Abstract
High-throughput sequencing (HTS) has quickly become a valuable tool for comparative genetics and genomics and is now regularly carried out in laboratories that are not connected to large sequencing centers. Here we describe an updated version of our protocol for constructing single- and paired-end Illumina sequencing libraries, beginning with purified genomic DNA. The present protocol can also be used for “multiplexing,” i.e. the analysis of several samples in a single flowcell lane by generating “barcoded” or “indexed” Illumina sequencing libraries in a way that is independent from Illumina-supported methods. To analyze sequencing results, we suggest several independent approaches but end users should be aware that this is a quickly evolving field and that currently many alignment (or “mapping”) and counting algorithms are being developed and tested.
Keywords: High-throughput sequencing, Solexa/illumina library, Reference genome, Paired-end library, De novo assembly, Barcoding
1. Introduction
High-throughput DNA sequencing (HTS) is now commonly used to determine the DNA sequence of closely related strains within one species, referred to as “re-sequencing” because a reference genome sequence had been previously determined by more traditional chain termination, or “Sanger,” sequencing. Single- or paired-end re-sequencing has proven useful for identification of SNPs, indels, rearrangements and differences in copy number compared to a reference genome (1–5). Paired-end high-throughput sequencing has also been used as the basis for de novo assembly of previously unknown genome sequences (6–9). De novo assembly from short paired-end reads is particularly feasible in filamentous fungi where small genomes (<100 Mb) are common (7, 9). In these cases, the coding portion of the genome is typically well represented, especially in species where the fraction of repetitive DNA is low (9) because of excision of repeats by recombination or mutation by RIP, respectively (10). The methods to construct high-throughput sequencing libraries with genomic DNA are essentially the same as those for DNA derived from chromatin immunoprecipitation (ChIP) or cDNA generated for transcriptome analyses as long as sufficient starting material can be produced.
Prior to preparation of a sequencing library, DNA must be isolated and purified (see http://www.illumina.com/support/documentation.ilmn; “Genomic DNA Sample Preparation Guide”). Here we begin with purified genomic DNA as a starting point (Fig. 1). We previously published a detailed protocol for the isolation of high-quality genomic DNA from Neurospora crassa (11), which we have now also used with Fusarium spp., Trichoderma spp., and Aspergillus nidulans. Genomic DNA, which should first be analyzed for contaminating levels of RNA and if necessary treated with RNase A (see Note 1), must be sheared to near-homogeneous fragment sizes. Three commonly used methods to generate small DNA fragments for HTS library generation are (1) digestion, (2) nebulization, and (3) sonication. For rapid whole-genome sequencing digestion is not suitable because of bias in enzyme recognition sites and enzyme activity. Nebulization has a tendency to generate heterogeneous DNA fragments and results in substantial loss of DNA by vaporization (12). Therefore, we describe here shearing of genomic DNA or whole cells by sonication with a Bioruptor, which allows us to reproducibly sonicate 24 samples at one time. We also provide an alternative protocol that describes sonication with a standard tip sonicator. In both cases the DNA is sheared almost randomly, generating double strand breaks with blunt ends, as well as 3′ - and 5′ -overhangs. The ends are made flush with a mixture of enzymes. A 3′ adenine overhang is added to make the DNA library compatible for ligation with Illumina sequencing adapters (Table 1), which all have 3′ thymine overhangs. After ligation, PCR primers that are compatible with the adapter sequences are used to amplify and enrich for successfully ligated DNA (see Note 2).
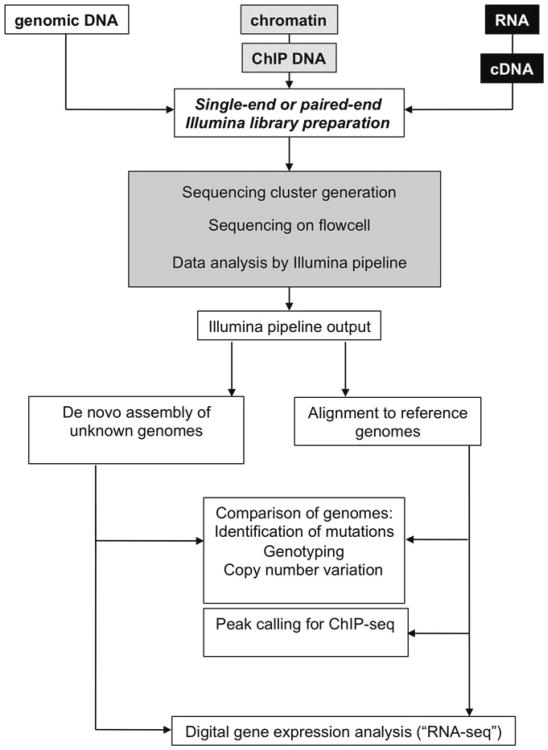
Fig. 1.
Workflow for rapid genome sequencing. Genomic DNA, ChIPed DNA, or cDNA prepared from RNA is ligated to single-end or paired-end adapters to generate a sequencing library. Sequencing centers generally perform the final “wet lab” (cluster generation and sequencing) and initial computational steps (running the Illumina pipeline) indicated in the gray box. This generates sequence files with quality scores, the Illumina output files, which are used as input for software programs that either assemble reads de novo (if the genome is unknown) or map reads to a reference genome. Reference genomes are necessary for efficient analysis of ChIP-seq data and for calling mutations, genotyping strains, or other applications. RNA-seq reads can either be assembled de novo, or mapped to a reference set of transcripts or a reference genome. More details on various analysis software can be found in the text.
Table 1. Oligonucleotide sequences for adapters or PCR primers supplied by Illumina, Inc. (copyright, 2008).
| Name | Sequence (5′−3′) |
|---|---|
| SE_P_Adapt1 | Phos-GATCGGAAGAGCTCGTATGCCGTCTTCTGCTTG |
| SE_Adapt2 | ACACTCTTTCCCTACACGACGCTCTTCC GATCT |
| SE_PCR1.1 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCT CTTCCGATCT |
| SE_PCR2.1 | CAAGCAGAAGACGGCATACGAGCTCTTCCGATCT |
| PE_P_Adapt1 | Phos-GATCGGAAGAGCGGTTCAGCAGGAATGCCGAG |
| PE_Adapt2 | ACACTCTTTCCCTACACGACGCTCTTCCGATCT |
| PE_PCR1.1 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTC TTCCGATCT |
| PE_PCR2.1 | CAAGCAGAAGACGGCATACGAGATCGGTCTCGGCATTCCTGCTGAACC GCTCTTCCGATCT |
The oligos listed here are used for single-end or paired-end library construction without barcodes. Oligos need not be highly purified (desalting is sufficient). The Adapt 1 primers need to be phosphorylated (“Phos,” and note “P” in adapter name) at the 5′ -most nucleotide. For more information see http://www.illumina.com/support/documentation.ilmn; “Illumina Adapter Sequences” (see Note 19). Oligonucleotide sequences © 2007–2011 Illumina, Inc. All rights reserved
To drastically reduce sequencing cost, several samples can be combined and sequenced in one lane (“multiplexing”). For a typical ∼40 Mb filamentous fungal genome, we routinely combine up to four samples in one lane of the Illumina GAIIx, or eight samples in the HiSeq 2000, which generally yield more than 30 or 250 million total reads per lane, respectively. Divided among four or eight equally loaded samples, this yields more than 7–40 million reads per sample, in our experience sufficient to call statistically significant ChIP-seq peaks and even single point mutations. For de novo assembly or transcriptome assembly we do not multiplex on this machine. On the Illumina HiSeq 2000 machine, 100–250 million reads are generated per lane, so additional samples can be multiplexed.
While Illumina advocates and supports an indexing method that does not reduce the read length (see http://www.illumina.com/support/documentation.ilmn; “Multiplexing Preparation Guide”), we often use adapters that have a four-nucleotide (nt) “barcode” to “index” each sample, based on an earlier 3-nt system (13). The barcode is sequenced as part of a normal read, thus yielding a unique four base identifier at the beginning of each read that is later used to sort or “parse” reads from the combined sample. This means that each read is reduced by 4 nt, e.g. from 58 to 54 nt on the HiSeq 2000 machine. Barcode sequences have been tested and optimized in various ways. We found problems with certain combinations of nucleotides that resulted in either higher than normal error rates or specific exclusion of some barcodes from cluster generation and/or sequencing, and thus started to balance the sequences of the adapters present in a single lane to yield even numbers of expected calls for all nucleotides in the first four cycles (one example set of four balanced barcodes would be 5′ -ACGT, 5′ -CGTA, 5′ -GTAC, and 5′ -TACG; Table 2). Because of the adapter design, the fifth base of each read is always a T.
Table 2. Adapter sequences for barcoding single-end and paired-end libraries.
| Name | Sequence (5′−3′) |
|---|---|
| SE_P_A1_ACGTT | ACGTAGATCGGAAGAGCTCGTATGCCGTCTTCTGCTTG |
| SE_P_A1_CGTAT | TACGAGATCGGAAGAGCTCGTATGCCGTCTTCTGCTTG |
| SE_P_A1_GTACT | GTACAGATCGGAAGAGCTCGTATGCCGTCTTCTGCTTG |
| SE_P_A1_TACGT | CGTAAGATCGGAAGAGCTCGTATGCCGTCTTCTGCTTG |
| SE_A2_ACGTT | ACACTCTTTCCCTACACGACGCTCTTCCGATCTACGTT |
| SE_A2_CGTAT | ACACTCTTTCCCTACACGACGCTCTTCCGATCTCGTAT |
| SE_A2_GTACT | ACACTCTTTCCCTACACGACGCTCTTCCGATCTGTACT |
| SE_A2_TACGT | ACACTCTTTCCCTACACGACGCTCTTCCGATCTTACGT |
| SE_P_A1_AGTCT | GACTAGATCGGAAGAGCTCGTATGCCGTCTTCTGCTTG |
| SE_P_A1_CTAGT | CTAGAGATCGGAAGAGCTCGTATGCCGTCTTCTGCTTG |
| SE_P_A1_GACTT | AGTCAGATCGGAAGAGCTCGTATGCCGTCTTCTGCTTG |
| SE_P_A1_TCGAT | TCGAAGATCGGAAGAGCTCGTATGCCGTCTTCTGCTTG |
| SE_A2_AGTCT | ACACTCTTTCCCTACACGACGCTCTTCCGATCTAGTCT |
| SE_A2_CTAGT | ACACTCTTTCCCTACACGACGCTCTTCCGATCTCTAGT |
| SE_A2_GACTT | ACACTCTTTCCCTACACGACGCTCTTCCGATCTGACTT |
| SE_A2_TCGAT | ACACTCTTTCCCTACACGACGCTCTTCCGATCTTCGAT |
| SE_P_A1_ATGCT | GCATAGATCGGAAGAGCTCGTATGCCGTCTTCTGCTTG |
| SE_P_A1_CATGT | CATGAGATCGGAAGAGCTCGTATGCCGTCTTCTGCTTG |
| SE_P_A1_GCATT | ATGCAGATCGGAAGAGCTCGTATGCCGTCTTCTGCTTG |
| SE_P_A1_TGCAT | TGCAAGATCGGAAGAGCTCGTATGCCGTCTTCTGCTTG |
| SE_A2_ATGCT | ACACTCTTTCCCTACACGACGCTCTTCCGATCTATGCT |
| SE_A2_CATGT | ACACTCTTTCCCTACACGACGCTCTTCCGATCTCATGT |
| SE_A2_GCATT | ACACTCTTTCCCTACACGACGCTCTTCCGATCT GCATT |
| SE_A2_TGCAT | ACACTCTTTCCCTACACGACGCTCTTCCGATCTTGCAT |
| SE_P_A1_ACTGT | CAGTAGATCGGAAGAGCTCGTATGCCGTCTTCTGCTTG |
| SE_P_A1_CTCAT | TGAGAGATCGGAAGAGCTCGTATGCCGTCTTCTGCTTG |
| SE_P_A1_GAGTT | ACTCAGATCGGAAGAGCTCGTATGCCGTCTTCTGCTTG |
| SE_P_A1_TGACT | GTCAAGATCGGAAGAGCTCGTATGCCGTCTTCTGCTTG |
| SE_A2_ACTGT | ACACTCTTTCCCTACACGACGCTCTTCCGATCTACTGT |
| SE_A2_CTCAT | ACACTCTTTCCCTACACGACGCTCTTCCGATCTCTCAT |
| SE_A2_GAGTT | ACACTCTTTCCCTACACGACGCTCTTCCGATCTGAGTT |
| SE_A2_TGACT | ACACTCTTTCCCTACACGACGCTCTTCCGATCTTGACT |
The oligos listed here are used for single-end or paired-end library construction with balanced barcodes. The same PCR primers as shown in Table 1 are used to amplify single-end or paired-end libraries. Oligos listed here have been used successfully (note that not all potential 5-nt barcodes are shown or have been tested yet). Oligos need not be highly purified (desalting is sufficient). All Adapt1 primers (“A1”) need to be phos-phorylated (note “P” in adapter name) at the 5′ -most nucleotide. These sequences are based on and contain the oligonucleotide sequences shown in Table 1, which are copyright of Illumina, Inc.; 2008. Note: Illumina does not support this type of barcoding and has a different system available for purchasing. The parsing and aligning of reads obtained with the barcodes shown here will require use of in-house scripts. Oligonucleotide sequences © 2007–2011 Illumina, Inc. All rights reserved. Derivative works created by Illumina customers are authorized for use with Illumina instruments and products only. All other uses are strictly prohibited.
There are few labs with access to their own HTS machines. In most cases sequencing libraries will be processed for sequencing by dedicated staff in sequencing centers or private companies. Thus we do not elaborate on the Illumina cluster generation and sequencing steps (Fig. 1). We wish to emphasize that investment in computational hardware and personnel “know-how” is essential to successfully undertake whole-genome sequencing approaches. Here, we do not go into details with regard to the downstream handling of sequencing data, for two reasons: (1) Currently the processing pipelines supplied by Illumina change frequently as throughput increases every few months and (2) new and drastically improved analysis tools become available or are being updated almost monthly. Thus, we provide only brief outlines for how to prepare for the flood of sequencing data but we do not recommend specific software. Instead we focus on the part of the sequencing that is most influenced by the end user of this technology, the preparation and troubleshooting of Illumina sequencing libraries.
2. Materials
2.1. Equipment to Determine DNA Concentration
Nanodrop spectrophotometer (NanoDrop products, Wilmington, USA).
Qubit fluorometer (Invitrogen, Carlsbad, USA).
2.2. Equipment to Shear DNA
Waterbath sonicator: Bioruptor UCD-200 (Diagenode, Liège, Belgium).
Tip sonicator: Branson 450 Sonifier (Branson Ultrasonics, Danbury, CT).
2.3. Processing of DNA
2.3.1. Shearing of DNA by Sonication
Genomic DNA.
TE Buffer: 10 mM Tris–HCl, 1 mM EDTA in sterile distilled H2O, pH 8.0. Stored at room temperature.
- Components of QIAquick PCR purification kit (Qiagen, Valencia, USA) or equivalent:
- PB buffer: 5.5 M guanidine hydrochloride, 20 mM Tris–HCl, pH 6.6. Store at room temperature.
- PE buffer: 20 mM NaCl, 2 mM Tris–HCl in 80% ethanol, pH 7.5. Store at room temperature.
- QIAquick columns.
- EB buffer: 10 mM Tris–HCl, pH 8.5. Store at room temperature.
2.3.2. End Repair of Sheared DNA Fragments (See Note 3)
Sheared genomic DNA.
10× T4 DNA ligase buffer: 500 mM Tris–HCl, 100 mM MgCl2, 100 mM dithiothreitol 10 mM ATP, pH 7.5 (New England Biolabs [NEB], Ipswich, USA) (see Note 4).
dNTPs (20 mM of each nucleotide, i.e. mix 20 μl each of 100 mM dATP, dCTP, dGTP, dTTP, and H2O).
T4 DNA polymerase, 3 units/ μL (NEB).
DNA polymerase I, large (Klenow) fragment, 5 units/μL (NEB).
T4 polynucleotide kinase, 10 units/μL (NEB).
Components of QIAquick purification kits (see Subheading 2.3.1, items a–d; Qiagen).
2.3.3. Addition of A-Overhang
End-repaired, sheared DNA.
10× NEBuffer 2: 500 mM NaCl, 100 mM Tris–HCl, 100 mM MgCl2, 10 mM dithiothreitol, pH 7.9 (NEB).
1 mM dATP.
DNA polymerase I, large (Klenow) fragment (3′ → 5′ exo-), 5 units/μL (NEB).
Components of QIAquick and MinElute purification kits (see Subheading 2.3.1, items a, b, d; Qiagen) or equivalent.
MinElute columns (Qiagen).
2.3.4. Ligation of Sequencing Adapters
A-tailed, end-repaired, sheared DNA.
Single-end (SE) and paired-end (PE) library adapters (Table 1) or SE barcode adapters (Table 2): mix 5 μM of each of two oligonucleotides in sterile distilled H2O. To anneal adapters, boil for 2 min (min) in a heat block and allow to cool for 30 min at room temperature. Store annealed adapters at −20°C.
T4 DNA ligase, 400 units/μL (NEB).
NuSieve GTG low melting temperature agarose (Lonza, Walkersville, USA).
TAE buffer: 40 mM Tris, 20 mM acetate, 1 mM EDTA. To make 50× stock, dissolve 242 g Tris base in water, add 57.1 mL glacial acetic acid, 100 mL of 500 mM EDTA (pH 8.0), and bring the final volume to 1 L. Store at 20°C.
Ethidium bromide solution (stocks of 10 mg/mL, use 5–10 μL stock for 100 mL of gel).
5× DNA gel loading buffer: 125 mg bromophenol blue, 125 mg xylene cyanol, 25 mL of 0.2 M EDTA (pH 8.0), 15 mL glycerol, 10 mL H2O.
DNA ladder able to resolve 100–1,000 bp (e.g., Qiagen GelPilot 1 kb Plus Ladder, #239095).
Glass microscope cover slips.
Components of QIAquick Gel Extraction and PCR purification kits (see Subheading 2.3.1, items b–d; Qiagen) or equivalent.
QG buffer: 5.5 M guanidine thiocyanate, 20 mM Tris–HCl, pH 6.6. Store at room temperature (Qiagen).
Isopropanol.
2.3.5. Amplification of Sequencing Library
DyNAzyme II DNA polymerase, 2 units/μL (Finnzymes, Vantaa, Finland).
10× buffer for DyNAzyme II DNA polymerase (Finnzymes): 100 mM Tris–HCl (pH 8.8), 15 mM MgCl2, 500 mM KCl, 1% Triton X-100.
Single-end (SE) and paired-end (PE) library PCR amplification primers (Table 1); DpnII sites are indicated in bold): mix 5 μM each in H2O. Store at −20°C.
2× Phusion Flash master mix: 1 unit/μL Phusion Flash High-Fidelity polymerase, 50 mM TAPS-HCl (pH 9.3), 100 mM KCl, 3 mM MgCl2, 2 mM β-mercaptoethanol, 400 μM of dATP, dCTP, dGTP, and dTTP (NEB).
2.3.6. Oligonucleotide Sequences
The two tables contain Illumina oligonucleotide sequences (Table 1; these oligonucleotide sequences are copyright of Illumina, Inc.; 2008) or barcode adapter sequences for construction of indexed single-end libraries (Table 2).
3. Methods
All procedures are carried out at room temperature unless otherwise noted.
3.1. Shearing of DNA by Sonication
Adjust the water bath temperature of the Bioruptor to 4°C prior to sonication (see Note 5).
Melt a 1% agarose gel and a 2% NuSieve low melting temperature agarose gel in TAE buffer and cool to 55°C. Add ethidium bromide to 0.5 μg/mL and pour in separate gel casting trays prior to sonication. When the gels are cool, place in separate electrophoresis chambers and cover with TAE buffer.
Dilute 5 μg purified genomic DNA into 100 μL TE buffer in an Eppendorf tube (recommended: VWR 87003-294; see Note 6).
Place the Eppendorf tube with DNA in the Bioruptor and sonicate on high for 20 min total with 30 s of sonication followed by 30 s of rest (T1 = 20 min, T2 = 30 s, T3 = 30 s).
Mix 10 μL of the sonicated DNA (0.5 μg) with 2 μL 5× DNA gel loading buffer and run on the 1% agarose gel to ensure the DNA has been sheared to less than 1,000 bp (Fig. 2). Sonicate for additional cycles if the DNA is inadequately sheared.
Mix the sheared DNA with five volumes of buffer PB and vortex to mix.
Apply the DNA sample to a QIAquick column and centrifuge at 18,000 × g in a bench top centrifuge for 1 min. Discard the flow-through.
Apply 750 μL buffer PE to the column and let rest for 5 min. Centrifuge at 18,000 × g in a bench top centrifuge for 1 min. Discard the flow-through and centrifuge at 18,000 × g for 2 min.
Immediately place the column in a sterile Eppendorf tube and apply 40.5 μL EB buffer to the center of the QIAquick membrane. Let rest for 1 min, then centrifuge at 18,000 × g in a bench top centrifuge for 1 min.
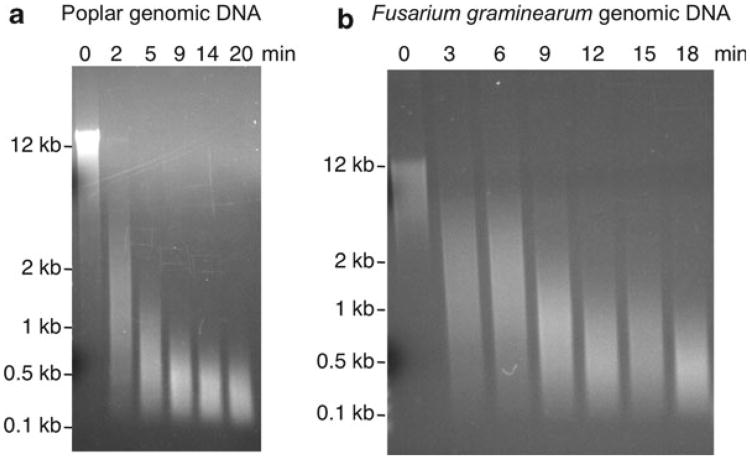
Fig. 2.
Shearing of DNA in the bioruptor and by tip sonication. (a) Shearing of poplar (Populus trichocarpa) genomic DNA in the bioruptor (5 μg DNA in TE buffer, 300 μL volume) on “high” setting with 30 s on/off cycles. The total time of sonication in minutes (min) is shown above the lanes in which ∼170 ng of DNA was loaded. (b) Shearing of Fusarium graminearum genomic DNA. Tissue was disrupted by bead beating for chromatin isolation, centrifuged and the supernatant sonicated in a bioruptor as described in (a), treated with RNAseA and run on a 1% agarose gel in TAE buffer.
3.2. End Repair of Sheared DNA Fragments
To the purified sheared DNA (from Subheading 3.1, step 9) add 5 μL 10× T4 DNA ligase buffer with 10 mM ATP, 1 μL 20 mM dNTPs, 2.5 μL T4 DNA polymerase, 0. 5 μL large DNA polymerase I (Klenow) fragment, and 2.5 μL T4 polynucleotide kinase. Incubate at room temperature for 30 min.
Stop the reaction by adding 250 μL buffer PB. Vortex to mix.
Apply the DNA sample to a QIAquick column and centrifuge at 18,000 × g in a bench top centrifuge for 1 min. Discard the flow-through.
Apply 750 μL buffer PE to the column and let rest for 5 min. Centrifuge at 18,000 × g in a bench top centrifuge for 1 min. Discard the flow-through and centrifuge at 18,000 × g for 2 min.
Immediately place the column in a sterile Eppendorf tube and apply 34 μL EB buffer to the center of the QIAquick membrane. Let rest for 1 min, then centrifuge at 18,000 × g in a bench top centrifuge for 1 min.
3.3. Addition of A-Overhang
To the purified sheared and end-repaired DNA (from Subheading 3.2, step 5), add 5 μL 10× NEBuffer 2, 10 μL 1 mM dATP, and 3 μL DNA polymerase I Klenow fragment (3′ → 5′ exo-). Incubate at 37°C for 30 min.
Stop the reaction by adding 250 μL buffer PB. Vortex to mix.
Apply the DNA sample to a MinElute column and centrifuge at 18,000 × g in a bench top centrifuge for 1 min. Discard the flow-through.
Apply 750 μL buffer PE to the column and let rest for 5 min. Centrifuge at 18,000 × g in a bench top centrifuge for 1 min. Discard the flow-through and centrifuge at 18,000 × g for 1 min.
Immediately place the column in a sterile Eppendorf tube and apply 17 μL EB buffer to the center of the MinElute membrane. Let rest for 1 min, then centrifuge at 18,000 × g in a bench top centrifuge for 1 min.
3.4. Ligation of Sequencing Adapters
To the purified sheared, end-repaired, and tailed DNA (from Subheading 3.3, step 5), add 2 μL 10× T4 ligase buffer with 10 mM dATP, 1 μL of 5 μM library adapters (non-barcoded single- or paired-end adapters, Table 1; barcoded single-end adapters, Table 2) and 1 μL T4 DNA ligase (see Note 7). Incubate at room temperature for 20 min.
Stop the reaction by mixing ligated DNA with 4 μL 5× DNA gel loading buffer.
Load the ligated DNA on the 2% NuSieve low melting temperature agarose gel. Use DNA ladder to resolve 100–1,000 bp and run for ∼45 min at 100 V (see Note 8).
View the gel on a UV transilluminator and excise a gel fragment between 250 and 350 bp with a clean glass cover slip (Fig. 3a; see Note 9).
Extract DNA with a QIAquick gel extraction kit: Weigh the gel slice and mix with three volumes of QG buffer in an Eppendorf tube. Melt the gel at 50°C for ∼10 min with vortexing every 3 min (see Note 10).
To increase recovery of small DNA fragments, add a volume of isopropanol equivalent to one gel slice volume, vortex to mix, and apply the sample to a QIAquick column. Centrifuge at 18,000 × g in a bench top centrifuge for 1 min. Discard the flow-through.
Apply 750 μL buffer PE to the column and let rest for 5 min. Centrifuge at 18,000 × g in a bench top centrifuge for 1 min. Repeat the PE wash. Discard the flow-through and centrifuge at 18,000 × g for 2 min (see Note 11).
Immediately place the column in a sterile Eppendorf tube and apply 50 μL EB buffer to the center of the QIAquick membrane. Let rest for 1 min, then centrifuge at 18,000 × g in a bench top centrifuge for 1 min. This constitutes the non-amplified Illumina sequencing library.
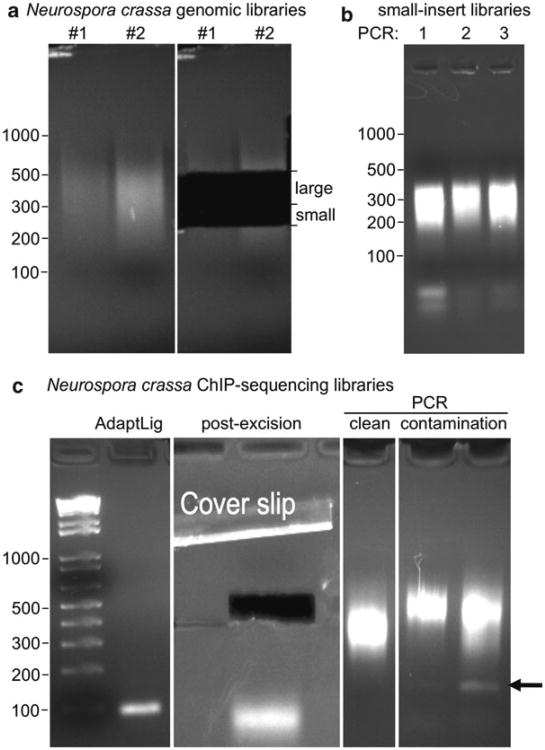
Fig. 3.
Preparation of genomic and ChIP paired-end sequencing libraries. (a) Neurospora crassa genomic DNA was sonicated with a tip sonicator (see Note 5), and libraries were generated as described. Two replicates were done in parallel and loaded on a 2% NuSieve agarose gel for DNA isolation (left panel). Two fractions were removed from the gel (250–325 bp, “small”; 325–500 bp, “large”). We do this to insure that we have library material for future PCRs even in cases when library amplifications fail or other unforeseen difficulties arise. (b) Three parallel PCR amplifications of the small insert libraries shown in (a) (5 μL of 35 μL were loaded). Note the presence of adapter and PCR primer bands. (c) Example of construction of ChIP-seq libraries. After ligation of adapters (“AdaptLig”) the library is invisible on the gel (but note the presence of adapter bands below 100 bp). After excision of the bands and extraction of DNA from the gel (Qiagen gel extraction kit), PCR reactions that are puri fi ed (Qiagen PCR puri fi cation kit) typically yield slightly smeary library bands (“clean”) but no bands <200 bp. It is important to adjust the adapter:DNA ratio to ∼10:1. In cases of great adapter excess, which can be a problem with ChIP-seq library generation, spurious bands are observed. Paired-end adapters run at ∼160 bp and single-end adapters run at ∼110 bp (“contamination,” see arrow). We would not subject the library in the right-most lane to sequencing as the amount of contamination present will significantly reduce the number of useable reads, because most reads will be adapter sequence instead of insert.
3.5. Amplification of Sequencing Library
Test a range of cycle numbers to determine the optimum number of PCR cycles to amplify the sequencing library (see Note 12).
- Mix 14.75 μL H2O, 1 μL library DNA, 2 μL 10× DyNAzyme buffer, 0.25 μL of 20 mM dNTPs, 1.5 m L of 5 μM library amplification primers, and 0.5 μL DyNAzyme (see Note 13) for each number of cycles to be tested and amplify using the following protocol on a thermocycler:
- 180 s at 94°C.
- 30 s at 94°C.
- 30 s at 60°C.
- 60 s at 72°C.
- Repeat (b) to (d) for 15, 18, or 21 cycles.
- 300 s at 72°C.
Melt a 2% agarose gel in TAE buffer and cool to 55°C. Add ethidium bromide to 0.5 μg/mL and pour in a gel casting tray. When the gel cools, place in an electrophoresis chambers and cover with TAE buffer.
Mix the PCR reactions with 4 m L of 5× DNA gel loading buffer and load into the 2% agarose gel wells next to a DNA ladder able to resolve 100–1,000 bp and run for 45 min at 100 V.
View the gel on a UV transilluminator. The appropriate number of cycles results in a just visible DNA smear of ∼50–100 bp greater than the size excised earlier (primers add 53 bp to the adapter ligated fragments; Notes 14 and 15).
After the appropriate number of cycles has been determined, amplify the DNA libraries in three separate reactions with 1–5 μL template DNA. Try to minimize the number of cycles necessary (Fig. 3b; Note 16).
- Mix 8.5 μL template DNA and H2O, 1.5 μL of 5 μM of each library amplification primers (SE_PCR1.1. and SE_PCR2.1. or PE_PCR1.1. and PE_PCR2.1.; Table 1) and 10 μL 2× Phusion Flash master mix. Amplify the library in three separate reactions using the following protocol on a Thermocycler (see Note 17):
- 30 s at 98°C.
- 10 s at 98°C.
- 30 s at 65°C.
- 30 s at 72°C.
- Repeat steps b–d for the number of cycles determined.
- 300 s at 72°C.
Pool the three reactions in an Eppendorf tube and add 300 μL buffer PB. Vortex to mix.
Apply the DNA sample to a QIAquick column and centrifuge at 18,000 × g in a bench top centrifuge for 1 min. Discard the flow-through.
Apply 750 μL buffer PE to the column and let rest for 5 min. Centrifuge at 18,000 × g in a bench top centrifuge for 1 min. Discard the flow-through and centrifuge at 18,000 × g for 2 min.
Immediately place the column in a sterile Eppendorf tube and apply 32 μL EB buffer to the center of the QIAquick membrane. Let rest for 1 min, then centrifuge at 18,000 × g in a bench top centrifuge for 1 min.
Melt a 2% agarose gel in TAE buffer and cool to 55°C. Add ethidium bromide to 0.5 μg/mL and pour in a gel casting tray. When the gel cools, place in an electrophoresis chamber and cover with TAE buffer.
Mix 5 μL of the purified PCR reactions with 5 μL of 1× DNA gel loading buffer and load into the 2% agarose gel wells next to a DNA ladder able to resolve 100–1,000 bp and run for 45 min at 100 V.
View the gel on a UV transilluminator. Only the library smear should be visible now, and no bands lower than 200 bp are expected (Figs. 3c and 4d).
Quantify the library by using a Qubit fluorometer or by quantitative PCR with the Illumina sequencing primers (see Note 18).
Dilute the sample in buffer EB to a concentration recommended by your core laboratory or sequencing company of choice.
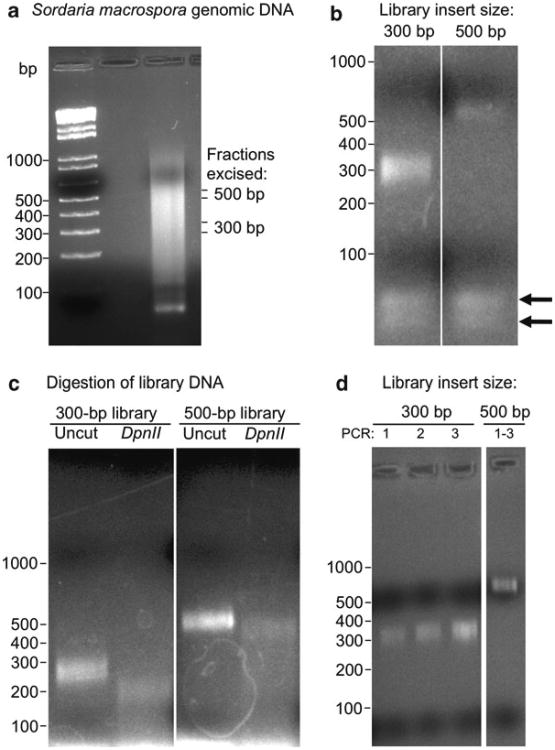
Fig. 4.
Preparation of paired-end sequencing libraries from Sordaria macrospora. (a) Genomic DNA (10 μg in 450 μL TE) was prepared as described in ref. (11) and sheared by sonication with a Branson 450 Sonifier with microtip (duty cycle 80%, output 1.2; five cycles of 10 s pulses, interrupted by 30 s rest on ice). Sheared DNA was concentrated (Qiagen PCR purification kit) and separated on a 1.2% agarose gel. Gel slices of ∼300 and ∼500 bp fragments were isolated and purified (Qiagen Gel extraction kit, see Subheading 3.4, step 5). (b) Paired-end libraries were constructed as described in the methods from the 300- and 500-bp fractions and amplified with 12 cycles of PCR, yielding barely visible bands. Note presence of adapters and PCR primers below the 100 bp marker. (c) Test for library complexity. One simple test to show that libraries contain predominantly genomic DNA and are not enriched for adapter dimers or other spurious overamplified bands is to digest the DyNAzyme-amplified libraries with DpnII. This cleaves most of the adapter off the inserts. If genomic DNA has been cloned it will result in a smear (similar to genomic DNA digested for Southern blots). If only low complexity samples have been cloned in the library, sharp banding is observed instead of the smear and the library is unsuitable for further processing. (d) Libraries were amplified with 18 cycles of PCR with Phusion Flash High-Fidelity polymerase and purified (Qiagen PCR purification kit; see three 300-bp library samples). We typically pool three independent 25 μmL PCR reactions before purification and run 5 μL of 35 μL to show that only expected bands are obtained. Note the absence of the adapters or PCR primers and primer dimers (compare to (b)).
3.6. Post-sequencing Data Analyses
Illumina software pipelines generate files of several formats, the most useful being “sequence.txt” files, which contain sequence and quality scores, and ELAND or bam files, which also contain the start and stop positions (or “coordinates”) specific reads map to in reference genomes (see http://www.illumina.com/support/documentation.ilmn). Pre-analysis of raw data directly from the sequencer is typically run by core facilities or companies who provide the actual sequencing services, and this relies on proprietary Illumina software. File types supported by software for post-sequencing analysis vary, and so does their input and output file types. For this reason, familiarity with the program manuals is required and they should be consulted frequently for updated information.
Older pipelines of Illumina Genome Analyzers generated a different set of files than the current pipeline (CASAVA 1.8). Illumina CASAVA 1.7, for example, still used with the Illumina GAIIx machines, relies on analysis by the Illumina GERALD package to generate a comprehensive file “sequence.txt” of all the reads and quality score. Files of the format “sequence.txt” contain each read in a 4-line format: header, read, header, quality score. The header has information about the location on the flow cell from which the read was collected, the read consists of base calls made by the software analysis of the images, and the quality score has information about the probability of a base call being wrong. In this pipeline, quality scores are in a phred scale 64 Illumina-type offset format. The newer version, CASAVA 1.8, which is required for the higher throughputs on HiSeq 2000 machines, has been changed in several ways including the available output formats. Now, archival bam files and a collection of compressed fastq files are the primary source of total sequence reads. CASAVA 1.8 also uses different, Sanger-type quality scores, which are in phred scale 33 offset in ASCII format.
Many freely available programs will take Illumina “sequence.txt” files as input, such as Bowtie (14) and Velvet (15). There are tools available to convert sequence.txt files into fastq and other desired formats, when necessary. Some, such as MAQ (16), have these scripts built into a comprehensive package. Similar tools for quality score conversion are available through the BioPerl project for the savvy programmer (http://www.bioperl.org), and the Java-based Vancouver Short Read Analysis package, which also converts between popular formats (http://vancouvershortr.sourceforge.net). When samples are not barcoded for multiplexing samples, the GERALD output of CASAVA 1.7, or the compressed fastq files of CASAVA 1.8 may be used directly as input for downstream applications.
This is slightly more challenging when several distinct samples are run in one lane of a flowcell (multiplexing), as “barcoded” or “indexed” samples. In this case, several files need to be generated, grouped by the barcodes. Multiplexed samples prepared with proprietary Illumina indexing kits are supported by the CASAVA 1.8 software and thus can easily be separated into different files according to the barcode; the barcode sequence is automatically removed in this process. The barcoding scheme we mention here, however, may require one additional concatenation step of the many CASAVA 1.8 output files to combine the contents of a lane into one single huge, unwieldy file. Alternatively, and likely more convenient, the many CASAVA 1.8 output files can be used as input for sorting by barcode, followed by concatenation of the appropriate files. Both approaches require additional scripts for parsing of reads according to barcodes and removal of the barcode sequences prior to read mapping. Some institutions may have in-house software available for sorting and removal of barcodes but there are also freely available tools, such as “NovoBarCode” (novoalign package from Novocraft Technologies; http://www.novocraft.com/main/index.php). The goal of the sequencing will determine subsequent steps and file formats or conversions necessary. Prior to undertaking an analysis project, perhaps even the sequencing itself, it is recommended that users become comfortable with the command line interface and basic scripting, and read program manuals available online. For most of the software packages user groups are available that can advise on specific problems. The Seqanswers forum (http://seqanswers.com) is particularly useful for keeping up to date with constantly changing formats and software.
3.6.1. De Novo Assembly Software
A variety of open source programs for de novo assembly of genomes are available and are being improved regularly. Some of the more popular programs have been compared on identical datasets and are reviewed elsewhere (17). Most rely on the construction of a De Bruijn graph and differ primarily in their error correction steps. The authors have used Velvet (15) and its many updates to assemble fungal genomes (9), but other state of the art short read assemblers including ABySS (18), QSRA (19, 20), SOAPdenovo (21), Ray (22), and others described in ref. (17) promise to improve the speed and accuracy of assembly from short reads.
3.6.2. Discovery of SNPs, Insertion/Deletions (Indels), and Genome Rearrangements for Mutation Mapping
Short read sequence alignment programs are useful for identifying single nucleotide polymorphisms (SNPs), indels, and rearrangements in organisms genetically similar to a reference sequence. The programs map each read to a reference genome and allow the user to specify a variety of options including the number of mismatches allowed per read and whether reads are mapped to a single match in the genome or all possible perfect matches in the reference genome if identical kmer repeats exist; kmer refers to a specific determined length, i.e. 36mer. If the algorithm uses quality score values it is essential that the user specify the appropriate quality score scale. Popular open source programs for read alignment include SOAP (23), MAQ (16), Bowtie (14), BWA (24), CASHX (25), and Stampy (26). While some programs such as MAQ include SNP and indel analysis, others, like Bowtie, require additional analysis by a package such as SAMtools (27) to call variants. We have used these approaches to find single-point mutations in several fungal genomes, for example, see (5). Deciding which software program to use for read alignment may depend on computational power available versus what is required, which depends on the size of your genome and amount of sequence data.
3.6.3. Analysis of ChIP-Sequencing and MeDIP-Sequencing Data
The analysis of ChIP-seq data begins with aligning reads to a reference genome using one of the software packages mentioned above. How regions of enrichment are determined depends on the target of immunoprecipitation. In some cases, such as centromere proteins with localization to defined regions of each chromosome, statistical analysis of enrichment levels is not crucial (28). In the case of transcription factors, however, enriched regions must be called with software that uses statistical tools to assign a confidence level to each of the identified peaks (29). Different software packages are designed specifically to analyze localized binding, e.g. by transcription factors, versus diffuse binding, e.g. by histones and their modifications. We have used our own scripts in the past to call enrichment peaks (28, 29) but have also used MACS (30), a package which uses binomial peak identification to build a model of transcription factor binding. Diffuse binding seen in ChIP of histone modifications or other chromatin proteins requires a package such as SICER (31) or RSEG (32), which identify domains of enrichment spanning multiple nucleosomes, rather than discrete peaks. A wide variety of open source peak finding programs exist, and new additions and updates are frequent. If a genome assembly and gene annotations are available, visual confirmation of identified peaks in a browser like IGV (http://www.broadinstitute.org/igv/), GenomeView (http://genomeview.org/), or gbrowse (http://gmod.org/wiki/GBrowse) can be used to refine the settings and adjustable parameters of the program.
3.6.4. Analysis of Transcriptomes
Transcriptome sequencing by analysis of cDNA, or “RNA-seq,” is performed to identify expressed genes from an organism with no reference genome, or in a quantitative way to identify differential expression between two or more conditions. If a reference genome is unavailable, RNA-seq reads must be assembled into contigs with Velvet (15) or SOAPdenovo (21), or any of the other de novo assemblers described above. If a reference genome is available, RNA-seq reads are mapped to the genome with an aligner that can find splice junctions. These include SOAP (23), Tophat (33), and BWA (24). Other programs are designed specifically to identify novel intron/exon junctions (20). Counting cDNA tags from different samples and replicate experiments to generate comparable datasets that give meaningful results is challenging. To quantify transcript levels and call differential expression between two samples, statistical tools have been developed, including DEseq (34), Cufflinks (35, 36), and GENEcounter (37) but this field is still very much in flux.
Acknowledgments
We thank Mark Dasenko, Chris Sullivan, Steve Drake, Matthew Peterson, and Scott Givan at the OSU CGRB core facility for assistance with Illumina sequencing, and Chris Sullivan, Jason Cumbie, Noah Fahlgren and Henry Priest for helpful discussions and sharing code. Work in our laboratory is supported by funds from the American Cancer Society (RSG-08-030-01-CCG), the National Institutes of Health (P01GM068087 and R01GM097637), and start-up funds from the OSU Computational and Genome Biology Initiative.
Footnotes
When generating libraries from pooled DNA samples (e.g., to do bulk segregant analyses) it is essential that an equal amount of DNA from each strain be used. If different amounts of RNA are present in different samples this will affect the measured amount of nucleic acids and underestimate the amount of DNA in some samples. Imbalances in the concentration of the DNA pool are sometimes significant enough to affect mutation mapping. While presence of RNA will reduce the effective DNA concentration this is not a problem with library construction of non-pooled samples.
DNA shearing, end repair, A-overhang addition, ligation, and the subsequent gel extraction should be performed in a single day for best ligation efficiency. Ligated DNA can be stored at −20°C and amplified by PCR at any time prior to Illumina sequencing cluster generation. The incubation steps prior to ligation are short so plan ahead and thaw buffers for the next reaction during the incubation so that the next step can be performed immediately.
Several manufacturers now offer kits that contain all or most enzymes required for library preparation (e.g., NEB #E6040, contains all required enzymes; NEB Quick-Blunting kit #E1201 or Epicentre End-It end-repair kit #ER0720, contain enzymes for blunting reactions). In our hands both kits and individual enzymes work equally well. We typically use enzymes from NEB but enzymes from other suppliers can be used. Compare units/μl (e.g., Invitrogen T4 ligase is typically less concentrated than NEB T4 ligase, so we use 2.5 μl of this enzyme).
Ligase buffer can be prepared fresh or premade buffers from several manufacturers can be used, e.g. 10× ligase buffer from NEB or 5× ligase buffer from Invitrogen: 250 mM Tris–HCl (pH 7.6), 50 mM MgCl2, 5 mM ATP, 5 mM DTT, 25% (w/v) polyethylene glycol (8000). No differences in library construction were observed when different buffers were used.
Instead of the Bioruptor, tip sonicators can be used, e.g. a Branson 450 sonifier equipped with a microtip. Genomic DNA is sonicated with five 10-s pulses with 30 s rest on ice between repeats (settings: output 1.2, duty cycle 80%).
The stiffness of the Eppendorf tube impacts shearing efficiency. Always use the same brand of tubes once a reliable Bioruptor protocol has been established. Volumes of 100–300 μL can be used in standard Eppendorf tubes in the Bioruptor.
One can measure the concentration of the tailed DNA with Invitrogen Picogreen kits. Typically, 1 μl of 5 μM adapters is used in a 25 μl reaction when starting with 5–10 μg of genomic DNA. In the case of chromatin immunoprecipitated (ChIPed) DNA we found that those adapter concentrations can be 1,000- to 10,000-fold too high and may contribute to spurious 160-bp bands that appear after library PCR (Fig. 3c). The goal is to have a ∼10:1 adapter:insert ratio.
To avoid cross contamination, leave an empty lane between each sample.
A non-UV transilluminator will result in less DNA degradation. We routinely use a standard low- or high-range UV transilluminator but expose the DNA for as short a time as possible. The size range of DNA insert will affect the standard deviation of insert size during library assembly.
Instead of weighing it is sufficient to use marks on the Eppendorf tubes to judge the volume after gel slices have been centrifuged. Melting the gel closer to room temperature will increase the yield of AT-rich DNA (12).
Two washes with PE buffer are necessary to get rid of all the salt from the QG buffer and to allow for measurement of DNA concentration with a nanodrop spectrophotometer.
Typically, 15–21 cycles of PCR are optimal for generation of libraries from genomic DNA. For ChIP-seq libraries up to 24 cycles are acceptable. The best genomic DNA libraries only require as few as 12 cycles while libraries made from dilute or poorly ligated DNA can require as many as 25 cycles to yield sufficient DNA. In general fewer cycles are better because less PCR bias will be introduced, resulting in fewer “clonal” reads (i.e. large numbers of reads that have identical start and stop coordinates when mapped to the genome sequence).
To test the library any Taq DNA polymerase can be used and we have not found large differences in overall amplification behavior. For the preparation of the library used for actual sequencing a high-fidelity and highly processive enzyme is preferred but Phusion Flash polymerase is not the only such enzyme available.
“Tightening” of the DNA smear with more cycles indicates over-amplification. DNA greater than 100 bp but lower than the excised library size should not be present. A band at ∼120 bp for single-end libraries and ∼160 bp for paired-end libraries indicates adapter-primer hybrids contaminating the library (Fig. 3c). Such libraries should not be sequenced. It is possible to amplify the libraries again and to isolate and gel purify fragments above the contaminating bands but often adapter primer multimers “hide” below the library bands and still result in significant contamination of sequencing runs with these nonspecific sequences. In addition, gel purification at this stage of the protocol often results in significant loss of library DNA.
Complexity of libraries can be assessed by digestion of test PCR fragments with DpnII, as this cleaves most of the adapter off the inserts (Fig. 4c). If predominantly genomic DNA has been cloned it will result in a smear similar to genomic DNA digested for Southern blots. Conversely, if mostly low complexity samples (i.e. adapters or small amounts of contaminants) have been cloned, sharp banding is observed instead of the smear and the library is unsuitable for further processing.
Ideally, doubling of the amount of template DNA is expected to reduce the number of cycles needed by one. For example, if 20 cycles with 1 μL of template is sufficient, 18 cycles with 4 μL of template should be similar and result in similar complexity of libraries. In practice this relationship is not exactly linear.
Increasing the initial denaturation step from 30 s to 3 min and each subsequent denaturation step from 10 to 80 s or decreasing the temperature ramp rate of the thermocycler to 2°C/s improves the yield of GC-rich amplicons. Similarly, addition of 2 M betaine also improves recovery of GC-rich amplicons. Decreasing the annealing temperature from 65 to 60°C slightly improves the yield of AT-rich amplicons (38).
Other quantification methods such as spectrophotometry (e.g., by Nanodrop or similar devices) are less accurate and typically result in overestimation of DNA concentration and production of significantly fewer clusters. Quantification by qPCR is most accurate since it directly measures the concentration of only DNA that is able to form clusters on the flowcell.
Results with in-house adapter and PCR primer mix will not be identical to results obtained with Illumina-purchased kits, as certain modifications are made to oligos in Illumina library preparation kits to improve performance.
The authors have no conflicting interests
References
- 1.Blumenstiel JP, Noll AC, Griffiths JA, et al. Identification of EMS-induced mutations in Drosophila melanogaster by whole-genome sequencing. Genetics. 2009;182:25–32. doi: 10.1534/genetics.109.101998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birkeland SR, Jin N, Ozdemir AC, et al. Discovery of mutations in Saccharomyces cerevisiae by pooled linkage analysis and whole-genome sequencing. Genetics. 2010;186:1127–1137. doi: 10.1534/genetics.110.123232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ehrenreich IM, Torabi N, Jia Y, et al. Dissection of genetically complex traits with extremely large pools of yeast segregants. Nature. 2010;464:1039–1042. doi: 10.1038/nature08923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wenger JW, Schwartz K, Sherlock G. Bulk segregant analysis by high-throughput sequencing reveals a novel xylose utilization gene from Saccharomyces cerevisiae. PLoS Genet. 2010;6:e1000942. doi: 10.1371/journal.pgen.1000942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pomraning KR, Smith KM, Freitag M. Bulk segregant analysis followed by high-throughput sequencing reveals the Neurospora cell cycle gene, ndc-1, to be allelic with the gene for ornithine decarboxylase, spe-1. Eukaryot Cell. 2011;10:724–733. doi: 10.1128/EC.00016-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reinhardt JA, Baltrus DA, Nishimura MT, et al. De novo assembly using low-coverage short read sequence data from the rice pathogen Pseudomonas syringae pv. oryzae. Genome Res. 2009;19:294–305. doi: 10.1101/gr.083311.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diguistini S, Liao NY, Platt D, et al. De novo genome sequence assembly of a filamentous fungus using Sanger, 454 and Illumina sequence data. Genome Biol. 2009;10:R94. doi: 10.1186/gb-2009-10-9-r94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li R, Fan W, Tian G, et al. The sequence and de novo assembly of the giant panda genome. Nature. 2010;463:311–317. doi: 10.1038/nature08696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nowrousian M, Stajich JE, Chu M, et al. De novo assembly of a 40 Mb eukaryotic genome from short sequence reads: Sordaria macrospora, a model organism for fungal morphogenesis. PLoS Genet. 2010;6:e1000891. doi: 10.1371/journal.pgen.1000891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pomraning KR, Connolly LR, Whalen JP, et al. Repeat-induced point mutation, DNA methylation and heterochromatin in Gibberella zeae (anamorph: Fusarium graminearum) In: Brown D, Proctor RH, editors. Fusarium genomics and molecular and cellular biology. Horizon Scientific Press; Norwich: 2011. [Google Scholar]
- 11.Pomraning KR, Smith KM, Freitag M. Genome-wide high throughput analysis of DNA methylation in eukaryotes. Methods. 2009;47:142–150. doi: 10.1016/j.ymeth.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 12.Quail MA, Kozarewa I, Smith F, et al. A large genome center's improvements to the Illumina sequencing system. Nat Methods. 2008;5:1005–1010. doi: 10.1038/nmeth.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cronn R, Liston A, Parks M, et al. Multiplex sequencing of plant chloroplast genomes using Solexa sequencing-by-synthesis technology. Nucleic Acids Res. 2008;36:e122. doi: 10.1093/nar/gkn502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Langmead B, Trapnell C, Pop M, et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin Y, Li J, Shen H, et al. Comparative studies of de novo assembly tools for next-generation sequencing technologies. Bioinformatics. 2011;27:2031–2037. doi: 10.1093/bioinformatics/btr319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simpson JT, Wong K, Jackman SD, et al. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Filichkin SA, Priest HD, Givan SA, et al. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 2010;20:45–58. doi: 10.1101/gr.093302.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bryant DW, Jr, Wong WK, Mockler TC. QSRA: a quality-value guided de novo short read assembler. BMC Bioinformatics. 2009;10:69. doi: 10.1186/1471-2105-10-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li R, Zhu H, Ruan J, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010;20:265–272. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boisvert S, Laviolette F, Corbeil J. Ray: simultaneous assembly of reads from a mix of high-throughput sequencing technologies. J Comput Biol. 2010;17:1519–1533. doi: 10.1089/cmb.2009.0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li R, Li Y, Kristiansen K, et al. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- 24.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fahlgren N, Sullivan CM, Kasschau KD, et al. Computational and analytical framework for small RNA profiling by high-throughput sequencing. RNA. 2009;15:992–1002. doi: 10.1261/rna.1473809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lunter G, Goodson M. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 2011;21:936–939. doi: 10.1101/gr.111120.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Handsaker B, Wysoker A, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith KM, Phatale PA, Sullivan CM, et al. Heterochromatin is required for normal distribution of Neurospora CenH3. Mol Cell Biol. 2011;31:2528–2542. doi: 10.1128/MCB.01285-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith KM, Sancar G, Dekhang R, et al. Transcription factors in light and circadian clock signaling networks revealed by genomewide mapping of direct targets for Neurospora White Collar Complex. Eukaryot Cell. 2010;9:1549–1556. doi: 10.1128/EC.00154-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, Liu T, Meyer CA, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zang C, Schones DE, Zeng C, et al. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song Q, Smith AD. Identifying dispersed epigenomic domains from ChIP-Seq data. Bioinformatics. 2011;27:870–871. doi: 10.1093/bioinformatics/btr030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh D, Orellana CF, Hu Y, et al. FDM: a graph-based statistical method to detect differential transcription using RNA-seq data. Bioinformatics. 2011;27:2633–2640. doi: 10.1093/bioinformatics/btr458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cumbie JS, Kimbrel JA, Di Y, et al. GENE-counter: a computational pipeline for the analysis of RNA-Seq data for gene expression differences. PLoS One. 2011;6:e25279. doi: 10.1371/journal.pone.0025279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aird D, Ross MG, Chen WS, et al. Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol. 2011;12:R18. doi: 10.1186/gb-2011-12-2-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]