | −0.18584 | .00696 | .13020 | .00610 |
| LOC647460 | ILMN_1729198 | No | −0.35417 | 0.30213 | .00702 | .02798 | .00670 |
| FAM69A | ILMN_1682577 | No | −0.39838 | 0.22979 | .00829 | .05197 | .00850 |
| C14orf80 | ILMN_1653553 | No | 0.53635 | 0.06719 | .00975 | .12307 | .01200 |
| ZNF684 | ILMN_1751393 | No | −0.26585 | 0.24363 | .01053 | .20257 | .01420 |
| CAMK1D | ILMN_1795561 | Yes | 0.19985 | −0.37048 | .01070 | .02572 | .01150 |
| GINS2 | ILMN_1809590 | Yes | 0.61090 | 0.18267 | .01118 | .00738 | .01220 |
| ACOT7 | ILMN_1740265 | No | 0.65108 | 0.07385 | .01519 | .02759 | .01630 |
| LOC647037 | ILMN_1761922 | No | −0.18321 | 0.31981 | .01595 | .08595 | .01530 |
| NUSAP1 | ILMN_1726720 | Yes | 0.53809 | 0.13494 | .01635 | .03178 | .01580 |
| THAP11 | ILMN_1780699 | No | 0.23107 | −0.27642 | .02019 | .05254 | .02370 |
| FAM108B1 | ILMN_1660440 | No | −0.38296 | 0.00115 | .02032 | .00046 | .02090 |
| BZW1 | ILMN_1793846 | No | −0.13509 | 0.33007 | .02387 | .05116 | .02280 |
| NXF1 | ILMN_2358652 | Yes | 0.11059 | −0.34754 | .02648 | .03248 | .02530 |
| PLA2G4C | ILMN_1810191 | No | 0.32896 | −0.08184 | .02707 | .01867 | .02710 |
| UGP2 | ILMN_2284181 | No | −0.29111 | 0.32012 | .02744 | .04307 | .03020 |
| CDC20 | ILMN_1663390 | Yes | 0.54947 | 0.14556 | .02830 | .02563 | .02590 |
| PLAGL2 | ILMN_1786601 | No | 0.44579 | −0.13340 | .02920 | .01518 | .03030 |
| CASZ1 | ILMN_1655191 | Yes | 0.59386 | 0.16751 | .03004 | .13843 | .03150 |
| CX3CR1 | ILMN_1745788 | No | 0.54696 | 0.04575 | .03028 | .06447 | .02790 |
| POLDIP3 | ILMN_1685845 | No | −0.46645 | −0.00050 | .03377 | .07222 | .03100 |
| ZBTB16 | ILMN_2402817 | No | −0.45215 | −0.08603 | .03660 | .00956 | .03660 |
| SLC25A43 | ILMN_1662097 | No | −0.28835 | 0.12678 | .03810 | .04372 | .04030 |
| ASPM | ILMN_1815184 | No | 0.42272 | −0.03327 | .03870 | .40102 | .04000 |
| NCOA6IP | ILMN_1651506 | Yes | 0.04771 | 0.40608 | .04086 | .00212 | .04030 |
| SLC8A3 | ILMN_1712023 | Yes | −0.09220 | −0.40295 | .04144 | .08910 | .04210 |
| GCLM | ILMN_1788547 | No | 0.13927 | −0.24781 | .04240 | .01150 | .04300 |
| CCNF | ILMN_1773119 | No | 0.57323 | 0.05194 | .04625 | .16164 | .04220 |
| IL18R1 | ILMN_1781700 | No | −0.33430 | −0.06447 | .04906 | .07070 | .05000 |
Abbreviations: ANOVA, analysis of variance; PSS, PTSD Symptom Scale; ~, described by.
List of 41 transcripts significant for the interaction between PSS score and rs9296158 after adjusting for age and sex. The transcripts were corrected for multiple testing using the permutation of regressor residuals test. The Pearson correlation coefficient between the transcript expression levels and the continuous PSS score is indicated across all individuals stratified by the rs9296158 A carrier status groups.
Illumina, San Diego, California.
Pathway Analysis
Analysis of gene expression data in the form of gene networks can provide a more comprehensive view of gene interactions underlying complex traits of interest.45 To investigate possible common components of regulatory pathways, the 41 transcripts were characterized using the BiblioSphere software.37 Pathway analysis of these 41 transcripts revealed a network that directly included 15 of the tested genes as well as associated transcription factors. These included CREB1, HDAC1, FOS, and STAT5B, which had previously been reported to be associated with PTSD symptoms42,43 (eFigure 4). In fact, STAT5B expression showed a significant correlation with FKBP5 expression in this data set, with a Pearson correlation of 0.25 and 2-tailed significance of P=.007.
Analysis of Transcription Factor Binding Sites
We next investigated the distribution of steroid hormone biding sites, including GR response elements, androgen receptor response elements, and progesterone receptor response elements, within the 32 transcripts regulated by the SNP×PTSD symptom interaction vs the 183 transcripts associated with PTSD symptom severity alone. For this, the JASPAR CORE database was probed to test for a possible overrepresentation of these steroid receptor response elements. Comparisons between the 183 transcripts identified using the PTSD symptom scores and 32 transcripts significant for the rs9296158×PSS score interaction revealed a highly significant overrepresentation of steroid response elements (GR response elements and androgen receptor response elements) among the 32 transcripts but not the 183 transcripts associated with PTSD symptom severity alone (eTable 4). No progesterone receptor response elements were predicted in any of the tested transcripts.
qPCR VALIDATION OF FKBP5 AND IL18R1 EXPRESSION
Because FKBP5 and IL18 had been reported in previous PTSD gene expression studies, we proceeded to validate FKBP5 and IL18R1 expression results in the 111 individuals from the microarray analysis using qPCR. For both FKBP5 and IL18R1, significant main effects for PSS score as well as significant interaction between PSS and rs9296158 were observed (FKBP5: PSS total score, P=.002552; PSS score×rs9296158, P=.006932; IL18R1: PSS total score, P =.012253; PSS score×rs9296158, P=.018549) in the qPCR data set.
qPCR REPLICATION OF FKBP5 AND IL18R1 EXPRESSION
Next, we proceeded to replicate our findings in an additional set of 98 individuals not used in the microarray analysis. We replicated the significant main effects for PSS score as well as the significant interaction between the 2 for FKBP5 and IL18R1 after adjusting for age and sex in the independent replication sample (FKBP5: PSS total score, P=.01967; PSS score×rs9296158, P=.00319; IL18R1: PSS total score, P=.03826; PSS score×rs9296158, P=.00408).
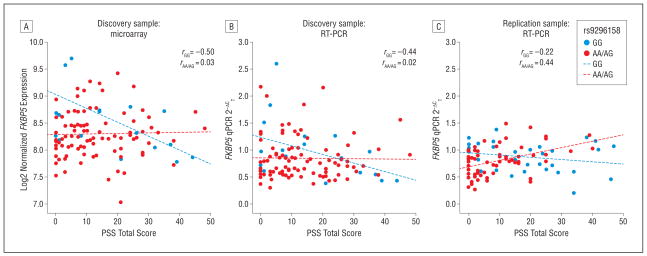
The genotype-dependent differences in correlation of FKBP5 messenger RNA levels and PSS score severity in the microarray data set, the qPCR validation, and the replication data set are depicted in Figure 2.
Figure 2.
Validation and replication of FKBP5 expression differences in posttraumatic stress disorder (PTSD) using quantitative polymerase chain reaction (qPCR). The PTSD Symptom Scale (PSS) total scores are plotted on the x-axis against the expression levels as measured by log2 expression for the microarray data and 2−ΔCT (change in crossing thresholds) for the qPCR validation and replication on the y-axis. The Pearson correlation coefficients for the rs9296158 A carrier status groups are shown. Post hoc genotype-dependent differences of the correlation coefficients were established by converting them into Fisher z scores.10 The z score differences between the 2 genotype groups were significant for the microarray expression (z=2.17; significant at P=.01), qPCR validation (z=2.12; significant at P=.05), and qPCR replication (z=2.87; significant at P=.01). RT-PCR indicates reverse transcriptase–polymerase chain reaction.
COMMENT
These data from endocrine measures as well as gene expression profiling suggest that the functional FKBP5 polymorphism rs9296158 is not only associated with an increased risk for PTSD following childhood trauma10,11 but might also define biologically distinct subtypes of this disorder. Endocrine measures and gene expression data were available in more than 200 traumatized individuals. In fact, we could replicate that only risk allele carriers of this SNP show the previously reported association of GR supersensitivity with PTSD.10,13 On the other hand, baseline cortisol levels were decreased in PTSD only in patients with the nonrisk GG genotype. This suggests that the inheritance of GR sensitivity–moderating FKBP5 polymorphisms10,22 can determine specific types of HPA-axis dysfunction within PTSD that were also reflected in gene-expression changes of a subset of GR-responsive genes.
A number of studies have reported both decreased baseline cortisol levels as well as increased GR sensitivity46 in patients with PTSD, although the findings are not uniformly consistent across studies.14,17,47 While we also observed these 2 endocrine features with PTSD, these findings are restricted to specific FKBP5 SNP genotype groups, which might be an explanation for some of the inconsistent findings in the literature. In this article, we replicated a relative GR resistance in nonaffected individuals carrying the risk allele, which is in line with our previous report.10 These data also support the previously reported decrease in negative feedback capacity in these individuals.23 Also, as previously reported, the genotype-dependent association with GR function seems to be state dependent, with increased GR sensitivity in risk allele carriers with PTSD. We are currently investigating the molecular basis of this state dependence and speculate that in addition to possible expression changes of other GR chaperones and other systems influencing the HPA axis, this might be mediated by epigenetic changes. Interestingly, the protective genotype, not the risk, was associated with decreasing baseline cortisol levels with increasing PTSD symptoms. In our sample, decreased baseline cortisol level and increasing GR sensitivity therefore do not seem to be strongly correlated, suggesting that other mechanisms besides efficiency of negative feedback of the HPA axis, possibly including stress reactivity, regulate baseline cortisol levels in PTSD. The possibility that these 2 measures are somewhat disjunct is also supported by the fact that while the finding of increased GR sensitivity is relatively consistent across studies, it is less so for baseline cortisol changes, with decreases, no changes, and increases reported in PTSD.16,48 A previous study had also reported that PTSD-related decreases in baseline cortisol level might be dependent on a functional GR polymorphism, a possible additional confounder in this association.49
The genotype-dependent endocrine differences with PTSD symptoms were also paralleled by genotype-dependent differences in whole-blood gene expression pattern, where expression levels of 183 transcripts were significantly associated with PTSD symptoms alone. Within these, none had been previously identified to be associated with PTSD. Adding the FKBP5 polymorphism to the model identified 32 transcripts not detected in the first model that showed a significant interaction between FKBP5 genotype and PTSD symptom severity. In fact, these transcripts showed strong genotype-dependent differences for the correlation of gene expression levels with PTSD symptom severity (eFigure 3), which likely has precluded their identification when analyzing the association with PTSD symptom severity alone. Among the top transcripts exhibiting the most significant gene expression interaction effects was FKBP5 itself. This is not surprising given the previously discussed influence of PTSD and FKBP5 on GR function and the fact that FKBP5 appears to be one of the most sensitive readouts of GR stimulation. FKBP5 has been shown to be one of the genes with the strongest induction by GR activation in a number of studies, including our own.50,51 Most notably, Yehuda and colleagues43 have also described a downregulation of FKBP5 gene expression in whole blood with PTSD, as we observed for rs9296158 GG carriers. Within these 32 genes, the IL18R1 transcript is also an interesting candidate. Zieker et al44 reported interleukin 18 to be differentially expressed in the peripheral blood of patients with PTSD, concordant with the differential regulation of its receptor IL18R1 in our cohort.
We also found that 15 of the genes regulated by the interaction between FKBP5 genotype and PTSD can be integrated into the same pathway (eFigure 4) using the BiblioSphere software.37 When expanding the annotation to include related transcription factors, we identified another 4 previously reported PTSD candidates: CREB1, FOS, HDAC1, and STAT5B42,43 to be part of this common regulatory pathway. We could confirm the significant correlation between expression levels of FKBP5 and STAT5B in this study, both of which had been previously shown to be regulated with PTSD by Yehuda et al.43 Our data suggest that a data mining approach could be informative in revealing coexpressed gene networks and might help identify transcripts that otherwise would be missed when not passing statistical thresholds of significance because of the subtle gene expression differences.
While we did identify a number of genes regulated by PTSD symptom severity itself, our data suggest that a network of genes is differentially regulated depending on FKBP5 genotype and related differences in GR sensitivity. In fact, there was a significant overrepresentation of GR response elements and androgen receptor response elements among these 32 transcripts significant for the interaction term compared with the 183 genes regulated by PTSD alone. Altogether, these findings suggest that the combined model of FKBP5 SNP genotype and PTSD symptoms seems most relevant for GR-dependent transcripts, while other predictive models might be necessary to capture differences in the expression of other gene classes.
While we were able to replicate some of the genes and pathways identified in previous studies, others could not be replicated. This is likely because of a number of reasons, including false-positive and false-negative associations in either study, differences in clinical study design, sample characteristics, type of microarrays, and statistical analysis.
A strength of our microarray gene expression study (n=111 in total) is the large sample size, with 41 individuals with a probable PTSD diagnosis, which, to our knowledge, is larger than any of the previously reported expression array studies for this disorder. In the overall sample, we investigated 75 individuals with a probable PTSD diagnosis. We acknowledge that our main PTSD definition is symptom driven; using the PSS might have included individuals with other anxiety disorders also showing high PSS scores.52 However, about 70% of our cohort had CAPS-based current PTSD diagnoses and these concorded well with the PSS scores (Pearson r=0.657, significant at a P=.01). The use of the PSS allowed us to include all individuals and to increase the power of the study by using a quantitative predictor vs using diagnostic cutoffs, a strategy now advocated by a number of investigators for biomarker discovery.42,53 A limitation of this highly traumatized sample is the heterogeneity regarding the timing of the trauma as well as the types of traumatic events. Despite this, our gene expression results appear to be comparable with other studies with much more defined index trauma, as described earlier.16,42
In this sample, all subjects had experienced at least 1 significant trauma; nonetheless, we noted significant differences in overall trauma exposure as well as the level of child abuse between individuals with high vs low PTSD symptoms. When correcting the analysis for adult trauma and child abuse exposure, about 50% of the associations with PTSD severity alone remained significant at a level of P=.05, indicating the importance of increased trauma exposure on gene expression, even when using traumatized individuals as controls (eTable 3). Differences in severity and timing of trauma exposure might also account for the slight differences in the PTSD×SNP interaction on FKBP5 messenger RNA expression in the discovery vs the replication sample (Figure 2 and eTable 1).
Independent replication is extremely important to assess the validity of gene expression findings, because more often than not specific changes reported for psychiatric disorders could not be replicated.54,55 In this study, we chose FKBP5 and IL18R1 for validation and replication, because they had previously been implicated in this disorder. We could, in fact, validate and replicate the association with genotype-dependent gene expression differences of both 2 transcripts using qPCR.
In summary, using a large sample of 219 traumatized individuals, we examined the effects of FKBP5 rs9296158 genotype, previously reported to increase the risk for PTSD with early trauma, on putative biomarkers for this disorder. In this study, we found that rs9296158 may be used to identify biologically different subtypes of PTSD in that the genotype groups differed with respect to PTSD-related changes in GR sensitivity. This was reflected in genotype- and PTSD-dependent differences in the expression of GR-dependent transcripts in whole blood. Our findings support the importance of using genetic markers and biological measures to increase the predictive validity of biomarkers in psychiatric disorders.56
Acknowledgments
Funding/Support: This work was supported by the Max Planck Society and National Institute of Mental Health grant MH071537. Dr Bradley receives grant support or has received awards from the American Foundation for Suicide Prevention and the American Psychoanalytic Association Psychoanalytic Research Fund. Within the last 3 years, Dr Ressler has received research funding support from the National Institute of Mental Health, the National Institute on Drug Abuse, the Burroughs Wellcome Foundation, and the National Alliance for Research on Schizophrenia and Depression. Dr Binder has current grant support from the National Institute of Mental Health and the Doris Duke Charitable Foundation.
Footnotes
Online-Only Material: The eFigures and eTables are available at http://www.archgenpsychiatry.com.
Financial Disclosure: Part of this work was funded by NeuroNova. Drs Binder, Menke, and Holsboer have filed a patent application as inventors of “Means and Methods for Diagnosing Predisposition for Treatment Emergent Suicidal Ideation (TESI)” (European application 08016477.5; international application PCT/EP2009/061575), and Drs Binder, Müller-Myhsok, and Holsboer have filed a patent application as inventors of “FKBP5: A Novel Target for Antidepressant Therapy” (international publication WO 2005/ 054500). Dr Holsboer is a founder and shareholder of Affectis Pharmaceuticals. Within the last 3 years, Dr Ressler has received research funding support from Lundbeck, and he has an unrelated role as cofounder of Extinction Pharmaceuticals for development of N-methyl-D-aspartate–based therapeutics. Dr Binder has current grant support from PharmaNeuroBoost.
References
- 1.Breslau N. The epidemiology of posttraumatic stress disorder: what is the extent of the problem? J Clin Psychiatry. 2001;62(suppl 17):16–22. [PubMed] [Google Scholar]
- 2.Kessler RC, Sonnega A, Bromet E, Hughes M, Nelson CB. Posttraumatic stress disorder in the National Comorbidity Survey. Arch Gen Psychiatry. 1995;52 (12):1048–1060. doi: 10.1001/archpsyc.1995.03950240066012. [DOI] [PubMed] [Google Scholar]
- 3.Koenen KC, Lyons MJ, Goldberg J, Simpson J, Williams WM, Toomey R, Eisen SA, True W, Tsuang MT. Co-twin control study of relationships among combat exposure, combat-related PTSD, and other mental disorders. J Trauma Stress. 2003;16(5):433–438. doi: 10.1023/A:1025786925483. [DOI] [PubMed] [Google Scholar]
- 4.Stein MB, Jang KL, Livesley WJ. Heritability of social anxiety-related concerns and personality characteristics: a twin study. J Nerv Ment Dis. 2002;190(4):219–224. doi: 10.1097/00005053-200204000-00002. [DOI] [PubMed] [Google Scholar]
- 5.True WR, Rice J, Eisen SA, Heath AC, Goldberg J, Lyons MJ, Nowak J. A twin study of genetic and environmental contributions to liability for posttraumatic stress symptoms. Arch Gen Psychiatry. 1993;50(4):257–264. doi: 10.1001/archpsyc.1993.01820160019002. [DOI] [PubMed] [Google Scholar]
- 6.Comings DE, Muhleman D, Gysin R. Dopamine D2 receptor (DRD2) gene and susceptibility to posttraumatic stress disorder: a study and replication. Biol Psychiatry. 1996;40(5):368–372. doi: 10.1016/0006-3223(95)00519-6. [DOI] [PubMed] [Google Scholar]
- 7.Gelernter J, Southwick S, Goodson S, Morgan A, Nagy L, Charney DS. No association between D2 dopamine receptor (DRD2) “A” system alleles, or DRD2 haplotypes, and posttraumatic stress disorder. Biol Psychiatry. 1999;45(5):620–625. doi: 10.1016/s0006-3223(98)00087-0. [DOI] [PubMed] [Google Scholar]
- 8.Segman RH, Cooper-Kazaz R, Macciardi F, Goltser T, Halfon Y, Dobroborski T, Shalev AY. Association between the dopamine transporter gene and posttraumatic stress disorder. Mol Psychiatry. 2002;7(8):903–907. doi: 10.1038/sj.mp.4001085. [DOI] [PubMed] [Google Scholar]
- 9.Koenen KC, Aiello AE, Bakshis E, Amstadter AB, Ruggiero KJ, Acierno R, Kilpatrick DG, Gelernter J, Galea S. Modification of the association between serotonin transporter genotype and risk of posttraumatic stress disorder in adults by county-level social environment. Am J Epidemiol. 2009;169(6):704–711. doi: 10.1093/aje/kwn397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Binder EB, Bradley RG, Liu W, Epstein MP, Deveau TC, Mercer KB, Tang Y, Gillespie CF, Heim CM, Nemeroff CB, Schwartz AC, Cubells JF, Ressler KJ. Association of FKBP5 polymorphisms and childhood abuse with risk of posttraumatic stress disorder symptoms in adults. JAMA. 2008;299(11):1291–1305. doi: 10.1001/jama.299.11.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie P, Kranzler HR, Poling J, Stein MB, Anton RF, Farrer LA, Gelernter J. Interaction of FKBP5 with childhood adversity on risk for post-traumatic stress disorder. Neuropsychopharmacology. 2010;35(8):1684–1692. doi: 10.1038/npp.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Kloet ER, Joëls M, Holsboer F. Stress and the brain: from adaptation to disease. Nat Rev Neurosci. 2005;6(6):463–475. doi: 10.1038/nrn1683. [DOI] [PubMed] [Google Scholar]
- 13.Yehuda R, Yang RK, Buchsbaum MS, Golier JA. Alterations in cortisol negative feedback inhibition as examined using the ACTH response to cortisol administration in PTSD. Psychoneuroendocrinology. 2006;31(4):447–451. doi: 10.1016/j.psyneuen.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 14.Mason JW, Giller EL, Kosten TR, Ostroff RB, Podd L. Urinary free-cortisol levels in posttraumatic stress disorder patients. J Nerv Ment Dis. 1986;174(3):145–149. doi: 10.1097/00005053-198603000-00003. [DOI] [PubMed] [Google Scholar]
- 15.Yehuda R. Neuroendocrine aspects of PTSD. Handb Exp Pharmacol. 2005;(169):371–403. doi: 10.1007/3-540-28082-0_13. [DOI] [PubMed] [Google Scholar]
- 16.Yehuda R, Bierer LM, Sarapas C, Makotkine I, Andrew R, Seckl JR. Cortisol metabolic predictors of response to psychotherapy for symptoms of PTSD in survivors of the World Trade Center attacks on September 11, 2001. Psychoneuroendocrinology. 2009;34(9):1304–1313. doi: 10.1016/j.psyneuen.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yehuda R, Southwick SM, Nussbaum G, Wahby V, Giller EL, Jr, Mason JW. Low urinary cortisol excretion in patients with posttraumatic stress disorder. J Nerv Ment Dis. 1990;178(6):366–369. doi: 10.1097/00005053-199006000-00004. [DOI] [PubMed] [Google Scholar]
- 18.Grad I, Picard D. The glucocorticoid responses are shaped by molecular chaperones. Mol Cell Endocrinol. 2007;275(1–2):2–12. doi: 10.1016/j.mce.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 19.Pratt WB, Toft DO. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr Rev. 1997;18(3):306–360. doi: 10.1210/edrv.18.3.0303. [DOI] [PubMed] [Google Scholar]
- 20.Scammell JG, Denny WB, Valentine DL, Smith DF. Overexpression of the FK506-binding immunophilin FKBP51 is the common cause of glucocorticoid resistance in three New World primates. Gen Comp Endocrinol. 2001;124(2):152–165. doi: 10.1006/gcen.2001.7696. [DOI] [PubMed] [Google Scholar]
- 21.Wochnik GM, Rüegg J, Abel GA, Schmidt U, Holsboer F, Rein T. FK506-binding proteins 51 and 52 differentially regulate dynein interaction and nuclear translocation of the glucocorticoid receptor in mammalian cells. J Biol Chem. 2005;280(6):4609–4616. doi: 10.1074/jbc.M407498200. [DOI] [PubMed] [Google Scholar]
- 22.Binder EB, Salyakina D, Lichtner P, Wochnik GM, Ising M, Pütz B, Papiol S, Seaman S, Lucae S, Kohli MA, Nickel T, Künzel HE, Fuchs B, Majer M, Pfennig A, Kern N, Brunner J, Modell S, Baghai T, Deiml T, Zill P, Bondy B, Rupprecht R, Messer T, Köhnlein O, Dabitz H, Brückl T, Müller N, Pfister H, Lieb R, Mueller JC, Lõhmussaar E, Strom TM, Bettecken T, Meitinger T, Uhr M, Rein T, Holsboer F, Muller-Myhsok B. Polymorphisms in FKBP5 are associated with increased recurrence of depressive episodes and rapid response to antidepressant treatment. Nat Genet. 2004;36(12):1319–1325. doi: 10.1038/ng1479. [DOI] [PubMed] [Google Scholar]
- 23.Ising M, Depping AM, Siebertz A, Lucae S, Unschuld PG, Kloiber S, Horstmann S, Uhr M, Müller-Myhsok B, Holsboer F. Polymorphisms in the FKBP5 gene region modulate recovery from psychosocial stress in healthy controls. Eur J Neurosci. 2008;28(2):389–398. doi: 10.1111/j.1460-9568.2008.06332.x. [DOI] [PubMed] [Google Scholar]
- 24.Gillespie CF, Bradley B, Mercer K, Smith AK, Conneely K, Gapen M, Weiss T, Schwartz AC, Cubells JF, Ressler KJ. Trauma exposure and stress-related disorders in inner city primary care patients. Gen Hosp Psychiatry. 2009;31(6):505–514. doi: 10.1016/j.genhosppsych.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gillespie CF, Phifer J, Bradley B, Ressler KJ. Risk and resilience: genetic and environmental influences on development of the stress response. Depress Anxiety. 2009;26(11):984–992. doi: 10.1002/da.20605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jovanovic T, Blanding NQ, Norrholm SD, Duncan E, Bradley B, Ressler KJ. Childhood abuse is associated with increased startle reactivity in adulthood. Depress Anxiety. 2009;26(11):1018–1026. doi: 10.1002/da.20599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, Light-foot S, Menzel W, Granzow M, Ragg T. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Foa EB, Tolin DF. Comparison of the PTSD Symptom Scale-Interview Version and the Clinician-Administered PTSD scale. J Trauma Stress. 2000;13(2):181–191. doi: 10.1023/A:1007781909213. [DOI] [PubMed] [Google Scholar]
- 29.Blake DD, Weathers FW, Nagy LM, Kaloupek DG, Klauminzer G, Charney DS, Keane TM. A clinician rating scale for assessing current and lifetime PTSD: the CAPS-1. Behav Therapist. 1990;13:187–188. [Google Scholar]
- 30.Beck AT, Ward CH, Mendelson M, Mock J, Erbaugh J. An inventory for measuring depression. Arch Gen Psychiatry. 1961;4:561–571. doi: 10.1001/archpsyc.1961.01710120031004. [DOI] [PubMed] [Google Scholar]
- 31.Hovens JE, Bramsen I, van der Ploeg HM, Reuling IE. Test-retest reliability of the trauma and life events self-report inventory. Psychol Rep. 2000;87(3 pt 1):750–752. doi: 10.2466/pr0.2000.87.3.750. [DOI] [PubMed] [Google Scholar]
- 32.Bernstein DP, Stein JA, Handelsman L. Predicting personality pathology among adult patients with substance use disorders: effects of childhood maltreatment. Addict Behav. 1998;23(6):855–868. doi: 10.1016/s0306-4603(98)00072-0. [DOI] [PubMed] [Google Scholar]
- 33.Kellogg SH, McHugh PF, Bell K, Schluger JH, Schluger RP, LaForge KS, Ho A, Kreek MJ. The Kreek-McHugh-Schluger-Kellogg Scale: a new, rapid method for quantifying substance abuse and its possible applications. Drug Alcohol Depend. 2003;69(2):137–150. doi: 10.1016/s0376-8716(02)00308-3. [DOI] [PubMed] [Google Scholar]
- 34.Huber W, von Heydebreck A, Sültmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18(suppl 1):S96–S104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- 35.Potter DM. A permutation test for inference in logistic regression with small- and moderate-sized data sets. Stat Med. 2005;24(5):693–708. doi: 10.1002/sim.1931. [DOI] [PubMed] [Google Scholar]
- 36.Wilcox RR, Keselman HJ, Muska J, Cribbie R. Repeated measures ANOVA: some new results on comparing trimmed means and means. Br J Math Stat Psychol. 2000;53(pt 1):69–82. doi: 10.1348/000711000159187. [DOI] [PubMed] [Google Scholar]
- 37.Krawetz S, Epple A, Scherf M. BiblioSphere: Hypothesis Generation in Regulatory Network Analysis. Bioinformatics for Systems Biology. New York, NY: Humana Press; 2009. pp. 401–412. [Google Scholar]
- 38.Scherf M, Epple A, Werner T. The next generation of literature analysis: integration of genomic analysis into text mining. Brief Bioinform. 2005;6(3):287–297. doi: 10.1093/bib/6.3.287. [DOI] [PubMed] [Google Scholar]
- 39.Sandelin A, Alkema W, Engström P, Wasserman WW, Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32(database issue):D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramakers C, Ruijter JM, Deprez RH, Moorman AF. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett. 2003;339(1):62–66. doi: 10.1016/s0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- 41.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 42.Segman RH, Shefi N, Goltser-Dubner T, Friedman N, Kaminski N, Shalev AY. Peripheral blood mononuclear cell gene expression profiles identify emergent post-traumatic stress disorder among trauma survivors. Mol Psychiatry. 2005;10(5):500–513. 425. doi: 10.1038/sj.mp.4001636. [DOI] [PubMed] [Google Scholar]
- 43.Yehuda R, Cai G, Golier JA, Sarapas C, Galea S, Ising M, Rein T, Schmeidler J, Müller-Myhsok B, Holsboer F, Buxbaum JD. Gene expression patterns associated with posttraumatic stress disorder following exposure to the World Trade Center attacks. Biol Psychiatry. 2009;66(7):708–711. doi: 10.1016/j.biopsych.2009.02.034. [DOI] [PubMed] [Google Scholar]
- 44.Zieker J, Zieker D, Jatzko A, Dietzsch J, Nieselt K, Schmitt A, Bertsch T, Fassbender K, Spanagel R, Northoff H, Gebicke-Haerter PJ. Differential gene expression in peripheral blood of patients suffering from post-traumatic stress disorder. Mol Psychiatry. 2007;12(2):116–118. doi: 10.1038/sj.mp.4001905. [DOI] [PubMed] [Google Scholar]
- 45.Rietkerk T, Boks MP, Sommer IE, de Jong S, Kahn RS, Ophoff RA. Network analysis of positional candidate genes of schizophrenia highlights myelin-related pathways. Mol Psychiatry. 2009;14(4):353–355. doi: 10.1038/mp.2008.86. [DOI] [PubMed] [Google Scholar]
- 46.Yehuda R. Advances in understanding neuroendocrine alterations in PTSD and their therapeutic implications. Ann N Y Acad Sci. 2006;1071:137–166. doi: 10.1196/annals.1364.012. [DOI] [PubMed] [Google Scholar]
- 47.Lemieux AM, Coe CL. Abuse-related posttraumatic stress disorder: evidence for chronic neuroendocrine activation in women. Psychosom Med. 1995;57(2):105–115. doi: 10.1097/00006842-199503000-00002. [DOI] [PubMed] [Google Scholar]
- 48.Yehuda R. Status of glucocorticoid alterations in post-traumatic stress disorder. Ann N Y Acad Sci. 2009;1179:56–69. doi: 10.1111/j.1749-6632.2009.04979.x. [DOI] [PubMed] [Google Scholar]
- 49.Bachmann AW, Sedgley TL, Jackson RV, Gibson JN, Young RM, Torpy DJ. Glucocorticoid receptor polymorphisms and post-traumatic stress disorder. Psychoneuroendocrinology. 2005;30(3):297–306. doi: 10.1016/j.psyneuen.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 50.Vermeer H, Hendriks-Stegeman BI, van Suylekom D, Rijkers GT, van Buul-Offers SC, Jansen M. An in vitro bioassay to determine individual sensitivity to glucocorticoids: induction of FKBP51 mRNA in peripheral blood mononuclear cells. Mol Cell Endocrinol. 2004;218(1–2):49–55. doi: 10.1016/j.mce.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 51.Woodruff PG, Boushey HA, Dolganov GM, Barker CS, Yang YH, Donnelly S, Ellwanger A, Sidhu SS, Dao-Pick TP, Pantoja C, Erle DJ, Yamamoto KR, Fahy JV. Genome-wide profiling identifies epithelial cell genes associated with asthma and with treatment response to corticosteroids. Proc Natl Acad Sci U S A. 2007;104(40):15858–15863. doi: 10.1073/pnas.0707413104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Engelhard IM, Arntz A, van den Hout MA. Low specificity of symptoms on the post-traumatic stress disorder (PTSD) symptom scale: a comparison of individuals with PTSD, individuals with other anxiety disorders and individuals without psychopathology. Br J Clin Psychol. 2007;46(pt 4):449–456. doi: 10.1348/014466507X206883. [DOI] [PubMed] [Google Scholar]
- 53.Segman RH, Shalev AY. Genetics of posttraumatic stress disorder. CNS Spectr. 2003;8(9):693–698. doi: 10.1017/s1092852900008889. [DOI] [PubMed] [Google Scholar]
- 54.Drago A, De Ronchi D, Serretti A. Incomplete coverage of candidate genes: a poorly considered bias. Curr Genomics. 2007;8(7):476–483. doi: 10.2174/138920207783591681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu FY, Hu WB, Liu L, Yu LH, Xi J, He XH, Zhu MR, Liu ZL, Xu YM. Lack of replication of a previously reported association between polymorphism in the 3′UTR of the alpha-synuclein gene and Parkinson’s disease in Chinese subjects. Neurosci Lett. 2010;479(1):31–33. doi: 10.1016/j.neulet.2010.05.022. [DOI] [PubMed] [Google Scholar]
- 56.Holsboer F. How can we realize the promise of personalized antidepressant medicines? Nat Rev Neurosci. 2008;9(8):638–646. doi: 10.1038/nrn2453. [DOI] [PubMed] [Google Scholar]