Abstract
Diets enriched in n-3 polyunsaturated fatty acids (n-3 PUFA) may protect against breast cancer but biochemical mechanisms are unclear. Our studies showed that the n-3 fatty acid, docosahexaenoic acid (DHA) up-regulated syndecan-1 (SDC-1) in human breast cancer cells and we tested the hypothesis that DHA-mediated up-regulation of SDC-1 induces apoptosis. DHA was delivered to MCF-7 cells by n-3 PUFA-enriched low density lipoproteins (LDL) or by albumin in the presence or absence of SDC-1 siRNA. The n-3 PUFA induced apoptosis, which was blocked by SDC-1 silencing. We also confirmed that SDC-1 up-regulation and apoptosis promotion by n-3 PUFA was mediated by PPARγ. Using a luciferase gene driven either by a PPAR response element (PPRE) or a DR-1 site present in the SDC-1 promoter, reporter activities were enhanced by n-3 LDL, DHA and PPARγ agonist, whereas activity of a luciferase gene placed downstream of a mutant DR-1 site was unresponsive. Co-transfection with dominant negative PPARγ DNA eliminated the increase in luciferase activity. These data provide strong evidence that SDC-1 is a molecular target of n-3 PUFA in human breast cancer cells through activation of PPARγ, and that n-3 PUFA-induced apoptosis is mediated by SDC-1. This provides a novel mechanism for the chemo-preventive effects of n-3 PUFA in breast cancer.
Introduction
Although still controversial, a number of human epidemiological studies have shown that a high dietary intake of fish is associated with a lower incidence of cancers including breast cancer (1–3). The omega-3 polyunsaturated fatty acids (n-3 PUFA) contained in fish oil are proposed to be the effectors of this protection. The two main PUFA constituents are eicosapentaenoic acid (EPA; 20:5n-3) and docosahexaenoic acid (DHA; 22:6n-3) which are converted by fish from alpha linolenic acid (αLNA; 18:3n-3) of ingested cold-water vegetation. Animal studies clearly support the idea that dietary supplementation with fish oil or its constituent fatty acids not only slows the growth of both xenograft (4, 5) and chemically-induced tumors (6, 7) but also increases sensitivity to chemotherapy (8, 9) and inhibits metastases (4, 5). DHA in particular was shown to be a potent enhancer of tumor cell chemosensitivity (10, 11). Biochemical mechanisms for the anti-tumor properties of n-3 PUFA are still unclear, but may include changes in eicosanoid metabolism, reduction in oncogene expression and modulation of lipid peroxidation [reviewed in (12, 13)].
It is known that fatty acids can regulate gene transcription. We have previously reported that the transmembrane heparan sulfate proteoglycan, syndecan-1 (SDC-1) is up-regulated by n-3 PUFA (14). SDC-1 has been shown to act as a tumor suppressor molecule by inhibition of cell growth and induction of apoptosis (15, 16). Proteolytic cleavage of the SDC-1 core protein at the cell surface generates a soluble ectodomain that retains anti-growth properties that depend on the presence of both core protein and heparan sulfate glycosaminoglycan chains (15, 16). Growth inhibition has been shown to require close proximity of SDC-1 ectodomain to the cell surface and to involve delay of cell G1-S progression and downregulation of cyclin D1 (16). Although there are inconsistent reports (17–19), the expression of SDC-1 is generally down-regulated in malignant tumors (20, 21), and lower levels of expression have been associated with high metastatic potential in malignant mesothelioma (22), invasive esophageal cancer (23), head and neck cancer (24), lung (25) and laryngeal cancer (26). SDC-1 expression was markedly reduced in squamous cell carcinoma and there was an inverse relationship between the level of SDC-1 expression and tumor aggressiveness (27). Furthermore, survival rate was decreased in patients with SDC-1 negative colorectal cancer (28) and was inversely associated with SDC-1 levels in gastric cancer (29). In vitro studies have shown that SDC-1 inhibits the invasion of tumor cells into type 1 collagen (30, 31) and that expression of SDC-1 in myeloma cells suppresses matrix metalloproteinase-9 (32). A reduction in SDC-1 promoted normal murine mammary epithelial-mesenchymal transition (31). Therefore, the decrease of SDC-1 expression may be an important step from tumorigenesis to the metastatic phenotype. However, there is little information on factors that regulate SDC-1.
Insight into a potential mechanism underlying the effects of n-3 PUFA on SDC-1 gene expression was suggested by the finding that the proximal promoter of the SDC-1 gene contains a specific sequence termed a DR-1 element (33). This sequence is recognized by several members of the nuclear hormone receptor superfamily including peroxisome proliferator-activated receptors (PPARs) that participate in activating the basic transcriptional machinery to regulate target genes. Fatty acids are known to bind and activate PPARs (34–36). There are three PPAR subtypes (α, β/δ, γ) with different tissue distributions and physiological involvements. Processes such as lipid and glucose homeostasis, inflammation, cell growth and differentiation, and apoptosis have all been shown as regulatory targets for the PPARs [reviewed in (37, 38)]. In human breast cancer cell lines the activation of PPARα (39) as well as PPARβ/δ (40) stimulated proliferation whereas ligands for PPARγ were growth inhibitory (41, 42).
In the human breast cancer cell line, MCF-7, n-3 PUFA not only inhibited cell proliferation and increased apoptosis but also promoted SDC-1 expression (14, 43). The SDC-1 response was mimicked by the PPARγ agonist, troglitazone and blocked by a PPARγ antagonist. Present studies were undertaken to demonstrate a novel pathway connecting n-3 PUFA, PPARγ regulation of SDC-1 and apoptosis. An important consideration in these studies was the delivery of n-3 PUFA to the cells by a method designed to model the in vivo process. Tumor cells obtain dietary PUFA by two major pathways: uptake of FA-albumin complexes that form after lipolysis of triglyceride-rich lipoproteins; and receptor mediated endocytosis of low density lipoproteins (LDL). Therefore, we have used LDL isolated from the plasma of monkeys fed a diet supplemented with fish oil to examine a mechanism for n-3 PUFA induced apoptosis.
Materials and Methods
Preparation of LDL and FA-bovine serum albumin (BSA) complexes
LDL were isolated from plasma of adult African Green (vervet) monkeys fed n-3 PUFA (fish oil) -enriched diets for 3–5 years. Diets, maintenance and clinical measurements of the animals are published (43). Animal care and procedures were approved by the Animal Care and Use Committee of Wake Forest University School of Medicine and animals were maintained by standard animal care procedures based on institutional guidelines. LDL isolation and characterization procedures have been previously described (43). To measure EPA and DHA concentrations in n-3 LDL preparations, 100 µl of LDL was spiked with 20.6 µg C17:0 (as Triheptadecanoate glyceride), then saponified, extracted and derivatized by standard procedures. LDL FA methyl esters were quantified by gas-liquid chromatography and EPA and DHA concentrations were calculated relative to C17:0. Based on these measurements, in all experiments when cells were treated with 100 µg/ml n-3 LDL, they were exposed to final concentrations of 52 µM EPA and 21µM DHA from the LDL. EPA and DHA were purchased as sodium salts (Sigma Chemical Company, St Louis, MO). FA-free BSA (Sigma) was prepared as a 125µM solution in Dulbecco’s modified Eagle medium (DMEM)/Ham’s F-12. FA salts were solublized to 600µM stocks in the BSA-media as described (4:1 ratio of FA:BSA), and stored in aliquots at −20 °C under argon (43). MS/MS analysis of the stock preparation of DHA showed <15% peroxidation products. With regard to DHA stability, over a 48 hour treatment of cells, the DHA (as percent of cell phospholipids) increased from 2.0 (time 0) to 3.7 (12 hours), to 5.7 (24 hours), and 3.5 (48 hours) indicating reasonable persistence over the time periods studied.
Preparation of SDC-1 ectodomain
The SDC-1 ectodomain expression construct was designed to encode a C-terminal polyhistidine fusion protein and was created using a two-step cloning process as follows. The SDC-1 ectodomain cDNA was amplified by PCR using the SDC-1 plasmid (OriGene Technologies, Inc., Rockville, MD) as the source of template, 5’-gcagaattcggcagcatgaggcgcgcggcgctct-3’ as the forward primer and 5’-gcaggatcctttcctgtccaggaggccctgtga-3’ as the reverse primer. The resulting cDNA was cut with BamHI and EcoRI restriction endonucleases and ligated into the pTcam4 expression plasmid (44). In the second round of PCR, 5’-gcaaagcttgaattcggcagcatgaggcgcgcg-3’ (forward) and 5’-gcaaagcttgaattcggcagcatgaggcgcgcg-3’ (reverse) primers were used to amplify the SDC-1 ectodomain cDNA from the modified pTcam4 plasmid. The amplified cDNA was treated with HindIII and XhoI and subsequently ligated into the pCEP4 (Invitrogen, Corp., Carlsbad, CA) expression plasmid. The resulting plasmid was transfected into 293-EBNA cells and stable expressing cells selected by the addition of 200 µg/ml hygromycin B. The cells were then amplified and grown to confluence in T175 flasks. Upon confluence, the culture media was replaced with serum-free DMEM. Media was then collected every 48 h and fresh media added back to the cells. Under these conditions, the transfected cells remain viable for several weeks allowing for the collection of conditioned media over an extended period of time. After collecting several liters of conditioned media, recombinant SDC-1 ectodomain was purified using the following procedure. Conditioned media was first concentrated using a Millipore Pellicon 2 tangential flow system (Millipore Corporation, Bedford, MA). Concentrated conditioned media was then subjected to anion exchange chromatography using an Amersham Biosciences ÄKTAexplorer chromatography system equipped with a 5 ml HiTrap Q column and bound SDC-1 ectodomain eluted with a linear gradient of 0–2 M sodium chloride in a buffer consisting of 20 mM Tris, 0.2 % CHAPS, pH 8.0.
Cell Culture
MCF-7 cell line was obtained from ATCC (Rockville, MD) and maintained in DMEM/F12 supplemented with 5% fetal bovine serum (FBS), 10mg/ml porcine insulin (Sigma), penicillin/streptomycin and L-glutamine at 37 °C in 5 % CO2. In experiments measuring mRNA, cleaved PARP levels and PPARγ protein synthesis, cells were seeded in 6-well plates at a density of 5 × 105 cells/well in growth medium. After 6 h, the medium was changed to DMEM/F12 with 0.5 % FBS for 18 h to up-regulate LDL receptors. PPARγ protein was determined after media supplementation with 100 µg/ml n-3 LDL, 30 µM DHA, 30 µM EPA or 5µM troglitazone (Cayman Chemical, Ann Arbor, MI) for 12 and 48 h.
Apoptosis assays
MCF-7 cells were seeded in 6-well plates at a density of 5 × 105 cells/well in DMEM/F12 with 0.5 % FBS for 18 h, and then exposed to n-3 LDL, DHA, EPA or troglitazone for 48 h, or to SDC-1 ectodomain for 72 h. The SDC-1 siRNA-transfected cells were also exposed to n-3 LDL, DHA, or troglitazone for 48 h. Apoptotic activity in adherent cells was determined by cleaved PARP using Western blots. Alternatively, apoptosis was measured with the Caspase-Glo 3/7 ®Assay in which 30 µl of Caspase-Glo 3/7 reagent was added to each well and incubated for 1 h at room temperature. Luminescence was measured by a Reporter Microplate Luminometer (Turner Biosystems). Treated cells were also stained with trypan blue (Invitrogen, Carlsbad, CA), and percent of dead cells were counted by hemocytometer.
Real-time PCR
RNA isolation and cDNA synthesis and real-time RT-PCR were as previously described (14). SDC-1 primers were 5’-ggagcaggacttcacctttg (forward) and 5’-ctcccagcacctctttcct (reverse). Peptidyl prolyl isomerase B (PPIB) housekeeping gene primers were 5’-gcccaaagtcaccgtcaa (forward) and 5’-tccgaagagaccaaagatcac (reverse). Care was taken to design primers so as to minimize amplification from any contaminating genomic DNA. The forward SDC-1 primer spans an exon/intron junction. The PPIB primers are on different exons separated by a ∼800 bp intron. Under the conditions of real-time PCR, we detected a single PPIB product of 82 bp by gel electrophoresis, and melt curve analyses performed at the end of the real-time PCR reproducibly show a single peak. SDC-1 data were normalized to the housekeeping control PPIB and are presented relative to control.
Western blot analysis
Cells were washed twice with ice-cold PBS and lysed for 10 min on ice; debris was then removed by centrifugation and equivalent amounts of protein were separated by 10 % SDS-PAGE and transferred onto PVDF membrane. The membranes were blocked with TBST (10 mM Tris-base, 100 mM NaCl, 0.1 % Tween 20, pH7.5) containing 5 % nonfat dry milk for 2 h at room temperature and then were washed three times with TBST for 5 min, and exposed to anti-PARP (Cell Signaling Technology, Beverly, MA), and anti-PPARγ (Abcam, Cambridge, UK) antibodies in TBST containing 3 % BSA at 4 °C overnight, followed by three washes with TBST, and were then incubated with horseradish peroxidase-conjugated secondary antibody for 1 hour, washed with TBST and developed using the Super Signal West Pico Kit (Super Signal West Pico, Pierce, Rockford, IL). The band densities on photographic films were analyzed using Quantity One software. For PARP cleavage measurement, band densities were normalized to β-actin and presented as the fractional change from control values.
SDC-1 siRNA transfection
Three SDC-1 siRNAs, #12432, 12527 and 142557 were purchased from Ambion. For transfection, 50 nM siRNA was added to 2.0 × 105 cells in 6-well plates using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) and DMEM/F12 medium lacking serum and antibiotics. Control cells were transfected with a negative control siRNA with no known mRNA target designed by Ambion (AM4624). At 6 h after transfection, each well was supplemented with 2 ml of complete growth medium. Forty eight h after transfection the medium was replaced with 2 ml of DMEM/F12 medium containing 0.5 % FBS and cells were treated with n-3 LDL (100 µg/ml), DHA (30 µM) and troglitazone (5 µM) for an additional 48 h. Cells were prepared for Western blot analysis as described above. Data are shown using SDC-1 siRNAs # 142557. Similar results were obtained with #12432, whereas #12527 failed to silence SDC-1 mRNA and did not inhibit n-3 PUFA-induced apoptosis.
Wild-type and mutant SDC-1 DR-1-luciferase constructs
The one copy reporter construct DR-1 from the SDC-1 promoter [pGL3-DR-1 wt] (33) was generated by annealing the oligonucleotides 5’-cggttcttcccctttgctctctcggccgtttccgctacacccgagct-3’ and 5’-cgggtgtagcggaaacggccgagagagcaaaggggaagaaccggtac-3’ before ligation into KpnI and SacI digested pGL3-promoter vector (Promega). The one copy mutant DR-1 reporter construct (pGL3-DR-1) was generated using the same method and the oligonucleotides 5’-cggttctAGTActGCcCTcactcggccgtttccgctacacccgagct-3’ and 5’-cgggtgtagcggaaacggccgagTgagGGCagTACTagaaccggtac-3’. Mutations are capitalized.
Transfection and luciferase assay
MCF-7 cells were co-transfected with the PPRE-TK-luciferase reporter plasmid and a control plasmid containing the lacZ gene or with pcDNA3-d/n PPARγ cDNA (L468/E471, kindly provided by Dr. V Chattergee, University of Cambridge, UK) and pGL3-luc-DR-1 (wild-type or mutant) using FuGENE 6 Transfection Reagent (Roche Diagnostics, Indianapolis, IN) according to manufacturer’s instructions. Transfected cells were incubated with 0, 100 µg/ml n-3 LDL, 30 µM DHA, 5 or 10 µM troglitazone for 8 or 24 h. Luciferase activities were measured using a Luciferase Assay Kit (Promega, Madison, WI) in a TD-20e luminometer (Turner Biosystems, Sunnyvale, CA). β-galactosidase activity was measured using β-Galactosidase Enzyme Assay Kit (Promega, Madison, WI) in SpectroMAX. Luciferase activities were normalized to β-galactosidase activity and are presented as the percentage of luciferase activity measured in the presence of stimulus, relative to the activity of control cells with no stimulation.
Data analysis
Data were analyzed by ANOVA and student’s t-test. The assays were carried out in triplicate and the averages are shown, together with SE.
Results
DHA induces apoptosis in MCF-7 cells
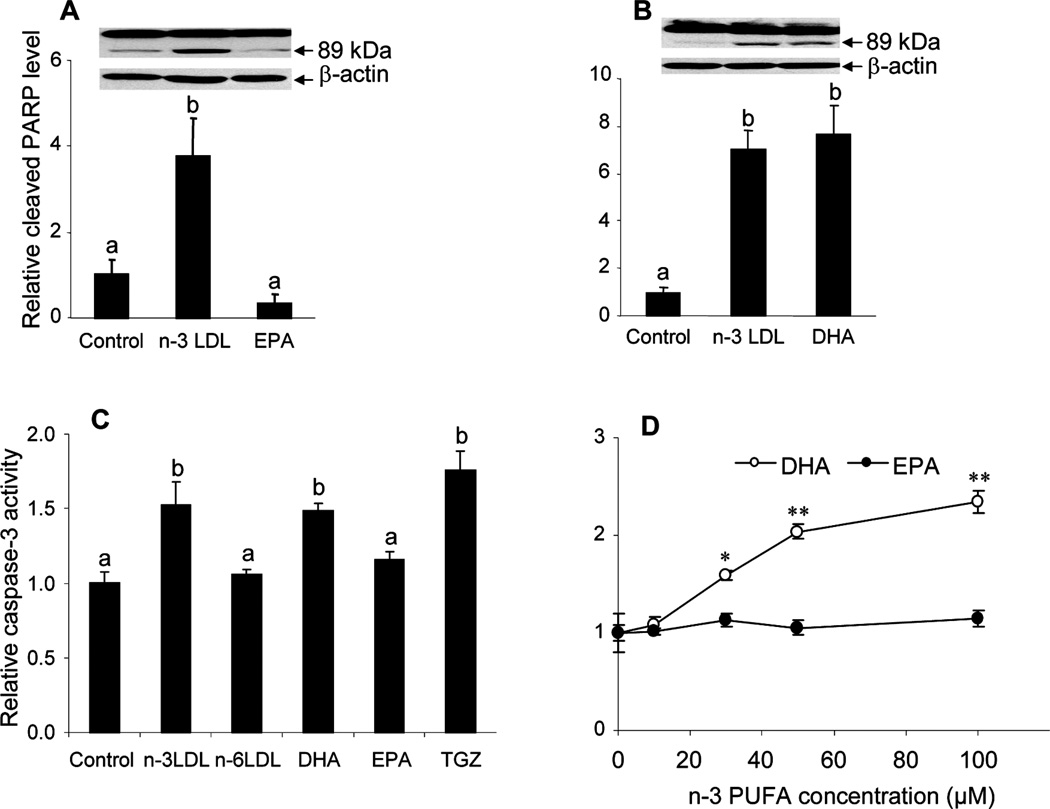
We have previously shown that n-3 PUFA-enriched LDL, but not n-6 PUFA -enriched LDL can induce apoptosis in MCF-7 cells (43). The n-3 LDL were shown to contain two predominant species of n-3 PUFA: EPA and DHA, and both were effectively delivered to the cells by the LDL. To clarify the role of the individual n-3 PUFA in apoptosis induction, MCF-7 cells were incubated with n-3 LDL, EPA-BSA or DHA-BSA for 48 h and apoptosis was measured by PARP cleavage. Consistent with previous observations, n-3 LDL caused a 4–7-fold increase in PARP cleavage product compared to control cells (figure 1A, B). EPA-BSA was not effective in induction of apoptosis (figure 1A) whereas a similar concentration of DHA-BSA resulted in an increase in PARP cleavage similar to that of n-3 LDL (figure 1B). Apoptosis was confirmed in cells treated with n-3 LDL and DHA by Caspase 3/7 -Glo® assay (figure 1C, D). As shown in Figure 1D, apoptosis induction by DHA was dose dependent, whereas no apoptotic effect was observed with EPA at similar doses for 48 h, or in cells treated for up to 72 h (not shown).
Figure 1. Effects of n-3 PUFA on apoptosis of MCF-7 cells.
Cells were pre-incubated in media containing 0.5 % FBS for 18 h and then media was supplemented with n-3 LDL (100 µg/ml), DHA or EPA (30 µM FA, 7.5 µM BSA) for 48 h. A and B. Measurement of apoptosis by Western blot identifying the 89 kDa PARP cleavage product and graphical presentation of intensity of 89 kDa band relative to β–actin. C, D. Measurement of apoptosis by Caspase 3/7-Glo® assay in MCF-7 cells treated for 48 h with n-3 LDL, n-6 LDL, DHA-BSA or EPA-BSA. In D, the 4:1 molar ratio of FA to BSA was maintained for each dose. Data are means ± S.E. of experiments performed in triplicate. Values with different letters are significantly different (P < 0.05), *P< 0.05, **P< 0.001.
SDC-1 induces apoptosis in MCF-7 cells
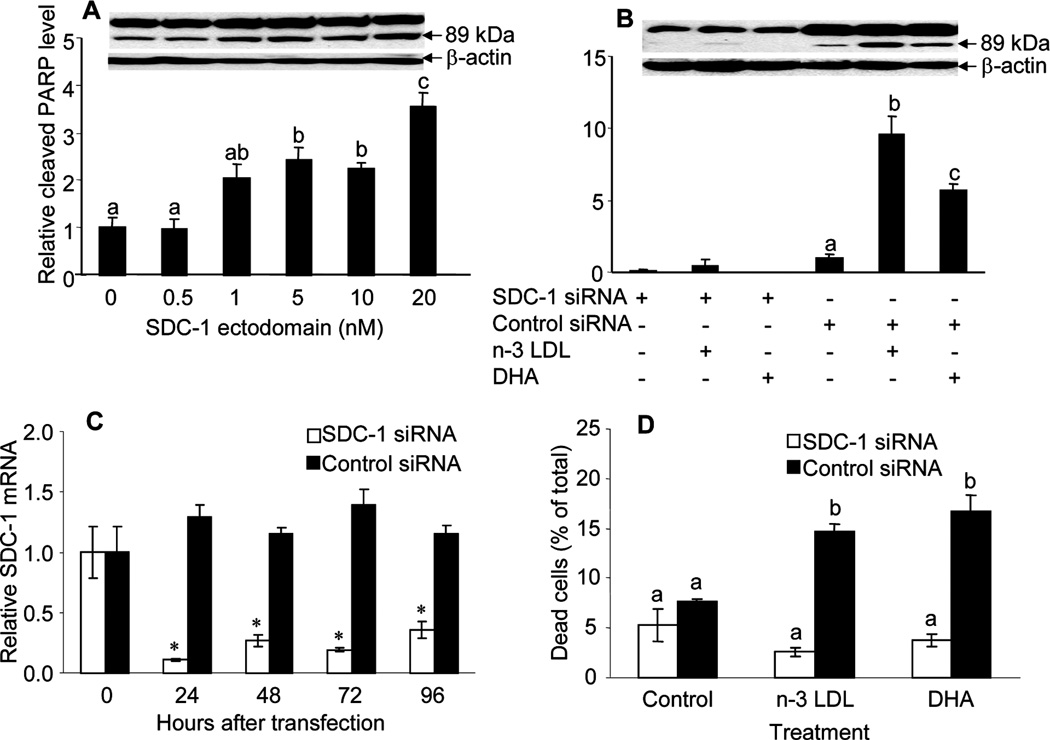
Consistent with the effect on apoptosis demonstrated in figure 1, previous studies have shown that both n-3 LDL and DHA-BSA significantly up-regulated the expression of SDC-1 in MCF-7 cells (14). To test the hypothesis that n-3 PUFA-induced apoptosis in MCF-7 cells is mediated by SDC-1, studies were first conducted to demonstrate that SDC-1 is a pro-apoptotic stimulant in this model system. Cells were cultured for 72 h in the presence of human recombinant SDC-1 ectodomain which resulted, as shown in figure 2A, in a dose dependent increase in apoptosis of the cells as measured by PARP cleavage. To confirm that SDC-1 is a mediator in n-3 LDL and DHA-induced apoptosis of MCF-7 cells, a SDC-1 siRNA was transfected into the cells to silence SDC-1 expression. This reagent effectively inhibited SDC-1 mRNA for up to 96 h after transfection (figure 2C). As shown in figure 2B, the SDC-1 siRNA blocked the ability of n-3 LDL and DHA to enhance PARP cleavage which was markedly stimulated in cells transfected with a control siRNA. It was also apparent that the level of intact PARP was different in cells treated with SDC-1 and control siRNA but further studies are required to determine the significance of this. In any case, the absolute as well as relative levels of PARP cleavage stimulated by n-3 PUFA were blocked by SDC-1 silencing and the ratio of cleaved to intact PARP was increased by n-3 LDL and DHA only in the presence of the non-targeting siRNA (0.26 ± 0.06 and 0.24 ± 0.05 for control cells, 0.52 ± 0.07 and 0.27 ± 0.05, p= 0.03 for n-3 LDL treated cells, 0.44 ± 0.06 and 0.13 ± 0.07, p= 0.02 for DHA-treated cells in the presence of non-targeting and SDC-1 siRNA respectively) Furthermore, a trypan blue exclusion assay confirmed that the SDC-1 siRNA inhibited n-3 PUFA-induced cell death (figure 2D).
Figure 2. SDC-1 induces apoptosis of MCF-7 cells.
A. Cells were incubated in media containing indicated concentrations of SDC-1 ectodomain for 72 h. B, Cells were transfected with SDC-1 siRNA or negative control siRNA for 48 h and then incubated with n-3 LDL (100 µg/ml) and DHA (30 µM FA, 7.5 µM BSA) for 48 h. Apoptosis was measured by PARP Western analysis (B). Data are presented relative to control and are means ±S.E. of three independent experiments performed in triplicate. C. MCF-7 cells were transfected with SDC-1 siRNA or negative control siRNA for the indicated times and SDC-1 mRNA was measured by real time RT-PCR. D. Cells transfected with SDC-1 siRNA or negative control siRNA were assayed for percentage cell death by trypan blue exclusion assay. Values with different letters are significantly different (P < 0.05), * P < 0.05.
PPARγ is a key factor in DHA or n-3 LDL up-regulation of SDC-1 expression
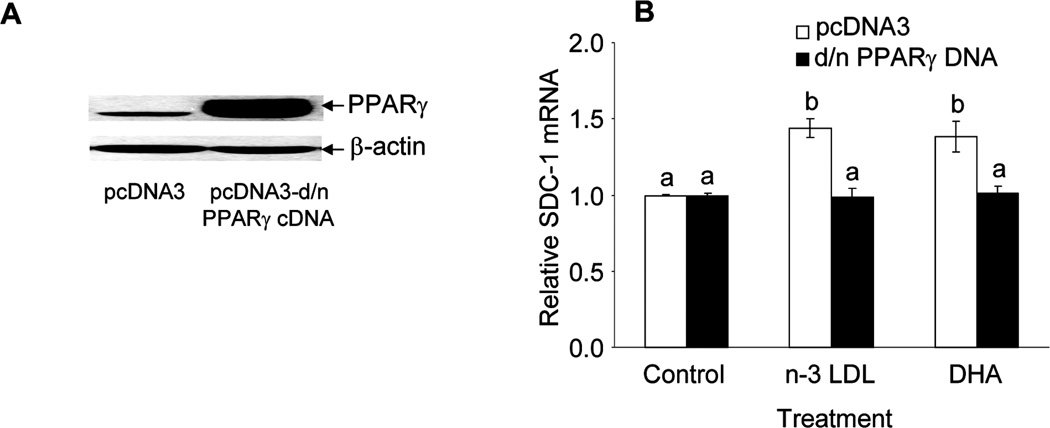
In previous studies we have shown that the PPARγ agonist troglitazone was an effective stimulator of SDC-1 expression, whereas the PPARδ agonist GW610742 had no effect. Moreover, the PPARγ antagonist GW259662 blocked the effect of n-3 LDL on SDC-1 expression (14). To directly implicate PPARγ in n-3 LDL and DHA regulation of SDC-1, MCF-7 cells were transfected with a d/n-PPARγ cDNA or its vector pcDNA3. Figure 3A shows a marked overexpression of PPARγ protein in the transfected cells. We then assessed the activity of the d/n- PPARγ construct by determining SDC-1 mRNA level after stimulation with n-3 LDL and DHA for 8 h (figure 3B). In cells transfected with the empty vector, both n-3 LDL and DHA up-regulated SDC-1 expression. No such response occurred in cells transfected with the vector encoding d/n- PPARγ cDNA. These data indicate that n-3 LDL and DHA up-regulated SDC-1 expression through PPARγ.
Figure 3. Dominant/negative PPARγ cDNA blocks the enhancement effect of n-3 LDL and DHA on SDC-1 expression.
MCF-7 cells were transfected with pcDNA3-d/n PPARγ cDNA or empty d/n PPARγ cDNA vector (pcDNA3) and (A) PPARγ protein was measured at 12 h by Western analysis. B. Transfected cells were treated with n-3 LDL (100 µg/ml) or DHA-BSA (30µ M FA, 7.5 µM BSA) for 8 h. Total RNA was isolated and SDC-1 mRNA was determined by real-time RT-PCR. Data are means ± S.E in triplicate experiments. Values with different letters are significantly different (P < 0.05).
PPARγ activation promotes apoptosis in MCF-7 cells
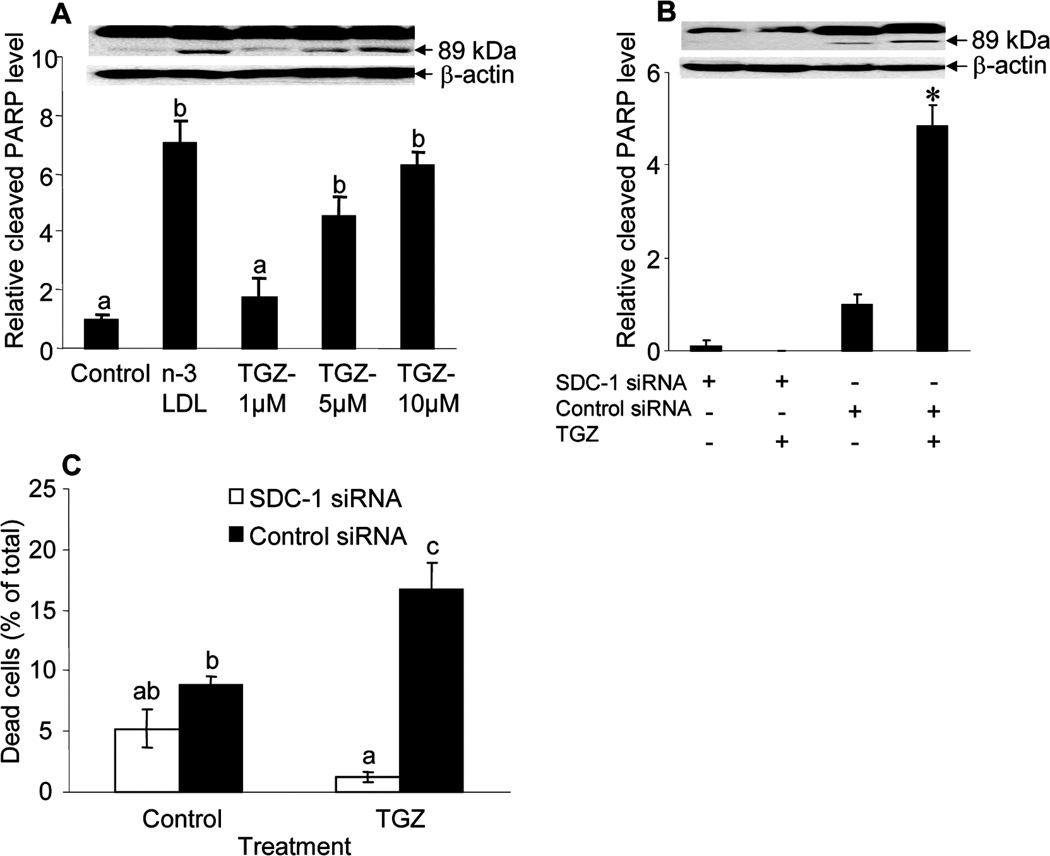
The observations thus far have indicated that n-3 LDL and DHA induce apoptosis of MCF-7 cells through up-regulation of SDC-1 in a PPARγ- activated transcriptional pathway. The next question addressed the ability of PPARγ to induce apoptosis in these cells. To examine this, cells were treated with the PPARγ agonist troglitazone and cleaved PARP was measured by Western blot analysis. As shown in figure 4A, a dose-dependent increase in cleavage product was observed in response to troglitazone treatment. Thus the PPARγ ligand has a similar effect to n-3 PUFA in promoting apoptosis of the breast cancer cells. To explore whether the activated PPARγ induced apoptosis of MCF-7 cells through SDC-1, the cells were transfected with SDC-1 or control siRNA and then were cultured in the presence of 5 µM troglitazone for 48 h (figure 4B, C). The apoptotic effect of troglitazone was inhibited when SDC-1 expression was silenced. These data indicate a critical involvement of SDC-1 in the promotion of apoptosis by n-3 PUFA, troglitazone, and the common target of these agents, PPARγ.
Figure 4. PPARγ-induced apoptosis of MCF-7 cells is eliminated by SDC-1 siRNA.
A. Cells were treated with 100 µg/ml of n-3 LDL and 1, 5 or 10 µM of PPARγ agonist troglitazone for 48 h, and apoptosis was measured by PARP Western analysis. B, C. Cells transfected with SDC-1 siRNA or negative control siRNA were incubated with 5 µM troglitazone for 48 h, and cleaved PARP levels were compared by Western analysis (B) or percentage cell death was measured by trypan blue exclusion assay (C). Data are means ± S.E. in triplicate experiments. Values with different letters are significantly different (P < 0.05), *P <0.05.
PPARγ is activated by n-3 PUFA and regulates SDC-1 transcription via the DR-1 site
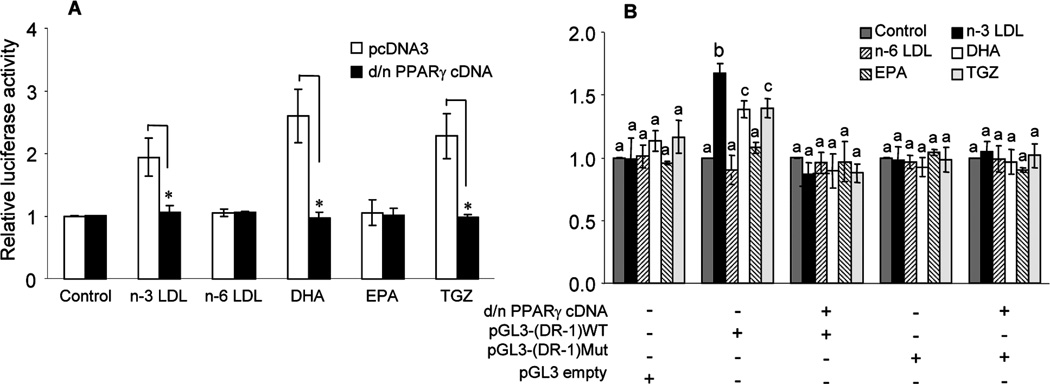
To further clarify the mechanism whereby the PPARγ pathway mediates n-3 PUFA up-regulation of SDC-1, effects on PPARγ expression were examined. Treatment with n-3 LDL and DHA had no effect on PPARγ mRNA level in the cells and furthermore, Western blot analysis showed that the level of PPARγ protein expression did not differ between control and n-3 PUFA treated cells or in cells treated with SDC-1 siRNA (not shown). Thus it appeared that activation rather than over-expression of PPARγ by n-3 PUFA resulted in up-regulation of SDC-1. To determine if n-3 LDL and DHA could activate a PPAR transcriptional activity, MCF-7 cells were co-transfected with a PPRE-TK-luciferase reporter plasmid and a control plasmid containing the lacZ gene. Luciferase activity was determined after treatment with n-3 LDL or n-6 LDL (100µg/ml), DHA or EPA (30µM) or troglitazone (5µM) as a positive control. DHA, n-3 LDL and troglitazone significantly increased luciferase activity indicating a PPAR response that was blocked by co-transfection with a d/n-PPARγ cDNA (figure 5A). To confirm the role of PPARγ in transcriptional regulation of SDC-1, the luciferase reporter gene was placed under the control of either the wild-type [pGL3-(DR-1) wt] or mutant [pGL3-(DR-1) mut] DR-1 element from the SDC-1 promoter. MCF-7 cells were co-transfected with wild-type or mutant SDC-1 DR-1-luciferase constructs and a d/n-PPARγ cDNA. The result showed that pGL3-(DR-1) wt-luciferase reporter was activated by n-3 LDL, DHA and troglitazone. Co-transfection of the same reporter with d/n-PPARγ DNA blocked luciferase activity (figure 5B). In addition, the transfected reporter gene containing mutant SDC-1 DR-1 site completely abolished the effect of n-3 LDL, DHA and troglitazone. No significant effect on luciferase activity was observed when the empty pGL3-promoter vector was transfected into MCF-7 cells and treated with n-3 LDL, DHA or troglitazone (figure 5B). These data establish SDC-1 as a molecular target for n-3 PUFA and PPARγ.
Figure 5. Activation of the human SDC-1 promoter by PPARγ activated by n-3 LDL or DHA.
A. Luciferase activity in MCF-7 cells co-transfected with luciferase driven by a PPAR response element (PPRE×3-tk-luciferase reporter) and a control plasmid containing the lacZ gene. Cells were transfected with pcDNA3-d/n PPARγ cDNA or empty d/n PPARγ cDNA vector (pcDNA3) and treated with n-3 LDL or n-6 LDL (100 µg/ml), DHA-BSA or EPA-BSA (30 µM FA, 7.5 µM BSA) or troglitazone (10 µM) and luciferase activity was measured at 24 h. B. Luciferase activity in MCF-7 cells co-transfected with a reporter construct containing the DR-1 from the SDC-1 promoter linked to a minimal promoter-luciferase reporter gene pGL3-(DR-1) wt or the same vector containing the mutant DR-1 [pGL3-(DR-1)mut], with or without the d/n PPARγ cDNA Following transfection, cells were treated with n-3 LDL or n-6 LDL (100 µg/ml), DHA-BSA or EPA-BSA (30 µM, 7.5 µM BSA) or troglitazone (10 µM) for 8 h. Luciferase activity values are normalized to β-galactosidase activity and presented as the percentage of luciferase activity measured in the presence of stimulus, relative to the activity of control cells with no stimulation. Basal activity was reduced approximately 2 fold by the d/n-PPARγ cDNA, but not changed by mutant DR-1. Values with different letters are significantly different (P < 0.05), *P <0.001.
Discussion
These studies have identified a novel pathway for the chemo-preventive properties of n-3 PUFA in human breast cancer cells. The most striking finding is that SDC-1, a key molecular target of n-3 LDL and DHA, is a critical player in the ability of n-3 PUFA to induce apoptosis in these cells. When SDC-1 expression was silenced by siRNA, the ability of n-3 PUFA to induce apoptosis was blocked. In addition, present studies have extended previous observations (14) by more clearly defining the role of PPARγ in the n-3 PUFA up-regulation of SDC-1 expression. The studies reported here delineate the pathway linking n-3 PUFA to cellular apoptosis as follows: the n-3 PUFA, DHA activates the nuclear transcription factor, PPARγ, which results in transcriptional up-regulation of the SDC-1 target gene, and the SDC-1 protein induces cell apoptosis.
Throughout these studies, we modeled two physiological processes of fatty acid delivery to the cells: by LDL and by albumin. Our previous studies have shown that LDL, isolated from the plasma of vervet monkeys fed a diet enriched in fish oil, contains two major n-3 PUFA species: EPA and DHA (43). Incubation of MCF-7 cells with this LDL showed that both EPA and DHA were efficiently delivered to the cells and detectable in cell phospholipids by 24 h (43) and as early as 8 h1. Proliferation of MCF-7 cells was inhibited by n-3 LDL, which also induced apoptosis in the cells (43). Neither of these effects was achieved by n-6 PUFA-enriched LDL. What was puzzling was that EPA-BSA was ineffective in apoptosis induction, a result confirmed by present studies. Evidence now indicates that by contrast, DHA-BSA is very effective in induction of apoptosis and that DHA is likely to be the apoptotic stimulus in the n-3 LDL.
DHA is synthesized from EPA by elongation and desaturation. However the conversion is not a straightforward one [for review, see (45)]. Specifically, EPA (20:5, n-3) is converted to docosapentaenoic acid (22:5, n-3) by elongase (Elovl)-5, and by Elovl-2 to 24:5, n-3. The next step requires desaturation of the 24:5 at the Δ6 position to produce 24:6, n-3. This product is translocated from the endoplasmic reticulum to the peroxisome where the β oxidation pathway exerts an acyl chain shortening of C2 to produce DHA (22:6, n-3). Although our previous studies have shown that there is some conversion of EPA to DHA in the MCF-7 cells, the inability of EPA to induce apoptosis suggests either that the pool of DHA derived from EPA may be less available to produce an apoptotic response in the cell or that a critical threshold level of DHA needed for activity cannot be achieved by this pathway. Our recent unpublished studies have shown that over a 48 hour treatment with EPA or DHA, the percent DHA in MCF-7 cell lipids changes from 1.8 at time 0 to 2.1 and 3.7 respectively at 48 h. It may be important to note that in humans, studies have consistently demonstrated that increased consumption of either EPA or its precursor α linolenic acid (18:3, n-3) does not result in a detectable increase in plasma DHA, thus highlighting the inefficiency of conversion in this pathway (46–48). Further studies are ongoing to clarify the metabolism of n-3 PUFA in relation to apoptosis in these cells. One likely possibility is that DHA is metabolized to a more active component that becomes a key player in the pathway that we have identified.
We specifically demonstrated effects of n-3 LDL and DHA on the level and function of SDC-1. DHA and n-3 LDL significantly up-regulated expression of SDC-1 in MCF-7 cells whereas EPA was ineffective. To connect the up-regulation of SDC-1 with apoptosis induction we showed that exogenous human recombinant SDC-1 ectodomain stimulated apoptosis in a dose-dependent manner. This is distinct from studies of Mali et al. (15), that showed cell growth inhibition but did not demonstrate apoptosis with a similar reagent. To demonstrate that SDC-1 was the target gene of n-3 LDL or DHA that was responsible for the pro-apoptotic effect, SDC-1 siRNA was transfected into the cells. SDC-1 gene silencing blocked the ability of n-3 LDL and DHA to promote apoptosis thus confirming a role for SDC-1 in n-3 PUFA-induced apoptosis in the breast cancer cells.
The present study verified SDC-1 as a novel PPARγ target gene. We had previously shown that PPARγ agonist troglitazone, DHA and n-3 LDL were effective stimulators of SDC-1 mRNA expression, whereas PPARδ agonist GW610742 had no effect. A PPARγ antagonist, GW259662, blocked the effect of n-3 LDL on SDC-1 expression (14). To confirm and further clarify the involvement of PPARγ in n-3 LDL and DHA regulation of SDC-1, we first utilized a dominant negative (d/n) - PPARγ cDNA which effectively eliminated the up-regulation of SDC-1 mRNA by n-3 LDL and DHA. Secondly, studies were undertaken to show that whereas the n-3 PUFA had no detectable effects on PPARγ message and protein levels, a significant increase in PPARγ activity was measured by reporter assays. To obtain direct evidence for a coupling mechanism between n-3 PUFA and PPARγ in SDC-1 gene up-regulation, we constructed a luciferase reporter gene under the control of either the wild-type [pGL3-(DR-1) wt] or mutant [pGL3-(DR-1) mut] DR-1 element from the SDC-1 promoter, and investigated whether the activity of PPARγ was enhanced by n-3 PUFA to regulate expression of SDC-1. DHA, n-3 LDL and troglitazone significantly increased activity of luciferase in cells transfected with wild-type SDC-1 DR-1-luciferase genes. Co-transfection of the same reporter with d/n-PPARγ cDNA inhibited activity and the transfected reporter construct containing a mutant SDC-1 DR-1 site was not responsive to n-3 LDL, DHA and troglitazone. DHA, n-3 LDL and troglitazone had no effect on luciferase activity in MCF-7 cells transfected with empty pGL3-promoter vector. Anisfeld et al (33), using a similar strategy, identified SDC-1 as a target gene for the farnesoid-X receptor (FXR) in hepatocytes. Although FXR is not expressed in breast epithelial cells, our data are consistent with those studies since the DR-1 element in the proximal promoter of SDC-1 is recognized by several nuclear hormone receptors including the PPARs. PPARγ is known to be directly activated by fatty acids to regulate gene expression (49, 50); however, our study is the first to demonstrate the functional coupling of an n-3 PUFA and PPARγ in up-regulation of SDC-1 and thence SDC-1-dependent apoptosis.
PPARγ ligands have been shown to induce apoptosis in a variety of tumor cells including human breast cancer cells (41). Thus, the dose-dependent increase in apoptosis of MCF-7 cells caused by troglitazone was not unexpected. The important new finding from our studies was that transfection of the cells with SDC-1 siRNA blocked apoptosis induction by both troglitazone and DHA or n-3 LDL. This indicates that the target gene, SDC-1 is required for the pro-apoptotic activity of PPARγ and that n-3 LDL and DHA promote apoptosis through a SDC-1 -dependent pathway.
In summary, the data presented in this work highlight a novel pathway in which n-3 LDL or DHA promote apoptosis of human breast cancer cells. Further studies are needed to delineate the pathway by which SDC-1 promotes apoptosis of the MCF-7 cells and to identify other participants in this process.
Acknowledgments
Financial support: Grant 05A071 from the American Institute for Cancer Research (IJE), R01CA115958 (IJE), P01CA106742 (IJE, JO’F) and R01CA114017 (IMB) from the National Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Abbreviations
- DHA
docosahexaenoic acid
- PPAR
Peroxisome Proliferator-activated Receptor
- d/n-PPARγ DNA
dominant/negative-PPARγ DNA
- EPA
eicosapentaenoic acid
- FA
fatty acid
- LDL
low density lipoproteins
- PARP
poly(ADP-ribose) polymerase
- PPRE
PPAR-response elements
- PUFA
polyunsaturated fatty acids
- SDC-1
syndecan-1
- TGZ
troglitazone
- BSA
bovine serum albumin
Footnotes
Sun H and Edwards IJ. Unpublished data.
References
- 1.Sasaki S, Horacsek M, Kesteloot H. An ecological study of the relationship between dietary fat intake and breast cancer mortality. Prev Med. 1993;22:187–202. doi: 10.1006/pmed.1993.1016. [DOI] [PubMed] [Google Scholar]
- 2.Kaizer L, Boyd NF, Kriukov V, Tritchler D. Fish consumption and breast cancer risk: an ecological study. Nutr Cancer. 1989;12:61–68. doi: 10.1080/01635588909514002. [DOI] [PubMed] [Google Scholar]
- 3.Hursting SD, Thornquist M, Henderson MM. Types of dietary fat and the incidence of cancer at five sites. Prev Med. 1990;19:242–253. doi: 10.1016/0091-7435(90)90025-f. [DOI] [PubMed] [Google Scholar]
- 4.Rose DP, Connolly JM. Effects of dietary omega-3 fatty acids on human breast cancer growth and metastases in nude mice. J Natl Cancer Inst. 1993;85:1743–1747. doi: 10.1093/jnci/85.21.1743. [DOI] [PubMed] [Google Scholar]
- 5.Rose DP, Connolly JM, Rayburn J, Coleman M. Influence of diets containing eicosapentaenoic or docosahexaenoic acid on growth and metastasis of breast cancer cells in nude mice. J Natl Cancer Inst. 1995;87:587–592. doi: 10.1093/jnci/87.8.587. [DOI] [PubMed] [Google Scholar]
- 6.Jurkowski JJ, Cave WT., Jr Dietary effects of menhaden oil on the growth and membrane lipid composition of rat mammary tumors. J Natl Cancer Inst. 1985;74:1145–1150. [PubMed] [Google Scholar]
- 7.Braden LM, Carroll KK. Dietary polyunsaturated fat in relation to mammary carcinogenesis in rats. Lipids. 1986;21:285–288. doi: 10.1007/BF02536414. [DOI] [PubMed] [Google Scholar]
- 8.Hardman WE, Avula CP, Fernandes G, Cameron IL. Three percent dietary fish oil concentrate increased efficacy of doxorubicin against MDA-MB 231 breast cancer xenografts. Clin Cancer Res. 2001;7:2041–2049. [PubMed] [Google Scholar]
- 9.Shao Y, Pardini L, Pardini RS. Dietary menhaden oil enhances mitomycin C antitumor activity toward human mammary carcinoma MX-1. Lipids. 1995;30:1035–1045. doi: 10.1007/BF02536289. [DOI] [PubMed] [Google Scholar]
- 10.Burns CP, North JA. Adriamycin transport and sensitivity in fatty acid-modified leukemia cells. Biochim Biophys Acta. 1986;888:10–17. doi: 10.1016/0167-4889(86)90064-9. [DOI] [PubMed] [Google Scholar]
- 11.Germain E, Chajes V, Cognault S, Lhuillery C, Bougnoux P. Enhancement of doxorubicin cytotoxicity by polyunsaturated fatty acids in the human breast tumor cell line MDA-MB-231: relationship to lipid peroxidation. Int J Cancer. 1998;75:578–583. doi: 10.1002/(sici)1097-0215(19980209)75:4<578::aid-ijc14>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 12.Hardman WE. Omega-3 fatty acids to augment cancer therapy. J Nutr. 2002;132:3508S–3512S. doi: 10.1093/jn/132.11.3508S. [DOI] [PubMed] [Google Scholar]
- 13.Stoll BA. N-3 fatty acids and lipid peroxidation in breast cancer inhibition. Br J Nutr. 2002;87:193–198. doi: 10.1079/BJNBJN2001512. [DOI] [PubMed] [Google Scholar]
- 14.Sun H, Berquin IM, Edwards IJ. Omega-3 polyunsaturated fatty acids regulate syndecan-1 expression in human breast cancer cells. Cancer Res. 2005;65:4442–4447. doi: 10.1158/0008-5472.CAN-04-4200. [DOI] [PubMed] [Google Scholar]
- 15.Mali M, Andtfolk H, Miettinen HM, Jalkanen M. Suppression of tumor cell growth by syndecan-1 ectodomain. J Biol Chem. 1994;269:27795–27798. [PubMed] [Google Scholar]
- 16.Dhodapkar MV, Abe E, Theus A, et al. Syndecan-1 is a multifunctional regulator of myeloma pathobiology: control of tumor cell survival, growth, and bone cell differentiation. Blood. 1998;91:2679–2688. [PubMed] [Google Scholar]
- 17.Barbareschi M, Maisonneuve P, Aldovini D, et al. High syndecan-1 expression in breast carcinoma is related to an aggressive phenotype and to poorer prognosis. Cancer. 2003;98:474–483. doi: 10.1002/cncr.11515. [DOI] [PubMed] [Google Scholar]
- 18.Tsanou E, Ioachim E, Briasoulis E, et al. Clinicopathological study of the expression of syndecan-1 in invasive breast carcinomas. correlation with extracellular matrix components. J Exp Clin Cancer Res. 2004;23:641–650. [PubMed] [Google Scholar]
- 19.Leivonen M, Lundin J, Nordling S, von Boguslawski K, Haglund C. Prognostic value of syndecan-1 expression in breast cancer. Oncology. 2004;67:11–18. doi: 10.1159/000080280. [DOI] [PubMed] [Google Scholar]
- 20.Inki P, Stenback F, Grenman S, Jalkanen M. Immunohistochemical localization of syndecan-1 in normal and pathological human uterine cervix. J Pathol. 1994;172:349–355. doi: 10.1002/path.1711720410. [DOI] [PubMed] [Google Scholar]
- 21.Mukunyadzi P, Sanderson RD, Fan CY, Smoller BR. The level of syndecan-1 expression is a distinguishing feature in behavior between keratoacanthoma and invasive cutaneous squamous cell carcinoma. Mod Pathol. 2002;15:45–49. doi: 10.1038/modpathol.3880488. [DOI] [PubMed] [Google Scholar]
- 22.Kumar-Singh S, Jacobs W, Dhaene K, et al. Syndecan-1 expression in malignant mesothelioma: correlation with cell differentiation, WT1 expression, and clinical outcome. J Pathol. 1998;186:300–305. doi: 10.1002/(SICI)1096-9896(1998110)186:3<300::AID-PATH180>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 23.Mikami S, Ohashi K, Usui Y, et al. Loss of syndecan-1 and increased expression of heparanase in invasive esophageal carcinomas. Jpn J Cancer Res. 2001;92:1062–1073. doi: 10.1111/j.1349-7006.2001.tb01061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anttonen A, Kajanti M, Heikkila P, Jalkanen M, Joensuu H. Syndecan-1 expression has prognostic significance in head and neck carcinoma. Br J Cancer. 1999;79:558–564. doi: 10.1038/sj.bjc.6690088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nackaerts K, Verbeken E, Deneffe G, Vanderschueren B, Demedts M, David G. Heparan sulfate proteoglycan expression in human lung-cancer cells. Int J Cancer. 1997;74:335–345. doi: 10.1002/(sici)1097-0215(19970620)74:3<335::aid-ijc18>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 26.Pulkkinen JO, Penttinen M, Jalkanen M, Klemi P, Grenman R. Syndecan-1: a new prognostic marker in laryngeal cancer. Acta Otolaryngol. 1997;117:312–315. doi: 10.3109/00016489709117794. [DOI] [PubMed] [Google Scholar]
- 27.Numa F, Hirabayashi K, Kawasaki K, et al. Syndecan-1 expression in cancer of the uterine cervix: association with lymph node metastasis. Int J Oncol. 2002;20:39–43. [PubMed] [Google Scholar]
- 28.Fujiya M, Watari J, Ashida T, et al. Reduced expression of syndecan-1 affects metastatic potential and clinical outcome in patients with colorectal cancer. Jpn J Cancer Res. 2001;92:1074–1081. doi: 10.1111/j.1349-7006.2001.tb01062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wiksten JP, Lundin J, Nordling S, Kokkola A, Haglund C. A prognostic value of syndecan-1 in gastric cancer. Anticancer Res. 2000;20:4905–4907. [PubMed] [Google Scholar]
- 30.Liebersbach BF, Sanderson RD. Expression of syndecan-1 inhibits cell invasion into type I collagen. J Biol Chem. 1994;269:20013–20019. [PubMed] [Google Scholar]
- 31.Kato M, Saunders S, Nguyen H, Bernfield M. Loss of cell surface syndecan-1 causes epithelia to transform into anchorage-independent mesenchyme-like cells. Mol Biol Cell. 1995;6:559–576. doi: 10.1091/mbc.6.5.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaushal GP, Xiong X, Athota AB, Rozypal TL, Sanderson RD, Kelly T. Syndecan-1 expression suppresses the level of myeloma matrix metalloproteinase-9. Br J Haematol. 1999;104:365–373. doi: 10.1046/j.1365-2141.1999.01180.x. [DOI] [PubMed] [Google Scholar]
- 33.Anisfeld AM, Kast-Woelbern HR, Meyer ME, et al. Syndecan-1 expression is regulated in an isoform-specific manner by the farnesoid-X receptor. J Biol Chem. 2003;278:20420–20428. doi: 10.1074/jbc.M302505200. [DOI] [PubMed] [Google Scholar]
- 34.Xu HE, Lambert MH, Montana VG, et al. Molecular recognition of fatty acids by peroxisome proliferator-activated receptors. Mol Cell. 1999;3:397–403. doi: 10.1016/s1097-2765(00)80467-0. [DOI] [PubMed] [Google Scholar]
- 35.Forman BM, Tontonoz P, Chen J, Brun RP, Spiegelman BM, Evans RM. 15-Deoxy-delta 12, 14-prostaglandin J2 is a ligand for the adipocyte determination factor PPAR gamma. Cell. 1995;83:803–812. doi: 10.1016/0092-8674(95)90193-0. [DOI] [PubMed] [Google Scholar]
- 36.Thoennes SR, Tate PL, Price TM, Kilgore MW. Differential transcriptional activation of peroxisome proliferator-activated receptor gamma by omega-3 and omega-6 fatty acids in MCF-7 cells. Mol Cell Endocrinol. 2000;160:67–73. doi: 10.1016/s0303-7207(99)00254-3. [DOI] [PubMed] [Google Scholar]
- 37.Kersten S, Desvergne B, Wahli W. Roles of PPARs in health and disease. Nature. 2000;405:421–424. doi: 10.1038/35013000. [DOI] [PubMed] [Google Scholar]
- 38.Lee CH, Olson P, Evans RM. Minireview: lipid metabolism, metabolic diseases, and peroxisome proliferator-activated receptors. Endocrinology. 2003;144:2201–2207. doi: 10.1210/en.2003-0288. [DOI] [PubMed] [Google Scholar]
- 39.Suchanek KM, May FJ, Lee WJ, Holman NA, Roberts-Thomson SJ. Peroxisome proliferator-activated receptor beta expression in human breast epithelial cell lines of tumorigenic and non-tumorigenic origin. Int J Biochem Cell Biol. 2002;34:1051–1058. doi: 10.1016/s1357-2725(02)00025-0. [DOI] [PubMed] [Google Scholar]
- 40.Stephen RL, Gustafsson MC, Jarvis M, et al. Activation of peroxisome proliferator-activated receptor delta stimulates the proliferation of human breast and prostate cancer cell lines. Cancer Res. 2004;64:3162–3170. doi: 10.1158/0008-5472.can-03-2760. [DOI] [PubMed] [Google Scholar]
- 41.Elstner E, Muller C, Koshizuka K, et al. Ligands for peroxisome proliferator-activated receptorgamma and retinoic acid receptor inhibit growth and induce apoptosis of human breast cancer cells in vitro and in BNX mice. Proc Natl Acad Sci U S A. 1998;95:8806–8811. doi: 10.1073/pnas.95.15.8806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mueller E, Sarraf P, Tontonoz P, et al. Terminal differentiation of human breast cancer through PPAR gamma. Mol Cell. 1998;1:465–470. doi: 10.1016/s1097-2765(00)80047-7. [DOI] [PubMed] [Google Scholar]
- 43.Edwards IJ, Berquin IM, Sun H, et al. Differential effects of delivery of omega-3 fatty acids to human cancer cells by low-density lipoproteins versus albumin. Clin Cancer Res. 2004;10:8275–8283. doi: 10.1158/1078-0432.CCR-04-1357. [DOI] [PubMed] [Google Scholar]
- 44.McQuillan DJ, Seo NS, Hocking AM, McQuillan CI. Recombinant expression of proteoglycans in mammalian cells. Utility and advantages of the vaccinia virus/T7 bacteriophage hybrid expression system. Methods Mol Biol. 2001;171:201–219. doi: 10.1385/1-59259-209-0:201. [DOI] [PubMed] [Google Scholar]
- 45.Sprecher H. The roles of anabolic and catabolic reactions in the synthesis and recycling of polyunsaturated fatty acids. Prostaglandins Leukot Essent Fatty Acids. 2002;67:79–83. doi: 10.1054/plef.2002.0402. [DOI] [PubMed] [Google Scholar]
- 46.Park Y, Harris WS. Omega-3 fatty acid supplementation accelerates chylomicron triglyceride clearance. J Lipid Res. 2003;44:455–463. doi: 10.1194/jlr.M200282-JLR200. [DOI] [PubMed] [Google Scholar]
- 47.Arterburn LM, Hall EB, Oken H. Distribution, interconversion, and dose response of n-3 fatty acids in humans. Am J Clin Nutr. 2006;83:1467S–1476S. doi: 10.1093/ajcn/83.6.1467S. [DOI] [PubMed] [Google Scholar]
- 48.Williams CM. Burdge GLong-chain n-3 PUFA: plant v. marine sources. Proc Nutr Soc. 2006;65:42–50. doi: 10.1079/pns2005473. [DOI] [PubMed] [Google Scholar]
- 49.Nagy L, Tontonoz P, Alvarez JG, Chen H, Evans RM. Oxidized LDL regulates macrophage gene expression through ligand activation of PPARgamma. Cell. 1998;93:229–240. doi: 10.1016/s0092-8674(00)81574-3. [DOI] [PubMed] [Google Scholar]
- 50.Kliewer SA, Sundseth SS, Jones SA, et al. Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors alpha and gamma. Proc Natl Acad Sci USA. 1997;94:4318–4323. doi: 10.1073/pnas.94.9.4318. [DOI] [PMC free article] [PubMed] [Google Scholar]