Abstract
We sought to identify novel pharmacogenetic markers associated with cardiovascular outcomes in patients with hypertension on antihypertensive therapy. We genotyped a 1:4 case:control cohort (n=1345) on the Illumina HumanCVD Beadchip from the International Verapamil SR-Trandolapril Study, where participants were randomized to a β blocker strategy or a calcium channel blocker strategy. Genome-spanning SNP × treatment interaction analyses of non-synonymous SNPs were conducted in white and Hispanic race/ethnic groups. Top hits from whites were tested in Hispanics for consistency. A genetic risk score was constructed from the top three signals and tested in the Nordic Diltiazem study. SIGLEC12 rs16982743 and A1BG rs893184 had a significant interaction with treatment strategy for adverse cardiovascular outcomes (International Verapamil SR-Trandolapril Study whites and Hispanics combined interaction P=0.0038, and 0.0036, respectively). A genetic risk score including rs16982743, rs893184 and rs4525 in F5, was significantly associated with treatment-related adverse cardiovascular outcomes in whites and Hispanics from the International Verapamil SR-Trandolapril Study and in the Nordic Diltiazem study (meta-analysis interaction P=2.39×10−5). In patients with a genetic risk score of zero or 1, calcium channel blocker treatment was associated with lower risk (OR (95% CI) = 0.60 (0.42-0.86)), and in those with a genetic risk score of 2-3, calcium channel blocker treatment was associated with higher risk, OR (95% CI) = 1.31 (1.08-1.59)). These results suggest cardiovascular outcomes may differ based on SIGLEC12, A1BG, F5 genotypes and antihypertensive treatment strategy. These specific genetic associations and our risk score provide insight into a potential approach to personalized antihypertensive treatment selection.
Keywords: Pharmacogenomics; Hypertension; antihypertensive agents; cardiovascular outcomes; genetic variation; beta-blockers, calcium channel blockers
INTRODUCTION
Hypertension (HTN) is the most common chronic disease in the United States, affecting about one third of the adult population, and is a major risk factor for acute myocardial infarction (MI), stroke, heart failure and renal failure.1 Numerous antihypertensive drugs are considered appropriate first-line therapy to lower blood pressure (BP) including diuretics, β-blockers (βB), calcium channel blockers (CCB), ACE inhibitors and angiotensin receptor blockers. These drugs are ultimately prescribed to prevent the long-term cardiovascular (CV) complications of HTN.2 However, there is great inter-patient variability in antihypertensive drug response, with only about 50% of patients achieving an adequate BP response to any one drug,3 and limited data available to guide treatment selection. Why patients respond differently to the same drug and why some patients experience adverse CV outcomes despite BP control while others do not, remains poorly understood. Pharmacogenomics aims to identify genetic markers that are associated with drug response, outcomes and adverse events.
In the past decade there have been many advances made in CV pharmacogenomics. Yet, to date, there have been few functional variants identified that associate with pharmacogenomic or treatment related CV outcomes in HTN patients. In order to discover variants that may be functional, we assessed non-synonymous SNPs (nsSNPs) from the Illumina HumanCVD chip4 for association with treatment related outcomes in the INternational VErapamil SR-Trandolapril STudy (INVEST), a HTN outcomes trial.5 Additionally, we replicated our findings in the NORdic DILtiazem (NORDIL) study, another HTN outcomes trial with similar treatment regimens. 6
METHODS
Study participants
The INVEST-GENEtic Substudy (INVEST GENES) collected DNA from 5,979 patients with HTN and coronary artery disease (CAD) enrolled in INVEST (www.clinicaltrials.gov, NCT00133692), residing in the United States and Puerto Rico. The genetic substudy was approved by the Institutional Review Board at the University of Florida; all patients provided voluntary, written informed consent; and all study procedures were in accordance with institutional guidelines and adhered to the principles of the Declaration of Helsinki and the U.S. Code of Federal Regulations for Protection of Human Subjects. The methods and results of INVEST have been previously published.5 Briefly, patients were randomized to a verapamil-SR (CCB) or an atenolol (βB) based treatment strategy with Hydrochlorothiazide (HCTZ) and trandolapril available as add-on treatment for BP control. The primary outcome (PO) was the first occurrence of all-cause death, nonfatal MI or nonfatal stroke. Secondary outcomes included the individual components of the PO. Overall, BP control and cardiovascular outcomes were similar between the two strategies.5
A nested case-control cohort was constructed from INVEST-GENES. Cases (n=269) were defined as those patients experiencing the PO. Each case was frequency matched to 4 controls (n=1076) based on age by decade, gender, and race/ethnicity. Patients in the case-control cohort were primarily classified as white (n=795), Hispanics (n=380), and black (n=170). The results presented here were limited to the white and Hispanic race/ethnic groups, as there is limited power in the black race/ethnic group.
DNA was collected on 4,200 Swedish subjects from NORDIL, a HTN outcomes trial comparing a CCB strategy (diltiazem), to conventional antihypertensive treatment utilizing diuretics, βBs, or both. The primary endpoint was fatal or nonfatal stroke, fatal or nonfatal MI, or other CV death, and information is available on all death events. The methods and results of NORDIL have been previously published.6
Additional details on the study designs and participants for INVEST and NORDIL are available in the online-only Data Supplement.
Genotyping and Quality Control
All 1345 INVEST patients in the nested case-control cohort were successfully genotyped on the Illumina HumanCVD (Cardiovascular Disease) Beadchip (Illumina, San Diego, CA), a gene-centric array containing ~50,000 SNPs in ~2,100 genes involved in cardiovascular, inflammatory, and metabolic processes.4 4,196 NORDIL subjects were successfully genotyped on the Illumina 610Quad genome-wide array (Illumina, San Diego, CA). Additional genotyping and quality control details are available in the online-only Data Supplement.
Statistical Analysis
This analysis was limited to nsSNPs on the HumanCVD Beadchip, with a minor allele frequency (MAF) ≥ 0.03, in order to identify markers with the greatest likelihood for functional effect and the greatest potential to replicate across race/ethnic groups in INVEST. Genome-spanning nsSNP × treatment interaction analyses were conducted by race/ethnic group under an additive genetic model using PLINK (http://pngu.mgh.harvard.edu/purcell/plink/).7 Analyses were adjusted for age, sex, principal components for ancestry, and history of MI, heart failure, and diabetes. The primary inference was made from the SNP × treatment interaction analysis in whites. The screening threshold was set at interaction P<0.05 in whites, based on the a priori hypothesis that nsSNPs are likely to be functional. SNPs meeting the screening P-value were examined in Hispanics for evidence of association. The significance threshold to move forward to our external replication cohort was P<0.05, with consistent direction of association. Thus, significant hits had a P<0.00125 (0.05 × 0.05 × 0.5). Gene regions were also examined, as patterns of linkage disequilibrium (LD) may vary between race/ethnic groups. For SNPs that showed consistent evidence of a pharmacogenomic (treatment interaction) association in both race/ethnic groups, whites and Hispanics were pooled together and a combined treatment interaction analysis was conducted. Adjusted odds ratios (OR) and 95% confidence intervals (CIs) for the occurrence of the PO was assessed by genotype using a logistic regression model, under both additive and dominant genetic models of inheritance, using SAS version 9.2 (Cary, NC). Pairwise measure of r2 and D’ of top hits were assessed using Haploview version 4.2.8
Based on the initial findings in the three groups (INVEST whites, INVEST Hispanics and NORDIL), the final genetic risk score was calculated from rs16982743, rs893184, and rs4525. One point was given for each genotype that conferred higher risk in the CCB arm/group versus the βB arm/group. The potential genetic risk scores ranged from 0 to 3 (one point each for SIGLEC12 rs16982743 GG (Gln/Gln), A1BG rs893184 GG (Arg/Arg), and F5 rs4525 AA (His/His)). The risk scores were dichotomized: low risk (0-1 points) and high risk (2-3 points), as only a small number of subjects had a score of 0 and 3, and they responded similarly to those with a score of 1 and 2, respectively. Adjusted ORs and 95% CIs for the occurrence of the PO were assessed by genetic risk score using a logistic regression model in SAS version 9.2 (Cary, NC). Secondary outcomes were also tested in INVEST with the final risk score. The significance level for the final risk score × treatment interaction in INVEST whites and Hispanics was P<0.05, and the significance level in NORDIL was a one-sided P<0.05, as we had a one-sided hypothesis for replication. Meta-analysis for the risk score was conducted using METAL and PLINK.7, 9 A weighted model was also tested but discarded because it was more complex but not more informative. Additional details of the statistical methods are available in the online-only Data Supplement.
RESULTS
Study Population and Baseline Characteristics
Baseline demographics and characteristics of the INVEST-GENES case-control cohort and NORDIL subjects are shown in Table 1. On average, INVEST subjects were older than NORDIL subjects. In addition, INVEST subjects had lower systolic BP (SBP) and diastolic BP (DBP) but this is likely explained by the fact that there was no washout of antihypertensive drugs in INVEST and 87.6% of INVEST subjects were treated at entry, while there was a washout in NORDIL (Table 1). Additionally the baseline demographics and characteristics of the INVEST-GENES case-control cohort and NORDIL subjects stratified by treatment group are shown in Table S1 (in the online-only Data Supplement).
Table 1. Baseline Demographics and Characteristics.
| Characteristic | INVEST Whites (n=795) |
INVEST Hispanics (n=380) |
NORDIL (n=4196) |
|---|---|---|---|
| Age, mean (SD) y | 70.8 ± 9.5 | 72.3 ± 8.8 | 60.1 ± 6.6 |
| Male | 415 (52.2) | 185 (48.7) | 2093 (49.9) |
| Primary Outcome Case | 159 (20.0) | 76 (20.0) | 376 (9.0) |
| BMI, mean (SD) kg/m2 | 28.6 ± 5.4 | 27.8 ± 4.6 | 28.2 ± 4.4 |
| SBP, mean (SD), mm Hg | 148.7 ± 17.5 | 148.1 ± 18.6 | 172.6 ± 18.9 |
| DBP, mean (SD), mm Hg | 81.7 ± 10.4 | 85.1 ± 10.1 | 103.1 ± 7.2 |
| Heart Rate, mean (SD), beats/min |
75.2 ± 9.3 | 73.6 ± 9.1 | N/A |
| History of | |||
| Diabetes | 153 (19.2) | 90 (23.7) | 342 (8.2) |
| Heart Failure (class I to III) | 50 (6.3) | 18 (4.7) | N/A |
| Myocardial Infarction | 280 (35.2) | 53 (13.9) | 101 (2.4) |
Values are presented as number (percentage) unless otherwise noted. SD: Standard Deviation. kg: kilograms. m: meters. SBP: systolic blood pressure. DBP: diastolic blood pressure. mm Hg: millimeters of Mercury. min: minute. N/A: characteristic not available in NORDIL genetic study
Genome-spanning Interaction Analysis and Between Race Consistency in INVEST
After SNP quality control procedures and applying a MAF cutoff of ≥0.03, there were 1,313 nsSNPs analyzed in the white race/ethnic group in INVEST. Sixty-five nsSNPs in the white race/ethnic group had a treatment interaction P<0.05. Two nsSNPs from whites showed evidence of consistent association in Hispanics: additive P<0.05 and consistent direction of association (Table 2, Figure S1). Allele counts and Hardy-Weinberg Equilibrium data are shown in Table S2.
Table 2. SNP-Treatment Interaction in INVEST.
| SNP | Chr | Position | Gene | Amino Acid Change |
Minor Allele |
Race | MAF Frequency |
Interaction P-value |
Combined Interaction P-value |
|---|---|---|---|---|---|---|---|---|---|
| rs16982743 | 19 | 56,696,715 | SIGLEC12 | Q29X | A (Stop) |
White Hisp |
0.179 0.195 |
0.0329 0.0215 |
0.0038 |
| rs893184 | 19 | 63,556,291 | A1BG | H52R | A (His) |
White Hisp |
0.046 0.101 |
0.0248 0.0310 |
0.0036 |
Chr: Chromosome. MAF: Minor Allele Frequency. Position: NCBI build 36 position.
The first SNP, rs16982743 in SIGLEC12 (sialic acid binding Ig-like lectin 12), results in a stop codon at amino acid (a.a.) position 29 (white Interaction P=0.0329, Hispanic Interaction P=0.0215, combined Interaction P=0.0038; Table 2, Figure S1A). In whites at rs16982743, stop codon carriers (A carriers) showed similar risk for the PO when treated with the CCB vs. βB strategy (OR (95% CI) = 0.78 (0.43-1.40), P=0.4015); whereas in those subjects with the Gln/Gln (G/G) genotype, the risk for PO was higher in the CCB strategy compared with the βB strategy (OR (95% CI) = 1.94 (1.21-3.14), P=0.0065, Interaction P=0.0159). There was a consistent association in Hispanics, so the white and Hispanic groups were pooled for a combined analysis, OR (95%CI) for stop codon carriers and Gln/Gln subjects were 0.82 (0.51-1.33) (P=0.4241), and 1.91 (1.29-2.83) (P=0.0013), respectively; Interaction P=0.0038; Figure S1A.
The second nsSNP that showed consistent pharmacogenomic association in whites and Hispanics was rs893184, located in A1BG (alpha-1-B glycoprotein), (white Interaction P=0.0248, Hispanic Interaction P=0.0310, combined Interaction P=0.0036; Table 2, Figure S1B). rs893184 causes a histidine (His) to arginine (Arg) substitution at a.a position 52 in A1BG. His carriers had a lower risk for the PO in the CCB strategy in whites and Hispanics, whereas Arg/Arg patients had a higher risk for the PO in the CCB strategy. After pooling the race/ethnic groups, the observed association was stronger (combined Interaction P=0.0036; Figure S1B).
LD was assessed between the two nsSNPs since they are located 6.85 Mb apart on chromosome 19. Overall there was very low LD between them, r2 values of 0 and 0.02 in INVEST whites and Hispanics, respectively; and D’ values of 0.03 and 0.24 in INVEST whites and Hispanics, respectively (Table S3).
Additionally, gene regions identified from the interaction analysis in whites were investigated in Hispanics. The F5-SELP-SELL-SELE region on chromosome 1 showed strong evidence of association in both whites and Hispanics: multiple nsSNPs associated in each race group. In this region, there were three independent nsSNPs with evidence of association in the white race/ethnic group (rs9332701, rs6131, and rs5361) (Figure S2) and two independent nsSNPs with evidence of association in the Hispanic race/ethnic group (rs4525, and rs6125) (Figure S3). Through LD assessment and haplotype interaction analysis, we discovered a two SNP haplotype spanning SELP and SELE, rs6125 and rs5361, was associated with treatment interaction in whites and replicated in Hispanics (white Interaction P=0.0097, Hispanic Interaction P=0.0028, Combined Interaction P= 0.0003, Table S4).
Genetic Risk Score and Replication in NORDIL
We built a genetic risk score in INVEST based on the two nsSNPs that showed evidence of association in both INVEST whites and INVEST Hispanics (rs16982743 and rs893184). Results from the treatment interaction analyses with the two nsSNP risk score were significant in whites (Interaction P=0.0012), Hispanics (Interaction P=0.0499), and combined (Interaction P=0.0004); with subjects with a low risk score (zero or one) more neutral to treatment strategy or trending towards favoring the CCB strategy (combined OR (95% CI) = 0.78 (0.50-1.22), P=0.2791) and subjects with a high risk score (two) experiencing significant risk in the CCB strategy (combined OR (95% CI) = 2.19 (1.45-3.33), P=0.0002). Since we also identified a region on chromosome 1 that showed consistent evidence of association in both INVEST whites and Hispanics, we evaluated a three nsSNP risk score including the two nsSNPs on chromosome 19, and adding each of the significant SNPs on chromosome 1. We also evaluated a model including the two nsSNPs on chromosome 19 with the significant haplotype from the chromosome 1 region. Four of the five 3-SNP models (including the chromosome 19 nsSNPs and then rs9332701, rs4525, rs6131, or rs6125) and the haplotype model showed equal or stronger evidence of association compared to the 2 SNP model (combined interaction P≤0.0004).
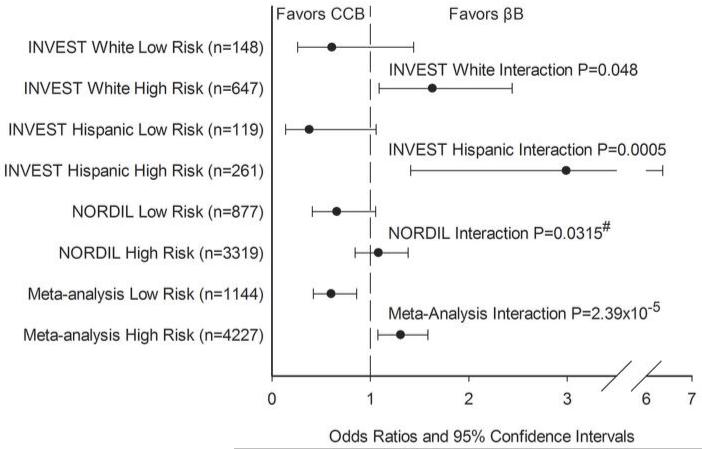
Next, we sought to replicate the genetic risk score models in NORDIL. We tested three of the four 3-SNP models above (the model with rs9332701 was not tested due to low rs9332701 MAF in NORDIL, and the haplotype model was not tested due to the fact that NORDIL data used imputed SNPs, thus construction of haplotypes was not appropriate). These data suggested that rs4525 (His865Arg) in F5 was the most informative nsSNP for the F5-SELP-SELL-SELE region across studies as part of a 3-SNP model (Table S5). The results of the treatment interaction analysis with the 3 SNP model (rs16982743, rs893184, and rs4525) genetic risk score relative to the PO for each of the three cohorts is shown in Figure 1. We observed consistent associations with the risk score in INVEST whites and Hispanics, with subjects with the low risk score (zero or one) being neutral to treatment strategy or trending towards favoring the CCB strategy, and subjects with the high risk score (two or three) being at significant risk in the CCB strategy (favoring the βB strategy). In NORDIL, these results replicated, with a significant risk score × treatment interaction (NORDIL Interaction P=0.0315, one sided P-value). When all three studies were combined in meta-analysis, the interaction P-value was more significant (Interaction P=2.39×10−5). In those with a low genetic risk score, CCB treatment was associated with lower risk of the PO (OR (95% CI) = 0.60 (0.42-0.86) P=6.51×10−3), and for those with a high genetic risk score, CCB treatment was associated with higher risk of the PO compared to βB treatment, OR (95% CI) = 1.31 (1.08-1.59, P=6.55×10−4), Figure 1).
Figure 1. Risk-Score pharmacogenomic association with the primary outcome.
Risk score calculated from SIGLEC12 rs16982743, A1BG rs893184, and F5 rs4525. One point was given for each genotype that conferred higher risk in the CCB arm/group versus the βB arm/group: rs16982743 GG (Gln/Gln), rs893184 GG (Arg/Arg), and rs4525 AA (His/His). Low Risk defined as a risk score = 0 or 1; High Risk defined as a risk score = 2 or 3.
#: One-sided P-value based on one-sided hypothesis in replication cohort.
Despite limited power, additional analyses of the risk score on secondary outcomes by genetic risk score in INVEST suggested that each element of the PO contributes (Figure S4). Both nonfatal MI, and all-cause death, appeared to contribute to the PO pharmacogenomic effect (white/Hispanic combined Interaction P = 0.0457, and 0.0026, respectively). Additionally, nonfatal stroke contributed to the PO pharmacogenomic effect in Hispanics (Interaction P=0.0404).
DISCUSSION
We performed a genome-spanning analysis of nsSNPs in cardiovascular, metabolic and inflammatory candidate genes to investigate genetic variants associated with differential treatment related outcomes in high risk HTN patients. Through this analysis, we identified three regions with a pharmacogenetic association with treatment related outcomes in white and Hispanic patients with HTN and CAD treated with a CCB versus βB strategy. In two of these regions a single SNP was associated in treatment interaction analysis: SIGLEC12 rs16982743 (Gln29Stop), and A1BG rs893184 (His54Arg). The third region on chromosome one, containing F5, SELP, SELL and SELE, showed evidence of association at multiple SNPs across INVEST whites and Hispanics. A risk score accounting for all three regions was significantly associated with treatment outcomes in INVEST whites, INVEST Hispanics and replicated in NORDIL.
This study represents the first pharmacogenetics study in HTN to identify and replicate genetic markers for differential treatment related outcomes across large clinical trials. This was likely aided by selecting SNPs that had associations in independent race/ethnic groups within INVEST for testing in a different clinical trial population. Additionally, the use of the genetic risk score enhanced the ability to replicate the finding in NORDIL. Risk scores have been more commonly used in disease genetics,10, 11 but their use in pharmacogenomics is valuable for future clinical translation. This is especially true for HTN pharmacogenomics, as often multiple signals are likely involved, as demonstrated in this study. Risk scores are typically constructed from prior association signals and tested in an independent population.10, 11 In this study, we constructed the score in the discovery population, but we have replicated the score in an independent population.
SIGLEC12, located at 19q13.4, encodes the sialic acid binding Ig-like lectin 12 gene. Siglecs are a family of single-pass, type I transmembrane proteins belonging to the immunoglobulin superfamily. Most siglecs are expressed on cells of the immune system or the hematopoietic system.12 SIGLEC12 is expressed on the epithelial cell surface and on some macrophages.12 It has been suggested that SIGLEC12 could be involved in the negative regulation of macrophage signaling by functioning as an inhibitory receptor.13 Additionally, other members of the Siglec family have been shown to be expressed abundantly on macrophages recruited during the pathogenesis of atherosclerosis.14 While there are no published data on the Gln29Stop SNP, premature stop codons are essentially always functional, and one this early in the protein would particularly be expected to be functional.
The Alpha 1B-Glycoprotein Precursor (A1BG) is also located at 19q13.4 (6.85 Mb away from SIGLEC12) and encodes a plasma glycoprotein with unknown function. The A1BG protein is a member of the immunoglobulin super family, and has been shown to form a complex with CRISP-3 (cysteine-rich secretory protein 3), a protein present in neutrophilic granulocytes that is thought to play a role in innate immunity.15
The last region identified from the treatment interaction analyses contained signals in three genes: F5, coagulation factor V; SELP, selectin P; and SELE, selectin E. Since we did not have a consistent signal across all three populations, it is likely we have not found the causal variant in this region. However, we did observe an association or trend in each group, and overall, rs4525 in F5 was the best representation of the signal. All three of these genes are located at chromosome 1q22-q25 and span approximately 220kb with SELL (selectin L) located between SELP and SELE. F5 encodes an essential cofactor in the blood coagulation cascade, participating as half of the complex that activates prothrombin to thrombin during the blood clotting process.16 SELP and SELE are members of the selectin family of cell adhesion molecules and both mediate leukocyte rolling, a process essential for the initiation of atherosclerosis.17 P-selectin is stored platelets and endothelial cells, and upon activation, redistributes to the plasma membrane, whereas, E-selectin is only expressed on endothelial cells.17
Prior genetic associations have been seen with F5, SELP and SELE and CVD. SNPs in F5 have been previously associated with DBP,18 CVD,19 and have shown a significant interaction with pravastatin in relation to CV events.20 Association studies have found that variants in SELP are associated with soluble and cell surface measures of P-selectin,21 coronary heart disease,21 and MI.22 Finally, variants in SELE have been associated with BP,23 essential HTN,24 CAD,25 stroke,26 and coronary heart disease in chlorthalidone (diuretic) treated patients from the GenHAT study.27
It is noteworthy that all three regions identified from a genome-spanning interrogation of nsSNPs are related to immune cell trafficking and/or are in the immunoglobulin superfamily. The functions of F5, P-selectin and E-selectin have been well characterized and variants in all three genes have been previously associated with CVD. Our data suggest that this region is also associated with treatment related CV outcomes in HTN patients. In addition, we observed evidence of an association with treatment related CV outcomes with variants in A1BG and SIGLEC12. While the exact functions of these genes are unknown, our results may hint that their functions are also related to the progression of the atherosclerotic environment in the vasculature. From our results, it is unclear whether the CCB strategy advanced this progression leading to increased risk in the genotypes identified, or if the βB strategy reduced this risk. We hypothesize that the latter would seem more plausible since it is more likely that one of the drugs is protective in a given genotype, versus a drug adding risk in a specific genotype.
Our study is not without limitations. We did not reach a Bonferroni significance level in our initial treatment interaction analysis of nsSNPs on the HumanCVD Beadchip, and we cannot discount the possibility of reaching these results by chance: with a P<0.00125 for 1313 nsSNPs analyzed in INVEST whites; we would expect 1.6 significant hits, and we found 2. However, our analysis only focused on nonsynonymous SNPs, giving them a higher probability of being functional variants. Additionally, consistent associations in both whites and Hispanics in INVEST reduces the risk of these representing a false positive. Replication of the risk score constructed from these SNPs in NORDIL further reduces the likelihood of this representing a chance finding. We also had reduced power in by race analyses. Still, we were able to identify three regions with evidence of association, and our genetic risk score constructed from these three regions was significant in INVEST whites, INVEST Hispanics, and NORDIL. Lastly, we may have missed true signals by the design of our study, since one of our replication cohorts had a different racial/ethnic background. Nevertheless, true functional variants should be functional across all populations and there are numerous examples for this within the pharmacogenomics literature.
PERSPECTIVES
There is great inter-patient variability in antihypertensive drug response, with only about 50% of patients responding to any one drug,3 and limited data available to guide treatment selection. Our study strived to identify novel genetic markers associated with treatment related outcomes in patients with HTN and CAD. We found that nsSNPs in A1BG, SIGLEC12 and the Selectin region are statistically associated with treatment related adverse CV outcomes in three independent hypertensive populations. By constructing a genetic risk score to account for all three regions, we were able to provide a potential approach for identifying which patients would benefit from a CCB strategy versus a βB strategy, in regards to treatment related adverse CV outcomes. These results require further investigation in order to clarify their role in HTN treatment, and to establish the mechanism through which they elicit their effect. However, the specific genetic associations and our risk score method provide insight into a potential approach to personalized antihypertensive treatment selection.
Supplementary Material
NOVELTY AND SIGNIFICANCE.
1) What is New?
Non-synonymous SNPs in SIGLEC12, A1BG, and the Selectin region are associated with antihypertensive treatment related adverse cardiovascular events.
2) What is Relevant?
There is great inter-patient variability in adverse outcomes related to antihypertensive treatment. This research identifies SNPs that may contribute to this variability and may help explain differential outcomes with a given HTN treatment based on genotype.
3) Summary
Non-synonymous SNPs in SIGLEC12, A1BG, and the Selectin region are associated with treatment related adverse cardiovascular outcomes in patients with HTN and CAD. A risk score accounting for the three regions provides a potential approach for identifying which patients would benefit from a CCB strategy versus a βB strategy.
ACKNOWLEDGEMENTS
We thank the INVEST site investigators and the INVEST-GENES participants. We thank Professor Thomas Hedner (Department of Clinical Pharmacology, Sahlgrenska Academy, Gotheburg, Sweden) and Professor Sverre Kjeldsen (Ullevaal University Hospital, University of Oslo, Oslo, Norway), who are investigators of the NORDIL study.
SOURCES OF FUNDING
This project was supported by NIH grants R01HL074730, U01GM074492, and NIH CTSA grant UL1RR092890, as well as grants from the University of Florida Opportunity Fund and Abbott Pharmaceuticals. INVEST was supported by the University of Florida and grants from BASF Pharma and Abbott Laboratories. Additional support for this work includes: NIH grants T32 DK007518 (CWM) and K23 HL086558 (RMCD). The NORDIL clinical study was supported by a grant from Pharmacia. The GWAS study of the NORDIL cohort was supported by the following grants: British Heart Foundation (BHF) Chair CH/98001 to AFD; BHF Programme RG/07/005/23633 to AFD, SP; BHF Special Project Grant SP/08/005/25115 to AFD, SP; European Union Ingenious HyperCare Consortium: Integrated Genomics, Clinical Research, and Care in Hypertension (grant number LSHM-C7-2006-037093). SP was supported by an intermediate research fellowship and a travel fellowship from the BHF (FS/05/095/19937; FS/10/016/28162).
Footnotes
CONFLICTS OF INTEREST/DISCLOSURES
Drs. Johnson, Pepine, Cooper-DeHoff, and Langaee have received funding from Abbott Laboratories.
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Roger VL, Go AS, Lloyd-Jones DM, Benjamin EJ, Berry JD, Borden WB, Bravata DM, Dai S, Ford ES, Fox CS, Fullerton HJ, Gillespie C, Hailpern SM, Heit JA, Howard VJ, Kissela BM, Kittner SJ, Lackland DT, Lichtman JH, Lisabeth LD, Makuc DM, Marcus GM, Marelli A, Matchar DB, Moy CS, Mozaffarian D, Mussolino ME, Nichol G, Paynter NP, Soliman EZ, Sorlie PD, Sotoodehnia N, Turan TN, Virani SS, Wong ND, Woo D, Turner MB. Heart disease and stroke statistics--2012 update: A report from the american heart association. Circulation. 2012;125:e2–e220. doi: 10.1161/CIR.0b013e31823ac046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chobanian AV, Bakris GL, Black HR, Cushman WC, Green LA, Izzo JL, Jr., Jones DW, Materson BJ, Oparil S, Wright JT, Jr., Roccella EJ. Seventh report of the joint national committee on prevention, detection, evaluation, and treatment of high blood pressure. ypertension. 2003;42:1206–1252. doi: 10.1161/01.HYP.0000107251.49515.c2. [DOI] [PubMed] [Google Scholar]
- 3.Materson BJ, Reda DJ, Cushman WC, Massie BM, Freis ED, Kochar MS, Hamburger RJ, Fye C, Lakshman R, Gottdiener J, Ramirez EA, Henderson WG. Single-drug therapy for hypertension in men. A comparison of six antihypertensive agents with placebo. The department of veterans affairs cooperative study group on antihypertensive agents. N Engl J Med. 1993;328:914–921. doi: 10.1056/NEJM199304013281303. [DOI] [PubMed] [Google Scholar]
- 4.Keating BJ, Tischfield S, Murray SS, Bhangale T, Price TS, Glessner JT, Galver L, Barrett JC, Grant SF, Farlow DN, Chandrupatla HR, Hansen M, Ajmal S, Papanicolaou GJ, Guo Y, Li M, Derohannessian S, de Bakker PI, Bailey SD, Montpetit A, Edmondson AC, Taylor K, Gai X, Wang SS, Fornage M, Shaikh T, Groop L, Boehnke M, Hall AS, Hattersley AT, Frackelton E, Patterson N, Chiang CW, Kim CE, Fabsitz RR, Ouwehand W, Price AL, Munroe P, Caulfield M, Drake T, Boerwinkle E, Reich D, Whitehead AS, Cappola TP, Samani NJ, Lusis AJ, Schadt E, Wilson JG, Koenig W, McCarthy MI, Kathiresan S, Gabriel SB, Hakonarson H, Anand SS, Reilly M, Engert JC, Nickerson DA, Rader DJ, Hirschhorn JN, Fitzgerald GA. Concept, design and implementation of a cardiovascular gene-centric 50 k snp array for large-scale genomic association studies. PLoS One. 2008;3:e3583. doi: 10.1371/journal.pone.0003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pepine CJ, Handberg EM, Cooper-DeHoff RM, Marks RG, Kowey P, Messerli FH, Mancia G, Cangiano JL, Garcia-Barreto D, Keltai M, Erdine S, Bristol HA, Kolb HR, Bakris GL, Cohen JD, Parmley WW. A calcium antagonist vs a non-calcium antagonist hypertension treatment strategy for patients with coronary artery disease. The international verapamil-trandolapril study (invest): A randomized controlled trial. JAMA. 2003;290:2805–2816. doi: 10.1001/jama.290.21.2805. [DOI] [PubMed] [Google Scholar]
- 6.Hansson L, Hedner T, Lund-Johansen P, Kjeldsen SE, Lindholm LH, Syvertsen JO, Lanke J, de Faire U, Dahlof B, Karlberg BE. Randomised trial of effects of calcium antagonists compared with diuretics and beta-blockers on cardiovascular morbidity and mortality in hypertension: The nordic diltiazem (nordil) study. Lancet. 2000;356:359–365. doi: 10.1016/s0140-6736(00)02526-5. [DOI] [PubMed] [Google Scholar]
- 7.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. Plink: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of ld and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 9.Willer CJ, Li Y, Abecasis GR. Metal: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meigs JB, Shrader P, Sullivan LM, McAteer JB, Fox CS, Dupuis J, Manning AK, Florez JC, Wilson PW, D’Agostino RB, Sr., Cupples LA. Genotype score in addition to common risk factors for prediction of type 2 diabetes. N Engl J Med. 2008;359:2208–2219. doi: 10.1056/NEJMoa0804742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ripatti S, Tikkanen E, Orho-Melander M, Havulinna AS, Silander K, Sharma A, Guiducci C, Perola M, Jula A, Sinisalo J, Lokki ML, Nieminen MS, Melander O, Salomaa V, Peltonen L, Kathiresan S. A multilocus genetic risk score for coronary heart disease: Case-control and prospective cohort analyses. Lancet. 2010;376:1393–1400. doi: 10.1016/S0140-6736(10)61267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitra N, Banda K, Altheide TK, Schaffer L, Johnson-Pais TL, Beuten J, Leach RJ, Angata T, Varki N, Varki A. Siglec12, a human-specific segregating (pseudo)gene, encodes a signaling molecule expressed in prostate carcinomas. J Biol Chem. 2011;286:23003–23011. doi: 10.1074/jbc.M111.244152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu Z, Lai CM, Maoui M, Banville D, Shen SH. Identification and characterization of s2v, a novel putative siglec that contains two v set ig-like domains and recruits protein-tyrosine phosphatases shps. J Biol Chem. 2001;276:23816–23824. doi: 10.1074/jbc.M102394200. [DOI] [PubMed] [Google Scholar]
- 14.Munday J, Floyd H, Crocker PR. Sialic acid binding receptors (siglecs) expressed by macrophages. J Leukoc Biol. 1999;66:705–711. doi: 10.1002/jlb.66.5.705. [DOI] [PubMed] [Google Scholar]
- 15.Udby L, Sorensen OE, Pass J, Johnsen AH, Behrendt N, Borregaard N, Kjeldsen L. Cysteine-rich secretory protein 3 is a ligand of alpha1b-glycoprotein in human plasma. Biochemistry. 2004;43:12877–12886. doi: 10.1021/bi048823e. [DOI] [PubMed] [Google Scholar]
- 16.Mann KG, Kalafatis M. Factor v: A combination of dr jekyll and mr hyde. Blood. 2003;101:20–30. doi: 10.1182/blood-2002-01-0290. [DOI] [PubMed] [Google Scholar]
- 17.Kansas GS. Selectins and their ligands: Current concepts and controversies. Blood. 1996;88:3259–3287. [PubMed] [Google Scholar]
- 18.Ehret GB, O’Connor AA, Weder A, Cooper RS, Chakravarti A. Follow-up of a major linkage peak on chromosome 1 reveals suggestive qtls associated with essential hypertension: Gennet study. Eur J Hum Genet. 2009;17:1650–1657. doi: 10.1038/ejhg.2009.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Auro K, Alanne M, Kristiansson K, Silander K, Kuulasmaa K, Salomaa V, Peltonen L, Perola M. Combined effects of thrombosis pathway gene variants predict cardiovascular events. PLoS Genet. 2007;3:e120. doi: 10.1371/journal.pgen.0030120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maitland-van der Zee AH, Peters BJ, Lynch AI, Boerwinkle E, Arnett DK, Cheng S, Davis BR, Leiendecker-Foster C, Ford CE, Eckfeldt JH. The effect of nine common polymorphisms in coagulation factor genes (f2, f5, f7, f12 and f13) on the effectiveness of statins: The genhat study. Pharmacogenet Genomics. 2009;19:338–344. doi: 10.1097/fpc.0b013e32832933b7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barbaux SC, Blankenberg S, Rupprecht HJ, Francomme C, Bickel C, Hafner G, Nicaud V, Meyer J, Cambien F, Tiret L. Association between p-selectin gene polymorphisms and soluble p-selectin levels and their relation to coronary artery disease. Arterioscler Thromb Vasc Biol. 2001;21:1668–1673. doi: 10.1161/hq1001.097022. [DOI] [PubMed] [Google Scholar]
- 22.Tregouet DA, Barbaux S, Escolano S, Tahri N, Golmard JL, Tiret L, Cambien F. Specific haplotypes of the p-selectin gene are associated with myocardial infarction. Hum Mol Genet. 2002;11:2015–2023. doi: 10.1093/hmg/11.17.2015. [DOI] [PubMed] [Google Scholar]
- 23.Sass C, Pallaud C, Zannad F, Visvikis S. Relationship between e-selectin l/f554 polymorphism and blood pressure in the stanislas cohort. Hum Genet. 2000;107:58–61. doi: 10.1007/s004390000325. [DOI] [PubMed] [Google Scholar]
- 24.Chang YP, Liu X, Kim JD, Ikeda MA, Layton MR, Weder AB, Cooper RS, Kardia SL, Rao DC, Hunt SC, Luke A, Boerwinkle E, Chakravarti A. Multiple genes for essential-hypertension susceptibility on chromosome 1q. Am J Hum Genet. 2007;80:253–264. doi: 10.1086/510918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wenzel K, Felix S, Kleber FX, Brachold R, Menke T, Schattke S, Schulte KL, Glaser C, Rohde K, Baumann G, Speer A. E-selectin polymorphism and atherosclerosis: An association study. Hum Mol Genet. 1994;3:1935–1937. doi: 10.1093/hmg/3.11.1935. [DOI] [PubMed] [Google Scholar]
- 26.Flex A, Gaetani E, Papaleo P, Straface G, Proia AS, Pecorini G, Tondi P, Pola P, Pola R. Proinflammatory genetic profiles in subjects with history of ischemic stroke. Stroke. 2004;35:2270–2275. doi: 10.1161/01.STR.0000140740.19421.fe. [DOI] [PubMed] [Google Scholar]
- 27.Lynch AI, Eckfeldt JH, Davis BR, Ford CE, Boerwinkle E, Leiendecker-Foster C, Arnett DK. Gene panels to help identify subgroups at high and low risk of coronary heart disease among those randomized to antihypertensive treatment: The genhat study. Pharmacogenet Genomics. 2012;22:355–366. doi: 10.1097/FPC.0b013e3283516ff8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.