Abstract
Vectors based on adeno-associated virus (AAV) are effective in gene delivery in vivo. Tissue-specific gene expression is often needed to minimize ectopic expression in unintended cells and undesirable consequences. Here we investigated if incorporation of target sequences of tissue-specific microRNA (miRNA) into AAV vectors could inhibit ectopic expression in tissues such as the liver and hematopoietic cells. First we inserted liver-specific miR-122 target sequences (miR-122T) into the 3′ untranslated region (UTR) of a number of AAV vectors. After intravenous delivery in mice, we found that 5 copies of the 20mer miR-122T reduced liver expression of luciferase by 50-fold and β-galactosidase (LacZ) by 70-fold. Five copies of miR-122T also reduced mRNA levels of a secretable protein (myostatin propeptide) from the AAV vector plasmid by 23–fold in the liver. However, gene expression in other tissues including the heart was not inhibited. Similarly, we inserted 4 copies of miR-142-3pT or miR-142-5pT, both hematopoietic lineage-specific, into the 3′ UTR of the AAV-luciferase vector. We wished to see if they could prolong transgene expression by inhibiting expression in antigen-presenting cells. However, in vivo luciferase gene expression in major tissues declined with time regardless of the miR-142 target sequences used. Quantitative analysis of the vector DNA in various tissues revealed that the decline of transgene expression in vivo was mainly due to promoter shut-off other than loss of AAV-transduced cells by immune destruction. Moreover, transgene expression was not detected in circulating mononuclear cells after delivering AAV9 vector with or without miR142T. These results demonstrate that live-specific miR-122 target sequence in AAV vectors was highly efficient in reducing liver expression, whereas hematopoietic miR-142 target sequences were ineffective in preventing decline of AAV vector gene expression in non-hematopoietic tissues resulted from promoter shut-off.
Introduction
Over the past decade, adeno-associated viruses (AAVs) have become vectors of choice for in vivo gene therapy of many genetic and chronic diseases such as hemophilia B1, Leber's congenital amaurosis2-4, Parkinson's disease5, hereditary muscular dystrophies6-8, and alpha-1 antitrypsin deficiency9, etc. The lack of pathogenicity of the wild-type virus, broad tissue tropism, and persistence of vector DNA have bolstered AAV's potential as a potent delivery vehicle for in vivo gene therapy applications10. The first in vivo demonstration of long-term and persistent transgene expression by AAV was achieved without apparent immune reaction11. Subsequently, more evidence showed that AAV could mediate successful and sustained transgene expression in small animal models6,12-15, large animal models16,17, and even human patients2,3,18 without noticeable side effects. However, host immune responses to the capsids or to the transgene products have been noticed in large animal studies and in human clinical trials. Several factors may influence the development of an immune response to AAV19, including vector serotypes, route of administration, vector doses, host species, the presence of preexisting immunity, and the specifications of the delivery cassette. In mice20 and non-human primates21, the AAV2 serotype elicits a stronger immune response than the AAV7 or AAV8 serotypes. With respect to route of administration, systemic delivery of AAV vectors often results in immune tolerance and is less likely to induce an immune reaction to the transgene than intramuscular delivery22,23. There is indication that the potential immune responses against AAV vectors are dose dependent24,25. Host species is relevant because large animals and humans have more complex and diverse immune systems than small rodents, making them more prone to developing an immune reaction to AAV26. Preexisting neutralizing antibodies will reduce and even block AAV-mediated gene transfer27.
The gene expression cassette, particularly the promoter and the nature of the transgene, will also affect immunogenicity. Tissue-specific promoters can reduce transgene expression in antigen-presenting cells and decrease the chances of developing an immune reaction to the transgene28. Additionally, selected transgenes can elicit a potent humoral and/or cellular immune response, as in the case of ovalbumin transgene29 and influenza virus hemagglutinin which leads to complete clearance of cells expressing the gene, while other transgenes such as beta-galactosidase elicit less immune response in normal mice30.
Numerous efforts have been made to achieve strong tissue-specific expression using tissue-specific promoters and enhancers 31. However, sometimes an enhancer from a different context could also lead to aberrant expression in undesirable tissues. For example, potent muscle-specific promoters with a strong myosin heavy chain (MHC) enhancer have the potential to generate leaky expression in the liver, a major depot for AAV vectors upon intravascular administration32,33. Thus, aberrant liver expression of transgenes may result in side-effects or even immune responses34, although hepatic gene transfer has been shown to induce immune tolerance to several protein antigens22.
MiRNAs , first discovered in Caenorhabditis elegans35, are considered an essential component of the gene expression regulatory networks. Evolutionarily conserved, miRNAs are small noncoding regulatory RNAs involved in RNA-mediated gene silencing at the post-transcriptional level36. At present, more than 500 human miRNAs have been identified37. Mature miRNA binds specific mRNAs through target sequences located in the 3′ UTR that are either partially or fully complementary to the miRNA. Previously, it was believed that the expression of the corresponding gene was silenced or reduced either by mRNA degradation (if fully complementary) or inhibition of translation (if partially complementary)38. More recently, it has been reported that miRNAs act predominately to reduce mRNA levels of nearly all targets39.Many miRNAs are expressed in a tissue-specific manner and play an important role in maintaining tissue-specific functions and differentiation40.
Our study is intended to improve controlled gene expression by exploiting post-transcriptional regulation via endogenous tissue-specific microRNA systems, thus, to inhibit transgene expression in undesired tissues by incorporating tissue-specific miRNA target sequences into the 3′untranslated region (UTR) of an AAV vector cassette. First, we investigated liver-specific miRNA target sequence miR-122T, since the liver is a major target organ by AAV. We found that miR-122T was able to effectively inhibited AAV gene expression in the liver. Subsequently, we also examined hematopoietic miRNA target sequences miR-142T, which could mediate inhibition of gene expression in hematopoietic lineage cells including antigen-presenting cells41. However, we found that the miR-142T sequence could not prevent decline of gene expression in non-hematopoietic tissues, where promoter shut-off, but not T-cell destruction of transgene-expression cells, had occurred.
Results
Liver-specific miRNA-122 target sequence in AAV vectors diminished transgene expression in liver
MiR-122, a 22-nucleotide miRNA, is derived from a liver-specific noncoding polyadenylated RNA transcribed from the gene hcr42. It is the most frequent miRNA isolated in the adult liver, representing approximately 70% of all cloned miRNAs there43. In order to test the efficacy of miR-122T in reducing unwanted AAV transgene expression in the liver, we synthesized either 3 or 5 copies of the target sequence (miRBase database accession number MI0008015) for incorporation into the 3′ UTR of the firefly luciferase gene in AAV-CB-Luc vector (Fig. 1).
Fig. 1.

AAV plasmid constructs containing or lacking the miR-122 target sequence. ITR: inverted terminal repeat; CB: chicken beta-actin promoter and cytomegalovirus enhancer; Luc: luciferase; En-MHC: myosin heavy chain enhancer; TNT: troponin promoter
We initially performed a hydrodynamic injection of naked plasmid DNA into mice (3 in each group) to determine the inhibitory effect of miR-122T. Hydrodynamic gene delivery via tail vein injection is an established and highly efficient procedure to deliver nucleic acids to the liver in small animals44. The mice were sacrificed one day after plasmid injection, and their liver tissues were carefully dissected for luciferase activity analysis and DNA copy number determination. The luciferase activities were normalized by both plasmid DNA copy number and protein concentration. We found that the plasmid containing 3 copies of the target sequence (3× miR-122T) caused a 10- to 50-fold reduction in luciferase expression, while the plasmid containing 5 copies (5× miR-122T) caused a 20- to 100-fold reduction, when compared to the AAV-CB-Luc control plasmid without the miR-122T sequence (Fig. 2a). To further confirm that the inhibitory effect was liver-specific, we transfected 293 cells, which did not express miR-122, using AAV-luciferase plasmids with or without miR-122T. No difference in luciferase activities was observed in 293 cells transfected with the above plasmids (Fig. 2b), suggesting that the inhibitory effect of miR-122T was liver-specific.
Fig. 2.

Luciferase activities after exposure to naked plasmid DNA containing the luciferase reporter gene and miR-122 target sequence in vivo and in vitro. a. Hydrodynamic injection of plasmids containing the miR-122 target sequence reduces luciferase expression in the liver of ICR mice (n=3). Incorporation of 3 copies reduces expression 10- to 50-fold (p<0.05), while incorporation of 5 copies reduces expression 20- to 100-fold (p<0.05). Luciferase activities were normalized by total protein concentration and DNA copy number. b. In vitro transfection of 293 cells (which do not contain microRNA-122) with plasmids containing or lacking the miR-122 target sequence results in no difference in luciferase expression, indicating that the effects of miR-122T are liver-specific (p>0.05). Luciferase activities were normalized by total protein concentration. The CB-GFP plasmid was used as a negative control. The transfection was done in triplicate wells. Error bars indicated standard deviation.
We subsequently packaged pAAV-CB-Luc and pAAV-CB-Luc-5xmiR-122T plasmids into AAV9, which was chosen for its superior ability to cross the blood vessel barrier and transduce cells in a broad range of tissues45,46. The AAV9-CB-Luc-5xmiR-122T vector and the control AAV9-CB-Luc without miR-122T were delivered into 6-week-old ICR mice via tail vein injection at a dose of 1×1012 vector genomes (vg) per mouse. The mice were sacrificed and tissues were harvested 3 weeks after vector delivery. Luciferase activities were analyzed for skeletal muscles, cardiac muscle, and the liver. As shown in Fig. 3, we did not observe difference in luciferase activities in the tibialis anterior (TA) muscles between the two groups. However, we did observe approximately 50-fold lower luciferase activities in the liver from mice injected with AAV9-CB-Luc-miR-122T (p<0.01, n=5) as compared with control vector AAV9-CB-Luc injected mice. Interestingly, we also observed significantly higher luciferase activities in cardiac muscle from mice injected with AAV9-CB-Luc-miR-122T (p<0.05, n=5) as compared with AAV9-CB-Luc injected mice (Fig. 3).
Fig. 3.
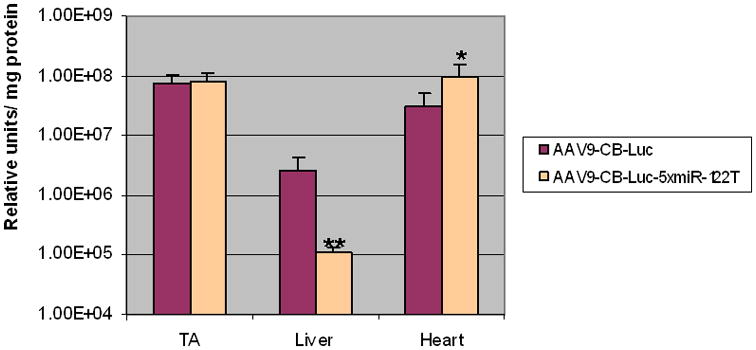
Reduction in liver expression of luciferase resulting from incorporation of the liver-specific miRNA target sequence miR-122T into AAV9 vectors containing the luciferase reporter gene. AAV9-CB-Luc vectors with or without the miR-122 target sequence were injected into the tail veins of 6-wk-old female ICR mice (1×1012 vg/mouse, n=5). After 3 weeks, there was no difference between luciferase expression of TA muscles, 50-fold lower expression in miR-122T liver (**, p<0.01), and significantly higher expression in miR-122T heart (*, p<0.05). Error bars indicated standard deviation.
To further evaluate the effect of miR-122T on reducing liver transgene expression, we examined a different reporter gene and promoter, thus, the LacZ gene and the en-MHC-TNT promoter, which contains the troponin T promoter (TNT) and the myosin heavy chain enhancer (en-MHC). The addition of en-MHC made the TNT promoter stronger in the heart, but it also made it very leaky in the liver. To reduce liver expression, we cloned 5 copies of miR-122T into the 3′ UTR of the LacZ gene driven by the en-MHC-TNT promoter (Fig. 1). The pAAV-en-MHC-TNT-LacZ and pAAV-en-MHC-TNT-LacZ-5xmiR-122T plasmids were separately packaged into AAV9 vectors. The vectors were then delivered into 6-week-old ICR mice via tail vein injection (3 × 1012 vg per mouse). The mice were sacrificed and their skeletal muscles, cardiac muscle, and liver were harvested 3 weeks after vector delivery. The tissues were cryosectioned and subjected to X-gal staining. We did not detect LacZ expression in the skeletal muscles from either group (data not shown), indicating that the en-MHC-TNT promoter, similar to its parental TNT promoter, was not active in the skeletal muscle. Surprisingly, the livers from mice injected with AAV9-en-MHC-TNT-LacZ showed very strong LacZ expression (Fig. 4a), which was not seen with the parental TNT promoter (Suppl. Fig. 1). On the other hand, the liver tissues from mice receiving AAV9-en-MHC-TNT-LacZ-5xmiR-122T showed only a few LacZ positive cells (Fig. 4a). Consistent with the luciferase reporter gene vector, higher LacZ expression in the heart was also observed from the cassette containing 5xmiR-122T (Compare Fig. 3 and Fig. 4a). Quantitative analysis of LacZ expression by the Chemiluminescent β-Galactosidase assay kit showed an approximately 70-fold decrease in LacZ expression in the liver in mice injected the vector containing the miR-122T, compared with those injected with vector without the miR-122T (Fig. 4b). Higher cardiac LacZ expression in the mice injected with AAV9-en-MHC-TNT-LacZ-5xmiR-122T was also observed (p=0.06, n=4). These data indicated that 5 copies of miR-122T into the 3′UTR of the AAV vector could effectively diminish liver expression.
Fig. 4.

Reduction in liver expression of LacZ mediated by miR-122T a. LacZ staining of liver and heart. 3 ×1012 vg of corresponding AAV9 vectors were delivered into 6-week-old ICR mice, which were sacrificed 3 weeks later. Liver tissues showed much weaker LacZ expression when exposed to the miR-122 target sequence. Heart tissues showed stronger LacZ expression when exposed to the miR-122 target sequence. scale bar: 100μm b. Quantitative analysis of LacZ expression of liver and heart. 3 weeks after injection. Liver tissues exposed to the miR-122 target sequence showed a 70-fold decrease in LacZ expression (**, p<0.001; n = 4). LacZ expression in heart tissues exposed to the miR-122 target sequence increased marginally over control (*, p=0.06). Error bars indicated standard deviation.
MicroRNA with perfect complementarity to its target mRNA usually causes direct mRNA cleavage39. The miR-122T utilized in this study was a fully complementary sequence to the mature miR-122. We expected that the reduction of liver transgene expression by miR-122T occur through mRNA degradation. To test this, we quantified the mRNA of the transgene by real-time PCR. Five copies of miR-122T was cloned into the 3′ UTR of the pAAV-CAG-dog-MPRO-Ig plasmid, which contains a large (900bp) intron (Fig. 5a). We chose to use this construct because we could differentiate the size of the real-time PCR product of cDNA of spliced mRNA from the backbone vector DNA, which was 900 bp longer. The forward and reverse primers were designed to span the intron region (Fig. 5a) with an expected RT-PCR product of 106 bp, while the vector DNA would be too large to be amplified by the qPCR program. The pAAV-CAG-dog-MPRO-Ig-5xmiR-122T plasmid was delivered into adult mice liver via hydrodynamic injection (three mice per group), while control mice received the AAV-CAG-dog-MPRO-Ig plasmid. The mice were sacrificed 24 hours after plasmid delivery, and their liver samples were harvested for RNA preparation. As shown in Fig. 5b, the transgene mRNA quantity was reduced by 96% in the presence of the 5xmiR-122T, suggesting that the inhibition of liver expression by miR-122T occurred through mRNA degradation.
Fig. 5.

Quantitative PCR analysis indicates that the reduction in liver expression mediated by the miR-122 target sequence occurs through mRNA degradation. a. Construction and schematic illustration of real-time PCR design. b. Relative quantification demonstrated a 96% reduction in mRNA expression mediated by the miR-122 target sequence (**, p<0.001). . The pAAV-CAG-dog-MPRO-Ig-5xmiR-122T plasmid was delivered to adult ICR mice via hydrodynamic injection, and the pAAV-CAG-dog-MPRO-Ig plasmid was delivered to the control mice (n=3 for each group). Error bars indicated standard deviation.
Examination of hematopoietic-specific miRNA target sequences for prolonged transgene expression in AAV vectors
One of the major hurdle for stable gene transfer in large animal models and human patients is the immune response against AAV vectors26 and transgenes47,48. Additionally, we have observed that some transgenes, such as luciferase, cannot retain long-term expression even in normal mice following systemic delivery of AAV vectors (unpublished data). We suspected that a cellular immune response against the luciferase might be developed during the course. In this study, we inserted hematopoietic-specific miRNA target sequences into the AAV vector in an attempt to down-regulate the transgene expression in immune cells, consequently prolong the transgene expression in desired tissues. The strategy of suppressing transgene expression in antigen-presenting cells has been successfully utilized in lentiviral vectors in order to avoid transgene-specific immunity49,50. Four copies of miR-142-3pT or 4 copies of miR-142-5pT were cloned into the 3′ UTR of the pAAV-CB-Luc plasmid, which contains the luciferase gene driven by the ubiquitous CB promoter (chicken beta-actin promoter and CMV enhancer) (Fig. 6a). AAV9 vectors containing the above reporters were produced. Since miRNAs are highly conserved, the miR-142-3p and miR-142-5p are identical among dog, mouse, and human, respectively37. Therefore, if this strategy works in one species, it is likely to work in others as well.
Fig. 6.

Incorporation of miR-142 target sequences into AAV vectors had no apparent effect on prolonging transgene expression in desired tissues. a. AAV-CB-Luc plasmid constructs containing or lacking hematopoietic miR-142 target sequences in the 3′ UTR. ITR: inverted terminal repeat; CB: chicken beta-actin promoter and cytomegalovirus enhancer; Luc: luciferase. b. Luciferase activity in both long-term and short term studies. There was a significant decline of luciferase expression in long-term studied tissues comparing with the short term counterparts (p<0.05). For the long-term study, AAV9-CB-Luc vectors with or without the miR-142 target sequences were injected into the tail veins of outbred ICR mice (2×1012 vg/mouse, n=6), and tissues were harvested three months later. For the short-term study, AAV9-CB-Luc vectors with or without the miR-142 target sequences were injected into the tail veins of C57BL/6 mice (1×1012 vg/mouse, n=3), and tissues were harvested two weeks later. c. Tissue vector copy numbers in both long-term and short-term studies. There was no significant differences in tissue vector copy numbers between short term and long-term studies (p>0.05), ranging from 10-80 copies per liver cell and 3-11 copies per nucleus for skeletal and cardiac muscles.
Three groups of AAV vectors (AAV9-CB-Luc, AAV9-CB-Luc-4xmiR-142-5pT, and AAV9-CB-Luc-4xmiR-142-3pT) were administered via tail vein injection to the outbred ICR mice at a dose of 2×1012 vg per mouse and 6 mice per group. All the mice were sacrificed 3 months after vector injection and their tissues (tibialis anterior muscle, upper limb muscle, cardiac muscle, and liver) were harvested and assayed for luciferase activities. As shown in Fig. 6 b, we did not observe a statistically significant difference in luciferase expression among all groups regardless the presence of the miRNA target sequences of hematopoietic lineage (p> 0.05, n=6). We also observed significant intra-group variability in luciferase expression with a marked decline of transgene expression in several mice (Fig. 6a) in all groups. To reveal short-term transgene expression among different groups, we performed similar experiments and sacrificed the mice two weeks after vector delivery. We noticed 10- to 100-fold greater luciferase expression in tissues examined in the short-term study as compared with the relatively long-term study (Fig. 6 b, p< 0.05), indicating a huge decline of long-term luciferase gene expression.
The decline of luciferase expression 3 months after vector delivery despite the presence of hematopoietic miRNA target sequences could be due to either immune clearance or promoter shut-off. To determine the cause of this phenomenon, we performed real-time PCR to determine vector copy numbers and histology with immunofluorescent staining against the T lymphocyte marker CD4. If the decline in the transgene was due to an immune destruction against the AAV vector transgene-positive cells, we would observe an equivalent loss of vector DNA copy numbers, along with mononuclear cell infiltration, and histopathology. Surprisingly, real-time PCR results revealed consistent AAV vector copy numbers in all mice studied for long-term groups, ranging from 10 to 80 copies per liver cell and 3 to 11 copies per nucleus for skeletal and cardiac muscles (Fig. 6 c). More importantly, the short-term study showed similar vector copy numbers to the long-term study (Fig. 6 c, p > 0.05, n=3 to 6.). The data indicated that the decline of transgene expression in the long-term study was not due to immune rejection of the vector-positive cells. In addition, H&E staining displayed normal tissue histology and no apparent mononuclear cell infiltration in any tissue examined. Immunofluorescent staining against mouse CD4 detected no significant CD4+ cells above the background control levels in all the tissues examined (Suppl. Fig. 2).
The intention of incorporating hematopoietic lineage-specific miRNA targeting sequence into AAV expression cassette in this study was to down regulate the transgene expression in antigene-presenting cells. To answer whether the transgene expressed in hematopoietic lineage cells, we analyzed the luciferase expression in mononuclear cells of blood circulation in different time point after delivering AAV9-CB-Luc vector with or without hematopoietic lineage-specific miRNA targeting sequence (three mice per group). AAV vectors with or without the hematopoietic miRNA targets (AAV9-CB-Luc, AAV9-CB-Luc-4xmiR-142-5pT, and AAV9-CB-Luc-4xmiR-142-3pT) were administered via tail vein injection in ICR mice at a dose of 2× 1012 vg per mouse. At 36 hours and 96 hours post vector delivery, blood was taken from each mouse via tail bleeding. Previous studies indicated that it would take as short as three days for the transgene to reach approximately one third of its peak level in vivo32. Mononuclear cells were isolated and subjected to luciferase activity analysis. The mononuclear cells isolated from different groups at various time point all displayed background level of luciferase expression (data not shown), suggesting hematopoietic lineage cells were poorly transduced by the AAV9 vectors.
From above data, we concluded that 1) there was no significant T-cell immune responses in tissues of vector-treated mice; 2) the hematopoietic miRNA target sequences failed to prolong luciferase expression, which declined 3 months post vector delivery; 3) promoter shut off, not immune rejection, was responsible for the decline of transgene expression in some individual mice. Incorporation of hematopoietic-specific miRNA-T in the 3′UTR of AAV9 vector was unable to help in this case.
Discussion
To develop a safer AAV vector, we intended to utilize endogenous miRNA machinery to provide improved control of transgene expression at the post-transcriptional level. Specifically, we incorporated multiple copies of the liver-specific miR-122 target sequences into the 3′ UTR of AAV expression cassette, for the purpose of decreasing unwanted liver expression. This strategy turned out to be highly effective. The reduction of luciferase expression mediated by 5xmiR-122T was 50-fold, while reduction of LacZ expression was over 70-fold. We did not observe any effects by miR-122T on transgene expression in the skeletal muscles. However, we did observe a slight but significant increase in cardiac muscle expression when AAV vectors containing 5xmiR-122T were used. The potential mechanism by which cardiac transduction was enhanced could be the presence of 5 copies of miR-122T in the 3′UTR of our vectors, which might increase mRNA stability in the cardiomyocytes or serve as a cardiac enhancer, since we did not notice similar phenomena in skeletal muscles. We further utilized quantitative PCR to demonstrate that mRNA degradation is the likely mechanism by which liver transgene expression was reduced.
One potential concern regarding this strategy is that a highly expressed transgene containing multiple binding sites for miR-122 could compete with endogenous targets for their cognate miRNA. However, this possibility is very unlikely for two reasons. First, the miRNA target sites are perfectly complementary, the miRISC will have a high catalytic rate, which reduces the likelihood of miR-122 from being titrated out41. Second, miR-122 is the most abundant miRNA in the liver, and possibly in the entire miRNA repertoire, making saturation improbable. Previous experiments have demonstrated that increasing concentrations of a perfectly complementary miRNA target site do not appreciably perturb miRNA regulation of an imperfect target41.
In this study, we also used the hematopoietic lineage miR-142-3p or miR-142-5p target sequences in AAV expression cassettes in an attempt to decrease transgene expression in antigen-presenting cells and prolong transgene expression in normal mice. These sequences were effective in lentiviral vectors for prolongation of transgene expression41,49,50. However, the presence of the hematopoietic lineage miRNA target sequence in the AAV-CB-Luc vectors was not effective in preventing marked long-term decline of luciferase expression in the tissues after systemic delivery. Further investigation on vector copy numbers in transduced tissues, histology, and lack of T-cell infiltration indicated that the decline in transgene expression was due to promoter shut-off. CMV promoter in AAV vector is prone to shut off in the liver in 2 to 3 weeks post intravenous gene transfer. In this study, we utilized the CB promoter (CMV enhancer and chicken beta-actin promoter) that was reported to render long-term (> 6 months) GFP gene expression in vivo in many tissues including the liver51,52. However, the same promoter driving the luciferase gene expression caused short-term expression in major tissues examined in this study using the mice. We have observed similar results in hamsters (data not shown). It is likely that the luciferase gene and the high vector dose (1012 vg per mouse, vs. 2 ×1011 v.g. per mouse for the GFP vector) both contributed to the promoter shut off, which was likely caused by the innate immunity in the liver, other than the T-cell immunity. Utilizing AAV vectors driven by tissue-specific promoters without viral regulatory elements and applying lower vector doses may prevent promoter shut-off in future experiments. Additionally, the barely detectable background levels of luciferase expression in mononuclear cells of AAV9-CB-Luc treated mice in our experiments indicates AAV9 , similar to AAV8 53, may not infect dendritic cells. It was also a possibility that the decline of the transgene expression was caused by activation of RNAi against the transgene, similar to RNAi against viral infection.
We cannot draw conclusions about the efficacy of hematopoietic miRNA target sequences in preventing an immune response against AAV-delivered transgenes because immune clearance was not the reason for the elimination of transgene expression in our experiments. A different species with a more complex immune system that is more prone to developing an immune response against AAV transgene expression, such as the dogs 47 and monkeys 48, should be utilized to answer this question. Nevertheless, our study is an important attempt toward exploiting endogenous miRNA regulatory machinery to further control transgene expression mediated by the AAV vector at the post-transcriptional levels.
Material and Methods
Plasmid construction and AAV virus production
To construct luciferase plasmids containing miRNA target sequences, multiple copies of different target sequences were synthesized and cloned into NheI and SacI sites positioned in the 3′ UTR of the AAV-CB-Luc plasmid31,54 (Fig. 1 and Fig. 6). Their sequences are listed as follows: Cfa-4xmiR-122T: CCA GAA GCT TGC TAGC caa aca cca ttg tca cac tcca TCAC caa aca cca ttg tca cac tcca GATATC caa aca cca ttg tca cac tcca TCAC caa aca cca ttg tca cac tcca AAGCTT GTAG. Cfa-4× miR-142-5-pT-S: CCA GAA GCT TGC TAGC agt agt gct ttc tac ttt atggg TCAC agt ag gct ttc tac ttt atggg GATATC agt agt gct ttc tac ttt atggg TCAC agt agt gct ttc tac ttt atggg AAG CTT GTAG. Cfa-4× miR-142-4×3pT-S: CCA GAA GCT TGC TAGC tcc ata aag tag gaa aca ctaca TCAC tcc ata aag tag gaa aca ctaca GATATC tcc ata aag tag gaa aca ctaca TCAC tcc ata aag tag gaa aca ctaca AAG CTT GTAG. The lower case letters indicate the target sequences, and the upper case letters are either linker sequences or restriction enzyme sites. To construct the pAAV-en-MHC-TNT-LacZ plasmid, the BamHI-SalI fragment of the troponin T (TNT) promoter (Gene bank M57905.1) was cloned into BamHI and SalI sites of the pAAV-CMV-LacZ plasmid to replace the CMV promoter, and the construction was named the pAAV-TNT-LacZ plasmid (Fig. 1). Then the EcoRI-HindIII fragment of the mouse α-myosin heavy chain complex enhancer (en-MHC) (Gene bank NW_001030560.1, from 1231272 to 1231460) was added in front of the TNT promoter of the pAAV-TNT-LacZ plasmid by cohesive ligation to form the pAAV-en-MHC-TNT-LacZ plasmid. Finally, the SalI-SphI fragment of the pAAV-CB-Luc-5xmiR-122T plasmid containing five copies of miR-122T and BGH (bovine growth hormone) polyA signal were cloned into SalI and SphI sites of the pAAV-en-MHC-TNT-LacZ plasmid to form the pAAV-en-MHC-TNT-LacZ-5xmiR-122T plasmid (Fig. 1).
The original AAV9 packaging plasmid was a generous gift from Dr. James Wilson and Dr. Guangping Gao 46. All of the AAV vectors were produced by the triple transfection method 55 and the viruses were purified by the second generation CsCl protocol that incorporates differential precipitation of AAV particles by polyethylene glycol (PEG)56. The virus titer was determined by both dot-blot and real-time PCR methods57,58.
Hydrodynamic naked plasmid injection and AAV vector delivery in vivo
All animal protocols were approved by the University of North Carolina at Chapel Hill Institutional Animal Care and Use Committee.
The detailed methods and mechanisms of hydrodynamic injection of plasmid DNA for delivery to liver cells have been described elsewhere44. Briefly, 50-100 ug of double CsCl purified DNA was dissolved in 8-10% body weight volume of PBS (in ml) for a given mouse. For example, if the mouse weight was 25 g, we would dissolve 50 ug of DNA in 2 to 2.5 ml of PBS. The total volume of PBS containing the DNA was then injected into the tail veins of adult ICR mice (6 to 8 weeks old) over approximately 5 seconds, using 3 ml syringes and 27G × ½ in PrecisionGlide™ Needles (BD, Cat # 305109). The mice were sacrificed 24 hours post-injection when the peak of gene expression was achieved, and the liver tissues were carefully dissected and quickly frozen in isopentane-liquid nitrogen slurry for future analysis.
We utilized outbred ICR mice for most of the experiments described in this manuscript. 6- to 8-week-old male ICR mice were purchased from Taconic (Hudson, New York). For intravenous injection, mice were first warmed under an infrared heat lamp (100 to 160 Watts) for 5 to 10 minutes to dilate the tail vein. The desired dilution of AAV was prepared in PBS, and the appropriate dose was administered to the tail vein of mice held in a restrainer without anesthesia using a 27G × ½ in PrecisionGlide™ Needle.
Real-time PCR analysis
Two real-time PCR assays were utilized in this study. To determine the mechanism underlying the reduction in liver expression mediated by miR-122T, relative quantification real-time PCR was used. Total RNA was extracted from liver tissues with TRIzol® Reagent from Invitrogen (Cat # 15596-026), and the first strand cDNA was synthesized utilizing the High-Capacity cDNA Reverse Transcription Kit with RNase Inhibitor from Applied Biosystems (Part # 4374966). For primers and probe designed for endogenous mRNA control, we utilized Gusb (glucuronidase, beta) which was purchased from Applied Biosystems (Cat # Mm00446953_m1). The primers and probe for pAAV-CAG-dog-MPRO-Ig molecules are listed as follows: RT-CAG(+): tct gac tga ccg cgt tactc; RT-CAG(−): ccg cgg tgg agc tca ag; RT-CAG probe: FAM-cat ttt ggc aaa gaa t-MGB.
To determine vector copy numbers in liver tissue, absolute quantification real-time PCR was used. Total DNA was extracted from liver tissues with the DNeasy Blood & Tissue Kit from QIAGEN (Cat # 69506, Valencia). The mouse glucagon gene was used as the endogenous control. The sequences for mouse glucagon primers and probes were as follows: Glucagon-real-F (mouse): AAG GGA CCT TTA CCA GTG ATG TG; Glucagon-real-R (mouse): ACT TAC TCT CGC CTT CCT CGG; Taqman mouse glucagon probe: FAM-cag caa agg aat tca-MGB. For the target AAV9 vectors, primers and probes were designed on the common BGH polyA sequence. Their sequences were as follows: BGH-F: AGC CTC GAC TGT GCC TTCTA; BGH-R: ATG CGA TGC AAT TTC CTCAT; BGH probe: FAM-TGC CAG CCA TCT GTTG. The copy number of delivered vector in a specific tissue per diploid cell was calculated as: (vector copy number/endogenous control) × 2.
Immunofluorescent staining against CD4
The snap-frozen tissues were cryo-thin-sectioned at 8 μm thickness. Then the slides were fixed with acetone for 10 minutes followed by three washes with PBS. Blocking was performed with 5% horse serum in PBS for 30 minutes. The rat anti-mouse CD4 monoclonal antibody (Pharmingen International, Cat # 01061D) was incubated at a 1:500 dilution in 1× PBS for one hour at room temperature. After three washes with 1× PBS, the secondary antibody Cy3-conjugated goat anti-rat IgG (Jackson ImmunoResearch, West Grove, PA, Cat # 706-165-1480) was incubated at a 1:500 ratio in 1× PBS. After another three washes with 1× PBS, the slides were incubated with 1× DAPI for 5 minutes to display the nucleus. Finally, the slides were mounted with the aqueous mounting medium Gel/Mount™ (Biomeda Corp Cat # M01, Foster City, CA).
X-gal staining for expression of LacZ and LacZ activity analysis
For X-gal staining, the cryo-thin-section slides were air-dried and fixed in X-gal fixative buffer containing 2% formaldehyde and 0.2% gluteraldehyde in PBS. After washing with PBS, the slides were subjected to X-gal staining buffer at 37°C overnight. The X-gal staining buffer consisted of 0.1 M phosphate buffer (pH 7.3) supplemented with 2 mM MgCl2, 5 mM potassium ferrocyanide (Sigma Cat # P-9287) and 5 mM potassium ferricyanide (Sigma Cat # P-8131). Before use, X-gal (5-bromo-4-chloro-3-indolyl-β-D-galactoside) was added at a final concentration of 1mg/ml 59.
To quantitate LacZ expression, we utilized the Galacto-Light Plus™ kit (Applied Biosystems, MA 01730. Cat # T1007). The manufacturer's protocol was strictly followed. The relative units of LacZ activity were normalized by total protein concentration, which was assessed with the Bio-Rad Protein Assay (Bio-Rad Laboratories, Inc, CA 94547 Cat # 500-0006).
Luciferase activity assay
Tissues (25-100 mg) were lysed and homogenized in luciferase lysis buffer (0.05% Triton X-100, 0.1 M Tris-HCl, pH 7.8, 2 mM EDTA). The homogenized lysates were extensively vortexed and spun down at 4°C for 2 minutes. The supernatant was utilized for luciferase activity analysis. For blood samples, the mononuclear cells were isolated using Ficoll-Paque density gradients. Blood was diluted 1:1 with phosphate buffered saline, and layered onto Ficoll-Paque (Sigma Diagnostics, Inc. Cat # Histopaque-1083) with the ratio of blood + PBS: Ficoll of 4:3. The blood was centrifuged at 1800 rpm for 35 min at room temperature. The lymphocyte layer (buffy coat) was removed and washed twice in PBS at 1200 rpm for 10 min each, following the same wash with media RPMI 1640. The cells were lysed in 200 ul of luciferase lysis buffer. Following two cycles of freeze-thaw in dry ice-ethanol bath, the homogenized lysates were extensively vortexed and spun down at 4°C for 2 minutes. Supernatant was utilized for luciferase activity assay. The analysis was performed according to a previously described protocol60, and the Luciferase Assay System (Cat # E1501) was purchased from Promega (Madison, WI).
Supplementary Material
Supplemental 1. The original TNT promoter displayed strong transgene expression in heart and slightly leaky expression in liver. 1× 1012 vg/ml of vectors were delivered into 6-week-old BL/6 mice, and the mice were sacrificed three weeks post delivery. Three animals were utilized in this group, and displayed are representative.
Supplemental 2. Histological analysis was performed to determine whether the decline in transgene expression was caused by an immune response to the vector, which would manifest as mononuclear cell infiltration and disturbed histology. Immunofluorescent staining against CD4 and histological examination showed normal morphology without apparent lymphocyte infiltration in the muscle and liver tissues, regardless of the presence of the miR-142 target sequence in the delivered vectors.
Acknowledgments
We thank Luke Roode (Division of Molecular Pharmacy, UNC Eshelman School of Pharmacy) for his critical reading of this manuscript. We appreciate James Wilson (Gene Therapy Center, University of Pennsylvania) and Guangping Gao (Gene Therapy Center, University of Massachusetts Medical School) for their generous gift of AAV9 packaging plasmid. This manuscript was supported by grant AR 56394, AR45967 (to X Xiao).
Footnotes
Disclosure Statement: No competing financial interests.
References
- 1.Manno CS, et al. AAV-mediated factor IX gene transfer to skeletal muscle in patients with severe hemophilia B. Blood. 2003;101:2963–2972. doi: 10.1182/blood-2002-10-3296. [DOI] [PubMed] [Google Scholar]
- 2.Maguire AM, et al. Age-dependent effects of RPE65 gene therapy for Leber's congenital amaurosis: a phase 1 dose-escalation trial. Lancet. 2009;374:1597–1605. doi: 10.1016/S0140-6736(09)61836-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maguire AM, et al. Safety and efficacy of gene transfer for Leber's congenital amaurosis. N Engl J Med. 2008;358:2240–2248. doi: 10.1056/NEJMoa0802315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hauswirth WW, et al. Treatment of leber congenital amaurosis due to RPE65 mutations by ocular subretinal injection of adeno-associated virus gene vector: short-term results of a phase I trial. Hum Gene Ther. 2008;19:979–990. doi: 10.1089/hum.2008.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaplitt MG, et al. Safety and tolerability of gene therapy with an adeno-associated virus (AAV) borne GAD gene for Parkinson's disease: an open label, phase I trial. Lancet. 2007;369:2097–2105. doi: 10.1016/S0140-6736(07)60982-9. [DOI] [PubMed] [Google Scholar]
- 6.Wang B, Li J, Xiao X. Adeno-associated virus vector carrying human minidystrophin genes effectively ameliorates muscular dystrophy in mdx mouse model. Proc Natl Acad Sci U S A. 2000;97:13714–13719. doi: 10.1073/pnas.240335297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yue Y, et al. A single intravenous injection of adeno-associated virus serotype-9 leads to whole body skeletal muscle transduction in dogs. Mol Ther. 2008;16:1944–1952. doi: 10.1038/mt.2008.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harper SQ, et al. Modular flexibility of dystrophin: implications for gene therapy of Duchenne muscular dystrophy. Nat Med. 2002;8:253–261. doi: 10.1038/nm0302-253. [DOI] [PubMed] [Google Scholar]
- 9.Brantly ML, et al. Sustained transgene expression despite T lymphocyte responses in a clinical trial of rAAV1-AAT gene therapy. Proc Natl Acad Sci U S A. 2009;106:16363–16368. doi: 10.1073/pnas.0904514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daya S, Berns KI. Gene therapy using adeno-associated virus vectors. Clin Microbiol Rev. 2008;21:583–593. doi: 10.1128/CMR.00008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiao X, Li J, Samulski RJ. Efficient long-term gene transfer into muscle tissue of immunocompetent mice by adeno-associated virus vector. J Virol. 1996;70:8098–8108. doi: 10.1128/jvi.70.11.8098-8108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirata R, Chamberlain J, Dong R, Russell DW. Targeted transgene insertion into human chromosomes by adeno-associated virus vectors. Nat Biotechnol. 2002;20:735–738. doi: 10.1038/nbt0702-735. [DOI] [PubMed] [Google Scholar]
- 13.Grimm D, et al. Preclinical in vivo evaluation of pseudotyped adeno-associated virus vectors for liver gene therapy. Blood. 2003;102:2412–2419. doi: 10.1182/blood-2003-02-0495. [DOI] [PubMed] [Google Scholar]
- 14.High KA. AAV-mediated gene transfer for hemophilia. Ann N Y Acad Sci. 2001;953:64–74. doi: 10.1111/j.1749-6632.2001.tb11361.x. [DOI] [PubMed] [Google Scholar]
- 15.Qiao C, et al. Amelioration of laminin-alpha2-deficient congenital muscular dystrophy by somatic gene transfer of miniagrin. Proc Natl Acad Sci U S A. 2005;102:11999–12004. doi: 10.1073/pnas.0502137102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang L, et al. Sustained correction of disease in naive and AAV2-pretreated hemophilia B dogs: AAV2/8-mediated, liver-directed gene therapy. Blood. 2005;105:3079–3086. doi: 10.1182/blood-2004-10-3867. [DOI] [PubMed] [Google Scholar]
- 17.Arruda VR, et al. Regional intravascular delivery of AAV-2-F.IX to skeletal muscle achieves long-term correction of hemophilia B in a large animal model. Blood. 2005;105:3458–3464. doi: 10.1182/blood-2004-07-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simonelli F, et al. Gene Therapy for Leber's Congenital Amaurosis is Safe and Effective Through 1.5 Years After Vector Administration. Mol Ther. 2009 doi: 10.1038/mt.2009.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vandenberghe LH, Wilson JM. AAV as an immunogen. Curr Gene Ther. 2007;7:325–333. doi: 10.2174/156652307782151416. [DOI] [PubMed] [Google Scholar]
- 20.Aiuti A, et al. Progress and prospects: gene therapy clinical trials (part 2) Gene Ther. 2007;14:1555–1563. doi: 10.1038/sj.gt.3303033. [DOI] [PubMed] [Google Scholar]
- 21.Vandenberghe LH, et al. Heparin binding directs activation of T cells against adeno-associated virus serotype 2 capsid. Nat Med. 2006;12:967–971. doi: 10.1038/nm1445. [DOI] [PubMed] [Google Scholar]
- 22.Martino AT, et al. Tolerance induction to cytoplasmic beta-galactosidase by hepatic AAV gene transfer: implications for antigen presentation and immunotoxicity. PLoS One. 2009;4:e6376. doi: 10.1371/journal.pone.0006376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao O, et al. Induction and role of regulatory CD4+CD25+ T cells in tolerance to the transgene product following hepatic in vivo gene transfer. Blood. 2007;110:1132–1140. doi: 10.1182/blood-2007-02-073304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L, et al. Cross-presentation of adeno-associated virus serotype 2 capsids activates cytotoxic T cells but does not render hepatocytes effective cytolytic targets. Hum Gene Ther. 2007;18:185–194. doi: 10.1089/hum.2007.001. [DOI] [PubMed] [Google Scholar]
- 25.Zhang TP, et al. Transgene expression levels and kinetics determine risk of humoral immune response modeled in factor IX knockout and missense mutant mice. Gene Ther. 2007;14:429–440. doi: 10.1038/sj.gt.3302881. [DOI] [PubMed] [Google Scholar]
- 26.Manno CS, et al. Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nat Med. 2006;12:342–347. doi: 10.1038/nm1358. [DOI] [PubMed] [Google Scholar]
- 27.Li H, et al. A preclinical animal model to assess the effect of pre-existing immunity on AAV-mediated gene transfer. Mol Ther. 2009;17:1215–1224. doi: 10.1038/mt.2009.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoshimura M, et al. AAV vector-mediated microdystrophin expression in a relatively small percentage of mdx myofibers improved the mdx phenotype. Mol Ther. 2004;10:821–828. doi: 10.1016/j.ymthe.2004.07.025. [DOI] [PubMed] [Google Scholar]
- 29.Brockstedt DG, et al. Induction of immunity to antigens expressed by recombinant adeno-associated virus depends on the route of administration. Clin Immunol. 1999;92:67–75. doi: 10.1006/clim.1999.4724. [DOI] [PubMed] [Google Scholar]
- 30.Sarukhan A, et al. Successful interference with cellular immune responses to immunogenic proteins encoded by recombinant viral vectors. J Virol. 2001;75:269–277. doi: 10.1128/JVI.75.1.269-277.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang B, et al. Construction and analysis of compact muscle-specific promoters for AAV vectors. Gene Ther. 2008;15:1489–1499. doi: 10.1038/gt.2008.104. [DOI] [PubMed] [Google Scholar]
- 32.Wang Z, et al. Adeno-associated virus serotype 8 efficiently delivers genes to muscle and heart. Nat Biotechnol. 2005;23:321–328. doi: 10.1038/nbt1073. [DOI] [PubMed] [Google Scholar]
- 33.Grimm D, et al. Liver transduction with recombinant adeno-associated virus is primarily restricted by capsid serotype not vector genotype. J Virol. 2006;80:426–439. doi: 10.1128/JVI.80.1.426-439.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, et al. AAV-mediated TRAIL gene expression driven by hTERT promoter suppressed human hepatocellular carcinoma growth in mice. Life Sci. 2008;82:1154–1161. doi: 10.1016/j.lfs.2008.03.023. [DOI] [PubMed] [Google Scholar]
- 35.Ambros V, et al. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr Biol. 2003;13:807–818. doi: 10.1016/s0960-9822(03)00287-2. [DOI] [PubMed] [Google Scholar]
- 36.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 37.Marquez RT, McCaffrey AP. Advances in microRNAs: implications for gene therapists. Hum Gene Ther. 2008;19:27–38. doi: 10.1089/hum.2007.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chekanova JA, Belostotsky DA. MicroRNAs and messenger RNA turnover. Methods Mol Biol. 2006;342:73–85. doi: 10.1385/1-59745-123-1:73. [DOI] [PubMed] [Google Scholar]
- 39.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mishra PJ, Merlino G. MicroRNA reexpression as differentiation therapy in cancer. J Clin Invest. 2009;119:2119–2123. doi: 10.1172/JCI40107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brown BD, et al. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat Biotechnol. 2007;25:1457–1467. doi: 10.1038/nbt1372. [DOI] [PubMed] [Google Scholar]
- 42.Chang J, et al. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1:106–113. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- 43.Girard M, et al. miR-122, a paradigm for the role of microRNAs in the liver. J Hepatol. 2008;48:648–656. doi: 10.1016/j.jhep.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 44.Herweijer H, Wolff JA. Gene Ther. 2006. Gene therapy progress and prospects: Hydrodynamic gene delivery. [DOI] [PubMed] [Google Scholar]
- 45.Inagaki K, et al. Robust systemic transduction with AAV9 vectors in mice: efficient global cardiac gene transfer superior to that of AAV8. Mol Ther. 2006;14:45–53. doi: 10.1016/j.ymthe.2006.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bish LT, et al. Adeno-associated virus (AAV) serotype 9 provides global cardiac gene transfer superior to AAV1, AAV6, AAV7, and AAV8 in the mouse and rat. Hum Gene Ther. 2008;19:1359–1368. doi: 10.1089/hum.2008.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Z, et al. Immunity to adeno-associated virus-mediated gene transfer in a random-bred canine model of Duchenne muscular dystrophy. Hum Gene Ther. 2007;18:18–26. doi: 10.1089/hum.2006.093. [DOI] [PubMed] [Google Scholar]
- 48.Gao G, et al. Adeno-associated virus-mediated gene transfer to nonhuman primate liver can elicit destructive transgene-specific T cell responses. Hum Gene Ther. 2009;20:930–942. doi: 10.1089/hum.2009.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brown BD, et al. Endogenous microRNA regulation suppresses transgene expression in hematopoietic lineages and enables stable gene transfer. Nat Med. 2006;12:585–591. doi: 10.1038/nm1398. [DOI] [PubMed] [Google Scholar]
- 50.Brown BD, et al. A microRNA-regulated lentiviral vector mediates stable correction of hemophilia B mice. Blood. 2007;110:4144–4152. doi: 10.1182/blood-2007-03-078493. [DOI] [PubMed] [Google Scholar]
- 51.Wang Z, et al. Rapid and highly efficient transduction by double-stranded adeno-associated virus vectors in vitro and in vivo. Gene Ther. 2003;10:2105–2111. doi: 10.1038/sj.gt.3302133. [DOI] [PubMed] [Google Scholar]
- 52.Xu L, et al. CMV-beta-actin promoter directs higher expression from an adeno-associated viral vector in the liver than the cytomegalovirus or elongation factor 1 alpha promoter and results in therapeutic levels of human factor X in mice. Hum Gene Ther. 2001;12:563–573. doi: 10.1089/104303401300042500. [DOI] [PubMed] [Google Scholar]
- 53.Lu Y, Song S. Distinct immune responses to transgene products from rAAV1 and rAAV8 vectors. Proc Natl Acad Sci U S A. 2009;106:17158–17162. doi: 10.1073/pnas.0909520106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang L, et al. A myocardium tropic adeno-associated virus (AAV) evolved by DNA shuffling and in vivo selection. Proc Natl Acad Sci U S A. 2009;106:3946–3951. doi: 10.1073/pnas.0813207106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiao X, Li J, Samulski RJ. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J Virol. 1998;72:2224–2232. doi: 10.1128/jvi.72.3.2224-2232.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ayuso E, et al. High AAV vector purity results in serotype- and tissue-independent enhancement of transduction efficiency. Gene Ther. 2009 doi: 10.1038/gt.2009.157. [DOI] [PubMed] [Google Scholar]
- 57.Rohr UP, et al. Quantitative real-time PCR for titration of infectious recombinant AAV-2 particles. J Virol Methods. 2005;127:40–45. doi: 10.1016/j.jviromet.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 58.Veldwijk MR, et al. Development and optimization of a real-time quantitative PCR-based method for the titration of AAV-2 vector stocks. Mol Ther. 2002;6:272–278. doi: 10.1006/mthe.2002.0659. [DOI] [PubMed] [Google Scholar]
- 59.Qiao C, et al. A novel gene expression control system and its use in stable, high-titer 293 cell-based adeno-associated virus packaging cell lines. J Virol. 2002;76:13015–13027. doi: 10.1128/JVI.76.24.13015-13027.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu CY, et al. A muscle-targeting peptide displayed on AAV2 improves muscle tropism on systemic delivery. Gene Ther. 2009;16:953–962. doi: 10.1038/gt.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental 1. The original TNT promoter displayed strong transgene expression in heart and slightly leaky expression in liver. 1× 1012 vg/ml of vectors were delivered into 6-week-old BL/6 mice, and the mice were sacrificed three weeks post delivery. Three animals were utilized in this group, and displayed are representative.
Supplemental 2. Histological analysis was performed to determine whether the decline in transgene expression was caused by an immune response to the vector, which would manifest as mononuclear cell infiltration and disturbed histology. Immunofluorescent staining against CD4 and histological examination showed normal morphology without apparent lymphocyte infiltration in the muscle and liver tissues, regardless of the presence of the miR-142 target sequence in the delivered vectors.


