Abstract
Most breast cancers at diagnosis are estrogen receptor (ER)-positive and depend on estrogen for growth and survival. Blocking estrogen biosynthesis by aromatase inhibitors (AI) has therefore become a first-line endocrine therapy for post-menopausal women with ER-positive breast cancers. Despite providing substantial improvements in patient outcome, AI resistance remains a major clinical challenge. The receptor tyrosine kinase RET and its co-receptor GFRα1 are upregulated in a subset of ER-positive breast cancers, and the RET ligand, glial-derived neurotrophic factor (GDNF) is upregulated by inflammatory cytokines. Here we report the findings of a multidisciplinary strategy to address the impact of GDNF-RET signaling in the response to AI treatment. In breast cancer cells in 2D and 3D culture, GDNF-mediated RET signaling is enhanced in a model of AI resistance. Further, GDNF-RET signaling promoted the survival of AI-resistant cells and elicited resistance in AI-sensitive cells. Both these effects were selectively reverted by the RET kinase inhibitor NVP-BBT594. Gene expression profiling in ER-positive cancers defined a proliferation-independent GDNF-response signature that prognosed poor patient outcome and, more importantly, predicted poor response to AI treatment with the development of resistance. We validated these findings by demonstrating increased RET protein expression levels in an independent cohort of AI-resistant patient specimens. Together, our results establish GDNF-RET signaling as a rational therapeutic target to combat or delay the onset of AI resistance in breast cancer.
Keywords: aromatase inhibitor, breast cancer, GDNF, resistance, RET
Introduction
Approximately 70% of breast tumors are positive for estrogen receptor alpha (ERα, called hereafter ER) expression, and the majority of these rely upon estrogen (E2)-mediated ER signaling for their growth. Endocrine therapy is the most common and effective treatment for this subset of breast cancers; targeting ER function by antagonizing binding of estrogens to the ER (selective ER modulators, e.g. tamoxifen), promoting ER degradation (selective ER downregulators, e.g. fulvestrant, also known as ICI182,780) or blocking estrogen biosynthesis (aromatase inhibitors, AIs) (1). AIs have become the first-line treatment choice for post-menopausal women with ER+ breast cancers (2). However, de novo or acquired AI resistance still limits their benefit for many patients. Several molecular mechanisms have been proposed to contribute to AI resistance. First, tumor cells can become hypersensitive to residual E2 and remain dependent on ER signaling for their growth (3). Of relevance for the current study, some ER+ breast cancer cells lines cultured long-term under E2 deprivation (LTED) display ER hypersensitivity to E2, thus modeling breast cancers that have developed resistance to AI treatment (4, 5). Second, tumor cells may escape the inhibitory effects of AIs by increasing ER activity independently of E2. This can result from EGFR, HER2 or IGF-IR overexpression (4, 6) leading to the activation of signaling cascades including the MAPK and PI3K/AKT pathways that promote ER phosphorylation, cell proliferation and cell survival (7).
These findings highlight the concept that combining AIs with therapies targeting signaling pathways that interact with ER is a strategy to enhance AI therapy response and prevent resistance, and have led to a number of combination therapy clinical trials. For example targeting of HER2 with trastuzumab or lapatinib in combination with the nonsteroidal AIs anastrozole or letrozole, respectively, has shown clinical benefit and improved outcome for metastatic breast cancer patients compared to treatment with AIs alone (8, 9). Further, the BOLERO-2 study reported recently that the mTOR inhibitor everolimus combined with the AI exemestane improved progression-free survival compared to exemestane alone in patients with ER+ advanced breast cancer previously treated with the AIs letrozole or anastrozole (10). However, despite the positive outcome of such trials, many patients fail to benefit from these combined therapeutic approaches. As a consequence there remains an urgent need to better understand the mechanisms of AI resistance, and to find and develop appropriate and more efficient therapeutic strategies.
Expression of the receptor tyrosine kinase RET (REarranged during Transfection) and its co-receptor GFRα1 (glycosyl phosphatidylinositol anchored GDNF family α-receptor-1) are low in normal breast but upregulated in a subset of ER+ breast cancers (11-13). Moreover, we have previously demonstrated that the RET ligand glial cell derived neurotrophic factor (GDNF) is upregulated by inflammatory cytokines and is expressed on infiltrating stromal fibroblasts and to a lesser extent by tumour cells in xenograft models (11). In RET+ ER+ breast cancer cells, GDNF stimulation results in an E2-independent increase in ER phosphorylation and transcriptional activity (13). However, little is known about the transcriptional program associated with GDNF-RET signaling in breast cancer cells or the relevance of this pathway to human disease. In particular, a role for GDNF-RET signaling in response and resistance to AI treatment has yet to be explored. In this study, we have identified a GDNF response gene set (RGS) with prognostic and predictive value in breast cancer, and demonstrate the utility of targeting GDNF-RET signaling in the context of AI treatment.
Material and Methods
Cell lines and assays
All cell lines were STR profiled in December 2012 by DNA Diagnostic Centre (DCC, London, UK). MCF7 cells used in the microarray experiments were maintained long-term in phenol red-free RPMI 1640 medium plus 10% dextran charcoal-treated fetal bovine serum (DCC), 1 nM E2 (Sigma), 2 mM L-glutamine, 50 U/ml penicillin and 50 μg/ml streptomycin. Long-term E2 deprived (LTED) cells were generated as previously described (4) by culturing cells in phenol red-free RPMI 1640 plus 10% DCC for a minimum of 20 weeks. MCF7, T47D and ZR75-1 cells were cultured over the same period in phenol red-free RPMI 1640 supplemented with 10% fetal bovine serum (FBS), 10 μg/ml insulin and 1 nM E2. MCF7 cells expressing full-length human aromatase (MCF7-2A) at clinically relevant levels or transfected the pBabeneo backbone (MCF7-neo) have been previously described (14). MCF7-2A and MCF7-neo cells were maintained in RPMI 1640 containing 10% FBS, 2 mM L-glutamine, and 1 mg/ml Geniticin/G418 (Invitrogen). For functional analysis, cells were E2-deprived for 3 days by culturing in phenol red-free RPMI 1640 supplemented with 10% DCC.
Cell based assays, siRNA transfection, immunohistochemistry, antibodies, quantitative real-time PCR (qRT-PCR) analysis were as described previously (11, 13) except the TaqMan probe sets (Applied Biosystem): RET (Hs00240887_m1), GFRA1 (Hs00237133_m1), ESR1 (Hs00174860_m1), PGR (Hs01556702_m1) and the following antibodies (Cell Signaling Technology): phospho-RET-Tyr905 (#3221), phospho-c-Jun (#2361) and c-Jun (#9165).
Generation of the GDNF-response gene set (GDNF-RGS)
MCF7 cells were E2-deprived for 3 days and then serum-starved overnight in the presence or absence of 100 nM ICI182,780 (Tocris Bioscience). The following day, cells were treated with GDNF (20 ng/ml) for 0, 4, 8, 24 and 48 hours in the presence or absence of ICI182,780. Triplicate samples from 3 independent experiments were hybridized onto whole genome HumanHT12_v3 Expression BeadChips (Illumina) by the Genomics Services Group, Wellcome Trust Centre for Human Genetics, Oxford. Data were extracted using BeadStudio (Illumina) software and were transformed and normalized using variance-stabilizing transformation and robust spline normalization method in the Lumi (2.6.0) package in R (http://www.bioconductor.org). Probes were discarded if they were not detected in any of the samples (detection p>0.01). Microarray data have been submitted to ArrayExpress database (E-MEXP-3662). To identify genes significantly regulated by GDNF treatment, a confidence score (CS) (15, 16) was calculated for each gene at each time point of GDNF treatment. CS was defined as the sum of individual scores given for fold change (FC), p-value (PV), expression level (EL), and present calls (PC).
Analysis of clinical datasets
Breast cancer subtypes in the NKI295 and Pawitan datasets were as reported by the authors. In TransBig dataset, the subtypes were retrieved and classify using PAM50 from ROCK (17). An unscaled GDNF-RGS score that recapitulates the degree of similarity to MCF7 cells upon GDNF treatment was generated as described previously (18). Thus, a high tumor GDNF-RGS score corresponds to a signature highly concordant with GDNF-activation in MCF7 cells.
Kaplan-Meier analysis and multivariate Cox proportional hazard regression analysis were carried out with the survival (2.36-12) and survplot (0.0.6) packages in R. GDNF-RGS positive and negative tumors were classified using the centroid Spearman correlation method with the nearest centroid >0.1 as described previously (19). The sample was not assigned if the correlation was ≤0.1.
Correlation of GDNF-RGS with response to letrozole
The cohort of patients treated with neoadjuvant letrozole has been described previously (20). Briefly, core biopsies from ER+ tumors were collected pre- and post-14 days letrozole treatment and subject to gene expression analysis. Response was classified based on tumor volume reduction after 3 months letrozole treatment as assessed by ultrasound. Patients with >50% reduction in tumor volume were considered responders. Follow-up data were available for 52 of the 58 patients. To examine the association between the GDNF-RGS, the log2 intensity (median-centered) of the 53 out of 67 GDNF-RGS genes available in the dataset were extracted using the ROCK database. When multiple probe sets mapped to the same gene, the one with the highest variance in the dataset was selected.
Statistical analysis
Statistical analysis was performed using GraphPad Prism Software as reported in the Figure legends and Results.
Results
GDNF-RET signaling is enhanced in an in vitro model of aromatase inhibitor (AI) resistance
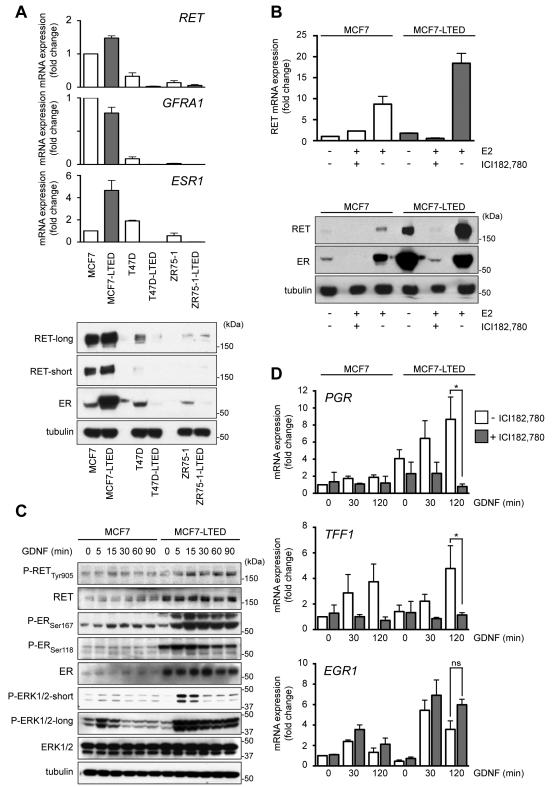
MCF7-LTED provide a widely accepted model of breast cancer cells that have developed resistance to aromatase inhibitor (AI) treatment (21). To investigate GDNF-RET signaling in the context of AI resistance, we monitored the time-dependent changes in RET, its co-receptor GFRA1 and ESR1 mRNA expression in MCF-7 cells and in two additional ER+ cell lines (T47D and ZR75-1) during E2 deprivation (Supplementary Fig. S1A). Compared to parental MCF7 cells, MCF7-LTED cells show a marked increase in RET and ER mRNA and protein expression (Fig. 1A). This increase in RET expression is comparable to that observed in an independent model of E2-deprived MCF7 cells (22). Compared to MCF7 cells, parental T47D cells express lower levels of RET (Fig. 1A) but retain responsiveness to GDNF/GFRα1 treatment (12). As previously reported (12), parental ZR75-1 have the lowest level of RET expression. By contrast to MCF7-LTED cells, T47D-LTED and ZR75-1-LTED cells have undetectable levels of ER, RET and GFRA1 (Fig. 1A) and do not respond to GDNF/GFRα1 treatment (Supplementary Fig. S1B). RET expression can be regulated by ER activation (12, 23) and in the in vitro cell models employed here, RET expression levels mirror the levels of ER expression (Fig. 1A). Moreover, when parental MCF7 cells are cultured in the presence of E2, RET expression is enhanced and this can be blocked by ICI182,780 treatment, that targets ER for proteasomal degradation (Fig. 1B). MCF7-LTED cells are hypersensitive to residual E2 (4, 5) and in the absence of exogenously added E2, a higher level of RET expression is observed (Fig. 1A and B). E2-mediated ER activation in these cells again results in increased RET expression that can be blocked by ICI182,780 treatment (Fig. 1B).
Figure 1.
Differential response to long-term E2 deprivation in ER+ breast cancer cells. A, top panel, qRT-PCR analysis for RET (n=4), GFRA1 (n=3) and ESR1 (n=3) in MCF7, T47D and ZR75-1 parental cells and their LTED derivatives. Data represent mean±SEM. Bottom panel, immunoblotting of total cell protein extracts. High and low exposures with RET antibody are shown. B, 3 day E2-deprived MCF7 and MCF7-LTED cells were serum-starved overnight and cultured for further 48 hours (top panel) or 96 hours (bottom panel) ± 10 pM E2 ± 100 nM ICI182,780. qRT-PCR analysis for RET (n=3). Data represents mean±SEM. C, 3 day E2-deprived MCF7 and MCF7-LTED cells were serum-starved overnight and stimulated with 20 ng/ml GDNF. D, 3 day E2-deprived MCF7 and MCF7-LTED cells were serum-starved overnight ± 1 μM ICI182,780 and stimulated with 20 ng/ml GDNF. qRT-PCR analysis for PGR (n=3), EGR1 (n=4), TFF1 (n=3). Data represent mean±SEM. Two-way ANOVA, Tukey-corrected, *p<0.05.
As the majority of the breast tumors that display endocrine therapy resistance retain ER expression, we employed MCF7-LTED cells to investigate GDNF-RET signaling in the context of AI resistance. The increased RET expression in MCF7-LTED cells is mirrored by enhanced GDNF-RET downstream signaling including increased ER activation monitored by Ser167 and Ser118 phosphorylation (24) (Fig. 1C) and transcriptional upregulation of E2-dependent genes TFF1 and PGR (Fig. 1D). Notably, the upregulation of TFF1 and PGR, but not that of the ER-independent gene EGR1, is inhibited in the presence of ICI 182,780. Consistent with their higher levels of RET expression and more sustained GDNF-induced RET signaling, MCF7-LTED cells show increased GDNF-induced transcriptional activation compared to MCF7 cells (Fig. 1D).
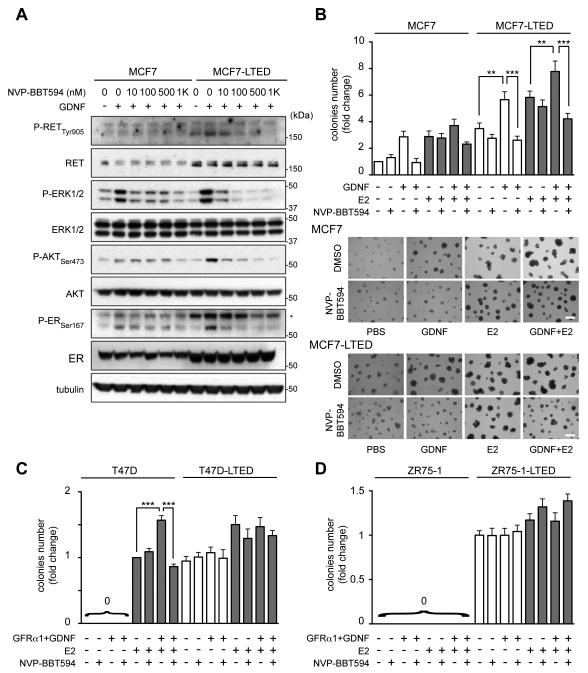
NVP-BBT594 impairs GDNF-RET signaling and GDNF-dependent growth of MCF7-LTED cells
Blocking RET with nM concentrations of the RET inhibitor NVP-BBT594 (Fig. 2A) or siRNA transfection (Supplementary Fig. S1C) demonstrates that GDNF signaling and ER phosphorylation are mediated solely via the RET receptor. Similarly, NVP-BBT594 blocks the GDNF-mediated enhancement of MCF7-LTED cell viability in 2D culture (not shown) and 3D colony formation (Fig. 2B). Compared to MCF7-LTED cells, parental MCF7 cells form smaller colonies in 3D culture but respond to GDNF and NVP-BBT594 treatment. The addition of 10 pM E2, to mimic the E2 level in post-menopausal patients that have relapsed on AI treatment and ceased AI therapy, increases 3D colony formation of both MCF7 and MCF7-LTED cells, and this effect is efficiently reverted by NVP-BBT594 (Fig. 2B). Parental T47D cells cultured in the absence of E2 and parental ZR75-1 cells, with or without E2, do not form colonies when cultured on Matrigel (Fig. 2C,D and Supplementary Fig. S1D). However, as previously reported (12) when parental T47D cells are cultured in presence of low level E2, GFRα1/GDNF stimulation results in increased 3D colony formation, which is significantly reverted by NVP-BBT594 (Figure 2C). Conversely, consistent with their low-level ER and RET expression (Fig. 1A,B), T47D-LTED and ZR75-1-LTED cells do not respond to GFRα1/GDNF stimulation and are minimally respond to the presence of E2 (Fig. 2C,D, Supplementary Fig. S1C). Importantly, NVP-BBT594 has no significant impact on T47D-LTED and ZR75-1-LTED 3D colony formation demonstrating that the effects observed in MCF7-LTED cells are due to selective RET inhibition by NVP-BBT594 rather than off-target toxicity.
Figure 2.
NVP-BBT594 impairs GDNF-RET signaling and GDNF-dependent growth in LTED cells that retain RET and ER expression. A, 3 day E2-deprived MCF7 and MCF7-LTED cells were serum-starved overnight and treated with NVP-BBT594 for 90 min followed by stimulation ± 20 ng/ml GDNF for 30 min. B-D, MCF7, T47D and ZR75.1 cells and LTED derivatives were plated on Matrigel ± GDNF (20 ng/ml), GFRα1 (100 ng/ml), NVP-BBT594 (100 nM) vehicle (DMSO) and E2 (10 pM). Colonies >200 μm in diameter was counted at day 7. Data represent the mean fold increase over control cells ± SEM, n=4. Data indicated as 0 denotes no colony growth. One-way ANOVA, Tukey-corrected, ** p<0.01, ***p<0.001. Representative images are shown. Scale bar, 400 μm.
GDNF-promoted AI resistance can be reverted by RET inhibition
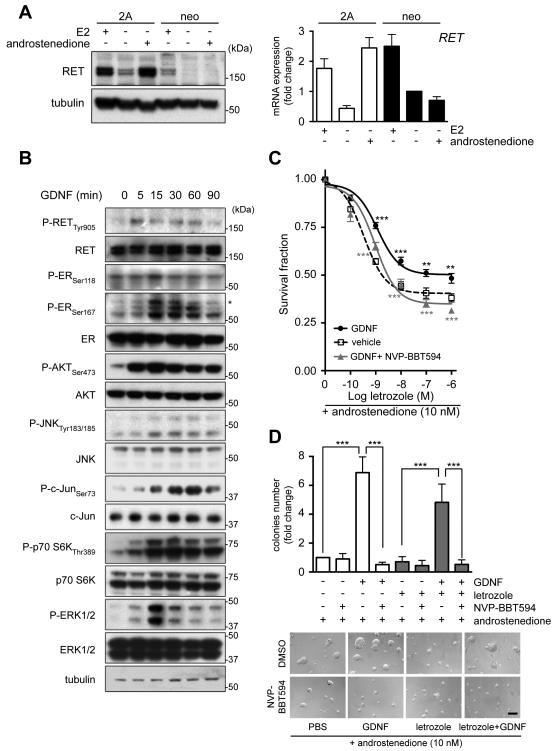
To assess further the effect of GDNF signaling in the response and adaptation to AI treatment we utilized MCF7 cells expressing aromatase enzyme (MCF7-2A) or the backbone vector (MCF7-neo). Treatment with androstenedione, which is converted into estrogens by aromatase, results in a concentration-dependent increase in MCF7-2A cell growth, but has no effect on MCF7-neo cells (Supplementary Fig. S2A). Conversely, increasing concentrations of letrozole, or the alternate AIs exemestane and anastrozole, in the presence of androstenedione impair MCF7-2A, but not MCF7-neo, cell survival (Supplementary Fig. S2B). Consistent with the known E2-mediated modulation of RET expression (see Fig. 1B) (12, 23), MCF7-2A cells show increased RET levels compared to MCF7-neo cells, and in both cell lines RET expression is reduced by E2 deprivation (Fig. 3A). Treatment with androstenedione restores basal RET mRNA and protein expression in the MCF7-2A cells but has no effect in MCF7-neo cells (Fig. 3A). Importantly, letrozole impairs androstenedione-induced RET expression both at mRNA and protein level in MCF7-2A cells (Supplementary Fig. S2C).
Figure 3.
NVP-BBT594 targets GDNF-RET signaling and sensitizes MCF7-2A cells to letrozole treatment. A, MCF7-2A or MCF7-neo cells were E2-deprived for 3 days with addition of 1 nM E2 or 10 nM androstenedione for the last 24 hours. Left panel, total cell protein extracts were subject to western blotting. Right panel, qRT-PCR analysis for RET (n=3). Data represents mean±SEM. B, MCF7-2A cells were E2-deprived for 3 days, serum-starved overnight and stimulated with 20 ng/ml GDNF. C, MCF7-2A cells in 2D culture were E2-deprived for 3 days and then cultured in the presence of 10 nM androstenedione in the presence of vehicle, 20 ng/ml GDNF or 20 ng/ml GDNF plus 100 nM NVP-BBT594 with the indicated concentration of letrozole for 6 days. Data represent mean survival fraction±SEM, n=3. Two-way ANOVA, Bonferroni-corrected, **p<0.01, ***p<0.001. Grey and black asterisks referred to statistical test between GDNF and GDNF+NVP-BBT594 and between untreated and GDNF treated samples, respectively. D, MCF7-2A cells were cultured on Matrigel in the presence of 10 nM androstenedione ± GDNF (20 ng/ml), NVP-BBT594 (100 nM), vehicle (DMSO) or letrozole (10 nM). Colonies >200 μm in diameter were counted at day 7. Data represent mean fold increase over control cells±SEM, n=3. One-way ANOVA, Bonferroni-corrected, ***p<0.001. Representative images are shown. Scale bar, 200 μm.
GDNF stimulation of AI sensitive MCF7-2A cells results in RET autophosphorylation, activation of ERK1/2 and AKT and enhanced ER phosphorylation (Fig. 3B). The GDNF-induced ER phosphorylation is abrogated by the mTOR inhibitor everolimus, partially blocked by PI3K/AKT and JNK inhibition but unaffected by MEK inhibition (Supplementary Fig. S2D). MCF7-2A cells show a high hormone dependency as evidenced by their inability to grow when deprived of androstenedione (Supplementary Figure S2A). Consequently for cell based assays, MCF7-2A cells were cultured with 10 nM androstenedione. GDNF administration significantly increases the resistance of MCF7-2A cells to letrozole (no GDNF, SF50 = 1.71 nM; plus GDNF, SF50 = 802 nM) (Fig. 3C), and the RET inhibitor NVP-BBT594 impairs GDNF-mediated RET downstream signaling (Supplementary Fig. S2E) and significantly enhances the antiproliferative effects of letrozole (SF50 = 2.9 nM) (Fig. 3C). Of note, the effect of GDNF on MCF7-2A cells is more pronounced when cells are cultured in 3D (Fig. 3D). In these experimental conditions that better mimic in vivo tumor growth, GDNF promotes colony formation both in the absence and presence of letrozole, while NVP-BBT594 completely abrogates this GDNF-induced resistance (Fig. 3D).
Identification of GDNF response genes in breast cancer cells
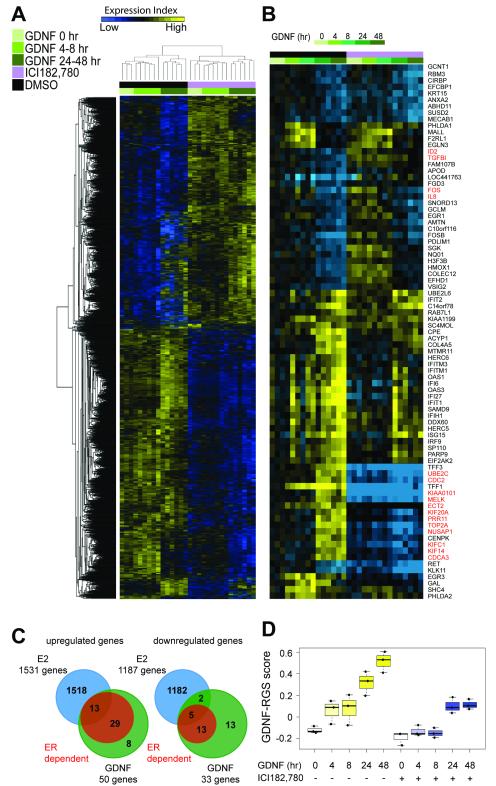
The preclinical in vitro models described here, together with our previous findings (13), suggest that increased RET expression and activation in ER+ breast cancers can promote resistance to endocrine therapy. However, the transcriptional program induced by GDNF-RET signaling in ER+ breast cancer cells and in particular the role of GDNF-induced ER-dependent versus ER-independent signaling in response to endocrine therapy is unknown. To address this, E2-deprived MCF7 cells were pretreated with or without ICI182,780 that targets ER for proteasomal degradation and thereby blocks expression of ER-dependent genes (Supplementary Fig. S3A). Cells were then GDNF stimulated for 0, 4, 8, 24 or 48 hours and RNA from 3 independent experiments was subject to gene expression profiling. Hierarchical cluster analysis shows that the samples divide into ICI182,780 treated and untreated groups and that within these two groups, the samples cluster according to early (4 - 8 hr) and late (24 - 48 hr) GDNF response (Fig. 4A). qRT-PCR of independent samples was used to validate the gene expression profiling and confirms that GDNF treatment induces transcriptional activation of ER-dependent genes TFF1 and TOP2A in MCF7 cells and that this activation is blocked by pretreatment with ICI 182,780. Conversely, the ER-independent genes ISG15 and PARP9 are upregulated in response to GDNF treatment both in the presence and absence of ICI 182,780 (Supplementary Fig. S3B).
Figure 4.
Identification of a GDNF transcriptional program in breast cancer cells. A, unsupervised hierarchical clustering of RNA transcripts in E2-deprived MCF7 cells treated with GDNF for 0, 4, 8, 24, and 48 hours ± ICI182,780 in three independent experiments. B, gene cluster analysis performed for the 83 genes significantly regulated by GDNF with a CS ≥11. In red are highlighted the 16 proliferation related genes removed to generate the 67 gene GDNF-response gene set (GDNF-RGS) (see Supplementary Fig. S4) C, comparison of genes upregulated (left) and downregulated (right) in E2-treated MCF7 cells (data from (25)) with genes regulated by GDNF in the absence of ICI182,780 (GDNF) and GDNF in the presence of ICI182,780 (ER-dependent). D, time course of GDNF-RGS score of the samples subject to gene expression profiling. Box and whisker plots represent median, 25 and 75 percentile values. Dots represent the three independent experiments.
Gene set enrichment analysis (GSEA) was applied to identify gene sets correlated with GDNF treatment in the presence or absence of ICI182,780 (Supplementary Table S1A). Many of the identified gene sets are related to response to serum, metabolic and apoptosis pathways, DNA damage and immune response pathways. Importantly, no correlation was found between the GDNF regulated genes in MCF7 cells and other growth factor response gene sets indicating that GDNF regulated genes do not have a substantial overlap with other growth factor signaling pathways.
To detect genes significantly regulated by GDNF a confidence score (CS) was calculated for each gene at each time point of GDNF treatment with a cut-off of ≥11.0, as reported previously (15, 16). 83 genes, 50 upregulated and 33 downregulated, were identified (Fig. 4B, C and Supplementary Table S1B). Gene ontology (GO) analysis revealed that a significant fraction of the GDNF regulated genes are functionally associated with immune system processes, apoptosis and response to stimulus (chemical, oxidative stress, biotic) (Supplementary Table S1C). A comparison of the gene expression profiles in the presence and absence of ICI182,780 revealed that 42 out of 50 (84%) and 18 out of 33 (54.5%) of GDNF upregulated and downregulated genes, respectively, were fully or partially dependent on ER (Fig. 4C). A comparison of GDNF-regulated genes with a comprehensive E2-regulated gene dataset (25) revealed that not all of the GDNF/ER-dependent genes are reported to be E2-dependent (Fig. 4C). This suggests that GDNF treatment can promote ER-mediated transcription of a subset of genes that are independent of the canonical E2 pathway involving estrogen response element sites. This is consistent with a previous study (26) that reported a subset of EGF-induced ER genomic targets that are distinct from those induced by E2 (see Discussion).
A proliferation-independent GDNF-response gene set (RGS) positivity correlates with poor clinical outcome
It is well established that proliferation related genes can dominate gene expression signatures, which de facto identify highly proliferative tumors (27). Consequently, the 83 GDNF-dependent gene list was robustly filtered (Supplementary Fig. S4). First, all potential proliferation related genes based on GO analysis were removed. Second, genes previously reported in two independent proliferation metagene signatures were removed (28, 29). Finally, the remaining 69 genes were correlated to Ki67 protein levels and TOP2A expression in a dataset of 81 ER+ breast cancers (30). Genes that showed rs>0.5 or rs<-0.5 were removed to generate the final GDNF-response gene set (RGS) comprising 67 proliferation-independent genes (Fig. 4B and Supplementary Table S1). The proliferation-independent GDNF-RGS is largely populated by GDNF late response genes and, as a result, the GDNF-RGS score is higher in the samples treated with GDNF for 24 - 48 hours (Fig. 4D). ICI182,780 has a significant impact on the GDNF-RGS score as 54 out of 67 of the GDNF-RGS genes are ER-dependent (Fig. 4C). However, samples that were GDNF-treated in presence of ICI182,780 for 24 - 48 hours show a higher GDNF-RGS score than untreated samples (without GDNF and without ICI182,780 pretreatment) indicating that GDNF-dependent ER-independent genes are accounted for within the GDNF-RGS score (Fig. 4D).
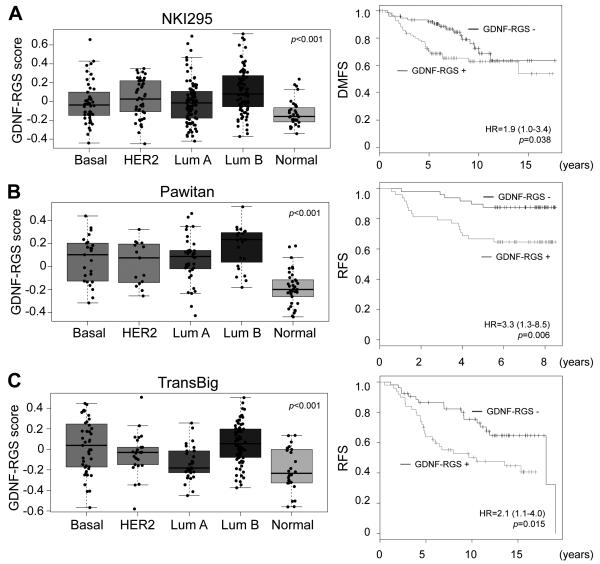
Breast cancers can be divided into molecular intrinsic subtypes that differ in their gene expression and clinical outcomes. Among the ER+ breast cancers, two subtypes can be distinguished: luminal A and luminal B (31). The GDNF-RGS score differs significantly between subtypes (ANOVA: p<0.001, Fig. 5A, B and C) with a Bonferroni-corrected analysis showing that GDNF-RGS score is higher in luminal B than in luminal A tumors in all 3 datasets analyzed (Fig. 5A p<0.001; Fig. 5B p<0.05; Fig. 5C p<0.001) (32-34). Further, analysis of 597 breast cancer samples from TGCA (http://cancergenome.nih.gov) using the ROCK database (17) shows that amongst the 5 intrinsic breast cancer subtypes, expression of the RET ligand GDNF is highest in the luminal B subtype (SAM score = 0.007, q-value = 0.02). Within ER+ breast cancers, luminal B tumors are characterized by poorer prognosis when compared to luminal A tumors (35) indicating that the GDNF-RGS and expression of GDNF associate with a clinically relevant molecular subtype of breast cancer. Consistent with this, using the nearest centroid method (19), GDNF-RGS positivity in ER+ cancers significantly associates with a decrease in distant metastases free survival (DMFS) (Fig. 5A -right panel) and in relapse free survival (RFS) (Fig 5B and C - right panels). Equivalent results are obtained when patients are stratified accordingly to their GDNF-RGS score (Supplementary Fig. S5). A significant association of GDNF-RGS positivity with decreased overall survival (OS), RFS or DMFS is also found if all cases are analyzed independently of their hormonal status (Supplementary Fig. S6). Importantly, multivariate Cox analysis reveals that the GDNF-RGS has an independent prognostic value in all three datasets analyzed (Table 1, Supplementary Fig. S6). Finally, these analyses indicate that patients bearing tumors characterized by active GDNF signaling have a significantly worse outcome either in the presence (NKI295, Pawitan) or absence (TransBig) of adjuvant treatment (Fig. 5, Table 1).
Figure 5.
GDNF-RGS correlates with the luminal B phenotype and poor prognosis in human breast cancers. A-C, left panels, box and whisker plots of GDNF-RGS scores by tumor subtype in the NKI295, Pawitan and TransBig datasets, respectively, showing significantly (one-way ANOVA, p<0.001) higher scores in luminal B breast cancer subgroup. Right panels, Kaplan-Meier analyses of the ER+ cases stratified by GDNF-RGS. Likelihood ratio test p-value and hazard ratio (HR) with 95% confidence interval are shown.
Table I.
Multivariate Cox proportional-hazard regression analyses of outcome according to the GDNF-RGS.
| Variable | HR | 95% CI | p |
|---|---|---|---|
| Van de Vijer et al. 2002 dataset (226 ER+ patients) DMFS | |||
| GDNF-RGS | 1.88 | 1.02-3.47 | 0.042 |
| Tumour size (>20 vs ≤ 20 mm) | 2.12 | 1.17-3.83 | 0.013 |
| Lymph node status | 0.64 | 0.35-1.15 | 0.135 |
| Age (>50 vs ≤ 50 yrs) | 0.63 | 0.26-1.49 | 0.291 |
| Pawitan et al. 2005 dataset (130 ER+ patients) RFS | |||
| GDNF-RGS | 2.95 | 1.07-8.06 | 0.035 |
| Grade (3 vs 2/1) | 1.36 | 0.54-3.46 | 0.514 |
| Tumour size (>20 vs ≤ 20 mm) | 2.57 | 0.96-6.85 | 0.060 |
| Lymph node status | 0.67 | 0.25-1.80 | 0.431 |
| Age (>50 vs ≤ 50 yrs) | 0.42 | 0.17-1.07 | 0.060 |
| PgR status | 0.75 | 0.24-2.36 | 0.485 |
| a Desmedt et al. 2007 dataset (134 ER+ patients) RFS | |||
| GDNF-RGS | 2.04 | 1.07-3.88 | 0.030 |
| Grade (3 vs 2/1) | 1.03 | 0.50-2.10 | 0.939 |
| Tumour size (>20 vs ≤ 20 mm) | 2.03 | 1.10-3.73 | 0.023 |
| Age (>50 vs ≤ 50 yrs) | 0.96 | 0.49-1.90 | 0.909 |
DMFS: distant metastasis free survival; RFS: relapse free survival.
No patients received systemic adjuvant therapy.
GDNF-RGS correlates with response to aromatase inhibitor treatment
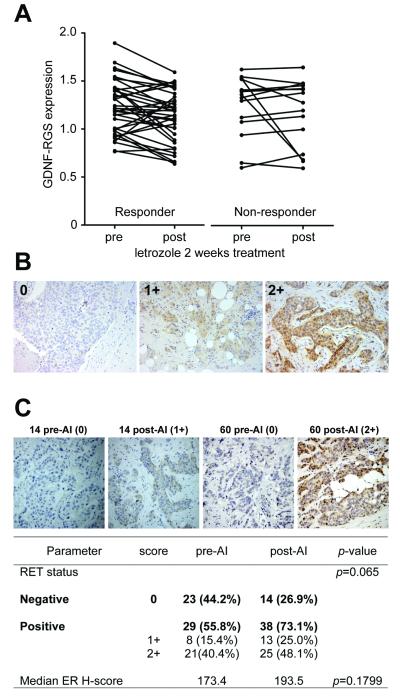
We next evaluated whether the GDNF-RGS correlates with response to AI treatment by retrieving gene expression data from biopsies of 52 ER+ breast cancers taken before and after 2 weeks of neoadjuvant letrozole treatment (20). The patients were subsequently divided into responder and non-responder groups defined by a >50% and <50% reduction, respectively, in tumor volume following a further 3 months of AI treatment. Pairwise comparison shows a significant decrease in GDNF-RGS score after 2 weeks of letrozole treatment in the responder cohort (p=0.009) but not in the non-responder cohort (p=0.804) (Fig. 6A).
Figure 6.
Correlation of GDNF-RGS and RET expression with response to aromatase inhibitors. A, changes in GDNF-RGS in 52 paired ER+ breast cancer samples pre- and post-2 week letrozole treatment (20). Responder patients (n=37) show a significant (Wilcoxon test, p=0.009) decrease in GDNF-RGS score. No significant change in GDNF-RGS was observed in the non-responder group (n=15). B, RET immunohistochemical staining of invasive breast cancers. Representative images of tumors scored as negative (0), moderate RET expression (1+) and strong RET expression (2+) are shown. C, RET expression was assessed in 52 paired primary tumor samples pre-aromatase inhibitor treatment (pre-AI) and recurrent/metastatic tumors following adjuvant AI treatment (post-AI) (37). Representative images of tumors pre-AI and post-AI are shown. Chi-square p-value for RET and paired t-test value for ER H-score (see Supplementary Fig. S8) is shown.
To validate these findings, we examined the effect of the nonsteroidal AI anastrozole on the GDNF-RGS score using gene expression data and Ki67 staining available for 69 paired ER+ tumors biopsies taken pre- and post-2 weeks of neoadjuvant anastrozole treatment (30). In such studies, a lack of response to AI treatment as monitored by a decrease in Ki67 staining has been shown to predict poor long-term disease outcome (1). The GDNF-RGS score does not correlate with levels of Ki67 in the pre-treatment samples (Supplementary Fig. S7A-C) consistent with the absence of proliferation related genes within the GDNF-RGS. However, this pre-treatment GDNF-RGS score shows a relatively weak but statistically significant correlation with the proportional two-week change in Ki67 (rs=-0.24, p=0.047) (Supplementary Fig. S7A). Furthermore, the change in GDNF-RGS score in the pre- and post-treatment samples positively correlates with post-treatment Ki67 (rs=0.32, p=0.007) and with the proportional two-week change in Ki67 (rs=0.35, p=0.0035) (Supplementary Fig. S7B,C). These data support the concept that GDNF-RET signaling plays an important role in the response and adaptation of breast cancer patients to AI treatment. Conversely, GDNF-RGS did not stratify for outcome in ER+ breast cancer patients that had exclusively received tamoxifen as adjuvant therapy (Supplementary Fig. S7D,E). This suggests that the different mechanism of action of AI and tamoxifen in breast cancer may influence the ER-dependent GDNF-mediated transcriptional profile.
RET expression increases in AI-resistant breast cancers
GDNF exerts its function as a ligand for the RET receptor tyrosine kinase (36). We have previously reported that RET expression is enhanced in primary tumors from patients who subsequently developed invasive recurrence after adjuvant tamoxifen treatment (13). To assess changes in RET protein expression in response to AI, we stained 52 paired samples of primary breast cancers and locally recurrent or metastatic tumors arising after adjuvant AI treatment (37) (Fig. 6B,C). RET expression is detected in 55.8% (29 out of 52) of the primary tumors. This percentage of RET-positive tumors is comparable to that reported for tamoxifen-resistant tumors (59.6%) and is significantly higher than found in a non-selected group of ER+ invasive breast cancers (37 out of 126; 29.4%) (13). This reflects the fact that the majority of patients in the AI treated cohort had previously received adjuvant tamoxifen treatment. After developing resistance to AI treatment, RET expression is detected in 73.1% (38 out of 52) with a borderline statistical significance (Chi-square, two-tailed: p=0.065) compared to the paired pre-treatment samples. As expected, ER expression was retained in the majority of post-AI treated samples with no significant change in ER H-score (Fig. 6C, Supplementary Figure S8).
DISCUSSION
In this study we highlight the GDNF-RET pathway as an important determinant of resistance to AI treatment in ER+ breast cancers. Crucially, direct inhibition of GDNF-RET signaling by the NVP-BBT594 compound in ER+ breast cancer cells enhances the sensitivity to AI treatment and reverts AI resistance. In addition, we have linked gene expression data derived from an in vitro experimental model to clinically relevant tumor samples. In particular, we have derived a proliferation-independent GDNF transcriptional profile and demonstrate that this correlates with worse prognosis and poor response to AI treatment in breast cancer patients.
A major concern in the generation of growth factor gene response signatures is that the signature can be dominated by proliferation related genes. As extensively reported, proliferation related gene signatures correlate with higher proliferating and higher grade tumors, and consequently with poor prognosis (27). GDNF is a weak mitogen for breast cancer cells (11, 12) but interrogation of the gene expression data shows that the GDNF induced transcriptional program included serum and other mitogen response pathways (Supplementary Table 1A, C). As a consequence, the initial list of 83 genes significantly regulated by GDNF with a CS ≥11 was subject to robust filtering to remove 16 proliferation related genes (Supplementary Fig. S4) to generate a 67 gene GDNF-RGS. The effectiveness of this approach is evidenced by the lack of correlation of the GDNF-RGS with levels of Ki67 staining and TOP2A expression in primary tumor samples (Supplementary Fig. S4). Further, the absence of proliferation genes within the GDNF-RGS has important implications for the observation that the GDNF-RGS associates with the luminal B subgroup of breast cancers Clinically, luminal B tumors have a poorer prognosis compared to luminal A tumors with an increased risk of early relapse with endocrine therapy and increased resistance to chemotherapy (31, 38). The data presented here suggest that the GDNF-mediated RET signaling in breast cancer cells triggers a transcriptional program associated with a more aggressive tumor phenotype independently of pro-mitogenic effects. Importantly, we demonstrate that the GDNF-RGS significantly correlated with a decrease in DMFS and in RFS (Fig. 5) in breast cancer patients.
Although a demonstration of prognostic value is of interest, more importantly this study revealed that a GDNF-RGS score is predictive for response to AI treatment in two independent studies (Fig. 6A and Supplementary Fig. S7A-C). Moreover, the correlation of the change in GDNF-RGS and the change in proliferation index of the tumors indicates that activation of GDNF signaling is also associated with the response to AI treatment. Clues as to the mechanism by which GDNF-RET signaling may promote the response and adaptation of breast cancers to AI treatment has come from taking a global approach to examine GDNF-RET signaling in breast cancer cells. First, this study has revealed that GDNF can promote both E2-independent activation of ER (Fig. 2 and 3) and a non-canonical ER transcriptional program (Fig. 4C). Promotion of a non-canonical ER cistrome has been reported in breast cancer cells following EGF stimulation in a process dependent on the transcription factor AP-1 (26). It is of note that GDNF-stimulation of MCF7-2A cells activates c-Jun (Fig. 3B), a key component of the AP-1 complex. Second, the GDNF-RGS is enriched with genes related to immune response pathways, in particular STAT1 target genes (Supplementary Table 1, Fig. 4B). STAT1 mediates the inflammatory response of interferon (IFN). IFN-related genes, such as ISG15, OAS1, IFI27 and OAS3 that are present in the GDNF-RGS, have been associated previously with radiation and chemotherapy resistance in breast and other cancers (39-41). Similarly, Dunbier and colleagues have identified an inflammatory gene expression signature associated with poor response to neoadjuvant AI treatment (30). In contrast to these reports, others have shown that an immune related signature is associated with better prognosis in triple negative and HER2+ breast cancers (28, 42) and that the presence of tumor-associated lymphocytes predicts good response to chemotherapy (43) and good clinical outcome in ER− cancers (44). These contrasting reports highlight the complex role of the immune system in different breast cancer subgroups and in the response to different therapeutic regimes. What is notable in this study is that the GDNF-RGS was derived from the MCF7 experimental model rather than from tumor specimens containing both tumor and stromal cells. This suggests that within the GDNF-RGS-positive tumors, the tumor cells are actively involved in the immune response. We have previously demonstrated that GDNF is secreted in response to pro-inflammatory cytokines by both tumor cells and stromal fibroblasts (11). This raises the possibility that GDNF-mediated upregulation of immune response pathways can reinforce GDNF signaling to promote cell survival in the AI resistant setting.
Despite the ability to identify breast cancer subsets, predict disease outcome and/or response to therapies (45, 46), there is still a lack of well defined targets causally associated with resistance to endocrine therapy that can be translated into the clinic. In recent years a number of studies have provided evidence that activation of growth factor signaling pathways could be a significant contributor to the luminal B phenotype of ER+ breast cancers. In particular ERBB2 and EGFR (4, 47), IGF-1R (48), FGFR (49) and more recently RET (13) and PDGFR (37) have been identified as potential targets in ER+ breast cancer (38). Given the plethora of growth factor signaling pathways that can impact upon ER, it has been considered desirable to target common downstream pathways in ER+ breast cancers. mTOR inhibitors have shown a beneficial effect on progression free survival when combined with AI therapy (10). However, as demonstrated here, GDNF activates multiple downstream pathways and, given the adaptability of tumor cells, targeting only one is likely to lead to compensatory upregulation of others. Such compensatory mechanisms have been well documented, for example, inhibition of mTOR in breast cancer cells results in enhanced IGF-1R signaling by abrogating a negative feedback loop (50). Similarly, we show that blockade of GDNF-induced MEK/ERK1/2 and PI3K signaling in MCF7-2A cells results in increased AKT and c-Jun phosphorylation, respectively (Supplementary Fig. S2).
Here we demonstrate, using a multidisciplinary approach, that GDNF-RET signaling is an important determinant of AI therapy response and resistance in ER+ breast cancers. The priority now is to determine whether RET inhibition is achievable in the clinical setting to prolong the efficacy of AIs in recurrent and/or metastatic disease and whether targeting growth factor signaling pathways in combination with an AI could prevent or delay the onset of AI resistance.
Supplementary Material
Acknowledgments
We thank Marjan Iravani and Zara Ghazoui for array samples processing and guidance on the gene expression analysis, Paola Francica for setting up the MCF7-2A model, Antony Fearns for help with the qRT-PCR analysis, Christine Desmedt (Institut Jules Bordet) and Jorge Reis-Filho for the helpful discussion on the gene expression data, Miguel Angel Pujana (IDIBELL, Barcelona) for providing the MCF7 long-term estrogen deprivation gene expression data and Novartis (NIBR, Basel) for supplying the NVP-BBT594 inhibitor.
Financial support: The work was funded by the Association of International Cancer Research (grant 09-0533 to IP-M and CMI) and by Breakthrough Breast Cancer (CMI). We acknowledge NHS funding to the NIHR Biomedical Research Centre.
Footnotes
Conflict of interest: MD and L-AM receive funding from AstraZeneca and Pfizer. The work presented here is not related to this funding. All authors declare no other conflicts of interest.
References
- 1.Forbes JF, Cuzick J, Buzdar A, Howell A, Tobias JS, Baum M. Effect of anastrozole and tamoxifen as adjuvant treatment for early-stage breast cancer: 100-month analysis of the ATAC trial. Lancet Oncol. 2008;9:45–53. doi: 10.1016/S1470-2045(07)70385-6. [DOI] [PubMed] [Google Scholar]
- 2.Winer EP, Hudis C, Burstein HJ, Wolff AC, Pritchard KI, Ingle JN, et al. American Society of Clinical Oncology technology assessment on the use of aromatase inhibitors as adjuvant therapy for postmenopausal women with hormone receptor-positive breast cancer: status report 2004. J Clin Oncol. 2005;23:619–29. doi: 10.1200/JCO.2005.09.121. [DOI] [PubMed] [Google Scholar]
- 3.Arpino G, Wiechmann L, Osborne CK, Schiff R. Crosstalk between the estrogen receptor and the HER tyrosine kinase receptor family: molecular mechanism and clinical implications for endocrine therapy resistance. Endocr Rev. 2008;29:217–33. doi: 10.1210/er.2006-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin LA, Farmer I, Johnston SR, Ali S, Marshall C, Dowsett M. Enhanced estrogen receptor (ER) alpha, ERBB2, and MAPK signal transduction pathways operate during the adaptation of MCF-7 cells to long term estrogen deprivation. J Biol Chem. 2003;278:30458–68. doi: 10.1074/jbc.M305226200. [DOI] [PubMed] [Google Scholar]
- 5.Martin LA, Pancholi S, Chan CM, Farmer I, Kimberley C, Dowsett M, et al. The anti-oestrogen ICI 182,780, but not tamoxifen, inhibits the growth of MCF-7 breast cancer cells refractory to long-term oestrogen deprivation through down-regulation of oestrogen receptor and IGF signalling. Endocr Relat Cancer. 2005;12:1017–36. doi: 10.1677/erc.1.00905. [DOI] [PubMed] [Google Scholar]
- 6.Stephen RL, Shaw LE, Larsen C, Corcoran D, Darbre PD. Insulin-like growth factor receptor levels are regulated by cell density and by long term estrogen deprivation in MCF7 human breast cancer cells. J Biol Chem. 2001;276:40080–6. doi: 10.1074/jbc.M105892200. [DOI] [PubMed] [Google Scholar]
- 7.Miller TW, Hennessy BT, Gonzalez-Angulo AM, Fox EM, Mills GB, Chen H, et al. Hyperactivation of phosphatidylinositol-3 kinase promotes escape from hormone dependence in estrogen receptor-positive human breast cancer. J Clin Invest. 2010;120:2406–13. doi: 10.1172/JCI41680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaufman B, Mackey JR, Clemens MR, Bapsy PP, Vaid A, Wardley A, et al. Trastuzumab plus anastrozole versus anastrozole alone for the treatment of postmenopausal women with human epidermal growth factor receptor 2-positive, hormone receptor-positive metastatic breast cancer: results from the randomized phase III TAnDEM study. J Clin Oncol. 2009;27:5529–37. doi: 10.1200/JCO.2008.20.6847. [DOI] [PubMed] [Google Scholar]
- 9.Johnston S, Pippen J, Jr., Pivot X, Lichinitser M, Sadeghi S, Dieras V, et al. Lapatinib combined with letrozole versus letrozole and placebo as first-line therapy for postmenopausal hormone receptor-positive metastatic breast cancer. J Clin Oncol. 2009;27:5538–46. doi: 10.1200/JCO.2009.23.3734. [DOI] [PubMed] [Google Scholar]
- 10.Baselga J, Campone M, Piccart M, Burris HA, 3rd, Rugo HS, Sahmoud T, et al. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N Engl J Med. 2012;366:520–9. doi: 10.1056/NEJMoa1109653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esseghir S, Todd SK, Hunt T, Poulsom R, Plaza-Menacho I, Reis-Filho JS, et al. A role for glial cell derived neurotrophic factor induced expression by inflammatory cytokines and RET/GFR alpha 1 receptor up-regulation in breast cancer. Cancer Res. 2007;67:11732–41. doi: 10.1158/0008-5472.CAN-07-2343. [DOI] [PubMed] [Google Scholar]
- 12.Boulay A, Breuleux M, Stephan C, Fux C, Brisken C, Fiche M, et al. The Ret receptor tyrosine kinase pathway functionally interacts with the ERalpha pathway in breast cancer. Cancer Res. 2008;68:3743–51. doi: 10.1158/0008-5472.CAN-07-5100. [DOI] [PubMed] [Google Scholar]
- 13.Plaza-Menacho I, Morandi A, Robertson D, Pancholi S, Drury S, Dowsett M, et al. Targeting the receptor tyrosine kinase RET sensitizes breast cancer cells to tamoxifen treatment and reveals a role for RET in endocrine resistance. Oncogene. 2010;29:4648–57. doi: 10.1038/onc.2010.209. [DOI] [PubMed] [Google Scholar]
- 14.Banerjee S, Zvelebil M, Furet P, Mueller-Vieira U, Evans DB, Dowsett M, et al. The vascular endothelial growth factor receptor inhibitor PTK787/ZK222584 inhibits aromatase. Cancer Res. 2009;69:4716–23. doi: 10.1158/0008-5472.CAN-08-4711. [DOI] [PubMed] [Google Scholar]
- 15.Frasor J, Danes JM, Komm B, Chang KC, Lyttle CR, Katzenellenbogen BS. Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology. 2003;144:4562–74. doi: 10.1210/en.2003-0567. [DOI] [PubMed] [Google Scholar]
- 16.Jelinsky SA, Harris HA, Brown EL, Flanagan K, Zhang X, Tunkey C, et al. Global transcription profiling of estrogen activity: estrogen receptor alpha regulates gene expression in the kidney. Endocrinology. 2003;144:701–10. doi: 10.1210/en.2002-220728. [DOI] [PubMed] [Google Scholar]
- 17.Sims D, Bursteinas B, Gao Q, Jain E, MacKay A, Mitsopoulos C, et al. ROCK: a breast cancer functional genomics resource. Breast Cancer Res Treat. 2010;124:567–72. doi: 10.1007/s10549-010-0945-5. [DOI] [PubMed] [Google Scholar]
- 18.Taube JH, Herschkowitz JI, Komurov K, Zhou AY, Gupta S, Yang J, et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci U S A. 2010;107:15449–54. doi: 10.1073/pnas.1004900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weigelt B, Mackay A, A’Hern R, Natrajan R, Tan DS, Dowsett M, et al. Breast cancer molecular profiling with single sample predictors: a retrospective analysis. Lancet Oncol. 2010;11:339–49. doi: 10.1016/S1470-2045(10)70008-5. [DOI] [PubMed] [Google Scholar]
- 20.Miller WR, Larionov AA, Renshaw L, Anderson TJ, White S, Murray J, et al. Changes in breast cancer transcriptional profiles after treatment with the aromatase inhibitor, letrozole. Pharmacogenet Genomics. 2007;17:813–26. doi: 10.1097/FPC.0b013e32820b853a. [DOI] [PubMed] [Google Scholar]
- 21.Johnston SR, Martin LA, Dowsett M. Life following aromatase inhibitors--where now for endocrine sequencing? Breast Cancer Res Treat. 2005;93(Suppl 1):S19–25. doi: 10.1007/s10549-005-9038-2. [DOI] [PubMed] [Google Scholar]
- 22.Jan R, Huang M, Lewis-Wambi J. Loss of pigment epithelium-derived factor: a novel mechanism for the development of endocrine resistance in breast cancer. Breast Cancer Res. 2012;14:R146. doi: 10.1186/bcr3356. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Stine ZE, McGaughey DM, Bessling SL, Li S, McCallion AS. Steroid hormone modulation of RET through two estrogen responsive enhancers in breast cancer. Hum Mol Genet. 2011;20:3746–56. doi: 10.1093/hmg/ddr291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ali S, Coombes RC. Endocrine-responsive breast cancer and strategies for combating resistance. Nat Rev Cancer. 2002;2:101–12. doi: 10.1038/nrc721. [DOI] [PubMed] [Google Scholar]
- 25.Creighton CJ, Cordero KE, Larios JM, Miller RS, Johnson MD, Chinnaiyan AM, et al. Genes regulated by estrogen in breast tumor cells in vitro are similarly regulated in vivo in tumor xenografts and human breast tumors. Genome Biol. 2006;7:R28. doi: 10.1186/gb-2006-7-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lupien M, Meyer CA, Bailey ST, Eeckhoute J, Cook J, Westerling T, et al. Growth factor stimulation induces a distinct ER(alpha) cistrome underlying breast cancer endocrine resistance. Genes Dev. 2010;24:2219–27. doi: 10.1101/gad.1944810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sotiriou C, Wirapati P, Loi S, Harris A, Fox S, Smeds J, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98:262–72. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 28.Desmedt C, Haibe-Kains B, Wirapati P, Buyse M, Larsimont D, Bontempi G, et al. Biological processes associated with breast cancer clinical outcome depend on the molecular subtypes. Clin Cancer Res. 2008;14:5158–65. doi: 10.1158/1078-0432.CCR-07-4756. [DOI] [PubMed] [Google Scholar]
- 29.Ghazoui Z, Buffa FM, Dunbier AK, Anderson H, Dexter T, Detre S, et al. Close and stable relationship between proliferation and a hypoxia metagene in aromatase inhibitor-treated ER-positive breast cancer. Clin Cancer Res. 2011;17:3005–12. doi: 10.1158/1078-0432.CCR-10-1704. [DOI] [PubMed] [Google Scholar]
- 30.Dunbier A, Ghazoui Z, Anderson H, Smith I, Dowsett M. Molecular Profiling of Aromatase Inhibitor-Treated Post-Menopausal Breast Tumours Identifies Determinants of Response. Cancer Res. 2010;70(24 Suppl) Abstract nr S2-5. [Google Scholar]
- 31.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Desmedt C, Piette F, Loi S, Wang Y, Lallemand F, Haibe-Kains B, et al. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res. 2007;13:3207–14. doi: 10.1158/1078-0432.CCR-06-2765. [DOI] [PubMed] [Google Scholar]
- 33.Pawitan Y, Bjohle J, Amler L, Borg AL, Egyhazi S, Hall P, et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res. 2005;7:R953–64. doi: 10.1186/bcr1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 35.Kennecke H, Yerushalmi R, Woods R, Cheang MC, Voduc D, Speers CH, et al. Metastatic behavior of breast cancer subtypes. J Clin Oncol. 2010;28:3271–7. doi: 10.1200/JCO.2009.25.9820. [DOI] [PubMed] [Google Scholar]
- 36.Morandi A, Plaza-Menacho I, Isacke CM. RET in breast cancer: functional and therapeutic implications. Trends Mol Med. 2011;17:149–57. doi: 10.1016/j.molmed.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 37.Weigel MT, Banerjee S, Arnedos M, Salter J, A’Hern R, Dowsett M, et al. Enhanced expression of the PDGFR/Abl signaling pathway in aromatase inhibitor-resistant breast cancer. Ann Oncol. 2012 doi: 10.1093/annonc/mds240. [DOI] [PubMed] [Google Scholar]
- 38.Tran B, Bedard PL. Luminal-B breast cancer and novel therapeutic targets. Breast Cancer Res. 2011;13:221. doi: 10.1186/bcr2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weichselbaum RR, Ishwaran H, Yoon T, Nuyten DS, Baker SW, Khodarev N, et al. An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proc Natl Acad Sci U S A. 2008;105:18490–5. doi: 10.1073/pnas.0809242105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duarte CW, Willey CD, Zhi D, Cui X, Harris JJ, Vaughan LK, et al. Expression signature of IFN/STAT1 signaling genes predicts poor survival outcome in glioblastoma multiforme in a subtype-specific manner. PLoS One. 2012;7:e29653. doi: 10.1371/journal.pone.0029653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khodarev NN, Roach P, Pitroda SP, Golden DW, Bhayani M, Shao MY, et al. STAT1 pathway mediates amplification of metastatic potential and resistance to therapy. PLoS One. 2009;4:e5821. doi: 10.1371/journal.pone.0005821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Teschendorff AE, Miremadi A, Pinder SE, Ellis IO, Caldas C. An immune response gene expression module identifies a good prognosis subtype in estrogen receptor negative breast cancer. Genome Biol. 2007;8:R157. doi: 10.1186/gb-2007-8-8-r157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Denkert C, Loibl S, Noske A, Roller M, Muller BM, Komor M, et al. Tumor-associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J Clin Oncol. 2010;28:105–13. doi: 10.1200/JCO.2009.23.7370. [DOI] [PubMed] [Google Scholar]
- 44.Mahmoud SM, Paish EC, Powe DG, Macmillan RD, Grainge MJ, Lee AH, et al. Tumor-infiltrating CD8+ lymphocytes predict clinical outcome in breast cancer. J Clin Oncol. 2011;29:1949–55. doi: 10.1200/JCO.2010.30.5037. [DOI] [PubMed] [Google Scholar]
- 45.van ’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 46.Chang JC, Wooten EC, Tsimelzon A, Hilsenbeck SG, Gutierrez MC, Elledge R, et al. Gene expression profiling for the prediction of therapeutic response to docetaxel in patients with breast cancer. Lancet. 2003;362:362–9. doi: 10.1016/S0140-6736(03)14023-8. [DOI] [PubMed] [Google Scholar]
- 47.Arpino G, Green SJ, Allred DC, Lew D, Martino S, Osborne CK, et al. HER-2 amplification, HER-1 expression, and tamoxifen response in estrogen receptor-positive metastatic breast cancer: a southwest oncology group study. Clin Cancer Res. 2004;10:5670–6. doi: 10.1158/1078-0432.CCR-04-0110. [DOI] [PubMed] [Google Scholar]
- 48.Atzori F, Traina TA, Ionta MT, Massidda B. Targeting insulin-like growth factor type 1 receptor in cancer therapy. Target Oncol. 2009;4:255–66. doi: 10.1007/s11523-009-0123-z. [DOI] [PubMed] [Google Scholar]
- 49.Hynes NE, Dey JH. Potential for targeting the fibroblast growth factor receptors in breast cancer. Cancer Res. 2010;70:5199–202. doi: 10.1158/0008-5472.CAN-10-0918. [DOI] [PubMed] [Google Scholar]
- 50.O’Reilly KE, Rojo F, She QB, Solit D, Mills GB, Smith D, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–8. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.