Abstract
Inhibition of the endocannabinoid receptor CB1 improves insulin sensitivity, lowers glycemia and slows atherosclerosis. We analyzed if common variants in the gene encoding CB1, CNR1, are associated with insulin resistance, risk of type 2 diabetes (T2D) or coronary heart disease (CHD).
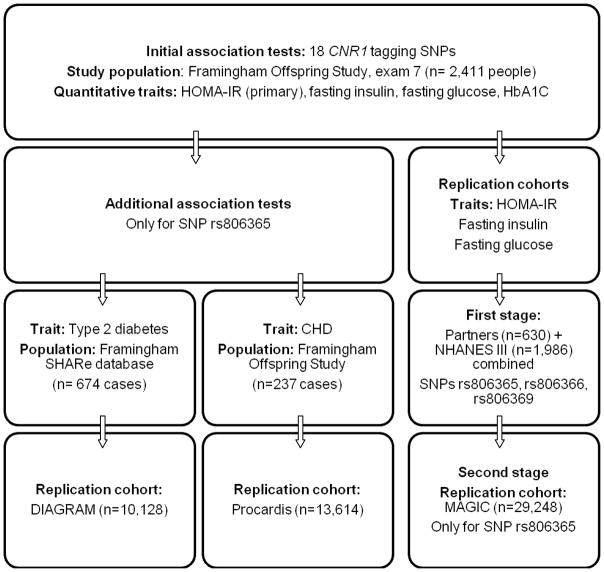
We studied 2,411 participants of the Framingham Offspring Study (mean age 60 years, 52% women) for quantitative traits and CHD, and the Framingham SHARe database for T2D risk. We genotyped 19 single nucleotide polymorphisms (SNPs) that tagged 85% (at r2=0.8) of common (>5%) CNR1 SNPs. Fasting blood glucose and insulin at the 7th (1999–2001) exam were collected. We used age-, sex-, BMI-adjusted models to test additive associations of genotype with HOMA-IR (linear mixed-effect models), T2D or CHD. To account for multiple tests of SNPs, we generated empirical P values. The C allele at SNP rs806365 (frequency, 57.4%), ~4.1kb 3′ from CNR1, was associated with increased HOMA-IR (n=2,261, beta=0.05 per C, empirical P=0.01), risk of T2D (674 cases, OR=1.19 per C, nominal P=0.01) and CHD (237 cases, HR=1.23 per C, nominal P=0.04). The association of rs806365 with HOMA-IR was replicated in a meta-analysis of two independent cohorts (NHANES III plus Partners Case-Control Diabetes Study; 2,540 white individuals, beta=0.037, nominal P=0.007), but not in the large MAGIC Consortium (n=29,248, nominal P=0.74). The association of rs806365 was not replicated either with T2D in DIAGRAM (n=10,128, nominal P=0.31), or with CHD in PROCARDIS (n=13,614, nominal P=0.37).
Although supported by initial results, we found no reproducible statistical association of common variation at CNR1 with insulin resistance, T2D or CHD.
Keywords: endocannabinoids, candidate genes, diabetes mellitus, insulin resistance, coronary heart disease
Introduction
Overweight, increased insulin resistance and subsequent progressive pancreatic beta cell relative insufficiency are determining factors for the development of type 2 diabetes mellitus (T2D) (1). Genetic background plays a role in the development of T2D (2), yet the contribution of variation in common single nucleotide polymorphisms (SNPs) has so far modestly explained the genetic basis of T2D (3,4) or improved risk prediction when added to validated clinical scores (5,6). Additional genetic contributors to T2D remain to be found.
The endocannabinoid system modulates body weight and insulin sensitivity (7–10). Endogenous cannabinoids (endocannabinoids) are lipid molecules that target the receptors CB1 and CB2 (11). CB1 is widely expressed in the brain, as well as in the liver, adipose tissue, gut, skeletal muscle and pancreas (12–14). In animal models, CB1 agonists lead to increased food intake, whereas CB1 inhibition reduce food intake and promote weight loss, thus reducing insulin resistance (15). Accordingly, therapy with rimonabant, a selective CB1 antagonist, is associated with significant weight loss, diminished waist circumference, improved cardiovascular risk profile and lower fasting insulin levels (16) and in the STRADIVARUS trial, with reduced coronary artery total atheroma volume by coronary intravascular ultrasonography (17). In T2D patients, rimonabant improves glycemic control, lowers insulin resistance and promotes weight loss (18–19).
We have previously tested the association of common genetic variation in or around CNR1, the gene that encodes CB1, with adiposity-related quantitative traits, finding no significant associations (20). Here, our main objective was to test the hypotheses that 1) common variants in the CNR1 gene are associated with insulin resistance and glucose-related quantitative traits in non-diabetic individuals and 2) insulin resistant-associated variants confer an increased risk for T2D and CHD. We studied the community-based, prospective Framingham Offspring Study, found one promising variant and sought replication of the finding in additional independent datasets.
Methods and Procedures
Primary association study population
The original prospective Framingham Heart Study was started in 1948 to study predisposing factors for cardiovascular disease. Participants are essentially all of white European ancestry. In 1971, the Framingham Offspring Study enrolled 5,124 offspring of the original cohort and the spouses of the offspring (21). Participants have been seen in the clinic about every 4 years (up to 7 exams) to receive a complete medical history and physical exam, including anthropometric measurement and blood sample collection. The data used for this study pertain to exam 7 (1998–2001). Genotypes were available for 2,792 participants.
For the analysis of quantitative continuous glycemic traits, we only included the 2,411 non-diabetic participants of the Framingham Offspring Study (1,389 unrelated and 1,022 related, that is, from 282 pedigrees with two or more members genotyped) for whom complete clinical and genotype data were available (Figure 1). To look for the association with risk of T2D as a binary trait, we conducted a case-cohort analysis using the FHS-SHARe database, that comprises the ~7,900 individuals from the three Framingham Heart Study cohorts (original, offspring and third generation) who have both genotype and follow-up data for diabetes onset at each exam (674 T2D cases) (22). Finally, only data from the Framingham Offspring Study were used to evaluate incidence of CHD (237 cases). All participants gave written informed consent. The study protocols were approved by the Institutional Review Board at the Boston University Medical Center.
Figure 1.
Flow-chart showing which databases have been used as primary association study populations or as replication cohorts for each trait. Framingham SHARe database includes the Framingham Heart Study original cohort, the Framingham Offspring Study and the Third Generation cohort. Framingham Heart Study, NHANES III and Partners databases contribute to MAGIC. SNP: single nucleotide polymorphism. CHD: Coronary heart disease.
Framingham outcome and control variables
Our main interest was to evaluate the association of genetic variation in or around CNR1 in the Framingham Offspring Study at exam 7 with insulin resistance reflected by the Homeostasis Model Assessment of Insulin Resistance (HOMA-IR=fasting insulin × fasting glucose/22.5) (23), as well as test for the association among common variation at CNR1 with risk of T2D and CHD. As secondary outcomes, we also analyzed fasting blood glucose levels, fasting insulin levels and glycated hemoglobin A1C at exam 7. We defined T2D as having a fasting plasma glucose ≥126 mg/dl (7.0 mmol/l) at any exam or diabetes treatment at any exam. To calculate body mass index (BMI) we divided weight in kilograms by height in meters squared. Coronary heart disease (CHD) was defined as according to established criteria (24).
Single-nucleotide polymorphism (SNP) selection
CNR1 is located on chromosome 6q15. We selected the 19 tagging SNPs in the region using a pair-wise approach to capture (with an r2=0.8) 100% of common variation (minor allele frequency (MAF) >5%) in the chromosomal region +20 kb 5′-upstream- and +10 kb 3′-downstream- of CNR1, using the Tagger function as implemented in Haploview (25), based on the Hap Map release 21a. We genotyped tag SNPs on an iPLEX Sequenom platform. Genotyped SNPs passed quality-control filters (MAF >5%; P>0.001 for deviation from Hardy-Weinberg equilibrium (HWE); minimal call rate: 94%, average call rate: 98%). The observed genotype frequencies for one SNP (rs6928813) deviated from HWE (P<0.001) and this SNP was not considered in our association analyses. After the present analysis was almost complete, an updated assembly of the CNR1 region was found to include a novel 20.5 kb segment in the 5′ region (26,27). However, given our wide initial tagging approach and the strong linkage disequilibrium in the region, the original 18 tag SNPs (after excluding rs6928813) still allowed for 85% coverage of CNR1 +10 kb downstream with r2=0.8.
Replication study populations
We looked for replication of association of CNR1 SNPs with HOMA-IR and T2D by de novo genotyping or in silico replication in several large independent cohorts. For associations with HOMA-IR we evaluated genotype data of the participants of the National Health and Nutrition Examination Survey III genetic cohort (NHANES-III) (28), as well as genotype data from the participants in the Partners Case-Control Diabetes Study from the Massachusetts General Hospital and the Brigham and Women’s Hospital (29). In NHANES-III, 1,986 of the subjects were white, did not have T2D and had available information. In the Partners Case-Control Diabetes Study, which had recruited 1,157 individuals, 630 subjects were white non-diabetic controls. We then meta-analyzed the replication results in whites (n=2,616, adding 1,986 from NHANES-III plus 630 from the Partners Study).
We sought additional confirmation of significant associations with HOMA-IR in the Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) data, in which the Framingham Offspring Study, the NHANES-III and the Partners datasets are included (30). For this study, MAGIC provided laboratory values information for up to 46,152 individuals. Framingham contributed data for 6,040 subjects to MAGIC, whereas both NHANES-III and Partners Study contributed to MAGIC with 1,219 and 635 subjects respectively.
We also searched for replication of the association with T2D risk in the Diabetes Genetics Replication and Meta-analysis (DIAGRAM) database (n=10,128) (31). Lastly, we had access to PROCARDIS (32), a large database designed to evaluate CHD, to look for replication of the association with CHD (n=13,614, including control data from the Wellcome Trust Case Control Consortium (WTCCC)).
Statistical methods
The values for HOMA-IR, fasting glucose, fasting insulin and HbA1C were logarithmically transformed for the regression analysis. In order to account for familial relationships, we used linear mixed-effect models as implemented in LMEKIN (version 1.1.0) in the R statistical programming environment (version 2.9.2) (33). As the main hypotheses, we tested the association of the SNPs around CNR1 with HOMA-IR. In predefined secondary analyses, we evaluated associations with fasting glucose levels, fasting insulin levels and HbA1C. For the meta-analysis of the NHANES and Partners datasets, we used METAL (34) to summarize the effects from linear regression models adjusted for sex, age and BMI.
To correct for multiple testing for each phenotype in the FHS sample, we used a simulation to determine the distribution of the minimum P value across all SNPs under the null hypothesis of no association. We simulated 1,000 null traits for our sample using SIMQTL in SOLAR with heritability set to 35% (35, 36). Each simulated trait was analyzed using linear mixed-effect models, and the minimum P values over all SNPs were recorded. The empirical P values correspond to the proportion of simulated minimum P values smaller or equal to the observed P values in our data. We set an empirical P value <0.05 to denote statistical significance. With our sample size of 2,516 individuals with genotyped DNA we had a statistical power of 87.8% at a significance level of 5% to identify SNPs explaining at least 0.5% of the variance of the quantitative traits, assuming an additive genetic model and a pair-wise correlation between the tested SNPs and a causal variant of r2 ≥ 0.8.
To test for associations with T2D, we built logistic regression models, adjusted for sex, age and BMI, to calculate odds ratios (ORs) in the SHARe case-cohort data. Using the same covariate adjustments for CHD, we used Cox proportional-hazards models using frailty for sibling correlation to estimate hazard ratios (HRs). The precise ascertainment in time for CHD events allowed for its analysis using Cox method. Since we planned to test one hypothesis (that the SNP associated with HOMA-IR was also associated with T2D or CHD), for these ORs and HRs, a nominal P value ≤ 0.05 defined statistical significance.
Results
Association of genetic variants in CNR1 with HOMA-IR
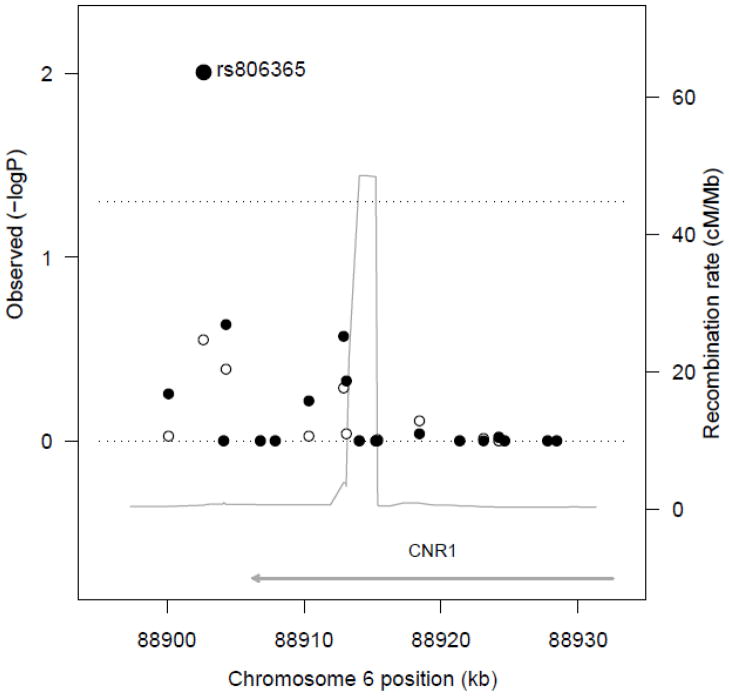
Our sample was a middle-aged cohort (mean 60.8±9.6 years), with about half women (52%) (Table 1). Online supplemental (OLS) Table S1 shows the characteristics and quality criteria for the tagging SNPs in the CNR1 gene. OLS Figure S1 depicts the updated CNR1 locus from 88,906,306 to 88,932,486 in chromosome 6q15 and its linkage disequilibrium structure in the CEU Hap Map population. The C allele of SNP rs806365 (frequency, 57.4%) (37), ~4.1 kb 3′ from CNR1, showed a significant association with increased HOMA-IR relative to T allele carriers in the BMI-adjusted model in the Framingham Offspring Study at exam 7 (n=2,261; beta=0.052 per C, empirical P=0.01; Figure 2 and Table 2). When waist circumference was substituted for BMI, results were unchanged. SNP rs806365 was not significantly associated with HOMA-IR in the non-BMI-adjusted model (empirical P=0.19), nor did it show any significant associations with fasting glucose levels or HbA1C (OLS Table S2). No other SNP was found to be significantly associated with HOMA-IR, fasting insulin, fasting glucose or HbA1C levels at an empirical P value <0.05 in the Framingham Offspring Study at exam 7. We observed that nominal significant or borderline associations between some of the SNPs and HOMA-IR in the tagging SNP analysis were clustered within a genomic segment whose boundaries were defined by rs806371 and rs10485171, 5′ to 3′ (Table 2). In order to fine map this signal, we built common (frequency >5%) haplotypes using five SNPs included in this genomic segment that showed nominally significant associations with HOMA-IR. Indeed, common haplotypes in this region of CNR1 have been previously associated with lipid traits (38). We found that the common haplotypes having the C allele for SNP rs806365 showed the strongest positive associations with HOMA-IR in our Framingham Offspring population, largely recapitulating our SNP analysis.
Table 1.
Characteristics of the Framingham study sample (n=2,411)
| Clinical variables | Mean ± standard deviation, or % |
|---|---|
| Age, years | 60.8 ± 9.6 |
| Women, % | 52.6% |
| Body mass index at exam 7, kg/m2 | 28.2 ± 5.4 |
| Diabetes Mellitus, % | 9.7% |
| Blood fasting glucose at exam 7, mg/dl | 105.5 ± 26.6 |
| Hemoglobin A1C at exam 7, % | 5.7 ± 1.0 |
| Fasting insulin levels at exam 7, μU/ml | 15.3 ± 11.0 |
| Homa-IR at exam 7, mmol/L × μU/ml | 4.2 ± 3.9 |
Figure 2.
Association among the CNR1 SNPs with HOMA-IR. The observed negative logarithm base 10 of the empirical P values for the association of CNR1 SNPs with HOMA-IR, without (white circles) and with BMI-adjustment (black circles) are plotted against genomic location. The recombination rate across the locus is shown by the thin grey line. The upper transverse dashed line represents an empirical P value = 0.05.
Table 2.
Association of tag SNPs at the CNR1 gene locus with insulin resistance raits at exam 7 a.
| SNP | SNP position | Risk allele (% frequency)/other (%) | HOMA-IR | FASTING INSULIN | ||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| B estimate (standard error) | Nominal P value | Empirical P value | B estimate (standard error) | Nominal P value | Empirical P value | |||
| rs10485171 | 88900109 | C (43.3)/T (56.7) | 0.029 (0.015) | 0.059 | 0.51 | 0.014 (0.013) | 0.28 | 0.97 |
| rs806365 | 88902668 | C (57.4)/T (42.6) | 0.052 (0.015) | 0.00061 | 0.011 | 0.035 (0.013) | 0.0076 | 0.096 |
| rs7766029 | 88904154 | C (52.9)/T (47.1) | 0.011 (0.015) | 0.47 | 1 | 0.0015 (0.013) | 0.91 | 1 |
| rs806366 | 88904308 | T (50.0)/C (50.0) | 0.037 (0.015) | 0.013 | 0.15 | 0.032 (0.013) | 0.013 | 0.15 |
| rs806368 | 88906819 | C (23.6)/T (76.4) | 0.014 (0.018) | 0.42 | 1 | 0.013 (0.016) | 0.42 | 1 |
| rs12720071 | 88907900 | A (89.3)/G (10.7) | 0.023 (0.026) | 0.37 | 0.99 | 0.017 (0.023) | 0.46 | 1 |
| rs1049353 | 88910354 | A (25.2)/G (74.8) | 0.033 (0.017) | 0.056 | 0.49 | 0.027 (0.015) | 0.069 | 0.56 |
| rs806369 | 88912897 | C (71.0)/T (29.0) | 0.038 (0.016) | 0.019 | 0.21 | 0.022 (0.014) | 0.11 | 0.75 |
| rs806371 | 88913082 | G (14.2)/T (85.8) | 0.046 (0.022) | 0.033 | 0.34 | 0.04 (0.019) | 0.035 | 0.36 |
| rs806374 | 88914039 | T (63.7)/C (36.3) | 0.0042 (0.015) | 0.78 | 1 | -9.1e-05 (0.013) | 0.99 | 1 |
| rs806375 | 88915240 | T (43.0)/A (57.0) | 0.011 (0.015) | 0.46 | 1 | 0.0021 (0.013) | 0.87 | 1 |
| rs806376 | 88915367 | C (48.3)/T (51.7) | 0.01 (0.015) | 0.5 | 1 | 0.0046 (0.013) | 0.73 | 1 |
| rs806380 | 88921372 | A (65.4)/G (34.6) | 0.0067 (0.015) | 0.67 | 1 | 0.0091 (0.013) | 0.5 | 1 |
| rs7752758 | 88923095 | G (10.5)/A (89.5) | 0.027 (0.024) | 0.26 | 0.96 | 0.018 (0.021) | 0.39 | 0.99 |
| rs12528858 | 88924207 | A (94.3)/G (5.7) | 0.04 (0.032) | 0.21 | 0.92 | 0.038 (0.028) | 0.18 | 0.9 |
| rs12205430 | 88924644 | T (79.0)/C (21.0) | 0.0075 (0.018) | 0.68 | 1 | 0.0067 (0.016) | 0.67 | 1 |
| rs6454673 | 88927768 | G (70.4)/A (29.6) | 0.0051 (0.016) | 0.75 | 1 | 0.0081 (0.014) | 0.56 | 1 |
| rs6914429 | 88928435 | C (11.4)/A (88.6) | 0.014 (0.024) | 0.55 | 1 | 0.0039 (0.021) | 0.85 | 1 |
Abbreviations: SNP = single nucleotide polymorphism, HOMA-IR: Homeostasis Model Assessment for Insulin Resistance. Results are derived from age-, sex-, BMI-adjusted regression models. SNPs position according to Hap Map data Rel 24/phase II Nov 08, dbSNP b126. SNP rs6928813 was not included in the statistical analysis, as its observed genotype frequencies deviated from Hardy–Weinberg equilibrium (HWE) (P<0.001).
The β estimates reflect the additive effect of the risk allele on the log-transformed outcome. Nominal P values are uncorrected for multiple testing. To determine the distribution of the minimum P value across all SNPs under the null hypothesis of no association, we simulated 1,000 traits for our sample, as an approach to correct for the multiple SNPs tested. The empirical P values correspond to the proportion of simulated minimum P values smaller or equal to the observed nominal P values in our data. An empirical P value <0.05 denotes statistical significance (see text for details).
Association of genetic variants in CNR1 with the risk of T2D
We tested for the association between SNP rs806365 with risk of T2D in the FHS-SHARe database. We detected a significant association for rs806365 with risk of T2D in the sex-, age-adjusted model (OR=1.15 [1.01–1.30] per C, nominal P=0.03) and also after further BMI-adjustment (OR=1.19 [1.04–1.35] per C, nominal P=0.01).
Association of genetic variants in CNR1 with coronary heart disease
In the sex-, age-adjusted proportional hazards model, the C allele of CNR1 rs806365 significantly conferred a higher risk of CHD in the Framingham Offspring Study (n=237 cases, HR=1.26 [1.02–1.55], nominal P=0.03). After further BMI-adjustment, the association remained significant (HR=1.23 [1.00–1.51], nominal P=0.04).
Tests of replication of associations with rs806365 in other cohorts
With these promising results for rs806365, we sought replication in other cohorts. In the meta-analysis of the NHANES-III and Partners data combined, SNP rs806365 was significantly associated with both HOMA-IR (n=2,540; beta=0.037, nominal P=0.007) and fasting insulin (n=2,553; beta=0.036, nominal P=0.004) in sex-, age-, BMI-adjusted models, supporting our initial finding in FHS (Table 3). We also evaluated rs806366 and rs806369 for HOMA-IR in these combined cohorts, since their empirical P values neared statistical significance in the FHS cohort. Nonetheless, the NHANES and Partners meta-analysis did not show significant associations for these SNPs (for HOMA-IR, nominal P values=0.5 and 0.27, respectively) (Table 3).
Table 3.
Summary of replication results for the association of the SNPs rs806365, rs806366 and rs806369 with insulin-resistance traits, fasting glucose and risk of diabetes in the following datasets: meta-analysis of the NHANES-III and Partners databases combined; MAGIC; and DIAGRAMa.
| HOMA-IR | Fasting glucose | Fasting insulin | Risk of T2D | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||
| SNP | Risk allele/other | N | β (SE) | P value | N | β (SE) | P value | N | β (SE) | P value | N | OR (CI) | P value |
| NHANES-PARTNERS | |||||||||||||
| rs806365 | C/T | 2,540 | 0.037 (0.014) | 0.007 | 2,551 | 0.020 (0.013) | 0.12 | 2,553 | 0.036 (0.013) | 0.004 | |||
| rs806366 | T/C | 2,573 | −0.009 (0.013) | 0.49 | 2,584 | −0.006 (0.013) | 0.66 | 2,586 | −0.011 (0.012) | 0.35 | |||
| rs806369 | T/C | 2,579 | 0.016 (0.014) | 0.27 | 2,590 | 0.005 (0.014) | 0.7 | 2,592 | 0.017 (0.013) | 0.19 | |||
|
| |||||||||||||
| MAGIC | |||||||||||||
| rs806365 | T/C | 29,248 | 0.001 (0.003) | 0.74 | 34,558 | 0.008 (0.004) | 0.07 | 29,561 | 0.001 (0.003) | 0.72 | |||
|
| |||||||||||||
| DIAGRAM | |||||||||||||
| rs806365 | T/C | 10,128 | 1.00 (0.94–1.07) | 0.31 | |||||||||
Abbreviations: SNP = single nucleotide polymorphism. HOMA-IR: Homeostasis Model Assessment for Insulin Resistance.
SE: standard error. T2D: type 2 diabetes. OR: odds ratio. CI: confidence interval.
NHANES: National Health and Nutrition Examination Survey III (28). Partners: Massachusetts General Hospital plus Brigham and Women’s Hospital case-control diabetes study database (29). MAGIC: The Meta-Analyses of Glucose and Insulin-related traits Consortium database (30). DIAGRAM: The Diabetes Genetics Replication and Meta-analysis (31).
All P values are nominal. Results from the meta-analysis of the NHANES III and Partners cohorts and from MAGIC are age-, sex-, BMI-adjusted, whereas those from DIAGRAM are not BMI-adjusted.
As results from both FHS and the NHANES-Partners meta-analysis consistently supported significant associations for SNP rs806365 with HOMA-IR, we searched for additional confirmation of the association between SNP rs806365 with HOMA-IR in the MAGIC dataset. We did not find a significant association of rs806365 with HOMA-IR (n=29,248; nominal P=0.74) or with fasting glucose levels (n=34,558; nominal P=0.07) in MAGIC in a sex-, age-, BMI-adjusted model (Table 3). Unfortunately, the phasing of haplotypes is not available in all data sets, which did not allow us to seek replication of the haplotype analyses results in our replication cohorts.
With regard to risk of T2D, the DIAGRAM database disclosed no statistically significant association with rs806365 (n=10,128, OR=1.0, nominal P=0.31), but these aggregate results were not BMI-adjusted (Table 3). P values for heterogeneity in MAGIC and DIAGRAM were 0.51 and 0.13, respectively.
Finally, the association of rs806365 with CHD in Framingham was not replicated in PROCARDIS (n=13,614; beta=0.025, nominal P=0.37).
Discussion
As a main finding, our study highlights the preliminary significant association of the SNP rs806365 near CNR1 with HOMA-IR, T2D and CHD in the Framingham Offspring Study. For these phenotypes we consistently found that the C allele, associated with higher levels of HOMA-IR, was also associated with increased risk for T2D. However, we acknowledge that rs806365 may not be the true functional variant that explains the significant association with the traits. For a complex trait like glucose homeostasis, the causative allele may actually be composed of a combination of common and rare variants (39).
The fact that the association between rs806365 with HOMA-IR was only significant after BMI-adjustment in Framingham would suggest that factors other than obesity may be playing a role to explain this association in our population. The associations with T2D and CHD in age, sex adjusted models remained significant before and after adjusting for BMI.
We replicated the association of rs806365 with HOMA-IR in a meta-analysis comprising the independent NHANES III and Partners cohorts, reinforcing the notion of its possible association with insulin resistance (40). Nonetheless, our efforts to confirm the results in the larger databases from the MAGIC, DIAGRAM and PROCARDIS consortia failed to disclose a significant association among CNR1 tagging SNPs with insulin resistance traits, risk of diabetes or CHD.
We performed this candidate-gene study of the CB1 cannabinoid receptor CNR1 gene to test the hypothesis of its association with insulin resistance-related traits. Previously described physiological facts supported the working hypothesis: firstly, mice null for the cannabinoid receptors have a lean phenotype even when they are fed with a high fat diet (41). Secondly, the cannabinoid receptors modulate glucose-induced calcium transient concentrations and insulin secretion by beta cells (8, 13). Thirdly, genetic variation may be associated with lower expression levels of CB1 in adipose tissues (42). This receptor regulation has been described within adipose tissues for SNP rs12720071 in CNR1, which is located ≈5.2 kb upstream from rs806365, yet in low linkage disequilibrium with it (r2=0.12). In spite of the reported CB1 regulator activity, we found no significant associations for rs12720071 with any trait in our cohort. And fourthly, accumulated evidence indicates that inhibition of CB1 activity with rimonabant is associated with a better glycemic control and diminished insulin resistance in treated patients (19).
Notwithstanding this background supporting the association of levels of function between CNR1 and both insulin secretion and resistance, there are few previous studies exploring the plausible association among common genetic variation in CNR1 and risk of diabetes or insulin resistance (38). Studies testing the association among genetic variants of the endocannabinoid gene and obesity have also shown conflicting results, since some groups (43,44) did not replicate results that had been reported earlier (45–48). The absence of an association with obesity in an earlier report by our group (20) did not preclude us from testing it for its association with T2D risk and insulin resistance, as a mutation in a gene might theoretically increase insulin resistance without having a discernible impact on obesity.
It is not clear why the positive replication results in the independent NHANES III and Partners databases were not reproduced in the larger MAGIC, DIAGRAM and PROCARDIS studies. The possibility of a falsely positive result in our cohort cannot be completely ruled out. Inclusion criteria in the different cohorts may vary. Genome-wide data are available in Framingham and there is no evidence of inflation of the type-I error for the traits analyzed due to population stratification (49). The NHANES III and Partners cohorts included population samples which were fairly similar to Framingham in terms of sample size and ancestry. Neither the publicly-available DIAGRAM nor PROCARDIS provided BMI-adjusted data. Although it might be argued that heterogeneity in larger databases can hamper the detection of true associations present in more homogeneous samples, the P values for heterogeneity for both DIAGRAM and MAGIC databases are not statistically significant. Thus, the negative evidence provided by the large consortia datasets does not allow to definitely prove or rule out a significant association among common genetic variation in or around CNR1 and insulin resistance or risk of T2D.
The large, unbiased community-based design, the standardized assessment of clinical covariates, the spectrum of insulin resistance and glucose-related traits and the comprehensive coverage of common genetic variation in and around the CNR1 gene initially gave robustness to our results. However, some limitations must be taken into account: our database is mainly comprised of white subjects of European ancestry, so that an attempt to extrapolate the results to other ethnicities must be done with caution. We focused on common genetic variants, and the coverage of the CNR1 region was not complete (85%); thus, we cannot definitely rule out that rarer genetic variants (minor allele frequency <5%) or additional tagging SNPs around this gene might be associated with insulin resistance-related traits. Finally, variants with very modest effects on the variance of the traits could have been missed, since we only had adequate power to detect genetic variants that explained >0.5% of the variance in the traits.
Conclusions
Although supported by initial results and biologically plausible, we did not find a reproducible statistical association of common variation at CNR1 with insulin resistance or T2D. Despite the lack of replication, a small significant effect for rs806365 on insulin resistance cannot be completely ruled out. The results remain consistent with the hypothesis, supported by other data, that genetic variation in the endocannabinoid system may have a modest effect on insulin resistance and related phenotypes.
Supplementary Material
Acknowledgments
This study was supported by: an investigator-initiated research grant from sanofi-aventis (JBM), the National Heart, Lung, and Blood Institute’s Framingham Heart Study (Contract No. N01-HC-25195), an American Diabetes Association Career Development Award (JBM), and the Boston University Linux Cluster for Genetic Analysis (LinGA) funded by the NIH-NCRR Shared Instrumentation grant (1S10RR163736-01A1). PROCARDIS was supported by the European Community Sixth Framework Program (LSHMCT-2007-037273), AstraZeneca, the Knut and Alice Wallenberg Foundation and the British Heart Foundation. PROCARDIS also made use of data generated by the WTCCC. A full list of the investigators who contributed to the generation of the WTCCC data, funding for which was provided by the Wellcome Trust under award 076113, is available from www.wtccc.org.uk. JMDMY is supported by a “Bolsa de Ampliación de Estudios” from the “Instituto de Salud Carlos III”, Madrid, Spain (2009/90071). JBM is also supported by NIDDK K24 DK080140. JCF is supported by a NIH Research Career Award K23 DK65978-04.
Footnotes
Disclosure statement
JBM currently has research grants from GlaxoSmithKline, and serves on a consultancy board for Interleukin Genetics.
References
- 1.Surampudi PN, John-Kalarickal J, Fonseca VA. Emerging concepts in the pathophysiology of type 2 diabetes mellitus. Mt Sinai J Med. 2009;76:216–226. doi: 10.1002/msj.20113. [DOI] [PubMed] [Google Scholar]
- 2.Permutt MA, Wasson J, Cox N. Genetic epidemiology of diabetes. J Clin Invest. 2005;115:1431–1439. doi: 10.1172/JCI24758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grant RW, Moore AF, Florez JC. Genetic architecture of type 2 diabetes: recent progress and clinical implications. Diabetes Care. 2009;32:1107–1114. doi: 10.2337/dc08-2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ruchat SM, Elks CE, Loos RJ, et al. association between insulin secretion, insulin sensitivity and type 2 diabetes susceptibility variants identified in genome-wide association studies. Acta Diabetol. 2009;46:217–226. doi: 10.1007/s00592-008-0080-5. [DOI] [PubMed] [Google Scholar]
- 5.Meigs JB, Shrader P, Sullivan LM, et al. Genotype score in addition to common risk factors for prediction of type 2 diabetes. N Engl J Med. 2008;359:2208–2219. doi: 10.1056/NEJMoa0804742. Erratum in: N Engl J Med 2009, 360, 648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Hoek M, Dehghan A, Witteman JC, et al. Predicting type 2 diabetes based on polymorphisms from genome-wide association studies: a population-based study. Diabetes. 2008;57:3129–3135. doi: 10.2337/db08-0425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engeli S, Bohnke J, Feldpausch M, et al. Activation of the peripheral endocannabinoid system in human obesity. Diabetes. 2005;54:2838–2843. doi: 10.2337/diabetes.54.10.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bermudez-Silva FJ, Suarez-Perez J, Nadal A, Rodriguez de Fonseca F. The role of the pancreatic endocannabinoid system in glucose metabolism. Best Pract Res Clin Endocrinol Metab. 2009;23:87–102. doi: 10.1016/j.beem.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 9.Cota D, Sandoval DA, Olivieri M, et al. Food intake-independent effects of CB1 antagonism on glucose and lipid metabolism. Obesity. 2009;17:1641–1645. doi: 10.1038/oby.2009.84. [DOI] [PubMed] [Google Scholar]
- 10.Motaghedi R, McGraw T. The CB1 endocannabinoid system modulates adipocyte insulin sensitivity. Obesity. 2008;16:1727–1734. doi: 10.1038/oby.2008.309. [DOI] [PubMed] [Google Scholar]
- 11.Howlett AC, Barth F, Bonner TI, et al. International Union of Pharmacology. XXVII. Classification of cannabinoid receptors. Pharmacol Rev. 2002;54:161–202. doi: 10.1124/pr.54.2.161. [DOI] [PubMed] [Google Scholar]
- 12.Matias I, Di Marzo V. Endocannabinoids and the control of energy balance. Trends Endocrinol Metabol. 2006;18:27–37. doi: 10.1016/j.tem.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 13.Tharp WG, Lee YH, Maple RL, Pratley RE. The cannabinoid CB1 receptor is expressed in pancreatic delta-cells. Biochem Biophys Res Commun. 2008;372:595–600. doi: 10.1016/j.bbrc.2008.05.077. [DOI] [PubMed] [Google Scholar]
- 14.Bermúdez-Silva FJ, Suárez J, Baixeras E, et al. Presence of functional cannabinoid receptors in human endocrine pancreas. Diabetologia. 2008;51:476–487. doi: 10.1007/s00125-007-0890-y. [DOI] [PubMed] [Google Scholar]
- 15.Chambers AP, Sharkey KA, Koopmans HS. Cannabinoid (CB1) receptor antagonist, AM 251, causes a sustained reduction of daily food intake in the rat. Physiology & Behavior. 2004;82:863–869. doi: 10.1016/j.physbeh.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 16.Després JP, Golay A, Sjöström L Rimonabant in Obesity-Lipids Study Group. Effects of rimonabant on metabolic risk factors in overweight patients with dyslipidemia. N Engl J Med. 2005;353:2121–2134. doi: 10.1056/NEJMoa044537. [DOI] [PubMed] [Google Scholar]
- 17.Nissen SE, Nicholls SJ, Wolski K, et al. Effect of rimonabant on progression of atherosclerosis in patients with abdominal obesity and coronary artery disease: the STRADIVARIUS randomized controlled trial. JAMA. 2008;299:1547–1560. doi: 10.1001/jama.299.13.1547. [DOI] [PubMed] [Google Scholar]
- 18.Scheen AJ, Finer N, Hollander P, Jensen MD, Van Gaal LF RIO-Diabetes Study Group. Efficacy and tolerability of rimonabant in overweight or obese patients with type 2 diabetes: a randomised controlled study. Lancet. 2006;368:1660–1672. doi: 10.1016/S0140-6736(06)69571-8. Erratum in: Lancet, 2006, 368, 1650. [DOI] [PubMed] [Google Scholar]
- 19.Rosenstock J, Hollander P, Chevalier S, Iranmanesh A SERENADE Study Group. SERENADE: the Study Evaluating Rimonabant Efficacy in Drug-naive Diabetic Patients: effects of monotherapy with rimonabant, the first selective CB1 receptor antagonist, on glycemic control, body weight, and lipid profile in drug-naive type 2 diabetes. Diabetes Care. 2008;31:2169–2176. doi: 10.2337/dc08-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lieb W, Manning AK, Florez JC, et al. Variants in the CNR1 and the FAAH Genes and Adiposity Traits in the Community. Obesity. 2009;17:755–760. doi: 10.1038/oby.2008.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kannel WB, Feinleib M, McNamara PM, Garrison RJ, Castelli WP. An investigation of coronary heart disease in families. The Framingham offspring study. Am J Epidemiol. 1979;110:281–290. doi: 10.1093/oxfordjournals.aje.a112813. [DOI] [PubMed] [Google Scholar]
- 22.Mailman MD, Feolo M, Jin Y, et al. The NCBI dbGaP database of genotypes and phenotypes. Nat Genet. 2007;39:1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- 24.Kannel WB, Wolf PA, Garrison RJ. The Framingham Study: An Epidemiological Investigation of cardiovascular Disease: Section 34: Some Risk Factors Related to the Annual Incidence of Cardiovascular Disease and Death Using Pooled Repeated Biennial Measurements: Framingham Heart Study, 30-Year-Follow-Up. Bethesda, Md: National Heart, Lung, and Blood Institute; 1987. [Google Scholar]
- 25.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 26.http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene&cmd=Retrieve&dopt=Graphics&list_uids=1268≠refseq
- 27.Zhang PW, Ishiguro H, Ohtsuki T, et al. Human cannabinoid receptor 1: 5′ exons, candidate regulatory regions, polymorphisms, haplotypes and association with polysubstance abuse. Mol Psychiatry. 2004;9:916–931. doi: 10.1038/sj.mp.4001560. [DOI] [PubMed] [Google Scholar]
- 28.Centers for Disease Control and Prevention. Plan and Operation of the Third National Health and Nutrition Examination Survey, 1988–94. Bethesda, Md: Centers for Disease Control and Prevention; 1994. http://www.cdc.gov/nchs/nhanes.htm. [Google Scholar]
- 29.Ai M, Otokozawa S, Schaefer EJ, et al. Glycated albumin and direct low density lipoprotein cholesterol levels in type 2 diabetes mellitus. Clin Chim Acta. 2009;406:71–74. doi: 10.1016/j.cca.2009.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeggini E, Weedon MN, Lindgren CM, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saxena R, Voight BF, Lyssenko V, et al. Diabetes Genetics Initiative of Broad Institute of Harvard and MIT Lund University, and Novartis Institutes of BioMedical Research. Genome-wide association analysis identifies loci for type-2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 32.PROCARDIS Consortium. A trio family study showing association of the lymphotoxin-alpha N26 (804A) allele with coronary artery disease. Eur J Hum Genet. 2004;12:770–774. doi: 10.1038/sj.ejhg.5201244. [DOI] [PubMed] [Google Scholar]
- 33.Lange K. Mathematical and Statistical Methods for Genetic Analysis. Springer-Verlag; New York: 1997. [Google Scholar]
- 34.METAL software. http://genome.sph.umich.edu/wiki/METAL_Program.
- 35.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poulsen P, Kyvik KO, Vaag A, Beck-Nielsen H. Heritability of type II (non-insulin-dependent) diabetes mellitus and abnormal glucose tolerance--a population-based twin study. Diabetologia. 1999;42:139–145. doi: 10.1007/s001250051131. [DOI] [PubMed] [Google Scholar]
- 37.NCBI. Single Nucleotide Polymorphism. http://www.ncbi.nlm.nih.gov/sites/entrez?db=snp&cmd=search&term=rs806365.
- 38.Baye TM, Zhang Y, Smith E, et al. Genetic variation in cannabinoid receptor 1 (CNR1) is associated with derangements in lipid homeostasis, independent of body mass index. Pharmacogenomics. 2008;9:1647–1656. doi: 10.2217/14622416.9.11.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goldstein DB. Common genetic variation and human traits. N Eng J Med. 2009;17:1696–1698. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- 40.Florez JC. Newly identified loci highlight beta cell dysfunction as a key cause of type 2 diabetes: where are the insulin resistance genes? Diabetologia. 2008;51:1100–1110. doi: 10.1007/s00125-008-1025-9. [DOI] [PubMed] [Google Scholar]
- 41.Ravinet-Trillou C, Delgorge C, Menet C, Arnone M, Soubrié P. CB1 cannabinoid receptor knockout in mice leads to leanness, resistance to diet-induced obesity and enhanced leptin sensitivity. Int J Obes Relat Metab Disord. 2004;28:640–648. doi: 10.1038/sj.ijo.0802583. [DOI] [PubMed] [Google Scholar]
- 42.Bordicchia M, Battistoni I, Mancinelli L, et al. Cannabinoid CB1 receptor expression in relation to visceral adipose depots, endocannabinoid levels, microvascular damage, and the presence of the CNR1 A3813G variant in humans. Metabolism. 2009;59:734–741. doi: 10.1016/j.metabol.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 43.Müller TD, Reichwald K, Wermter AK, et al. No evidence for an involvement of variants in the cannabinoid receptor gene (CNR1) in obesity in German children and adolescents. Mol Genet Metab. 2007;90:429–434. doi: 10.1016/j.ymgme.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 44.Löfgren P, Sjölin E, Wåhlen K, Hoffstedt J. Human adipose tissue cannabinoid receptor 1 gene expression is not related to fat cell function or adiponectin level. J Clin Endocrinol Metab. 2007;92:1555–1559. doi: 10.1210/jc.2006-2240. [DOI] [PubMed] [Google Scholar]
- 45.Benzinou M, Chèvre JC, Ward KJ, et al. Endocannabinoid receptor 1 gene variations increase risk for obesity and modulate body mass index in European populations. Hum Mol Genet. 2008;17:1916–1921. doi: 10.1093/hmg/ddn089. [DOI] [PubMed] [Google Scholar]
- 46.Peeters A, Beckers S, Mertens I, Van Hul W, Van Gaal L. The G1422A variant of the cannabinoid receptor gene (CNR1) is associated with abdominal adiposity in obese men. Endocrine. 2007;31:138–141. doi: 10.1007/s12020-007-0022-y. [DOI] [PubMed] [Google Scholar]
- 47.Russo P, Strazzullo P, Cappuccio FP, et al. Genetic variations at the endocannabinoid type 1 receptor gene (CNR1) are associated with obesity phenotypes in men. J Clin Endocrinol Metab. 2007;92:2382–2386. doi: 10.1210/jc.2006-2523. [DOI] [PubMed] [Google Scholar]
- 48.Jaeger JP, Mattevi VS, Callegari-Jacques SM, Hutz MH. Cannabinoid type-1 receptor gene polymorphisms are associated with central obesity in a Southern Brazilian population. Dis Markers. 2008;25:67–74. doi: 10.1155/2008/841490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dupuis J, Langenberg C, Prokopenko I, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–116. doi: 10.1038/ng.520. Erratum in: Nat Genet 2010, 42, 464. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.