Abstract
Methods to determine neurochemical concentrations in small samples of tissue are needed to map interactions among neurotransmitters. In particular, correlating physiological measurements of neurotransmitter release and the tissue content in a small region would be valuable. HPLC is the standard method for tissue content analysis but it requires microliter samples and the detector often varies by the class of compound being quantified; thus detecting molecules from different classes can be difficult. In this paper, we develop capillary electrophoresis with fast-scan cyclic voltammetry detection (CE-FSCV) for analysis of dopamine, serotonin, and adenosine content in tissue punches from rat brain slices. Using field-amplified sample stacking, the limit of detection was 5 nM for dopamine, 10 nM for serotonin, and 50 nM for adenosine. Neurotransmitters could be measured from a tissue punch as small as 7 µg (7 nL) of tissue, three orders of magnitude smaller than a typical HPLC sample. Tissue content analysis of punches in successive slices through the striatum revealed higher dopamine but lower adenosine content in the anterior striatum. Stimulated dopamine release was measured in a brain slice, then a tissue punch collected from the recording region. Dopamine content and release had a correlation coefficient of 0.71, which indicates much of the variance in stimulated release is due to variance in tissue content. CE-FSCV should facilitate measurements of tissue content in nanoliter samples, leading to a better understanding of how diseases or drugs affect dopamine, serotonin, and adenosine content.
Introduction
Tissue content studies are popular in neuroscience to correlate intraneuronal stores of neurochemicals with behavior, physiological measurements, or disease. Typically, a brain is harvested from an animal after an experiment and the region of interest is dissected out. Separations are performed to analyze the compounds of interest because brain samples are highly complex. High performance liquid chromatography (HPLC) is the most popular method and the detector can be UV-Vis,1,2 fluorescence,3–5 mass spectrometry,6 or electrochemical detection.7,8 In order to correlate tissue content with measurements of neurotransmitter release, tissue content should be determined in the small portion of the brain from which recordings were collected.9 While more recent use of smaller columns and smaller particles with HPLC has led to better mass sensitivity, most HPLC studies still utilize at least microliter sample volumes.8 Capillary electrophoresis (CE) has very low mass sensitivity and can be used with nanoliter volume samples.10 In addition, rapid separations are feasible with CE because high voltages can be applied to short capillaries and separation efficiency is not dependent on column length, as it is in HPLC.11 To identify neurotransmitters, CE is coupled to a variety of detectors, including fluorescence,12–14 mass spectrometry,15 and electrochemical detection.16,17
Electrochemical detection is popular for detecting biogenic amines, such a dopamine, and their metabolites, which are easily oxidizable. For purines, detection in biological samples is primarily achieved using HPLC with fluorescence,3 mass spectrometry,6 or UV detection.1,18 Adenosine is electroactive,19,20 but has rarely been detected electrochemically after separations due to its high oxidation potential.21 Other purines are electroactive as well, including guanine and uric acid.20,22 The ability to quantitate both adenosine and dopamine would be useful because adenosine and dopamine receptors are colocalized23 and adenosine modulates dopamine neurotransmission.24
Only a few tissue content studies have simultaneously detected catecholamines and adenosine in biological samples1,2,5,6 and most required two different detectors. For example, electrochemical detection was used for catecholamines and UV-Vis for adenosine detection after HPLC separation.2 Low concentrations of adenosine cannot be monitored in the samples because the set-up requires splitting the sample and UV is not very sensitive. HPLC with electrochemical detection and fluorescence detection has been adopted to detect dopamine and adenosine, respectively, in rat phenochromocytoma PC12 cells after two kinds of sample treatments.5 However, fluorescence detection is not ideal because it requires derivatization and a long reaction time. Kennedy’s lab recently derivatized microdialysis samples with benzoyl chloride and detected compounds including dopamine and adenosine using HPLC-MS detection.6 A single method to detect adenosine and dopamine using electrochemical detection has not been developed but would facilitate measurements of small amounts of neurotransmitters without derivatization.
Our lab has recently developed capillary electrophoresis with fast-scan cyclic voltammetry (FSCV) detection to quantitate dopamine, serotonin, octopamine and tyramine content in a single fruit fly central nervous system.16 The advantage of FSCV is that it provides a cyclic voltammogram chemical signature of the molecule being detected, which is not provided with typical amperometric detection. Samples as small as 7 µg can be analyzed using sample stacking to preconcentrate the analyte injected. In this study, we further developed CE-FSCV to determine serotonin, dopamine, and adenosine tissue content from small tissue punches from single brain slices. Thus, we expanded the CE-FSCV technique from positively charged monoamine neurotransmitters to include neutral compounds and have shown that rat brain slices can be analyzed, which have a different pH and sample matrix than Drosophila samples. The ability to quantitate serotonin, dopamine, and adenosine in brain slices permits measurements of their interactions and allows a better correlation of physiological measurements, such as stimulated release, with tissue content.
Methods
Chemicals
Serotonin, dopamine, norepinephrine, isoproterenol, adenosine, tryptophan, and guanine were purchased from Sigma (St. Louis, MO), and 10 mM stock solutions prepared in 0.1 M perchloric acid. An artificial cerebral spinal fluid (aCSF) solution was used in brain dissections: 126 mM NaCl, 2.5 mM KCl, 1.2 mM NaH2PO4, 2.4 mM CaCl2 (dihydrate),1.2 M MgCl2 (hexahydrate), 25 mM NaHCO3, 11 mM glucose, and 15 mM tris(hydroxymethyl) aminomethane, with the pH adjusted to 7.4 (all from Fisher, Suwanee, GA)25. The separation buffer was 150 mM NaH2PO4 with 1 mM β–cyclodextrin (pH 4.0). The detection cell buffer was 100 mM NaH2PO4 (pH 6.5). Buffer solutions for CE were filtered with a 0.2 µm nylon filter (Alltech, Deerfield, IL).
Instrumentation and Data Acquisition
The CE with end column fast-scan cyclic voltammetry was built in house, as previously reported.16 Briefly, the separation capillary was 39–42 cm long (11 µm i.d., 148 µm o.d., Polymicro Technologies, Phoenix, AZ). The detection end of the capillary was etched to 20–30 µm o.d. with 25% hydrogen fluoride for 3 h to decouple the separation current from the detector. Between biological samples, the capillary was rinsed with separation buffer for 5 min.
Positive high voltage (15 kV) was applied at the injection end of the capillary through a platinum wire in the buffer reservoir using a DC power supply (Spellman, Plainview, NY) and ground was stainless steel tubing attached to the detection cell. The end of the capillary positioned 15 ± 2 µm from the microelectrode. The working electrode was a 34.5 ± 2.5 µm diameter carbon-fiber disk microelectrode (Specialty Materials, Lowell, MA). The reference electrode was a 250 µm diameter chloridized silver wire placed about 50 µm from the working electrode. A cross-flow detection buffer flowed slowly (0.5 mL/min) to flush the area around the electrode and prevent analytes from accumulating.
Detection was performed with FSCV with a two-electrode configuration using a Dagan ChemClamp potentiostat (Dagan, Minneapolis, MN) with a custom-modified headstage. The data acquisition software and hardware were the same as previously described26. The electrode was scanned from −0.4 to 1.5 V and back at 400 V/s every 100 ms. Tissue content was calculated using the ratio of the peak height for the tissue sample and the internal standard. To correct for injection bias, this amount was then divided by the ratio of the peak height for the internal standard in the tissue sample and the standard sample.
Brain slice experiments
Adult, male Sprague-Dawley rats (250–350 g) purchased from Charles River were housed in a vivarium and given food and water ab libitum. All experiments were approved by the Animal Care and Use Committee of the University of Virginia.
Rats were anesthetized with isoflurane (1 mL/100 g rat weight) and decapitated. The brain was removed within 2 minutes and placed in 0–5 °C artificial cerebral spinal fluid (aCSF) for 2–3 minutes. The aCSF was bubbled with a mixture of 95% oxygen and 5% carbon dioxide (carbogen) for 30 minutes prior to introduction of the brain tissue. Slices were made with a vibratome (Leica VT1000S, Bannockburn, IL), with a slicing speed of 3 mm/s and a vibration frequency of 8. Tissue samples were homogenized in 10 µL (400 µm thick slices) or 2 µL (100 µm thick slices) of ice-cold 5 mM perchloric acid containing 0.3 µM isoproterenol as the internal standard. The brain sample was broken with a metal wire and then sonicated for 60 s with a tissue sonicator. The homogenates were centrifuged at 11,000 rpm for 5 min at room temperature and filtered through a 0.22-µm PVDF membrane centrifugal filter (Millipore Co., Bedford, MA, USA). The filtrate of 3 µL was analyzed by capillary electrophoresis by directly placing the capillary into the centrifuge tube and performing an electrokinetic injection. Multiple injections (2– 3) can be made from the same sample.
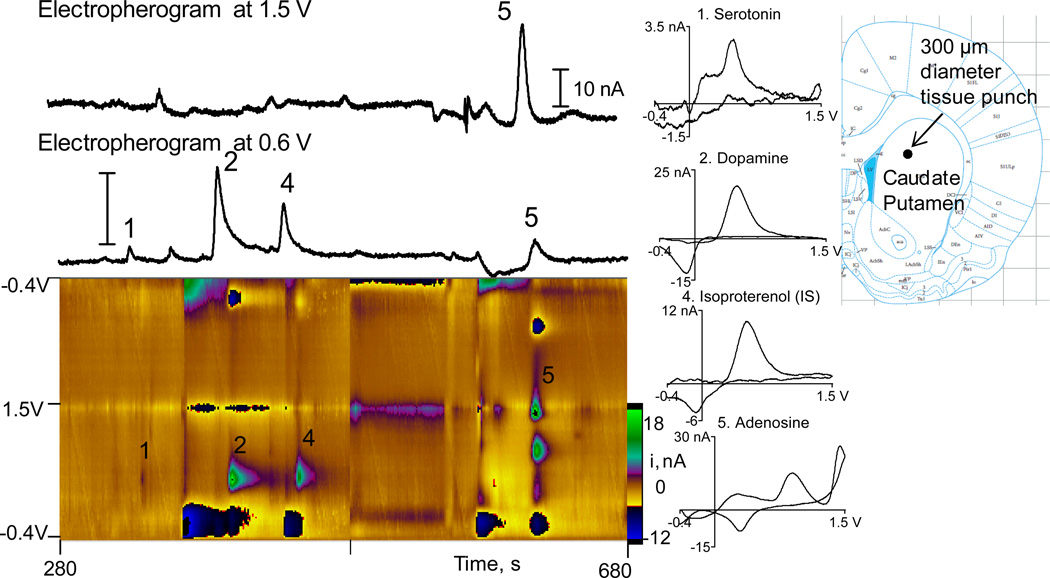
For the characterization of neurotransmitter detection in brain slice punches, 400 µm thick coronal slices were taken from the caudate-putamen of a chilled sliced. The tissue was never frozen and tissue was easily removed from the punch using compressed air. Punches 300 µm in diameter (Zivic Instruments, Pittsburgh PA) were collected in an area of the dorsal-medial caudate-putamen, corresponding to the following coordinates: 1.9 mm anterior, 4.5 mm ventral, 1.8 mm medial. Sample size limitations were tested in the prefrontal cortex using 300 µm punches in 100 µm thick slices at 2.52 mm anterior, −3.4 mm ventral, 0.4 mm medial. To correlate stimulated dopamine release and tissue content, the electrode was implanted in dorsal-medial caudate-putamen and stimulated release was measured. Then a tissue punch was collected, centered around the indentations left where the electrode was implanted. The slices were stimulated with a single, biphasic pulse, 4 ms long, and an amplitude of 300 µA using a BSI-950 Biphasic Stimulus Isolator (Dagan), which is typical for dopamine measurements.27 In order to test regional differences in the caudate-putamen, four sequential 400 µm-thick coronal slices were made starting at coordinates of +2.5 mm AP and ending at 0.9 mm AP.
Statistics
Error bars are standard error of the mean (SEM). Statistics were performed in GraphPad Prism (La Jolla, CA). Two-tailed student’s t tests were used to compare the means from two groups, while an ANOVA was used to compare 3 or more groups. Results were considered statistically significant at the 95% confidence level.
Results and Discussion
Detection of neurotransmitters in a tissue punch from a single brain slice requires a sensitive and selective measurement technique. CE-FSCV allows rapid separation of sub-picomolar amounts of neurotransmitters with the added benefit of cyclic voltammograms to aid in identification of analytes during detection.16 We used a waveform of −0.4 to 1.5 V for fast-scan cyclic voltammetry to facilitate both biogenic amine and purine detection.20
Separation of dopamine, serotonin and adenosine
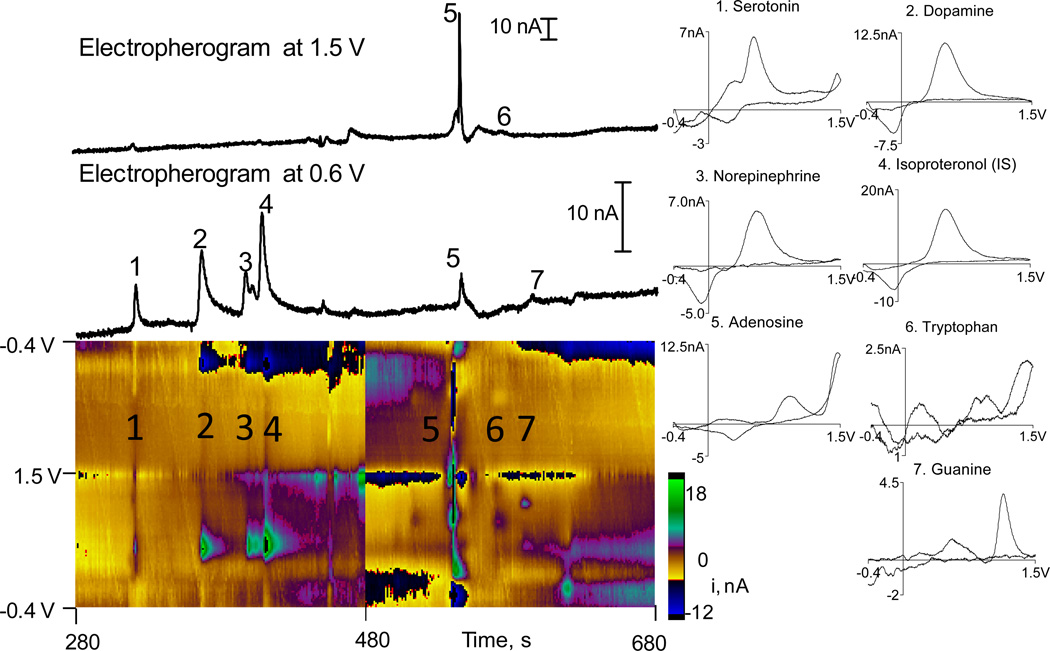
Fig. 1 shows detection of biologically relevant standards: 100 nM serotonin, 100 nM dopamine, 100 nM norepinephrine, 500 nM adenosine, 500 nM tryptophan, and 500 nM guanine. Isoproterenol was added as an internal standard at 200 nM. The color plot shows all the data, with applied voltage on the y-axis, time on the x-axis and the color representing current. Compounds are numbered in the order they migrate. The color plots are background subtracted for each 4 minute data file and a new file began at 480 s, immediately following the previous data file so that no data was lost. The background-subtracted cyclic voltammograms on the right for each compound facilitate identification of the analytes. Above the color plots are electropherograms which plot current changes at two different potentials: 0.6 V and 1.5 V. These are equivalent of taking a line scan across the color plot at these two potentials. Traditional electropherograms for amperometric detection would include every peak because any compound with an oxidation potential lower than the holding potential would be oxidized. Our traces only show a peak in the current over time trace only if there is current at that specific voltage in the cyclic voltammogram. Thus, not every compound appears in every trace.
Figure 1.
Separation of standards. The peaks in the electropherograms and color plot are numbered and those numbers correspond to the cyclic voltammograms. Serotonin, dopamine, and norepinephrine are 100 nM, isoproterenol 200 nM, and adenosine, tryptophan and guanine are 500 nM. The color plot shows all the data, with applied voltage on the yaxis, time on the x-axis and current in false color. Color plots were background subtracted for each data file and a new data file starts at 480 s. The electropherograms are horizontal traces through the color plot at specific potentials, 1.5 V and 0.6 V. Because different compounds have different oxidation peaks, they do not all appear in one electropherogram. Cyclic voltammograms are taken at each peak and help confirm the identity of the analyte.
For repeated injections of the same standard, relative standard deviations of peak heights were less than 7%. With standards, the LOD was 10 ± 3 nM for serotonin, 5 ± 3 nM for dopamine, and 50 ± 20 nM for adenosine (n=4). The error for these figures is larger because of the different sensitivity of electrodes used on different days.
For this separation, we chose to separate cationic neurotransmitters dopamine, serotonin, and norepinephrine, the purines adenosine and guanine, and the electroactive amino acid tryptophan. Isoproterenol was used as an internal standard and is an isopropyl modified analog of norepinephrine, so it has the same cyclic voltammogram as norepinephrine. Serotonin migrates first, followed by dopamine, norepinephrine and isoproterenol. All of those compounds are positively charged at the separation pH so their fast migration was expected. For compounds with similar CVs (dopamine, norepinephrine and isoproterenol), peaks were identified by spiking the samples with each compound. Adenosine, tryptophan and guanine are uncharged and migrated later. Although tryptophan and guanine are hard to identify in the two electropherogram traces because their main oxidation peaks are 0.95 V and 1.15V, respectively, they are clearly identified in the color plot and CVs. Adenosine is also clearly identified and has a very prominent secondary oxidation and reduction peak with this type of electrode compared to other types of carbon fibers.20
CE-FSCV was used to analyze sub-pmol amounts of material with detection limits in the low nM range for dopamine and serotonin. FSCV does not have as low of detection limits as amperometry because the noise from fast scanning is greater, so field-amplified sample stacking was used to maximize the amount of sample injected.16 Sample stacking arises during electrokinetic injection where the sample has a low conductivity compared to the electrophoresis buffer and the samples stack at beginning of the capillary when a voltage is applied.28 Because the voltage drop over the capillary is negative, positively charged analytes are preferentially loaded, neutral molecules like adenosine can be injected, but fewer negatively charged molecules, such as ATP, would be injected. Similarly, anionic metabolites such as DOPAC and 5-HIAA were not detected. Thus, while the technique is not optimal for analysis of anionic metabolites for dopamine and serotonin or establishing metabolite ratios, CE-FSCV does facilitate detection of small amounts of neurotransmitters and purines simultaneously in very small amounts of tissue.
Separations of tissue punch samples from brain slices
Figure 2 shows a separation from a tissue punch from a brain slice in the rat caudate-putamen. The slice was 400 µm thick and a 300 µm o.d. tissue punch was used to collect the sample. The color plot shows serotonin, dopamine and adenosine are detected in the slice. Norepinephrine is present at low levels in the striatum and it was not detected, which indicates it was under our detection limits. Norepinephrine levels are known to be low in the striatum29. The peak migration times are a bit delayed compared to Fig. 1, due to a different capillary length or differences in electroosmotic flow, but the peak order is useful for determining the identity. The cyclic voltammograms confirm peak identity so identification is possible even when migration times are slightly shifted. Other peaks are present, especially before adenosine, which is expected given the complexity of the sample. Using the current from the standards run after the tissue sample, the amount of each analyte was calculated for each sample. The amounts were then corrected for any injection errors by dividing by the ratio of the peak height of the internal standard in the rat sample and the standard. The total amount in the sample was calculated to be 0.44 pmol serotonin, 1.8 pmol dopamine, and 11.1 pmol adenosine.
Figure 2.
Separation of a tissue punch sample from a representative rat caudate putamen slice. The tissue punch was 300 µm o.d. and the slice was 400 µm thick. Color plots were background subtracted for each data file and a new data file starts at 480 s. The amount of analyte in this sample is 0.44 pmol serotonin, 1.8 pmol dopamine, and 11.1 pmol adenosine. Isoproterenol was the internal standard.
Tissue content is usually reported in terms of ng per mg of wet tissue or ng per mg of protein. The volume of a tissue punch from a 400 µm thick slice using a 300 µm punch was 28 nL, which corresponds to 29 µg of tissue. Because this is too small to be accurately weighed, the weight was calculated using the volume measurements and the known density of brain tissue (1.04 mg/mL30). In addition, the amount of tissue was too low to partition the sample to determine protein content.31. Thus, values are reported in terms of mg of wet tissue and Table 1 compares values for our results and other studies.
Table 1.
Literature reports of tissue content in the striatum.
| Reference | DA (pmol/mg) | 5-HT (pmol/mg) | Aden (pmol/mg) | DA/5HT | Aden/DA |
|---|---|---|---|---|---|
| 39 | 143 | 6.8 | N.A. | 6.6 | N.A |
| 40 | 47.7 | 1.4 | N.A. | 29.8 | N.A. |
| 41 | 28.0 | 0.96 | N.A. | 25.3 | N.A. |
| 42 | 52.5 | 8.8 | N.A. | 5.2 | N.A. |
| 43 | 65.5 | 3.6 | N.A. | 25.1 | N.A. |
| 44 | 52.2 | 21.6 | N.A. | 2.2 | N.A. |
| 1 | 50.0 | 2.7 | 641 | 17.9 | 19.9 |
| 33 | N.A. | N.A. | 618 | N.A. | N.A. |
| Fang (this work) | 56.0 | 13.4 | 492 | 4.2 | 8.8 |
N.A. means value was not reported. DA is dopamine, 5-HT is serotonin, and Aden is adenosine.
The amounts of dopamine and serotonin in the striatum measured by CE-FSCV fall within the range of values reported in the literature (Table 1). Our dopamine values were in the middle of previously reported values for the striatum while our values for serotonin were higher than many studies, although within the overall range. FSCV at carbon-fiber microelectrodes is particularly sensitive to serotonin,32 but this should not result in higher values, as concentrations were determined from external standards and normalizing data to an internal standard, isoproternol. Serotonin fouling was also only a minimal issue, as standards run after a tissue sample had about 90 % of the signal as standards run before the sample, indicating the electrode was maintaining sensitivity. The electrode was used for an entire day without repolishing and fouling effects were insignificant. The ratio of dopamine to serotonin in the striatum is also in line with other experiments, although those ratios can vary by an order of magnitude (Table 1). Our experiments were performed on a specific portion of the striatum so it is possible that the tissue content and ratios might be different than other portions of the striatum or the overall average if the whole region was tested. The overall agreement in content demonstrates CE-FSCV is a valid method for tissue content analysis.
Fewer studies have examined adenosine tissue content in brain samples. Because adenosine is a metabolite of ATP, the main energy molecule, it is not surprising that it is present in greater amounts than dopamine and serotonin, neurotransmitters which are specifically synthesized. Betto et al used HPLC with UV detection for adenosine and electrochemical detection for dopamine to determine a tissue content of 640 ± 213 pmol adenosine/mg tissue.1 Akula et al found an adenosine concentration of 619 pmol/mg tissue in the striatum with HPLC-UV.33 These values are slightly higher, but in a similar range, to our value of 490 ± 160 pmol adenosine/mg tissue. Betto et al found a ratio of adenosine to dopamine of about 19 but our ratio of adenosine to dopamine was 9, lower than their value. This difference may be due to discrete sampling in the dorsal-medial striatum while their samples encompassed the entire striatum.
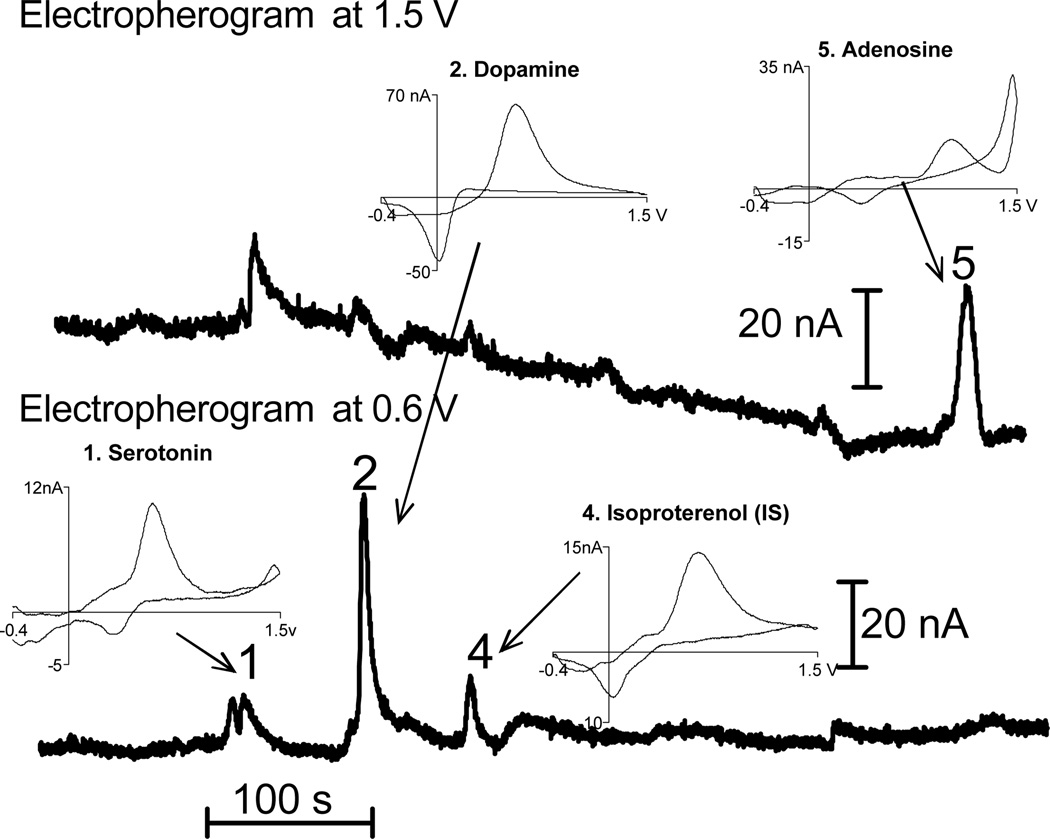
One hundred µm thick slices were analyzed to demonstrate that CE-FSCV can quantitate very small samples (Fig. 3). Tissue punch samples from the prefrontal cortex were examined to show that this analysis can be done in other brain regions. Serotonin, dopamine, and adenosine were detected, although the tissue sample was only 7.1 nL. The peaks are evident and the compounds are easily quantitated. The amount of analyte in this sample is 0.082 pmol serotonin, 0.12 pmol of dopamine, and 4.3 pmol of adenosine, which corresponds to tissue content values of 11 pmol/mg for serotonin, 23 pmol/mg for dopamine, 602 pmol/mg for adenosine, respectively. These contents were similar to tissue punches obtained from 400 µm slices which contained on average 13 pmol/mg of serotonin, 25 pmol/mg of dopamine and 686 pmol/mg of adenosine in the prefrontal cortex. The content, normalized for the amount of tissue, is the same in 100 µm and 400 µm thick slices indicating that decreasing the volume of the slice does not affect the measurement. This experiment is proof of principle that small regions could be studied with CE-FSCV, for example discrete portions of the nucleus accumbens or hippocampus, or tissue samples from smaller organisms such as mice.
Figure 3.
Tissue content of a punch from a 100 µm thick slice. A 300 µm tissue punch was obtained from a 100 µm thick slice of the frontal cortex. Serotonin, dopamine, and adenosine can all be clearly seen in this sample, which is only 7 µg. Isoproterenol was the internal standard. The amount of analyte in this tissue punch is 0.082 pmol of serotonin, 0.12 pmol of dopamine, and 4.3 pmol of adenosine.
Regional differences in dopamine, serotonin, and adenosine content
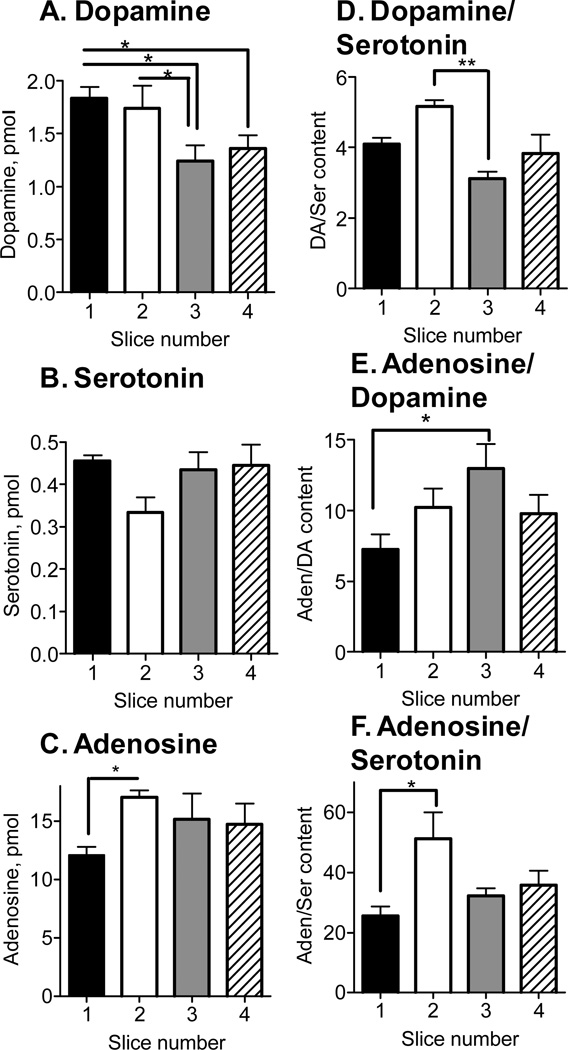
To determine whether there was a regional difference in tissue content in the caudate-putamen, slices were collected from the rostral to the caudal portion of the brain. The first slice was 2.5 mm before bregma and 4 sequential, 400 µm thick slices were collected and punches were taken from the dorsal-medial region. The left column of Figure 4 (A–C) shows average absolute tissue content for each analyte. With averaged data, there was a significant difference in dopamine and adenosine between slices with only a significant difference between slice 1 and slices 3 and 4, as well as slice 2 and 3, for dopamine (1-way ANOVA with paired Bonferonni post-test). This indicates a general trend for higher dopamine in the anterior portion of the brain. There were no significant differences for serotonin. For adenosine, tissue content was significantly lower in the first slice compared to the second slice (1-way ANOVA with paired Bonferonni post-test).
Figure 4.
Dopamine, serotonin, and adenosine content in the dorsal-medial portion of the caudate-putamen. Four sequential, 400 µm slices were taken, starting at +2.5 mm AP (slice 1) and ending at +0.9 AP (slice 4). The left column (A–C) shows the absolute amount of each compound found in a 300 µm o.d. tissue punch. Statistics are one-way ANOVA followed by paired Bonferonni post-test (n=4). The right column (D–F) shows ratios of analytes. Statistics are one-way ANOVA followed by paired Bonferonni post-test (n=4) (*, p<0.05).
Another way to analyze the data collected from slices taken from front to back in the caudate is to examine the ratios of the different neurochemicals in each slice. After calculating the ratios for each slice and then averaging, the first slice of the caudate has about 4 times as much dopamine as serotonin and there is a significant difference between slice 2 and 3 (Fig. 4D). For all the slices, there was on average about 10 times more adenosine than dopamine and there was a significant difference in the adenosine/dopamine ratio between slices 1 and 3 (Fig. 4E). The greatest difference in concentration was seen for adenosine and serotonin where on average, for all the slices, there was 37-times more adenosine than serotonin (Fig. 4F). Slices 1 and 2 had a significantly different adenosine to serotonin ratio. Thus significant differences were differentiated although there were not clear, substantial progressions from front to back in the caudate.
Regional differences were found within the striatum for dopamine and adenosine. The higher values of dopamine in the anterior caudate agree well with literature.34,35 Regional variations in adenosine have not been studied in the striatum but the results showed higher amounts of adenosine in the posterior caudate, the opposite trend of dopamine. In all slices, there was an order of magnitude more adenosine than serotonin or dopamine which shows higher concentrations of this neuromodulator than neurotransmitters. The striatum is known to be heterogeneous, displaying a patch-matrix organization,36 and have regional differences in neurotransmitter receptors37 and synthesis38 so the ability to collect and analyze samples from discrete slices and discrete regions will facilitate tissue content analysis of specific subregions.
Correlating stimulated release and tissue content
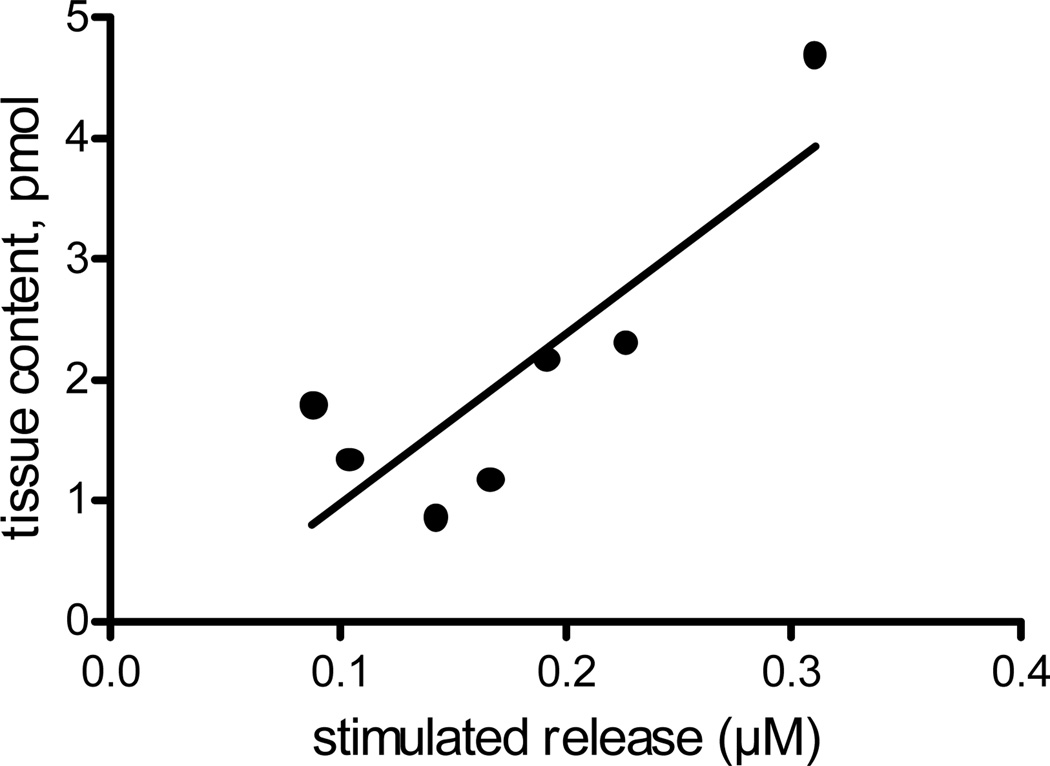
One reason to quantify neurotransmitter content from tissue punches from slices is to understand the relationship between the intraneuronal stores of neurotransmitters and neuronal release. To correlate stimulated dopamine release and tissue content, we electrically stimulated a caudate-putamen slice in 3 different places, by lowering the electrode into the slice between stimulations. A 300 µm tissue punch was collected from the area where electrochemical measurements were performed and the dopamine content measured with CE-FSCV. Taking 3 measurements of stimulated release at different depths allowed an average of release throughout that area of the slice. For 7 slices, the correlation coefficient for tissue content and stimulated release (Fig. 5) is 0.71, which indicates that 71 % of the variance in the stimulated release can be explained by variance in the tissue content.
Figure 5.
Correlation of stimulated dopamine release and tissue content. Stimulated dopamine release was measured in the caudate-putamen in a 400 µm coronal slice. The electrode was moved down three times to sample from top to bottom and those values averaged to get a stimulated release value. The electrode was removed and a 300 µm tissue punch obtained from the area where stimulated release was taken. The correlation coefficient is R2=0.71 (n=7).
Stimulated dopamine release correlated with tissue content very well, considering that the microelectrode only samples from a small space compared to the overall volume of the 300 µm diameter punch. The data show that release is not just a function of local environment, such as how close the electrode is located to release sites, but also is correlated to intraneuronal stores. The measurements of tissue content were similar to the previous experiments where no electrochemical experiments were performed, thus the electrochemical measurements did not appear to damage the tissue or reduce content. Thus, CE-FSCV should be a useful method to study the correlation of stimulated release and tissue content from a specific area of a brain slice.
Conclusions
In this work, we demonstrated that CE-FSCV can be used to quantitate dopamine, serotonin and adenosine from small volume samples such as tissue punches from brain slices. This is the first study to quantitate both biogenic amines and adenosine in the same tissue sample. Tissue content can be quantitated from as small as 7 µg of tissue. Stimulated dopamine release was also correlated with dopamine tissue content in a tissue sample taken directly around the electrode. Previous studies had evaluated monoamines in Drosophila nerve cords, which have an extracellular pH of 6.5 and a different sample matrix. The mammalian extracellular pH is 7.4, thus this study shows CE-FSCV can be used with a variety of tissue samples. The extension of CE-FSCV to quantitate adenosine also demonstrates that neutral compounds can be measured, opening up many more potential analytes for future studies. CE-FSCV allows codetermination of biogenic amines and adenosine in a tissue punch from single brain slice and will facilitate studies of how disease states affect content of neurotransmitters and neuromodulators.
Acknowledgments
This work was funded by the National Institute of Health R01NS076875.
References
- 1.Betto P, Popoli P, Ricciarello G, Caporali MG, Antonini R. J. Chromatogr. B Biomed. Appl. 1994;662:21–25. doi: 10.1016/0378-4347(94)00385-8. [DOI] [PubMed] [Google Scholar]
- 2.Echizen H, Itoh R, Ishizaki T. Clin. Chem. 1989;35:64–68. [PubMed] [Google Scholar]
- 3.Wojcik WJ, Neff NH. J. Neurochem. 1982;39:280–282. doi: 10.1111/j.1471-4159.1982.tb04736.x. [DOI] [PubMed] [Google Scholar]
- 4.Yoshitake T, Fujino K, Kehr J, Ishida J, Nohta H, Yamaguchi M. Anal Biochem. 2003;312:125–133. doi: 10.1016/s0003-2697(02)00435-9. [DOI] [PubMed] [Google Scholar]
- 5.Yamboliev IA, Smyth LM, Durnin L, Dai Y, Mutafova-Yambolieva VN. Eur. J. Neurosci. 2009;30:756–768. doi: 10.1111/j.1460-9568.2009.06869.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song P, Mabrouk OS, Hershey ND, Kennedy RT. Anal. Chem. 2012;84:412–419. doi: 10.1021/ac202794q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colebrooke RE, Humby T, Lynch PJ, McGowan DP, Xia J, Emson PC. Eur. J. Neurosci. 2006;24:2622–2630. doi: 10.1111/j.1460-9568.2006.05143.x. [DOI] [PubMed] [Google Scholar]
- 8.Nguyen AT, Aerts T, Van DD, De Deyn PP. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2010;878:3003–3014. doi: 10.1016/j.jchromb.2010.09.019. [DOI] [PubMed] [Google Scholar]
- 9.Howard CD, Keefe KA, Garris PA, Daberkow DP. J. Neurochem. 2011;118:668–676. doi: 10.1111/j.1471-4159.2011.07342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rossell S, Gonzalez LE, Hernandez L. Journal of Chromatography B-Analytical Technologies in the Biomedical and Life Sciences. 2003;784:385–393. doi: 10.1016/s1570-0232(02)00826-7. [DOI] [PubMed] [Google Scholar]
- 11.Kennedy RT, Watson CJ, Haskins WE, Powell DH, Strecker RE. Curr. Opin. Chem. Biol. 2002;6:659–665. doi: 10.1016/s1367-5931(02)00373-3. [DOI] [PubMed] [Google Scholar]
- 12.Benturquia N, Couderc F, Sauvinet V, Orset C, Parrot S, Bayle C, Renaud B, Denoroy L. Electrophoresis. 2005;26:1071–1079. doi: 10.1002/elps.200410150. [DOI] [PubMed] [Google Scholar]
- 13.Liu L, Li Q, Li N, Ling J, Liu R, Wang Y, Sun L, Chen XH, Bi K. J. Sep. Sci. 2011;34:1198–1204. doi: 10.1002/jssc.201000799. [DOI] [PubMed] [Google Scholar]
- 14.Park YH, Zhang X, Rubakhin SS, Sweedler JV. Anal. Chem. 1999;71:4997–5002. doi: 10.1021/ac990659r. [DOI] [PubMed] [Google Scholar]
- 15.Moini M, Schultz CL, Mahmood H. Anal. Chem. 2003;75:6282–6287. doi: 10.1021/ac034708i. [DOI] [PubMed] [Google Scholar]
- 16.Fang HF, Vickrey TL, Venton BJ. Anal. Chem. 2011;83:2258–2264. doi: 10.1021/ac103092z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ream PJ, Suljak SW, Ewing AG, Han KA. Anal. Chem. 2003;75:3972–3978. doi: 10.1021/ac034219i. [DOI] [PubMed] [Google Scholar]
- 18.Bowser MT, Kennedy RT. Electrophoresis. 2001;22:3668–3676. doi: 10.1002/1522-2683(200109)22:17<3668::AID-ELPS3668>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 19.Dryhurst G. Talanta. 1972;19:769–778. doi: 10.1016/0039-9140(72)80004-3. [DOI] [PubMed] [Google Scholar]
- 20.Swamy BEK, Venton BJ. Anal. Chem. 2007;79:744–750. doi: 10.1021/ac061820i. [DOI] [PubMed] [Google Scholar]
- 21.Henderson RJ, Jr, Griffin CA. J. Chromatogr. 1981;226:202–207. doi: 10.1016/s0378-4347(00)84222-x. [DOI] [PubMed] [Google Scholar]
- 22.Brajter-Toth A, El-Nour KA, Cavalheiro ET, Bravo R. Anal. Chem. 2000;72:1576–1584. doi: 10.1021/ac9906680. [DOI] [PubMed] [Google Scholar]
- 23.Ferre S, Ciruela F, Canals M, Marcellino D, Burgueno J, Casado V, Hillion J, Torvinen M, Fanelli F, de Benedetti P, Goldberg SR, Bouvier M, Fuxe K, Agnati LF, Lluis C, Franco R, Woods A. Parkinsonism & Related Disorders. 2004;10:265–271. doi: 10.1016/j.parkreldis.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 24.Okada M, Mizuno K, Kaneko S. Neurosci Lett. 1996;212:53–56. doi: 10.1016/0304-3940(96)12780-4. [DOI] [PubMed] [Google Scholar]
- 25.Pajski ML, Venton BJ. ACS Chem Neurosci. 2010;1:775–787. doi: 10.1021/cn100037d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heien MLAV, Phillips PEM, Stuber GD, Seipel AT, Wightman RM. Analyst. 2003;128:1413–1419. doi: 10.1039/b307024g. [DOI] [PubMed] [Google Scholar]
- 27.Jones SR, Garris PA, Kilts CD, Wightman RM. J. Neurochem. 1995;64:2581–2589. doi: 10.1046/j.1471-4159.1995.64062581.x. [DOI] [PubMed] [Google Scholar]
- 28.Cao CX, He YZ, Li M, Qian YT, Gao MF, Ge LH, Zhou SL, Yang L, Qu QS. Anal. Chem. 2002;74:4167–4174. doi: 10.1021/ac0201880. [DOI] [PubMed] [Google Scholar]
- 29.Kitanaka N, Kitanaka J, Takemura M. Eur. J. Pharmacol. 2003;474:63–70. doi: 10.1016/s0014-2999(03)02015-6. [DOI] [PubMed] [Google Scholar]
- 30.DiResta GR, Lee JB, Arbit E. J. Neurosci. Methods. 1991;39:245–251. doi: 10.1016/0165-0270(91)90103-7. [DOI] [PubMed] [Google Scholar]
- 31.Salvatore MF, Fisher B, Surgener SP, Gerhardt GA, Rouault T. Mol. Brain Res. 2005;139:341–347. doi: 10.1016/j.molbrainres.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 32.Hashemi P, Dankoski EC, Petrovic J, Keithley RB, Wightman RM. Anal Chem. 2009;81:9462–9471. doi: 10.1021/ac9018846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akula KK, Kaur M, Bishnoi M, Kulkarni SK. J. Sep. Sci. 2008;31:3139–3147. doi: 10.1002/jssc.200800316. [DOI] [PubMed] [Google Scholar]
- 34.Patel J, Trout SJ, Kruk ZL. Naunyn Schmiedebergs Arch. Pharmacol. 1992;346:267–276. doi: 10.1007/BF00173539. [DOI] [PubMed] [Google Scholar]
- 35.Strong R, Samorajski T, Gottesfeld Z. J. Neurochem. 1982;39:831–836. doi: 10.1111/j.1471-4159.1982.tb07967.x. [DOI] [PubMed] [Google Scholar]
- 36.Gerfen CR, Herkenham M, Thibault J. J. Neurosci. 1987;7:3915–3934. doi: 10.1523/JNEUROSCI.07-12-03915.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joyce JN, Loeschen SK, Marshall JF. Brain Res. 1985;338:209–218. doi: 10.1016/0006-8993(85)90149-0. [DOI] [PubMed] [Google Scholar]
- 38.Graybiel AM, Hirsch EC, Agid YA. Proc. Natl Acad. Sci. USA. 1987;84:303–307. doi: 10.1073/pnas.84.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moreira CM, Masson S, Carvalho MC, Brandao ML. Brain Res. Bull. 2007;71:466–474. doi: 10.1016/j.brainresbull.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 40.Hebert MA, Gerhardt GA. Brain Res. 1998;797:42–54. doi: 10.1016/s0006-8993(98)00370-9. [DOI] [PubMed] [Google Scholar]
- 41.Muneoka K, Kuwagata M, Ogawa T, Shioda S. Life Sci. 2010;87:738–742. doi: 10.1016/j.lfs.2010.10.016. [DOI] [PubMed] [Google Scholar]
- 42.Bavaresco CS, Ben J, Chiarani F, Netto CA, Wyse AT. Pharmacol. Biochem. Behav. 2008;90:594–597. doi: 10.1016/j.pbb.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 43.Matthews K, Dalley JW, Matthews C, Tsai TH, Robbins TW. Synapse. 2001;40:1–10. doi: 10.1002/1098-2396(200104)40:1<1::AID-SYN1020>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 44.Brenes JC, Fornaguera J. Behav. Brain Res. 2009;198:199–205. doi: 10.1016/j.bbr.2008.10.036. [DOI] [PubMed] [Google Scholar]