Abstract
Bacillus thuringiensis subsp. israelensis produces three Cry toxins (Cry4Aa, Cry4Ba and Cry11Aa) that are active against Aedes aegypti larvae. The identification of the rate-limiting binding steps of Cry toxins that are used for insect control in the field, such as those of B. thuringiensis subsp. israelensis, should provide targets for improving insecticides against important insect pests. Previous studies showed that Cry11Aa binds to cadherin receptor fragment CR7–11 (cadherin repeats 7–11) with high affinity. Binding to cadherin has been proposed to facilitate Cry toxin oligomer formation. In the present study, we show that Cry4Ba binds to CR7–11 with 9-fold lower binding affinity compared with Cry11Aa. Oligomerization assays showed that Cry4Ba is capable of forming oligomers when proteolytically activated in vitro in the absence of the CR7–11 fragment in contrast with Cry11Aa that formed oligomers only in the presence of CR7–11. Pore-formation assays in planar lipid bilayers showed that Cry4Ba oligomers were proficient in opening ion channels. Finally, silencing the cadherin gene by dsRNA (double-stranded RNA) showed that silenced larvae were more tolerant to Cry11Aa in contrast with Cry4Ba, which showed similar toxic levels to those of control larvae. These findings show that cadherin binding is not a limiting step for Cry4Ba toxicity to A. aegypti larvae.
Keywords: Aedes aegypti, Bacillus thuringiensis, cadherin, Cry toxin, pore-forming toxin, receptor binding
INTRODUCTION
Bacillus thuringiensis pore-forming toxins are valuable tools for the control of insect pests in agriculture and also against mosquitoes that are vectors of human diseases [1]. For the control of mosquitoes, B. thuringiensis subsp. israelensis has been used worldwide for the control of Aedes aegypti, and certain Culex and Anopheles species that are vectors of important diseases such as dengue fever and malaria [2]. Upon sporulation, B. thurigiensis subsp. israelensis produces crystal inclusions that are formed by at least three structurally related Cry toxins (Cry11Aa, Cry4Aa and Cry4Ba) and a non-related Cyt toxin (Cyt1Aa).
The three B. thurigiensis subsp. israelensis Cry toxins belong to a family of 3d (three-domain)-Cry toxins that have members that are active against insect larvae from different insect orders such as Lepidoptera, Coleoptera and Diptera. The 3d-Cry toxins have three discrete structural domains. Domain I is a seven α-helix bundle implicated in membrane insertion, toxin oligomerization and pore formation. Domain II is a β-prism of three antiparallel β-sheets packed around a hydrophobic core with exposed loop regions involved in recognition of insect midgut proteins, and domain III is a β-sandwich of two antiparallel β-sheets also involved in midgut protein recognition. Since domains II and III are implicated in binding to larvae insect gut proteins, they largely mediate insect specificity [1].
Cry toxins are widely believed to exert their toxicity by forming pores in the epithelia of insect midgut cells bursting them by osmotic lysis. To do so, Cry toxins rely on binding to specific proteins present in the surface of epithelium midgut cells of the target insects. The identification of the rate-limiting binding steps of Cry toxins that are used for insect control in the field should provide targets for improving insecticides against important insect pests, identifying resistant gene alleles and succeeding in insect-resistant-managing strategies. In lepidopteran insects, the Cry1A toxin-binding proteins identified include cadherin proteins, GPI (glycosylphophatidylinositol)-anchored APNs (aminopeptidases N) and GPI-anchored ALPs (alkaline phosphatases) [1,3]. In order to insert into the cell membrane, Cry1A toxins undergo a sequential binding interaction with the receptors mentioned above [4,5]. First Cry1A crystal inclusions are solubilized due to high pH present in the larval midgut lumen and the 130 kDa Cry1A protoxins are then proteolytically activated by larval midgut proteases, yielding 60 kDa monomers that have the three-dimensional structure described above. Cry1A monomers bind to highly abundant APN and ALP molecules by domain II and domain III residues. The binding affinity of the Cry1A monomer with GPI proteins is in the 100–200 nM range [5]. Binding to GPI-anchored proteins is believed to concentrate the Cry1A toxin in the apical surface of insect midgut cells, where Cry1A then binds with high affinity to cadherin (Kd of 1 nM) through exposed loops of domain II [6,7]. Insect cadherins that bind Cry toxins are composed of three domains: an ectodomain formed by 11–12 CRs (cadherin repeats), a transmembrane domain and an intracellular domain [8]. The binding interaction with cadherin facilitates further processing of the Cry1A toxin by an unidentified protease clipping out helix α-1, facilitating the formation of toxin oligomers of 250 kDa [9]. The Cry1A oligomers bind again to APN and ALP with high affinity (Kd of 0.6–1 nM) through domain II residues [4,5,10]. Binding to APN or ALP has been proposed to facilitate the insertion of the pre-pore oligomer into the insect membrane, forming pores that burst the insect midgut cells [11].
In the case of the dipteran A. aegypti, similar Cry toxin-binding midgut proteins have been identified such as cadherin, APN and ALP [12–15], suggesting that the mode of action of Cry toxins in mosquitoes is conserved [12]. Regarding the A. aegypti cadherin, it was shown that Cry11Aa binds with high affinity (Kd of 16.7 nM) to a fragment containing the membrane-proximal part of the ectodomain of this receptor molecule (CR7–11) [13]. Binding competition experiments showed that Cry4Aa and Cry11Ba, produced by B. thuringiensis subsp. jegathesen, competed with Cry11Aa for binding to this cadherin fragment showing that these toxins share the same cadherin-binding sites. In contrast, Cry4Ba did not compete with Cry11Aa for binding to CR7–11, indicating that Cry4Ba may not rely on binding to cadherin for toxicity [13].
In the present study, we characterized the binding of Cry4Ba to A. aegypti CR7–11. Our data show that Cry4Ba binds this cadherin fragment with at least 9-fold lower affinity in comparison with the binding affinity of Cry11Aa for this fragment. We show that oligomerization of Cry11Aa depends on its interaction with cadherin. In contrast, Cry4Ba readily forms oligomers in vitro after activation with proteases in the absence of the cadherin protein. The Cry4Ba oligomers produced stable ion channels with high probability of being open in black lipid bilayers indicating their functionality. Finally, our results suggest that Cry4Ba is a Cry toxin that does not require cadherin binding to induce toxicity in A. aegypti larvae, since cadherin-silenced larvae are not more tolerant to Cry4Ba toxin in contrast with Cry11Aa where cadherin-silenced larvae became more tolerant to this toxin. Although cadherin binding has been shown to be an important binding step of different Cry toxins, the results of the present study show that it is not a limiting step in the toxicity of Cry4Ba to A. aegypti.
EXPERIMENTAL
Growth of B. thuringiensis strains and production of Cry4Ba and Cry11Aa crystal inclusions
For the production of Cry4Ba and Cry11Aa crystals, acrystalliferous B. thuringiensis strains containing plasmids pHT618 [16] or pGC6 [17] respectively were cultured for 3 days at 29 °C and 200 rev./min in nutrient broth sporulation medium supplemented with erythromycin (10 μg/ml). Spores and inclusion bodies produced by the B. thuringiensis strains were harvested and washed three times with 0.3 M NaCl and 0.01 M EDTA, pH 8.0.
Expression of CR7–11
The Escherichia coli M15 (pREP4) strain containing the expression plasmid pQE30G10 (cloned in pQE30 series vector, Qiagen) for CR7–11 has been described previously [13]. For protein expression, the gene construct was induced by addition of 1 mM IPTG (isopropyl β-D-thiogalactopyranoside) and protein was produced as inclusion bodies. Inclusion bodies were solubilized in 8 M urea. The N-terminally His6-tagged recombinant protein was purified using Ni-NTA (Ni2+ -nitrilotriacetate) resin (Qiagen) under denaturing conditions and resolved by SDS/PAGE (10% gels). The CR7–11 protein was dialysed against PBS.
ELISA
ELISA 96-well plates (Costar) were incubated for 12 h at 4 °C with 2.5 μg of Cry4Ba or Cry11Aa in 50 mM NaHCO3 (pH 9.6), followed by five washes with PBS and 0.2 % Tween 20. The plates were then incubated with PBS, 0.5 % gelatin (Bio-Rad Laboratories) and 0.2 % Tween 20 for 1 h at 37 °C and washed five times with PBS and 0.1 % Tween 20. The ELISA plates were incubated with different concentrations of CR7–11 for 2 h at 37 °C and washed again with PBS and 0.2 % Tween 20. The CR7–11 fragment that bound Cry4Ba was revealed with a 1:5000 dilution of horseradish-peroxidase-conjugated anti-His antibody (Qiagen) for 1 h and washed three times with PBS and 0.1 % Tween 20. Finally, plates were incubated with 2 mM o-phenylenediamine and 0.7 % H2O2 in 0.1 M sodium phosphate (pH 5). The enzymatic reaction was stopped with 6 M HCl and the absorbance was read at 490 nm with a microplate reader from Molecular Devices. The data presented are the result of four repetitions of each CR7–11 concentration shown in Figure 1.
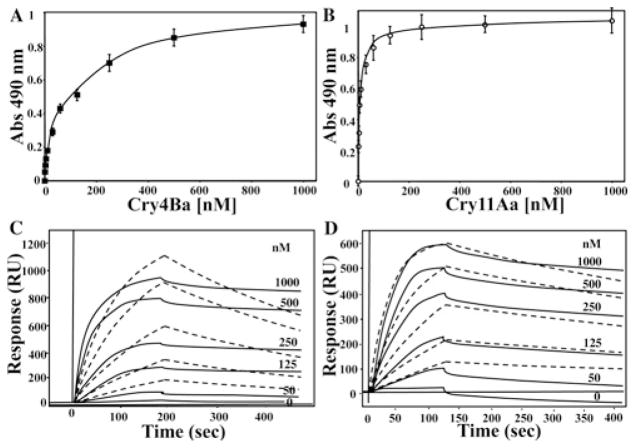
Figure 1. Cry4Ba binds with lower affinity than Cry11Aa to A. aegypti CR7–11.
Cry4Ba (A) or Cry11Aa (B) binding to CR7–11 analysed by ELISA saturation binding assays. ELISA binding assays were performed by adding 2.5 μg of Cry4Ba or Cry11Aa to ELISA plates and then incubating with different concentrations of CR7–11 recombinant protein. Results are means ± S.D. of four repetitions. SPR binding analyses of Cry4Ba (C) or Cry11Aa (D) were performed by immobilizing Cry toxins using conventional amine coupling. Sensograms of serial doubling dilutions of CR7–11 are shown as continuous lines, whereas broken lines represent curves generated by a one-site binding model. RU, response units.
Biosensor (SPR) analysis of Cry4Ba and Cry11Aa affinities to CR7–11
SPR (surface plasmon resonance) measurements were performed using a SensiQ instrument (ICx Nomadics). The running buffer for all experiments, HBS (Hepes-buffered saline) (pH 7.4), containing 10 mM Hepes, 150 mM NaCl and 0.005 % Tween 20, was freshly prepared, filtered (pore size of 0.22 mm) and degassed. Cry4Ba and Cry11Aa toxins were immobilized on to a carboxy-functionalized sensorchip (ICx Nomadics) by conventional amine coupling at densities less than 1500 RU (response units). The flow cells were blocked with a 5 min injection of 1 M ethanolamine at a flow rate of 10 ml/min. The analyte (CR7–11) was injected at a flow rate of 25 ml/min. Serial doubling dilutions of CR7–11 were analysed, and the surface was regenerated with a 1 min injection of 20 mM NaOH. Triplicate injections of each toxin concentration were carried out. The data were analysed using the SensiQ Software Qdat version B.02. This software employs non-linear regression and the Levenberg–Marquadt algorithm to fit experimental data to a binding interaction model that defines the interaction.
Preparation of SUVs (small unilamellar vesicles)
SUVs were prepared as described previously [18]. Briefly, egg-yolk PC (phosphatidylcholine) (Avanti Polar Lipids), cholesterol (Avanti Polar Lipids) and stearylamine (Sigma) from chloroform stocks, were mixed in glass vials in a 10:3:1 (by vol.) proportion at a final concentration of 2.6 mmol and dried by gentle argon flow evaporation followed by overnight storage under vacuum to remove residual chloroform. Lipids were then hydrated in 650 ml of 10 mM Ches [2-(N-cyclohexylamino)ethanesulfonic acid] (pH 9) and 150 mM KCl, or with 10 mM Hepes (pH 7) and 150 mM KCl by 30 min of incubation followed by vortex-mixing. The lipid sample was sonicated twice for 2 min in a Branson 1200 bath sonicator to prepare SUVs. SUVs were used within 2–3 days upon their preparation.
Oligomer-formation assays
For Cry11Aa, oligomer formation was performed as described previously [18]. For these assays, 2.5 μg of soluble Cry11Aa protoxin was incubated in a 100 ml final volume with 250 mM SUVs and 2 %(w/w) trypsin, in the presence or absence of 2.5 μg of CR7–11 protein fragment. For Cry4Ba, a similar activation procedure was performed: 2.5 μg of soluble Cry4Ba protoxin was activated under the same conditions, but with 10 % (w/w) chymotrypsin. The mixture was incubated for 1 h at 37 °C, and 1 mM PMSF was added to stop the reaction. The membrane fraction was separated by centrifugation at 90 000 rev./min for 1 h at 4 °C using a TLA-100 rotor (Beckman). The samples were analysed by SDS/PAGE (10 % gels) and Western blotting using polyclonal anti-Cry11Aa or anti-Cry4Ba antibodies.
PLB (planar lipid bilayer) experiments
For pore-formation assays, membrane fractions containing Cry4Ba oligomer (starting with 2.5 μg of soluble Cry4Ba protoxin for toxin activation as explained above) were suspended in 50 ml of buffer containing 5 mM Hepes (pH 7.4) and 10 mM KCl. PLBs were made by the Müeller and Rudin method as described previously [19], with egg-derived PC (20 mg/ml in n-decane). Typical bilayer capacitance values were between 250 and 350 pF. Buffer solution (5 mM Hepes, pH 7.4, and 300 mM KCl) was added to the cis compartment, whereas in the trans compartment, the buffer solution was modified to contain 10 mM KCl. The different KCl concentrations in the cis and trans compartments facilitate membrane vesicles fusion to the bilayer by adding 15 ml of the vesicle suspension containing Cry4Ba oligomer to the cis compartment, whereas that in the trans compartment was held at virtual ground. All experiments were performed at room temperature (25 °C). Single channel currents were recorded with a Dagan 3900A patch-clamp amplifier. Currents were filtered at 200 or 500 Hz, digitized online at 1 or 2 kHz respectively and analysed on a personal computer using a Digidata 1200 interface and Axotape and pClamp software (Axon Instruments).
RNAi (RNA interference) assays
Two fragments for the 5′- and 3′-end of the cloned A. aegypti cadherin gene were obtained using the cloned cadherin gene as template. The 507 bp 5′fragment was obtained using 5′-AATACCGGAGCATGAGGATG-3′and 5′-ACACCGTTCTTGCCACTTTC-3′ as the sense and antisense primers respectively, whereas the 3′ 781 bp fragment was obtained using 5′-TGTGATAGTGGATCGGTTTGAG-3′ and 5′-TTCTTCGTTTTCGGTGAAATCT-3′ as the sense and antisense primers respectively. As control dsRNA (double-stranded RNA), a LacZ fragment of 900 bp was amplified with appropriate primers. The fragments were cloned into pLitmus28i (HiScribe™, New England Biolabs) vector containing two T7 promoters flanking the multi-cloning site. These promoters enabled amplification of the cloned fragment by PCR using a T7 oligonucleotide. The PCR products were purified with QIAquik PCR purification kit protocol (Qiagen). In vitro transcription of both DNA strands of the insert was performed with T7 RNA polymerase using the HiScribe™ RNAi transcription kit (New England Biolabs) following the manufacturer’s instructions, yielding dsRNA.
RNAi experiments were performed as described previously [20]. A total of 200 neonate A. aegypti larvae in 10 ml of dechlorinated water were fed for 16 h with 200 μg of dsRNA (1 μg per larva) previously encapsulated with Effectene transfection reagent (Qiagen). For encapsulating the dsRNA with Effectene, 200 μg of dsRNA in a final volume of 4 ml of DNA-condensation buffer (EC buffer, as indicated in the Effectene transfection reagent handbook) were mixed for 10 s by vortex agitation with 0.8 ml of Enhancer buffer and incubated for 5 min at room temperature. Then the sample was mixed with 1.3 ml of Effectene by vortex agitation (10 s) and incubated for 10 min at room temperature. This sample was diluted with dechlorinated water to a final volume of 10 ml. After dsRNA feeding, the mosquito larvae were transferred to clean water and fed on a regular diet until they reached fourth instar for bioassays or for gut dissection to be analysed by quantitative real-time PCR.
To quantify cadherin transcript levels in dsRNA-treated and untreated larvae, total RNA was isolated form the dissected midguts and then reverse-transcribed using oligo(dT) and SuperScript II (Invitrogen). To reduce genomic contamination, primers that flank a predicted exon–exon junction were used. qPCR (quantitative PCR) was performed on ABI 7700 (Applied Biosystems) using Brilliant SYBR Green QPCR Master Mix (Qiagen). The fold changes between transcript levels in silenced and normal larvae was calculated by the comparative threshold cycle (CT) method.
Insect bioassays
For synergistic toxicity assays, ten early fourth-instar A. aegypti larvae reared at 28 °C and a 12 h light/12 h dark cycle were placed in 2 ml of dechlorinated water with a 10–15 % lethal concentration of Cry11Aa or Cry4Ba (75 ng/ml for both toxins) and 10- or 25-fold higher concentration of CR7–11 as described previously (n = 4) [21]. The concentration of protein in the CR7–11 inclusion bodies was determined after SDS/PAGE of different quantities of CR7–11 and comparing with known BSA protein concentrations on the same gel as described in [21]. Mortality was analysed after 24 h.
For dsRNA-treated larvae, bioassays were performed using 20 larvae in 200 ml of dechlorinated water that contained individual toxin concentrations at the LC50 level (100 ng/ml for Cry4Ba and 250 ng/ml for Cry11Aa). Larval mortality was analysed after 24 h.
RESULTS
Binding of Cry4Ba and Cry11Aa to cadherin receptor
Previous work indicated that Cry4Ba did not compete with Cry11Aa for binding to CR7–11, suggesting that either Cry4Ba does not bind to this cadherin fragment or it binds with a much lower affinity than Cry11Aa [13]. The binding affinity of Cry11Aa to CR7–11 was determined previously by ELISA binding assays showing a Kd value of 16.7 nM [13]. To analyse the binding of Cry4Ba to CR7–11, a binding saturation curve of Cry4Ba to cadherin CR7–11 produced in E. coli was obtained from an ELISA binding assay. As control, we determined also the binding of Cry11Aa to CR7–11. Figure 1(A) shows that Cry4Ba binds to CR7–11 in a saturable way with lower binding affinity (Kd of 134.9 nM) than that of Cry11Aa (Kd of 14.8 nM). To determine the binding affinities in real time, the binding of Cry4Ba and Cry11Aa to CR7–11 was determined by SPR analyses showing that CR7–11 bound both immobilized Cry4Ba and Cry11Aa (Figure 1B and Table 1). The overall affinity for CR7–11 binding to Cry4Ba (Kd of 154 nM) or to Cry11Aa (Kd of 17 nM) was estimated from the kon and koff values obtained from the binding curve (Figure 1B and Table 1). These results show that Cry4Ba binds CR7–11 with 9-fold lower affinity than Cry11Aa.
Table 1.
Binding kinetics of CR7–11 to Cry4Ba and Cry11Aa
| Toxin | kon (×10 −5 M −1 · s −1) | koff (×103 s −1) | Kd (nM) |
|---|---|---|---|
| Cry4Ba | 0.132 | 2.034 | 154 |
| Cry11Aa | 1.470 | 2.521 | 17 |
kon is the association rate constant; koff is the dissociation rate constant; Kd is the apparent affinity (koff/kon).
Oligomer formation by Cry11Aa and Cry4Ba
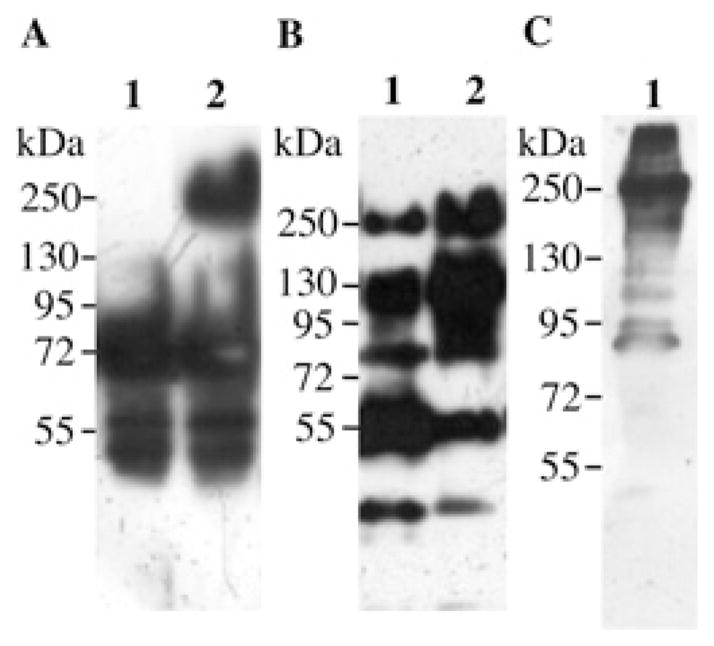
Previous work showed that Cry11Aa, when proteolytically activated in the presence of Cyt1Aa, was capable of forming oligomers in vitro that were proficient on pore-formation activity, suggesting that Cyt1Aa may function as a receptor of Cry11Aa, inducing oligomerization of the toxins such as cadherin receptors in lepidopteran insects [18]. However, oligomerization of Cry11Aa when activated in the presence of A. aegypti cadherin or CR7–11 fragment has not been analysed previously. Figure 2(A) shows that Cry11Aa forms oligomers of 250 kDa when activated by protease treatment in the presence of the CR7–11 fragment and lipid vesicles (lane 2). In contrast, Cry11Aa did not oligomerize in the absence of CR7–11 (Figure 2A, lane 1). We also analysed the formation of Cry4Ba oligomers performing a similar experiment in the presence or absence of CR7–11 and lipid vesicles. Figures 2(B) and 2(C) (lanes 1) show that Cry4Ba oligomers were formed even when activated in the absence of CR7–11.
Figure 2. In vitro oligomer formation of Cry11Aa and Cry4Ba.
Cry11Aa (A) or Cry4Ba (B and C) were activated by protease treatment with or without CR7–11, and oligomer formation was revealed by Western blotting with anti-Cry11Aa or anti-Cry4Ba polyclonal antibodies. (A) Proteolytic activation of Cry11Aa without (lane 1) or with (lane 2) CR7–11 in the assay. (B) Proteolytic activation of Cry4Ba without (lane 1) or with (lane 2) CR7–11 in the assay. (C) Cry4Ba oligomer association with membrane vesicles after recovering the membrane pellet by centrifugation (lane 1). Molecular masses are indicated in kDa.
Pore formation of Cry4Ba oligomers
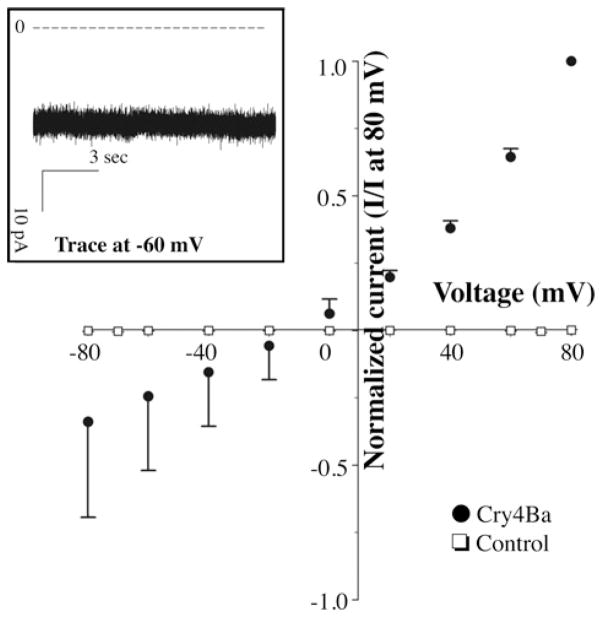
To determine the functionality of Cry4Ba oligomers that were formed in the absence of CR7–11, their capacity to induce ion channel activity was determined in black lipid bilayers. Cry4Ba protoxin was activated by protease treatment in the presence of lipid vesicles, but in the absence of CR7–11 peptide. Lipid vesicles containing Cry4Ba oligomers were recovered by centrifugation and the presence of Cry4Ba oligomer in the lipid vesicles was revealed by Western blot analysis (Figure 2C). The Cry4Ba-containing vesicles were then fused to a lipid bilayer, and ion channel activity was recorded as described in the Experimental section. Figure 3 shows that the Cry4Ba oligomer response involved the formation of ion channels with high open probability as described previously for Cry1Ab oligomers [19].
Figure 3. Cry4Ba oligomers induce ionic channels with high open probability in black lipid bilayers.
Current–voltage (I–V ) relationship of the macroscopic currents induced by the oligomeric Cry4Ba toxin measured under asymmetrical 300 mM cis and 10 nM trans KCl conditions. The inset shows a representative trace of the currents induced by Cry4Ba oligomer.
Toxicity of Cry4Ba and Cry11Aa in the presence of CR7–11
Previous work has shown that cadherin fragments containing Cry-binding sites are able to enhance toxicity of Cry toxins in different insect orders [21–23]. In lepidopteran insects, this synergistic effect correlated with enhanced oligomer formation [24]. To determine the effect of A. aegypti CR7–11 on the toxicity of Cry4Ba and Cry11Aa toxins, toxicity assays were performed with a 10 % lethal concentration of each toxin (75 ng/ml) in the presence of CR7–11 fragment as described in the Experimental section. Figure 4 shows that Cry11Aa toxicity was enhanced 2.7-fold in the presence of a 10-fold excess of CR7–11. In contrast, there was no change in Cry4Ba toxicity with the same molar excess of CR7–11. However, a 25-fold excess of CR7–11 enhanced 3.5-fold Cry11Aa toxicity, whereas a 2.5-fold increased Cy4Ba toxicity was observed under these conditions (Figure 4).
Figure 4. Effect of CR7–11 on Cry4Ba and Cry11Aa toxicity.
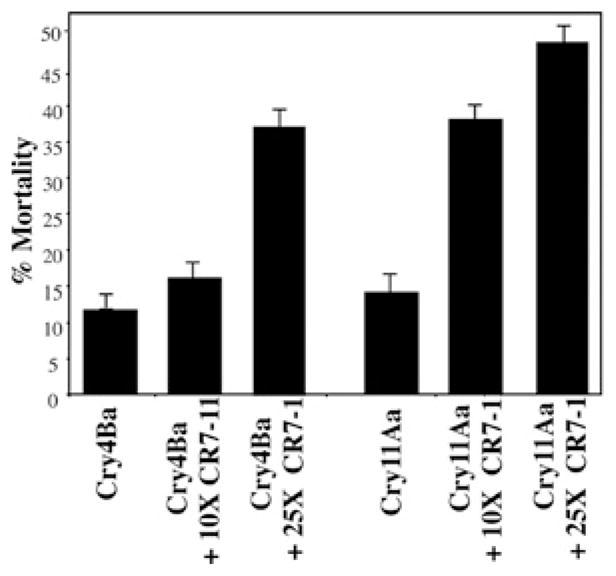
Ten fourth-instar larvae were exposed to 75 ng/ml Cry11Aa or Cry4Ba with or without a 10- or 25-fold higher concentration of CR7–11. Results are means ± S.D. of three repetitions.
Toxicity of Cry11Aa and Cry4Ba to A. aegypti cadherin-silenced larvae
In order to determine whether cadherin binding is a limiting step in the toxicity of Cry11Aa and Cry4Ba, we decided to test the toxicity of Cry11Aa and Cry4Ba to A. aegypti larvae that were silenced for the cadherin transcript by feeding with cadherin dsRNA. Previous work showed efficient gene silencing in A. aegypti larvae by feeding with dsRNA molecules encapsulated in Effectene liposomes [20]. Two dsRNA molecules from the cadherin gene of 507 and 781 bp were produced by in vitro transcription using T7 polymerase as described in the Experimental section. These dsRNA molecules were encapsulated using Effectene and then used to feed neonate larvae. Transcript levels of cadherin RNA were analysed by qPCR in silenced fourth-instar larvae. Figure 5(A) shows that both fragments reduced the cadherin transcript 2–3-fold in comparison with non-silenced larvae. Silenced larvae were exposed to either Cry4Ba or Cry11Aa at non-saturated concentrations (100 and 250 ng/ml respectively) that showed a 70 % mortality to determine the effect of reduced cadherin transcript on the toxicity of these Cry toxins. Figure 5(B) shows that silenced larvae (lanes 1 and 2) become more tolerant to Cry11Aa (30 % mortality) in contrast with Cry4Ba that showed the same toxic levels to silenced or control larvae (Figure 5C). As a control, a dsRNA of LacZ gene fragment was used to feed A. aegypti larvae and toxicity to Cry11Aa or Cry4Ba was analysed under the same conditions, showing a minor effect on toxicity of Cry11Aa and no effect on toxicity of Cry4Ba (Figures 5B and 5C, bars 4).
Figure 5. Silencing of A. aegypti cadherin gene and its effect on toxicity of Cry11Aa and Cry4Ba.
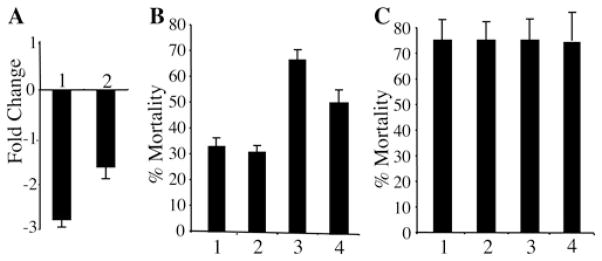
(A) Cadherin transcript levels determined by qPCR of silenced larvae with the cadherin dsRNA of 781 (1) or 587 bp (2) compared with non-silenced larvae. (B) Percentage of Cry11Aa mortality (250 ng/ml) of silenced larvae using cadherin dsRNA of 781 bp (1) or 587 bp (2) compared with non-silenced larvae (3) and with a control of silenced larvae with LacZ dsRNA (4) (n = 6). (C) Percentage of mortality of silenced larvae with cadherin dsRNA of 781 bp (1) or 587 bp (2) compared with non-silenced larvae (3) and to a control of silenced larvae with LacZ dsRNA (4) treated with Cry4Ba (100 ng/ml). Results are means ± S.D. of six repetitions.
DISCUSSION
The mode of action of three 3d-Cry toxins seems to be conserved in different insect orders. Cry toxin-binding cadherin proteins have been described in lepidopteran, dipteran and coleopteran insects [3]. It has been shown that binding to cadherin facilitates oligomer formation of Cry1A and Cry3A toxins, active against lepidopteran and coleopteran larvae respectively, when activated in vitro by protease treatment [9,25]. In the present study, we show that binding to A. aegypti (dipteran) cadherin fragment facilitates the formation of Cry11Aa oligomers after protease treatment, indicating that cadherin binding fulfils the same role in the three different insect orders, i.e. to facilitate additional proteolytic removal of helix α-1 resulting in the formation of a pre-pore oligomer that is responsible for toxin membrane insertion and pore formation. Nevertheless, the results of the present study indicate that Cry4Ba has 9-fold lower binding affinity to A. aegypti cadherin fragment compared with Cry11Aa explaining previous observations that Cry4Ba was not able to compete with Cry11Aa for binding to CR7–11 [13]. It is important to point out that Cry4Ba is slightly more toxic (LC50 = 100 ng/ml) than Cry11Aa (LC50 = 250 ng/ml), regardless of the lower affinity of Cry4Ba to the A. aegypti cadherin. This result suggests that Cry4Ba does not rely on binding to cadherin for A. aegypti toxicity either because it relies on binding to another as yet unidentified midgut molecule for oligomer formation or because it can form functional oligomers just by protease activation. In the lepidopteran Lymantria dispar, Cry1Ac toxin does not bind to cadherin, but instead relies on the binding to a different midgut protein, an anionic glycoconjugate (BTR270) [26].
In the present study, we have shown that Cry4Ba is capable of forming oligomers that are proficient in pore-formation activity when proteolytically activated in vitro in the absence of the cadherin protein in contrast with Cry11Aa that formed oligomers only when treated with proteases in the presence of CR7–11 fragment. Thus this result suggests that Cry4Ba does not rely on another receptor for oligomer formation since it can form oligomers in vitro without receptor interaction. Previous work showed that Cry4Ba is capable of forming oligomers in vitro [27]. Additionally, when the three-dimensional crystal structure of Cry4Ba was solved, it revealed that the Cry4Ba domain I helices α1-α2b were absent owing to proteolysis during the crystallization process, resulting in the formation of a crystal structure composed of three monomers that make inter-monomer contacts through domain I helices α-3, α-4 and α-6 [28]. The Cry4Ba oligomer structure was later proposed to represent the pre-pore oligomer and was used to model the structure of the related Cry4Aa when inserted into the membrane [29]. These results suggest that Cy4Ba, in contrast with Cry11Aa, is capable of forming the pre-pore oligomer without the need for cadherin or other midgut molecule interactions.
The mechanism of action of Cry toxins also involves other protein receptors such as GPI-anchored receptors (APN or ALP) [1]. The principal role of GPI-anchored receptors is to bind the pre-pore oligomer and to facilitate its insertion into the membrane [11]. It was reported recently that expression of an A. aegypti GPI-anchored ALP isoform (mALP) in Sf9 insect cells resulted in Cry4Ba binding to the transfected cells inducing susceptibility to this toxin [30]. These results support those of the present study that Cry4Ba does not rely on cadherin binding to exert its toxic effect, since expression of mALP alone, which would bind the Cry4Ba pre-pore, was sufficient to confer Sf9 susceptibility to the toxin [29]. It is important to mention that other GPI-anchored Cry1A-binding molecules such as APNs from different insects have been expressed in insect cell lines but failed to confer susceptibility to Cry1A toxins (reviewed in [3]), probably due to the fact that Cry1A toxins do not form oligomers in the absence of cadherin interaction [11].
In the three insect orders considered, cadherin fragments containing Cry toxin-binding sites enhance the toxicity of the corresponding Cry protein and, in the case of Cry1Ab, this enhanced toxicity correlated with enhanced oligomer formation [21–24]. To gain in vivo evidence that Cry4Ba does not rely on cadherin binding to exert its toxicity to A. aegypti larvae, toxicity assays of Cry11Aa and Cry4Ba were performed in the presence of CR7–11. Our results show that CR7–11 enhanced Cry11Aa toxicity more efficiently than the toxicity of Cry4Ba, since lower amounts of CR7–11 were sufficient to observe enhanced Cry11Aa toxicity in contrast with Cry4Ba that required higher amounts of CR7–11 to observe an effect on its toxicity (Figure 4). These results could be explained by the differences in binding affinities of Cry11Aa and Cry4Ba to CR7–11. A previous study showed that a similar protein fragment containing CR9-11 from Anopheles gambiae cadherin enhanced Cry4Ba toxicity to both A. gambiae and A. aegypti [21]. Interestingly, the A. gambiae cadherin fragment was shown to bind Cry4Ba with high affinity (Kd of 13 nM) in contrast with at least 11-fold lower binding affinity of Cry4Ba to the A. aegypti CR7–11 (Kd of 154 nM). Overall, these results might suggest that Cry4Ba is capable of forming the pre-pore oligomer in the absence of cadherin binding, but a high-affinity binding site (such as A. gambiae CR9–11) could still enhance Cry4Ba oligomer formation and toxicity. This remains to be determined.
Although cadherin binding has been shown to be an important binding step of different Cry toxins, the results of the present study show that it is not a limiting step in the toxicity of Cry4Ba to A. aegypti, since cadherin-silenced larvae were as sensitive to Cry4Ba as control larvae in contrast with Cry11Aa that showed reduced toxicity to cadherin-silenced larvae. Identifying the rate-limiting binding steps of Cry toxins that are used for insect control in the field should provide targets for improving insecticides against important insect pests, identifying resistant gene alleles and succeeding in insect-resistant-managing strategies.
Acknowledgments
We thank Lizbeth Cabrera for technical assistance.
FUNDING
Research was funded in part by the National Institutes of Health (NIH) [grant number 1R01 AI066014], Dirección General Asuntos del Personal Académico (DGAPA)/Universidad Nacional Autónoma de México (UNAM) [grant numbers IN218608 and IN210208-N] and Consejo Nacional de Ciencia y Tecnología (CONACyT) [grant number 81639].
Abbreviations used
- 3d
three-domain
- ALP
alkaline phosphatase
- APN
aminopeptidase N
- CR
cadherin repeat
- dsRNA
double-stranded RNA
- GPI
glycosylphophatidylinositol
- PC
phosphatidylcholine
- PLB
planar lipid bilayer
- qPCR
quantitative PCR
- RNAi
RNA interference
- SPR
surface plasmon resonance
- SUV
small unilamellar vesicle
Footnotes
AUTHOR CONTRIBUTION
Claudia Rodríguez-Almazán performed experiments (ELISA binding assays of Cry11Aa and Cry4Ba to CR7–11, final oligomerization assays of Cry11Aa and Cry4Ba, the synergism of CR7–11 on Cry11Aa and Cry4Ba toxicity). Esmeralda Reyes performed experiments (initial binding assays of Cry toxins to Cry4Ba, initial oligomerization assays of Cry4Ba, prepared Cry4Ba membrane oligomer in lipid vesicles for pore-formation assays). Fernando Zuñiga-Navarrete performed experiments (SPR binding assays of Cry4Ba and Cry11Aa to CR7–11). Carlos Muñoz-Garay carried out pore-formation assays of Cry4Ba oligomer in black lipid bilayers. Isabel Gómez performed SPR binding assays of Cry4Ba and Cry11Aa to CR7–11. Amy Evans performed experiments (cadherin gene-silencing assays). Supaporn Likitvivatanavong performed experiments (cloned the two cadherin gene fragments for dsRNA synthesis and also participated in the cadherin gene-silencing assays). Alejandra Bravo designed experiments, and contributed to materials and artwork. Sarjeet Gill designed experiments and contributed to materials. Mario Soberón designed experiments, contributed to materials and wrote the paper.
References
- 1.Bravo A, Gill SS, Soberón M. Mode of action of Bacillus thuringiensis Cry and Cyt toxins and their potential for insect control. Toxicon. 2007;49:423–435. doi: 10.1016/j.toxicon.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soberón M, Fernandez LE, Pérez C, Bravo A. Mode of action of mosquitocidal Bacillus thuringiensis toxins. Toxicon. 2007;49:597–600. doi: 10.1016/j.toxicon.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 3.Pigott CR, Ellar DJ. Role of receptors in Bacillus thuringiensis crystal toxin activity. Microbiol Mol Biol Rev. 2007;71:255–281. doi: 10.1128/MMBR.00034-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pacheco S, Gomez I, Arenas I, Saab-Rincon G, Rodríguez-Almazan C, Gill SS, Bravo A, Soberón M. Domain II loop 3 of Bacillus thuringiensis Cry1Ab toxin is involved in a “ping–pong” binding mechanism with Manduca sexta aminopetidase-N and cadherin receptors. J Biol Chem. 2009;284:32750–32757. doi: 10.1074/jbc.M109.024968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arenas I, Bravo A, Soberón M, Gómez I. Role of alkaline phosphatase from Manduca sexta in the mechanism of action of Bacillus thuringiensis Cry1Ab toxin. J Biol Chem. 2010;285:12497–12503. doi: 10.1074/jbc.M109.085266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gómez I, Dean DH, Bravo A, Soberón M. Molecular basis for Bacillus thuringiensis Cry1Ab toxin specificity: two structural determinants in the Manduca sexta Bt-R1 receptor interact with loops α-8 and 2 in domain II of Cy1Ab toxin. Biochemistry. 2003;42:10482–10489. doi: 10.1021/bi034440p. [DOI] [PubMed] [Google Scholar]
- 7.Gómez I, Arenas I, Benitez I, Miranda-Rios J, Becerril B, Grande R, Almagro JC, Bravo A, Soberón M. Specific epitopes of Domains II and III of Bacillus thuringiensis Cry1Ab toxin involved in the sequential interaction with cadherin and aminopeptidase-N receptors in Manduca sexta. J Biol Chem. 2006;281:34032–34039. doi: 10.1074/jbc.M604721200. [DOI] [PubMed] [Google Scholar]
- 8.Bel Y, Escriche B. Common genomic structure for the Lepidoptera cadherin-like genes. Gene. 2006;381:71–80. doi: 10.1016/j.gene.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 9.Gómez I, Sánchez J, Miranda R, Bravo A, Soberón M. Cadherin-like receptor binding facilitates proteolytic cleavage of helix α-1 in domain I and oligomer pre-pore formation of Bacillus thuringiensis Cry1Ab toxin. FEBS Lett. 2002;513:242–246. doi: 10.1016/s0014-5793(02)02321-9. [DOI] [PubMed] [Google Scholar]
- 10.Bravo A, Gómez I, Conde J, Muñoz-Garay C, Sánchez J, Miranda R, Zhuang M, Gill SS, Soberón M. Oligomerization triggers binding of a Bacillus thuringiensis Cry1Ab pore-forming toxin to aminopeptidase N receptor leading to insertion into membrane microdomains. Biochim Biophys Acta. 2004;1667:38–46. doi: 10.1016/j.bbamem.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 11.Pardo-López L, Gómez I, Rausell C, Sánchez J, Soberón M, Bravo A. Structural changes of the Cry1Ac oligomeric pre-pore from Bacillus thuringiensis induced by N-acetylgalactosamine facilitates toxin membrane insertion. Biochemistry. 2006;45:10329–10336. doi: 10.1021/bi060297z. [DOI] [PubMed] [Google Scholar]
- 12.Likitvivatanavong S, Chen J, Evans AE, Bravo A, Soberón M, Gill SS. Role of cadherin, alkaline phosphatase and aminopeptidase N as receptors of Cry11Ba toxin from Bacillus thuringiensis jegathesan in Aedes aegypti. J Agric Food Chem. 2011;59:2829–2838. doi: 10.1128/AEM.01852-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J, Aimanova KG, Fernandez LE, Bravo A, Soberón M, Gill SS. Aedes aegypti cadherin serves as a putative receptor of the Cry11Aa toxin from Bacillus thuringiensis subsp. israelensis. Biochem J. 2009;424:191–200. doi: 10.1042/BJ20090730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernandez LE, Aimanova KG, Gill SS, Bravo A, Soberón M. A GPI-anchored alkaline phosphatase is a functional midgut receptor of Cry11Aa toxin in Aedes aegypti larvae. Biochem J. 2006;394:77–84. doi: 10.1042/BJ20051517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen J, Aimanova KG, Pan S, Gill SS. Identification and characterization of Aedes aegypti aminopeptidase N as a putative receptor of Bacillus thuringiensis Cry11A toxin. Insect Biochem Mol Biol. 2009;39:688–699. doi: 10.1016/j.ibmb.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delecluse A, Poncet S, Klier A, Rappoport G. Expression of cryIVA and cryIVB genes, independently or in combination, in a crystal-negative strain of Bacillus thuringiensis subsp. israelensis. Appl Environ Microbiol. 1993;59:3922–3927. doi: 10.1128/aem.59.11.3922-3927.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang C, Yu YM, Dai SM, Law SK, Gill SS. High-level cryIVD and cytA gene expression in Bacillus thuringiensis does not require the 20-kilodalton protein, and the coexpressed gene products are synergistic in their toxicity to mosquitoes. Appl Environ Microbiol. 1993;59:815–821. doi: 10.1128/aem.59.3.815-821.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pérez C, Muñoz-Garay CC, Portugal L, Sánchez J, Gill SS, Soberón M, Bravo A. Bacillus thuringiensis subsp. israelensis Cyt1Aa enhances activity of Cry11Aa toxin by facilitating the formation of a pre-pore oligomeric structure. Cell Microbiol. 2007;9:2931–2937. doi: 10.1111/j.1462-5822.2007.01007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rausell C, Muñoz-Garay C, Miranda-CassoLuengo R, Gómez I, Rudiño-Piñera E, Soberón M, Bravo A. Tryptophan spectroscopy studies and black lipid bilayer analysis indicate that the oligomeric structure of Cry1Ab toxin from Bacillus thuringiensis is the membrane-insertion intermediate. Biochemistry. 2004;43:166–174. doi: 10.1021/bi035527d. [DOI] [PubMed] [Google Scholar]
- 20.Cancino-Rodezno A, Alexander C, Villaseñor R, Pacheco S, Porta H, Pauchet Y, Gill SS, Soberón M, Bravo A. The mitogen-activated protein kinase p38 pathway is involved in insect defense against Cry toxins from Bacillus thuringiensis. Insect Biochem Mol Biol. 2010;40:58–63. doi: 10.1016/j.ibmb.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park Y, Hua G, Abdullah MA, Rahman K, Adang MJ. Cadherin fragments from Anopheles gambiae synergize Bacillus thuringiensis Cry4Ba’s toxicity against Aedes aegypti larvae. Appl Environ Microbiol. 2009;75:7280–7282. doi: 10.1128/AEM.01870-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park Y, Abdullah MA, Taylor MD, Rahman K, Adang MJ. Enhancement of Bacillus thuringiensis Cry3Aa and Cry3Bb toxicities to coleopteran larvae by a toxin-binding fragment of an insect cadherin. Appl Environ Microbiol. 2009;75:3086–3092. doi: 10.1128/AEM.00268-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J, Hua G, Jurat-Fuentes JL, Abdullah MA, Adang MJ. Synergism of Bacillus thuringiensis toxins by a fragment of a toxin-binding cadherin. Proc Natl Acad Sci USA. 2007;104:13901–13906. doi: 10.1073/pnas.0706011104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pacheco S, Gomez I, Gill SS, Bravo A, Soberón M. Enhancement of insecticidal activity of Bacillus thuringiensis Cry1A toxins by fragments of a toxin-binding cadherin correlates with oligomer formation. Peptides. 2009;30:583–588. doi: 10.1016/j.peptides.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fabrick J, Oppert C, Lorenzen MD, Morris K, Oppert B, Jurat-Fuentes JL. A novel Tenebrio molitor cadherin is a functional receptor for Bacillus thuringiensis Cry3Aa toxin. J Biol Chem. 2009;284:18401–18410. doi: 10.1074/jbc.M109.001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valaitis AP. Localization of Bacillus thuringiensis Cry1A toxin-binding molecules in gypsy moth larval gut using fluorescence microscopy. J Invertebr Pathol. 2011;108:69–75. doi: 10.1016/j.jip.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 27.Likitvivatanavong S, Katzenmeier G, Angsuthanasombat C. Asn183 in α5 is essential for oligomerisation and toxicity of the Bacillus thuringiensis Cry4Ba toxin. Arch Biochem Biophys. 2006;445:46–55. doi: 10.1016/j.abb.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 28.Boonserm P, Davis P, Ellar DJ, Li J. Crystal structure of the mosquito-larvicidal toxin Cry4Ba and its biological implications. J Mol Biol. 2005;348:363–382. doi: 10.1016/j.jmb.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 29.Taveecharoenkool T, Angsuthanasombat C, Kantchanawarin C. Combined molecular dynamics and continuum solvent studies of the pre-pore Cry4Aa trimer suggest its stability in solution and how it may form a pore. PMC Biophys. 2010;3:1–16. doi: 10.1186/1757-5036-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dechklar M, Tiewsiri K, Angsuthanasombat C, Pootankit K. Functional expression in insect cells of glycosylphosphatidylinositol-linked alkaline phosphatase from Aedes aegypti larval midgut: a Bacillus thuringiensis Cry4Ba toxin receptor. Insect Biochem Mol Biol. 2011;41:159–166. doi: 10.1016/j.ibmb.2010.11.006. [DOI] [PubMed] [Google Scholar]