Abstract
A large number of common disorders, including cancer, have complex genetic traits, with multiple genetic and environmental components contributing to susceptibility. A literature search revealed that even among several meta-analyses, there were ambiguous results and conclusions. In the current study, we conducted a thorough meta-analysis gathering the published meta-analysis studies previously reported to correlate any random effect or predictive value of genome variations in certain genes for various types of cancer. The overall analysis was initially aimed to result in associations (1) among genes which when mutated lead to different types of cancer (e.g. common metabolic pathways) and (2) between groups of genes and types of cancer. We have meta-analysed 150 meta-analysis articles which included 4,474 studies, 2,452,510 cases and 3,091,626 controls (5,544,136 individuals in total) including various racial groups and other population groups (native Americans, Latinos, Aborigines, etc.). Our results were not only consistent with previously published literature but also depicted novel correlations of genes with new cancer types. Our analysis revealed a total of 17 gene-disease pairs that are affected and generated gene/disease clusters, many of which proved to be independent of the criteria used, which suggests that these clusters are biologically meaningful.
Keywords: Cancer, Meta-analysis, Gene, Association, Interaction, Single-nucleotide polymorphism, Alleles, Clustering
Introduction
Cancer is the result of a complicated process that involves the accumulation of both genetic and epigenetic alterations in various genes [1]. The somatic genetic alterations in cancer include point mutations, small insertion/deletion events, translocations, copy number changes and loss of heterozygosity [2]. These changes either augment the action and/or expression of an oncoprotein or silence tumour suppressor genes. Single-nucleotide polymorphism (SNP) is the most common form of genetic variation in the human genome. Although common SNPs for disease prediction are not ready for widespread use [3], recent genome-wide association studies (GWASs) using high-throughput techniques have identified regions of the genome that contain SNPs with alleles that are associated with increased risk for cancer such as FGFR2 in breast cancer [4-7].
The knowledge on gene mutations that predispose tumour initiation or tumour development and progress will give an advantage in cancer patients' treatment. Despite the complexity and variability of cancer genome, numerous studies have examined the correlation of genome variation with cancer development and progression [8]. However, ambiguous results have been generated from the attempt to link genome variants with cancer prediction or detection. A literature search revealed that even among several meta-analyses, there were unclear results and conclusions.
We have, therefore, conducted a thorough meta-analysis of meta-analysis studies previously reported to correlate the random effect or predictive value of genome variations in certain genes for various types of cancer. The aim of the overall analysis was the detection of correlations (1) among genes whose mutation might lead to different types of cancer (e.g. common metabolic pathways) and (2) between groups of genes and types of cancer.
Methods
We performed a thorough field synopsis by studying published meta-analysis studies involving the association of various types of cancer with SNPs located in certain genomic regions. For each published meta-analysis included in our study, we also investigated the number of patients (cases) and controls, date, type of study, study group details (e.g. gender, race, age, etc.), measures included, allele and genotype frequency and also the outcome of each study, i.e. if there was an association or not, the interactions noticed in each of these studies, etc.
We have meta-analysed 150 meta-analysis articles (Additional file 1), which included 4,474 studies, 2,452,510 cases and 3,091,626 controls (5,544,136 individuals in total). The meta-analyses that have been meta-analysed included various racial groups, e.g. Caucasians, Far Eastern populations (Asian, Chinese, Japanese, Korean, etc.), African-American and other population groups (native Americans, Latinos, Aborigines, etc.). Three types of studies were included: (1) pooled analysis, (2) GWAS and (2) other studies, e.g. search in published reports. Collected data consisted of a list of genes, genomic variants and diseases with a known genotype-phenotype association (whether or not a given variation has an impact on susceptibility to a given disease). The principle of our study was to use data mining techniques to find groups (referred to as clusters hereafter) of genes or diseases that behave similarly according to related data. Such groupings will make it possible to find different cancer types susceptible to similar genotypes as well as different genes associated to similar cancer types. Furthermore, our approach would facilitate predicting whether susceptibility to one type of cancer may be indicative of predisposition to another cancer type. Moreover, the association between a group of genes and a given phenotype may suggest that these genes interact or belong to the same biochemical pathway. In order to allow data mining analysis, genotype-phenotype associations had to be classified within a fixed set of categories, i.e. yes/small yes/may/no. Moreover, genes or diseases with fewer than two entries were not considered in our analysis since their clustering would not be meaningful.
Then, data were processed using a state-of-the-art general purpose clustering tool, CLUTO [9]. Data analysis consisted in finding the tightest and most reliable groupings. Since CLUTO offers a wide range of methods, and many different scoring schemes can be used to estimate similarity between genotypes or phenotypes, cluster reliability was assessed by their robustness to clustering criteria (details are provided in Additional file 1). As a consequence, each putative association has been qualified as either ‘highly consistent’ or ‘moderately consistent’. The biological significance of those clusters was, first, evaluated using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) [10,11], a biological database and web resource of known and predicted protein-protein interactions. The STRING database contains information from numerous sources, including experimental data, computational prediction methods and public text collections. It is widely accessible, and it is regularly updated. Second, literature research was performed to complete this initial evaluation.
Results and discussion
In this study, we performed a meta-analysis of published meta-analysis studies to investigate possible correlations among genes and SNPs and various types of cancer, as well as among gene-gene and/or gene-environmental interactions. Furthermore, an advanced literature research was applied in order to evaluate our results obtained from our meta-analysis. Our data were not only consistent with previously published literature but we have also depicted novel correlations of genes with new types of cancer. Our analysis showed a total of ten cancer-related genes that are affected (Table 1).
Table 1.
Summary of genes and SNPs identified by meta-analysis to be positively correlated with various cancers
|
Gene |
Cancer type |
SNPs |
References |
Supporting references |
|
|---|---|---|---|---|---|
| rs number | Other name | ||||
|
ERCC2 |
BC |
rs13181 |
p.K715Q |
[12,13] |
[14-17] |
|
ERCC2 |
BC |
rs1799793 |
p.D312N |
[12,18] |
[14-16] |
|
ERCC2 |
LC |
rs13181 |
p.K751Q |
[19,20] |
[17,21,22] |
|
ERCC2 |
LC |
rs1799793 |
p.D312N |
[23] |
[17,21,22] |
|
CCND1 |
BC |
rs603965 |
c.870G>A |
[24] |
[25-31] |
|
CYP2E1 |
CRC |
rs3813867 |
NA |
[32] |
[32-41]a |
|
CYP2E1 |
HNC |
rs3813867 |
NA |
[42,43] |
[44] |
|
CYP2E1 |
HNC |
rs6413432 |
NA |
[42] |
[44] |
|
GSTP1 |
CRC |
rs1695 |
p.I105V |
[45] |
[39,46-55] |
|
IL6 |
BC |
rs1800795 |
c.-174G>C |
[56,57] |
|
|
MTHFR |
GC |
rs1801131 |
c.1298A>C |
[58] |
[59,60]b |
|
MTHFR |
BC |
rs1801131 |
c.677C>T, c.1298A>C |
[61,62] |
[63,64] |
|
SOD2 |
BC |
rs4880 |
p.V16A, p.A9V |
[62,65,66] |
|
|
TGFB1 |
BC |
rs1800469 |
NA |
[67-69] |
|
|
TGFB1 |
BC |
rs1800470 |
NA |
[67,70-73] |
|
|
TGFB1 |
BC |
rs1982073 |
NA |
[74] |
[64,75-77] |
|
TP53 |
BC |
rs1042522 |
p.R72P |
[78,79] |
[80-94] |
|
TP53 |
UBC |
rs1042522 |
p.R72P |
[95] |
[96-100] |
|
TP53 |
CRC |
rs1042522 |
p.R72P |
[78,101-103] |
[104-108] |
|
TP53 |
CRC |
rs17878362 |
NA |
[78] |
[104-108] |
|
TP53 |
EC |
rs1042522 |
p.R72P |
[109,110] |
[111] |
|
TP53 |
LC |
rs1042522 |
p.R72P |
[78] |
[112-117] |
|
TP53 |
LC |
rs17878362 |
NA |
[78] |
[112-117] |
| VEGFA | BC | rs3025039, rs699947 | c.936C>T, c.-2578C>A | [20,45,118,119] | [120] |
These findings are supported by the published literature. aFor a different SNP (rs1329149); bfor c.677C>T and c.1298A>C. NA not available.
Correlation of SNPs' genes with various types of cancer
The association highlighted by our meta-analysis between the CYP2E1 gene and colorectal cancer (CRC), head and neck cancer (HNC) and liver cell carcinoma (LLC) is supported by published data [33-39,44,121]. An additional literature search to evaluate our initial results revealed novel correlations of the gene combination CYP2E1 and GSTM1 with prostate cancer (PC) susceptibility, lung cancer (LC) and bladder cancer (UBC) as shown in Table 2[126-128]. A similar correlation was found in CRC using a knockdown model [32,40,41]. Studies not only confirm the possibility of association between the CCND1 gene and breast cancer (BC) [25] but also suggest involvement with squamous cell carcinoma (SCC), oesophageal cancer (EC), oral cancer (OC) and malignant glioma (MG), as arisen from the interaction between the CCND1 and CCND3 genes [26,122-124]. This is further corroborated in mouse model studies that show association of CCND1 with BC [25,27-31,153] and PC [125].
Table 2.
Summary of genes and SNPs identified by further literature search as positively correlated with various cancers
|
Gene |
Cancer type |
SNPs |
References |
|
|---|---|---|---|---|
| rs number | Other name | |||
|
CCND1 |
OC |
rs603965 |
c.870G>A |
[26,122-124] |
|
CCND1 |
PC |
rs603965 |
c.870G>A |
[125] |
|
CYP2E1 |
PC |
NA |
NA |
[126] |
|
CYP2E1 |
LC |
NA |
NA |
[127] |
|
CYP2E1 |
UBC |
NA |
NA |
[128] |
|
CYP2E1 |
OC |
NA |
NA |
[40] |
|
ERCC2 |
OC |
rs1799793, rs13181 |
p.D312N, p.K751Q |
[23] |
|
ERCC2 |
HNC |
rs1799793, rs13181 |
p.D312N, p.K751Q |
[129-131] |
|
GSTP1 |
PC |
rs1695 |
p.I105V |
[126,128,132,133] |
|
MTHFR |
BCC |
rs1801131 |
c.677C>T, c.1298A>C |
[134] |
|
MTHFR |
ALL |
rs1801131 |
c.677C>T, c.1298A>C |
[59,135,136] |
|
MTHFR |
LC |
rs1801131 |
c.677C>T, c.1298A>C |
[137] |
|
MTHFR |
UBC |
rs1801131 |
c.677C>T, c.1298A>C |
[138] |
|
MTHFR |
CC |
rs1801131 |
c.677C>T, c.1298A>C |
[139] |
|
MTHFR |
NHL |
rs1801131 |
c.677C>T, c.1298A>C |
[140,141] |
|
MTHFR |
HNC |
rs1801131 |
c.677C>T, c.1298A>C |
[142] |
|
TGFB1 |
GC |
rs1982073 |
c.+29C>T |
[143] |
|
TGFB1 |
LC |
rs1982073 |
c.+29C>T |
[144] |
|
TGFB1 |
PC |
rs1982073 |
c.+29C>T |
[145] |
|
TGFB1 |
PC |
rs1982073 |
c.+29C>T |
[146] |
|
TGFB1 |
CRC |
rs1982073 |
c.+29C>T |
[147] |
|
TP53 |
EmCa |
rs1042522/rs17878362 |
p.R72P |
[148] |
|
TP53 |
PC |
rs1042522/rs17878362 |
p.R72P |
[114,149] |
|
TP53 |
OVCa |
rs1042522/rs17878362 |
p.R72P |
[150] |
|
TP53 |
GC |
rs1042522/rs17878362 |
p.R72P |
[151] |
| TP53 | OC | rs1042522/rs17878362 | p.R72P | [152] |
NA not available.
Moreover, as far as the ERCC2 is concerned along with the association of ERCC1 gene with BC and LC which is already confirmed [14-17,21,22], we have also identified from our further literature search on humans the existence of an association with OC [26] and with HNC [129-131]. There were no similar mouse studies that could confirm or overrule our findings.
Our findings regarding the GSTP1 gene are confirmed by the published literature [39,46-55]. Furthermore, we have noticed an association with PC derived from the combination of GSTM1 and CYP1A1[126,128,132,133]. Likewise, previous experimental evidence supports the association we found between the MTHFR gene and BC, basal cell carcinoma (BCC) [63,134] and gastric cancer (GC) [59,60]. An association was also found between MTHFR gene with other types of cancer, such as acute lymphoblastic leukaemia (ALL) [135,136,154], LC [137], UBC coming from interaction between CTH and GSTM1[138], CRC [139], non-Hodgkin's lymphoma (NHL) [140,141], BC [64] and HNC [142]. Specifically, in the case of NHL, the gene combination of MTHFR and TYMS might influence the susceptibility to NHL[140,141].
Concerning TGFB1, apart from the BC [64] that was confirmed from the results of our further literature search on humans and on mouse model [75,76], we have noticed also the following associations with gastric dysplasia, LC, pancreatic cancer (PanC) and BC [77,143-146]. Also, an association of TGFB1 with CRC was found using a mouse model [147].
In addition for TP53 gene, we have observed in the results of our meta-analysis that it is associated with BC, UBC, CRC, EC and LC [80-87,96-100,104-108,111-113,149]. We have observed also that TP53 gene might be associated with OC [88,148], too. Concerning the literature research on knockout mice, we have confirmed the associations with BC [89-94] and LC [114-117], and we have found also associations with ovarian cancer (OVCa) [150], GC [151] and OC [152]. Moreover for the VEGFA gene, based on further literature TGFB1 research, we have confirmed the association with BC [120], but we had not found any other evidence supporting the association with other types of cancer.
Correlations between groups of genes and various types of cancer
We have examined and confirmed the highly consistent gene clustering results over further literature search via STRING. Our search revealed additional types of cancer, except from the types that we have studied in our meta-analysis that seems to be related with pair of genes. STRING database reports binding interaction between GSTP1 and GSTM1 genes, activating interaction between MMP2 and EGF genes, between VEGFA and IL1B genes and between MMP-9 and IL8 genes (Table 3). The application of our machine learning method has highlighted that those pair of genes have similar association profiles and, therefore, might be involved in the same pathways. The genes that do not appear in the associations do not probably correlate with the presence of a certain type of cancer.
Table 3.
Putative gene-gene associations with various cancer types
|
Gene associations |
Considered phenotypes |
Comments |
STRING confirmation |
Literature confirmation |
|
|---|---|---|---|---|---|
| Gene 1 | Gene 2 | ||||
|
GSTP1 |
GSTM1 |
4 |
|
Binding interaction |
[Reference]: study type |
|
TGFB1 |
IL6 |
5 |
4 of 5 based on ‘yes’ |
|
|
|
MMP2 |
EGF |
3 |
Based on ‘yes’ |
Activating interaction |
|
|
VEGFA |
IL1B |
2 |
|
Activating interaction |
|
|
MMP9 |
IL8 |
4 |
Based on ‘may’ |
Activating interaction |
KEGG: same process |
| MMP1 | MMP3 | 5 | Based on ‘may’ | ||
First, in our meta-analyses, we observed that the interaction between IL6 and TGFB1 genes was associated to the following types of cancer: BC, CRC, GC, LC and PC as shown in Table 4. Although further literature search on humans could not validate our highly consistent results, we discovered that these interactions are associated to additional types of cancer, such as HNC [187], CRC [158], renal cancer (RC), small cell lung cancer [188], malignant melanoma (MM) [189-192] and OVCa [193]. Additionally, regarding our further research on the interaction between IL6 and TGFB1 genes on mouse models, we have confirmed our initial results principally for BC [155-157] and PC [159] and have noticed associations with epithelial cancer [194], skin tumour [195], LC [196], OVCa and cervical cancer (CC) [197,198] and HNSCC [199]. Second, we found that the interaction between MMP-2 and EGF was associated with LC, BC and GC (Table 4). Subsequently with a further literature search, we confirmed the association with BC osteolysis [163,164] and also found new associations with EC [200], LC, RC and PC [162]. Furthermore, in some cases, we have observed the association of the aforementioned genes with OSCC [201]. In this study, EGF induced MMP-1 expression that is required for type I collagen degradation. In addition, MMP-1 is also associated with human papillomavirus [202] and BC [165].
Table 4.
Summary of gene-gene interactions and the corresponding SNPs in these genes
|
Gene 1 |
Gene 2 |
Cancer type |
SNP's gene 1 |
SNP's gene 2 |
References (gene 1) |
References (gene 2) |
Supporting references |
||
|---|---|---|---|---|---|---|---|---|---|
| rs number | Other name | rs number | Other name | ||||||
|
IL6 |
TGFB1 |
BC |
rs1800795 |
c.-174G>C |
rs1800469, rs1800470 |
c.-509C>T, p.T29C |
[56] |
[67-70,72-74] |
[155-157] |
|
IL6 |
TGFB1 |
CRC |
rs1800795 |
c.-174G>C |
rs1800470 |
p.T29C |
[57] |
[71] |
[158] |
|
IL6 |
TGFB1 |
GC |
rs1800795 |
c.-174G>C |
rs1800470 |
p.T29C |
[57] |
[71] |
|
|
IL6 |
TGFB1 |
LC |
rs1800795 |
c.-174G>C |
rs1800470 |
p.T29C |
[57] |
[71] |
|
|
IL6 |
TGFB1 |
PC |
rs1800795 |
c.-174G>C |
rs1800470 |
p.T29C |
[57] |
[71] |
[159] |
|
MMP2 |
EGF |
LC |
rs2438650 |
c.-1306C>T |
rs4444903 |
c.61A>G |
[160] |
[161] |
[162] |
|
MMP2 |
EGF |
BC |
rs2438650 |
c.-1306C>T |
rs4444903 |
c.61A>G |
[160] |
[161] |
[163-165] |
|
MMP2 |
EGF |
GC |
rs2438650 |
c.-1306C>T |
rs4444903 |
c.61A>G |
[160] |
[161] |
|
|
VEGFA |
IL1B |
BC |
rs3025039 |
c.936C>T |
rs114327 |
NA |
[166-169] |
[170] |
[171] |
|
VEGFA |
IL1B |
BC |
rs699947 |
c.-2578C>A |
rs1143634 |
NA |
[172] |
[170] |
[171] |
|
VEGFA |
IL1B |
BC |
NA |
NA |
rs16944 |
NA |
NA |
[170] |
[171] |
|
VEGFA |
IL1B |
GC |
rs3025039 |
c.936C>T |
rs3087258 |
NA |
[45] |
[173] |
|
|
VEGFA |
IL1B |
GC |
rs699947 |
c.-2578C>A |
NA |
IL1B-31-ami |
[95] |
[173] |
|
|
MMP9 |
IL8 |
BC |
rs3918242 |
c.-1562C>T |
rs4073 |
c.-251A>T |
[160] |
[174] |
[171] |
|
MMP9 |
IL8 |
CRC |
rs3918242 |
c.-1562C>T |
rs4073 |
c.-251A>T |
[160] |
[174] |
|
|
MMP9 |
IL8 |
GC |
rs3918242 |
c.-1562C>T |
rs4073 |
c.-251A>T |
[160] |
[175] |
|
|
MMP9 |
IL8 |
LC |
rs3918242 |
c.-1562C>T |
rs4073 |
c.-251A>T |
[160] |
[174] |
|
|
MMP1 |
MMP3 |
BC |
rs1799750 |
c.-1607 1G>2G |
rs3025058 |
c.-1171 5A>6A |
[176] |
[176] |
|
|
MMP1 |
MMP3 |
CRC |
rs1799750 |
c.-1607 1G>2G |
rs3025058 |
c.-1171 5A>6A |
[176] |
[176] |
|
|
MMP1 |
MMP3 |
HNC |
rs1799750 |
c.-1607 1G>2G |
rs3025058 |
c.-1171 5A>6A |
[176] |
[176] |
|
|
MMP1 |
MMP3 |
LC |
rs1799750 |
c.-1607 1G>2G |
rs3025058 |
c.-1171 5A>6A |
[176] |
[176] |
[177,178] |
|
MMP1 |
MMP3 |
OVCa |
rs1799750 |
c.-1607 1G>2G |
rs3025058 |
c.-1171 5A>6A |
[176] |
[176] |
|
|
GSTP1 |
GSTM1 |
CRC |
rs1695 |
p.I105V |
rs1065411 |
GSTM1 present/null |
[45] |
[179] |
|
|
GSTP1 |
GSTM1 |
BC |
rs1695 |
p.I105V |
rs1065412 |
GSTM1 present/null |
[180] |
[181] |
[182,183] |
|
GSTP1 |
GSTM1 |
OVCa |
rs1695 |
p.I105V |
rs1065413 |
GSTM1 present/null |
[184] |
[184] |
|
| GSTP1 | GSTM1 | UBC | rs1695 | p.I105V | rs1065414 | GSTM1 present/null | [185] | [186] | |
These were identified in our meta-analysis. Their correlation with various cancer types is also shown. NA not available.
Another interesting interaction that was revealed from our analysis was between the VEGFA and IL1B genes that were associated with BC and GC (Table 4). After proceeding with a further literature search, we have not found similar results - except from one report [171] - but we have identified additional associations with HNC, ALL, laryngeal carcinoma and MM [203-206]. For MMP-9 and IL8 interaction, there was no study confirming our initial results for BC, CRC and GC on neither humans nor mouse models. We have observed though that there was evidence for an association with nasopharyngeal carcinoma [171], LC [177,178] and UBC [207]. Similarly, we could not find any study that could support the interactions between MMP-1 and MMP-3 and GSTP1 with GSTM1, although two studies confirmed that GSTP1 and GSTM1 interactions could be associated with BC [182,183] (Table 4).
Indications from further literature search on human models revealed associations for MMP-1 and MMP-3 with types of cancer such as BCC, metatypical cancer of the skin [208], colorectal adenoma and RC [209,210], and for GSTP1 and GSTM1, endometrial cancer (EmCa) [211], LC [212], multiple myeloma (observed no significant association to prostatic adenoma and adenocarcinoma) [213], PC [133,214], ALL [215], chronic myeloid leukaemia [216] and PanC [217].
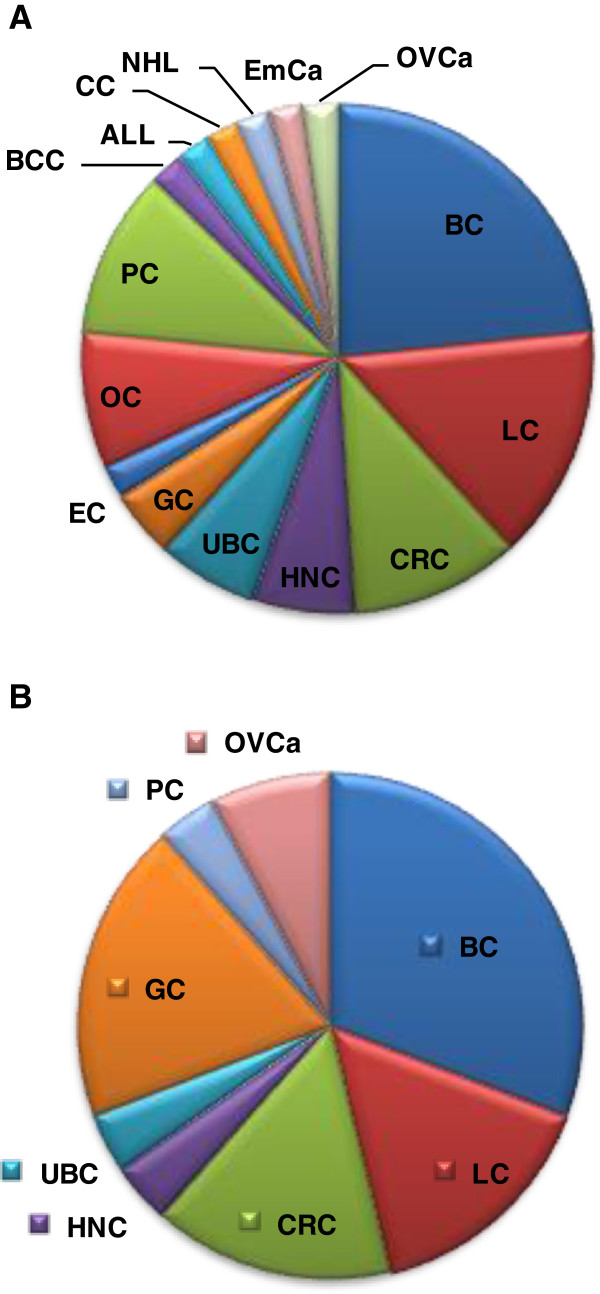
We have then attempted to depict the various types of cancers according to the number of SNPs and genes and/or gene clusters found from our meta-analysis to be meaningfully associated with certain cancer types. Our data indicate that BC is correlated more often than the other types of cancer both with the number of SNPs (Figure 1A) as well as with the number of genes or gene clusters (Figure 1B). This observation underlies the heterogeneity of BC, indicating that it is, most likely, not a single disease but a spectrum of related disease states.
Figure 1.
The distribution of various cancer types. According to (A) the number of SNPs per cancer type and (B) the number of genes or gene correlations per cancer type. By extrapolating the data in Tables 1, 2, 3 and 4, it seems that the number of genome variations and genes is profoundly bigger in BC, probably indicating that this type of cancer is not a single disease but, most likely, a spectrum of related disease states.
Conclusions
In essence, our meta-analysis study generated clusters of genes and diseases, many of which proved to be independent of the criteria used, which suggests that these clusters are most likely biologically meaningful. Preliminary study of some clusters and of our results shows that indeed these genes interact. As regards the associations, with a further literature analysis on human and mouse models, we have also found meaningful gene associations related to other cancer types not previously reported in the literature, an observation that warrants further investigation.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
ZL carried out the data collection, result analysis and participated in the manuscript preparation. EG participated in the manuscript preparation and data analysis. MF participated in the result and statistical analysis and manuscript revision. EK participated in the data collection and manuscript revision. JCN carried out the result and statistical analysis and participated in the manuscript preparation. HPK participated in the manuscript preparation. GPP participated in the design of the study, data analysis and manuscript preparation. CP conceived of the study, participated in its design and coordination as well as manuscript preparation. All authors read and approved for the final manuscript.
Supplementary Material
Genes and cancer types included in this meta-analysis.
Contributor Information
Zoi Lanara, Email: zoi_lanara@hotmail.com.
Efstathia Giannopoulou, Email: giannop@upatras.gr.
Marta Fullen, Email: m.fullen@kingston.ac.uk.
Evangelos Kostantinopoulos, Email: ekonstantinopoulos@yahoo.gr.
Jean-Christophe Nebel, Email: J.Nebel@kingston.ac.uk.
Haralabos P Kalofonos, Email: kalofonos@upatras.gr.
George P Patrinos, Email: gpatrinos@upatras.gr.
Cristiana Pavlidis, Email: chpavlidou@upatras.gr.
Acknowledgements
This study was conducted to fulfil the requirements of an undergraduate thesis, jointly with the Universities of Trieste, Italy and Patras, Greece. This work was partly funded by the University of Patras research budget and a European Commission grant (GEN2PHEN; FP7-200754) to GPP.
References
- Lea IA, Jackson MA, Li X, Bailey S, Peddada SD, Dunnick JK. Genetic pathways and mutation profiles of human cancers: site- and exposure-specific patterns. Carcinogenesis. 2007;28(9):1851–1858. doi: 10.1093/carcin/bgm176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutt A, Beroukhim R. Single nucleotide polymorphism array analysis of cancer. Curr Opin Oncol. 2007;19(1):43–49. doi: 10.1097/CCO.0b013e328011a8c1. [DOI] [PubMed] [Google Scholar]
- Chung CC, Chanock SJ. Current status of genome-wide association studies in cancer. Hum Genet. 2011;130(1):59–78. doi: 10.1007/s00439-011-1030-9. [DOI] [PubMed] [Google Scholar]
- Rae JM, Skaar TC, Hilsenbeck SG, Oesterreich S. The role of single nucleotide polymorphisms in breast cancer metastasis. Breast Cancer Res. 2008;10(1):301. doi: 10.1186/bcr1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teraoka SN, Bernstein JL, Reiner AS, Haile RW, Bernstein L, Lynch CF, Malone KE, Stovall M, Capanu M, Liang X, Smith SA, Mychaleckyj J, Hou X, Mellemkjaer L, Boice JD Jr, Siniard A, Duggan D, Thomas DC. WECARE Study Collaborative Group, Concannon P. Single nucleotide polymorphisms associated with risk for contralateral breast cancer in the women's environment, cancer, and radiation epidemiology (WECARE) study. Breast Cancer Res. 2011;13(6):R114. doi: 10.1186/bcr3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaki K, Liu ET. Structural mutations in cancer: mechanistic and functional insights. Trends Genet. 2012;28(11):550–559. doi: 10.1016/j.tig.2012.07.002. [DOI] [PubMed] [Google Scholar]
- You JS, Jones PA. Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell. 2012;22(1):9–20. doi: 10.1016/j.ccr.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Lazaro M. A new view of carcinogenesis and an alternative approach to cancer therapy. Mol Med. 2010;16(3–4):144–153. doi: 10.2119/molmed.2009.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen MD, Deshpande MS, Karypis G, Johnson J, Crow JA, Retzel EF. wCLUTO: a Web-enabled clustering toolkit. Plant Physiol. 2003;133(2):510–516. doi: 10.1104/pp.103.024885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, Jensen LJ, von Mering C. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39(Database issue):D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Search Tool for the Retrieval of Interacting Genes/Proteins. http://www.string-db.org.
- Jiang Z, Li C, Xu Y, Cai S, Wang X. Associations between XPD polymorphisms and risk of breast cancer: a meta-analysis. Breast Cancer Res Treat. 2010;123(1):203–212. doi: 10.1007/s10549-010-0751-0. [DOI] [PubMed] [Google Scholar]
- Qiu LX, Yao L, Zhang J, Zhu XD, Zhao XM, Xue K, Mao C, Chen B, Zhan P, Yuan H, Hu X-C. XPD Lys751Gln polymorphism and breast cancer susceptibility: a meta-analysis involving 28,709 subjects. Breast Cancer Res Treat. 2010;124(1):229–235. doi: 10.1007/s10549-010-0813-3. [DOI] [PubMed] [Google Scholar]
- Wang HC, Liu CS, Wang CH, Tsai RY, Tsai CW, Wang RF, Chang CH, Chen YS, Chiu CF, Bau DT, Huang CY. Significant association of XPD Asp312Asn polymorphism with breast cancer in Taiwanese patients. Chin J Physiol. 2010;53(2):130–135. doi: 10.4077/cjp.2010.amk005. [DOI] [PubMed] [Google Scholar]
- Han W, Kim KY, Yang SJ, Noh DY, Kang D, Kwack K. SNP-SNP interactions between DNA repair genes were associated with breast cancer risk in a Korean population. Cancer. 2012;118(3):594–602. doi: 10.1002/cncr.26220. [DOI] [PubMed] [Google Scholar]
- Hussien YM, Gharib AF, Awad HA, Karam RA, Elsawy WH. Impact of DNA repair genes polymorphism (XPD and XRCC1) on the risk of breast cancer in Egyptian female patients. Mol Biol Rep. 2012;39(2):1895–1901. doi: 10.1007/s11033-011-0935-7. [DOI] [PubMed] [Google Scholar]
- Yin J, Vogel U, Wang C, Liang D, Ma Y, Wang H, Yue L, Liu D, Ma J, Sun X. Hapmap-based evaluation of ERCC2, PPP1R13L, and ERCC1 and lung cancer risk in a Chinese population. Environ Mol Mutagen. 2012;53(3):239–245. doi: 10.1002/em.21681. [DOI] [PubMed] [Google Scholar]
- Yao L, Qiu LX, Yu L, Yang Z, Yu XJ, Zhong Y, Hu XC. The association between ERCC2 Asp312Asn polymorphism and breast cancer risk: a meta-analysis involving 22,766 subjects. Breast Cancer Res Treat. 2010;123(1):227–231. doi: 10.1007/s10549-010-0754-x. [DOI] [PubMed] [Google Scholar]
- Zhang J, Gu SY, Zhang P, Jia Z, Chang JH. ERCC2 Lys751Gln polymorphism is associated with lung cancer among Caucasians. Eur J Cancer. 2010;46(13):2479–2484. doi: 10.1016/j.ejca.2010.05.008. [DOI] [PubMed] [Google Scholar]
- Zhan P, Wang Q, Wei SZ, Wang J, Qian Q, Yu LK, Song Y. ERCC2/XPD Lys751Gln and Asp312Asn gene polymorphism and lung cancer risk: a meta-analysis involving 22 case–control studies. J Thorac Oncol. 2010;5(9):1337–1345. doi: 10.1097/JTO.0b013e3181e7fe2a. [DOI] [PubMed] [Google Scholar]
- Yin J, Vogel U, Ma Y, Qi R, Wang H, Yue L, Liang D, Wang C, Li X, Song T. HapMap-based study of a region encompassing ERCC1 and ERCC2 related to lung cancer susceptibility in a Chinese population. Mutat Res. 2011;713(1–2):1–7. doi: 10.1016/j.mrfmmm.2011.05.003. [DOI] [PubMed] [Google Scholar]
- Christiani DC. ERCC2/XPD polymorphisms and lung cancer risk. J Thorac Oncol. 2011;6(1):233. doi: 10.1097/JTO.0b013e318200e058. author reply 233–235. [DOI] [PubMed] [Google Scholar]
- Zhang J, Qiu LX, Leaw SJ, Hu XC, Chang JH. The association between XPD Asp312Asn polymorphism and lung cancer risk: a meta-analysis including 16,949 subjects. Med Oncol. 2011;28(3):655–660. doi: 10.1007/s12032-010-9501-8. [DOI] [PubMed] [Google Scholar]
- Sergentanis TN, Economopoulos KP. Cyclin D1 G870A polymorphism and breast cancer risk: a meta-analysis comprising 9,911 cases and 11,171 controls. Mol Biol Rep. 2011;38(8):4955–4963. doi: 10.1007/s11033-010-0639-4. [DOI] [PubMed] [Google Scholar]
- Millar EK, Dean JL, McNeil CM, O'Toole SA, Henshall SM, Tran T, Lin J, Quong A, Comstock CE, Witkiewicz A, Musgrove EA, Rui H, Lemarchand L, Setiawan VW, Haiman CA, Knudsen KE, Sutherland RL, Knudsen ES. Cyclin D1b protein expression in breast cancer is independent of cyclin D1a and associated with poor disease outcome. Oncogene. 2009;28(15):1812–1820. doi: 10.1038/onc.2009.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita H, Mori S, Tanda N, Nakayama K, Kanzaki A, Sato A, Morikawa H, Motegi K, Takebayashi Y, Fukumoto M. Loss of heterozygosity of nucleotide excision repair factors in sporadic oral squamous cell carcinoma using microdissected tissue. Oncol Rep. 2001;8(5):1133–1138. doi: 10.3892/or.8.5.1133. [DOI] [PubMed] [Google Scholar]
- Ghosh-Choudhury N, Ghosh-Choudhury G, Celeste A, Ghosh PM, Moyer M, Abboud SL, Kreisberg J. Bone morphogenetic protein-2 induces cyclin kinase inhibitor p21 and hypophosphorylation of retinoblastoma protein in estradiol-treated MCF-7 human breast cancer cells. Biochim Biophys Acta. 2000;1497(2):186–196. doi: 10.1016/s0167-4889(00)00060-4. [DOI] [PubMed] [Google Scholar]
- Musgrove EA, Hui R, Sweeney KJ, Watts CK, Sutherland RL. Cyclins and breast cancer. J Mammary Gland Biol Neoplasia. 1996;1(2):153–162. doi: 10.1007/BF02013639. [DOI] [PubMed] [Google Scholar]
- Sutherland RL, Hamilton JA, Sweeney KJ, Watts CK, Musgrove EA. Expression and regulation of cyclin genes in breast cancer. Acta Oncol. 1995;34(5):651–656. doi: 10.3109/02841869509094043. [DOI] [PubMed] [Google Scholar]
- Taneja P, Frazier DP, Kendig RD, Maglic D, Sugiyama T, Kai F, Taneja NK, Inoue K. MMTV mouse models and the diagnostic values of MMTV-like sequences in human breast cancer. Expert Rev Mol Diagn. 2009;9(5):423–440. doi: 10.1586/ERM.09.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Ionescu-Tiba V, Burns K, Gadd M, Zukerberg L, Louis DN, Sgroi D, Schmidt EV. The role of the cyclin D1-dependent kinases in ErbB2-mediated breast cancer. Am J Pathol. 2004;164(3):1031–1038. doi: 10.1016/S0002-9440(10)63190-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou GW, Hu J, Li Q. CYP2E1 PstI/RsaI polymorphism and colorectal cancer risk: a meta-analysis. World J Gastroenterol. 2010;16(23):2949–2953. doi: 10.3748/wjg.v16.i23.2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva TD, Felipe AV, Pimenta CA, Barao K, Forones NM. CYP2E1 RsaI and 96-bp insertion genetic polymorphisms associated with risk for colorectal cancer. Genet Mol Res. 2012;11(3):3138–3145. doi: 10.4238/2012.September.3.2. [DOI] [PubMed] [Google Scholar]
- Sameer AS, Nissar S, Qadri Q, Alam S, Baba SM, Siddiqi MA. Role of CYP2E1 genotypes in susceptibility to colorectal cancer in the Kashmiri population. Hum Genomics. 2011;5(6):530–537. doi: 10.1186/1479-7364-5-6-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Zhou Y, Zhou Z, Liu J, Yuan X, Matsuo K, Takezaki T, Tajima K, Cao J. A novel polymorphism rs1329149 of CYP2E1 and a known polymorphism rs671 of ALDH2 of alcohol metabolizing enzymes are associated with colorectal cancer in a southwestern Chinese population. Cancer Epidemiol Biomarkers Prev. 2009;18(9):2522–2527. doi: 10.1158/1055-9965.EPI-09-0398. [DOI] [PubMed] [Google Scholar]
- Kury S, Buecher B, Robiou-du-Pont S, Scoul C, Sebille V, Colman H, Le Houerou C, Le Neel T, Bourdon J, Faroux R, Ollivry J, Lafraise B, Chupin LD, Bézieau S. Combinations of cytochrome P450 gene polymorphisms enhancing the risk for sporadic colorectal cancer related to red meat consumption. Cancer Epidemiol Biomarkers Prev. 2007;16(7):1460–1467. doi: 10.1158/1055-9965.EPI-07-0236. [DOI] [PubMed] [Google Scholar]
- Chen K, Jin MJ, Fan CH, Song L, Jiang QT, Yu WP, Ma XY, Yao KY. A case–control study on the association between genetic polymorphisms of metabolic enzymes and the risk of colorectal cancer. Zhonghua Liu Xing Bing Xue Za Zhi. 2005;26(9):659–664. [PubMed] [Google Scholar]
- van der Logt EM, Bergevoet SM, Roelofs HM, Te Morsche RH, Dijk Y, Wobbes T, Nagengast FM, Peters WH. Role of epoxide hydrolase, NAD(P)H:quinone oxidoreductase, cytochrome P450 2E1 or alcohol dehydrogenase genotypes in susceptibility to colorectal cancer. Mutat Res. 2006;593(1–2):39–49. doi: 10.1016/j.mrfmmm.2005.06.018. [DOI] [PubMed] [Google Scholar]
- Landi S, Gemignani F, Moreno V, Gioia-Patricola L, Chabrier A, Guino E, Navarro M, de Oca J, Capella G, Canzian F. A comprehensive analysis of phase I and phase II metabolism gene polymorphisms and risk of colorectal cancer. Pharmacogenet Genomics. 2005;15(8):535–546. doi: 10.1097/01.fpc.0000165904.48994.3d. [DOI] [PubMed] [Google Scholar]
- Cotterchio M, Boucher BA, Manno M, Gallinger S, Okey AB, Harper PA. Red meat intake, doneness, polymorphisms in genes that encode carcinogen-metabolizing enzymes, and colorectal cancer risk. Cancer Epidemiol Biomarkers Prev. 2008;17(11):3098–3107. doi: 10.1158/1055-9965.EPI-08-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Huang XF. The signal pathways in azoxymethane-induced colon cancer and preventive implications. Cancer Biol Ther. 2009;8(14):1313–1317. doi: 10.4161/cbt.8.14.8983. [DOI] [PubMed] [Google Scholar]
- Tang K, Li Y, Zhang Z, Gu Y, Xiong Y, Feng G, He L, Qin S. The PstI/RsaI and DraI polymorphisms of CYP2E1 and head and neck cancer risk: a meta-analysis based on 21 case–control studies. BMC Cancer. 2010;10:575. doi: 10.1186/1471-2407-10-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Yu X, Du Y. Meta-analyses of the effect of cytochrome P450 2E1 gene polymorphism on the risk of head and neck cancer. Mol Biol Rep. 2011;38(4):2409–2416. doi: 10.1007/s11033-010-0375-9. [DOI] [PubMed] [Google Scholar]
- Garcia SM, Curioni OA, de Carvalho MB, Gattas GJ. Polymorphisms in alcohol metabolizing genes and the risk of head and neck cancer in a Brazilian population. Alcohol Alcohol. 2010;45(1):6–12. doi: 10.1093/alcalc/agp078. [DOI] [PubMed] [Google Scholar]
- Economopoulos KP, Sergentanis TN. GSTM1, GSTT1, GSTP1, GSTA1 and colorectal cancer risk: a comprehensive meta-analysis. Eur J Cancer. 2010;46(9):1617–1631. doi: 10.1016/j.ejca.2010.02.009. [DOI] [PubMed] [Google Scholar]
- Ebrahimkhani S, Asgharian AM, Nourinaier B, Ebrahimkhani K, Vali N, Abbasi F, Zali MR. Association of GSTM1, GSTT1, GSTP1 and CYP2E1 single nucleotide polymorphisms with colorectal cancer in Iran. Pathol Oncol Res. 2012;18(3):651–656. doi: 10.1007/s12253-011-9490-8. [DOI] [PubMed] [Google Scholar]
- Sameer AS, Qadri Q, Siddiqi MA. GSTP1 I105V polymorphism and susceptibility to colorectal cancer in Kashmiri population. DNA Cell Biol. 2012;31(1):74–79. doi: 10.1089/dna.2011.1297. [DOI] [PubMed] [Google Scholar]
- Wang J, Joshi AD, Corral R, Siegmund KD, Marchand LL, Martinez ME, Haile RW, Ahnen DJ, Sandler RS, Lance P, Stern MC. Carcinogen metabolism genes, red meat and poultry intake, and colorectal cancer risk. Int J Cancer. 2012;130(8):1898–1907. doi: 10.1002/ijc.26199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainz J, Rudolph A, Hein R, Hoffmeister M, Buch S, von Schonfels W, Hampe J, Schafmayer C, Volzke H, Frank B, Brenner H, Försti A, Hemminki K, Chang-Claude J. Association of genetic polymorphisms in ESR2, HSD17B1, ABCB1, and SHBG genes with colorectal cancer risk. Endocr Relat Cancer. 2011;18(2):265–276. doi: 10.1530/ERC-10-0264. [DOI] [PubMed] [Google Scholar]
- Jones BA, Christensen AR, Wise JP Sr, Yu H. Glutathione S-transferase polymorphisms and survival in African-American and white colorectal cancer patients. Cancer Epidemiol. 2009;33(3–4):249–256. doi: 10.1016/j.canep.2009.08.004. [DOI] [PubMed] [Google Scholar]
- Skjelbred CF, Saebo M, Hjartaker A, Grotmol T, Hansteen IL, Tveit KM, Hoff G, Kure EH. Meat, vegetables and genetic polymorphisms and the risk of colorectal carcinomas and adenomas. BMC Cancer. 2007;7:228. doi: 10.1186/1471-2407-7-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talseth BA, Meldrum C, Suchy J, Kurzawski G, Lubinski J, Scott RJ. Genetic polymorphisms in xenobiotic clearance genes and their influence on disease expression in hereditary nonpolyposis colorectal cancer patients. Cancer Epidemiol Biomarkers Prev. 2006;15(11):2307–2310. doi: 10.1158/1055-9965.EPI-06-0040. [DOI] [PubMed] [Google Scholar]
- Romero RZ, Morales R, Garcia F, Huarriz M, Bandres E, De la Haba J, Gomez A, Aranda E, Garcia-Foncillas J. Potential application of GSTT1-null genotype in predicting toxicity associated to 5-fluouracil irinotecan and leucovorin regimen in advanced stage colorectal cancer patients. Oncol Rep. 2006;16(3):497–503. [PubMed] [Google Scholar]
- Probst-Hensch NM, Sun CL, Van Den Berg D, Ceschi M, Koh WP, Yu MC. The effect of the cyclin D1 (CCND1) A870G polymorphism on colorectal cancer risk is modified by glutathione-S-transferase polymorphisms and isothiocyanate intake in the Singapore Chinese health study. Carcinogenesis. 2006;27(12):2475–2482. doi: 10.1093/carcin/bgl116. [DOI] [PubMed] [Google Scholar]
- Gaustadnes M, Orntoft TF, Jensen JL, Torring N. Validation of the use of DNA pools and primer extension in association studies of sporadic colorectal cancer for selection of candidate SNPs. Hum Mutat. 2006;27(2):187–194. doi: 10.1002/humu.20248. [DOI] [PubMed] [Google Scholar]
- Yu KD, Di GH, Fan L, Chen AX, Yang C, Shao ZM. Lack of an association between a functional polymorphism in the interleukin-6 gene promoter and breast cancer risk: a meta-analysis involving 25,703 subjects. Breast Cancer Res Treat. 2010;122(2):483–488. doi: 10.1007/s10549-009-0706-5. [DOI] [PubMed] [Google Scholar]
- Xu B, Niu XB, Wang ZD, Cheng W, Tong N, Mi YY, Min ZC, Tao J, Li PC, Zhang W, Wu HF, Zhang ZD, Wang ZJ, Hua LX, Feng NH, Wang XR. IL-6–174G>C polymorphism and cancer risk: a meta-analysis involving 29,377 cases and 37,739 controls. Mol Biol Rep. 2011;38(4):2589–2596. doi: 10.1007/s11033-010-0399-1. [DOI] [PubMed] [Google Scholar]
- Dong X, Wu J, Liang P, Li J, Yuan L, Liu X. Methylenetetrahydrofolate reductase C677T and A1298C polymorphisms and gastric cancer: a meta-analysis. Arch Med Res. 2010;41(2):125–133. doi: 10.1016/j.arcmed.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Wang Z, Chen JQ, Liu JL, Qin XG, Huang Y. Polymorphisms in ERCC1, GSTs, TS and MTHFR predict clinical outcomes of gastric cancer patients treated with platinum/5-Fu-based chemotherapy: a systematic review. BMC Gastroenterol. 2012;12:137. doi: 10.1186/1471-230X-12-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balassiano K, Lima S, Jenab M, Overvad K, Tjonneland A, Boutron-Ruault MC, Clavel-Chapelon F, Canzian F, Kaaks R, Boeing H, Meidtner K, Trichopoulou A, Laglou P, Vineis P, Panico S, Palli D, Grioni S, Tumino R, Lund E, Bueno-de-Mesquita HB, Numans ME, Peeters PH, Ramon Quirós J, Sánchez MJ, Navarro C, Ardanaz E, Dorronsoro M, Hallmans G, Stenling R. et al. Aberrant DNA methylation of cancer-associated genes in gastric cancer in the European Prospective Investigation into Cancer and Nutrition (EPIC-EURGAST) Cancer Lett. 2011;311(1):85–95. doi: 10.1016/j.canlet.2011.06.038. [DOI] [PubMed] [Google Scholar]
- Qi X, Ma X, Yang X, Fan L, Zhang Y, Zhang F, Chen L, Zhou Y, Jiang J. Methylenetetrahydrofolate reductase polymorphisms and breast cancer risk: a meta-analysis from 41 studies with 16,480 cases and 22,388 controls. Breast Cancer Res Treat. 2010;123(2):499–506. doi: 10.1007/s10549-010-0773-7. [DOI] [PubMed] [Google Scholar]
- Qiu LX, Zhang J, Li WH, Zhang QL, Yu H, Wang BY, Wang LP, Wang JL, Wang HJ, Liu XJ, Luo ZG, Wu XH. Lack of association between methylenetetrahydrofolate reductase gene A1298C polymorphism and breast cancer susceptibility. Mol Biol Rep. 2011;38(4):2295–2299. doi: 10.1007/s11033-010-0361-2. [DOI] [PubMed] [Google Scholar]
- Perel'muter VM, Zav'ialova MV, Vtorushin SV, Slonimskaia EM, Kritskaia NG, Garbukov E, Litviakov NV, Stakheeva MN, Babyshkina NN, Malinovskaia EA, Denisov EV, Grigor'eva ES, Nazarenko MS, Sennikov SV, Goreva EP, Kozlov VA, Voevoda MI, Maksimov VN, Beliavskaia VA, Cherdyntseva NV. Genetic and clinical and pathological characteristics of breast cancer in premenopausal and postmenopausal women. Adv Gerontol. 2008;21(4):643–653. [PubMed] [Google Scholar]
- Stevens VL, McCullough ML, Pavluck AL, Talbot JT, Feigelson HS, Thun MJ, Calle EE. Association of polymorphisms in one-carbon metabolism genes and postmenopausal breast cancer incidence. Cancer Epidemiol Biomarkers Prev. 2007;16(6):1140–1147. doi: 10.1158/1055-9965.EPI-06-1037. [DOI] [PubMed] [Google Scholar]
- Qiu LX, Mao C, Yao L, Yu KD, Zhan P, Chen B, Liu HG, Yuan H, Zhang J, Xue K, Hu XC. XRCC3 5′-UTR and IVS5-14 polymorphisms and breast cancer susceptibility: a meta-analysis. Breast Cancer Res Treat. 2010;122(2):489–493. doi: 10.1007/s10549-009-0726-1. [DOI] [PubMed] [Google Scholar]
- Chen Y, Pei J. Possible risk modifications in the association between MnSOD Ala-9Val polymorphism and breast cancer risk: subgroup analysis and evidence-based sample size calculation for a future trial. Breast Cancer Res Treat. 2011;125(2):495–504. doi: 10.1007/s10549-010-0978-9. [DOI] [PubMed] [Google Scholar]
- Qi X, Zhang F, Yang X, Fan L, Zhang Y, Chen L, Zhou Y, Chen X, Zhong L, Jiang J. Transforming growth factor-beta1 polymorphisms and breast cancer risk: a meta-analysis based on 27 case–control studies. Breast Cancer Res Treat. 2010;122(1):273–279. doi: 10.1007/s10549-010-0847-6. [DOI] [PubMed] [Google Scholar]
- Huang Y, Hao Y, Li B, Xie J, Qian J, Chao C, Yu L. Lack of significant association between TGF-beta1-590C/T polymorphism and breast cancer risk: a meta-analysis. Med Oncol. 2011;28(2):424–428. doi: 10.1007/s12032-010-9491-6. [DOI] [PubMed] [Google Scholar]
- Woo SU, Park KH, Woo OH, Yang DS, Kim AR, Lee ES, Lee JB, Kim YH, Kim JS, Seo JH. Association of a TGF-beta1 gene -509 C/T polymorphism with breast cancer risk: a meta-analysis. Breast Cancer Res Treat. 2010;124(2):481–485. doi: 10.1007/s10549-010-0871-6. [DOI] [PubMed] [Google Scholar]
- Gu D, Zhuang L, Huang H, Cao P, Wang D, Tang J, Chen J. TGFB1 T29C polymorphism and breast cancer risk: a meta-analysis based on 10,417 cases and 11,455 controls. Breast Cancer Res Treat. 2010;123(3):857–861. doi: 10.1007/s10549-010-0766-6. [DOI] [PubMed] [Google Scholar]
- Wei BB, Xi B, Wang R, Bai JM, Chang JK, Zhang YY, Yoneda R, Su JT, Hua LX. TGFbeta1 T29C polymorphism and cancer risk: a meta-analysis based on 40 case–control studies. Cancer Genet Cytogenet. 2010;196(1):68–75. doi: 10.1016/j.cancergencyto.2009.09.016. [DOI] [PubMed] [Google Scholar]
- Ma X, Chen C, Xiong H, Li Y. Transforming growth factorbeta1 L10P variant plays an active role on the breast cancer susceptibility in Caucasian: evidence from 10,392 cases and 11,697 controls. Breast Cancer Res Treat. 2010;124(2):453–457. doi: 10.1007/s10549-010-0843-x. [DOI] [PubMed] [Google Scholar]
- Huang Y, Li B, Qian J, Xie J, Yu L. TGF-beta1 29T/C polymorphism and breast cancer risk: a meta-analysis involving 25,996 subjects. Breast Cancer Res Treat. 2010;123(3):863–868. doi: 10.1007/s10549-010-0796-0. [DOI] [PubMed] [Google Scholar]
- Qiu LX, Yao L, Mao C, Chen B, Zhan P, Xue K, Zhang J, Yuan H, Hu XC. TGFB1 L10P polymorphism is associated with breast cancer susceptibility: evidence from a meta-analysis involving 47,817 subjects. Breast Cancer Res Treat. 2010;123(2):563–567. doi: 10.1007/s10549-010-0781-7. [DOI] [PubMed] [Google Scholar]
- Radisky DC, Hartmann LC. Mammary involution and breast cancer risk: transgenic models and clinical studies. J Mammary Gland Biol Neoplasia. 2009;14(2):181–191. doi: 10.1007/s10911-009-9123-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuelten CH, Busch JI, Tang B, Flanders KC, Oshima A, Sutton E, Karpova TS, Roberts AB, Wakefield LM, Niederhuber JE. Transient tumor-fibroblast interactions increase tumor cell malignancy by a TGF-Beta mediated mechanism in a mouse xenograft model of breast cancer. PLoS One. 2010;5(3):e9832. doi: 10.1371/journal.pone.0009832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W. Genetic polymorphisms in the transforming growth factor-beta signaling pathways and breast cancer risk and survival. Methods Mol Biol. 2009;472:265–277. doi: 10.1007/978-1-60327-492-0_11. [DOI] [PubMed] [Google Scholar]
- Hu Z, Li X, Qu X, He Y, Ring BZ, Song E, Su L. Intron 3 16 bp duplication polymorphism of TP53 contributes to cancer susceptibility: a meta-analysis. Carcinogenesis. 2010;31(4):643–647. doi: 10.1093/carcin/bgq018. [DOI] [PubMed] [Google Scholar]
- Ma Y, Yang J, Liu Z, Zhang P, Yang Z, Wang Y, Qin H. No significant association between the TP53 codon 72 polymorphism and breast cancer risk: a meta-analysis of 21 studies involving 24,063 subjects. Breast Cancer Res Treat. 2011;125(1):201–205. doi: 10.1007/s10549-010-0920-1. [DOI] [PubMed] [Google Scholar]
- Chunder N, Mandal S, Roy A, Roychoudhury S, Panda CK. Differential association of BRCA1 and BRCA2 genes with some breast cancer-associated genes in early and late onset breast tumors. Ann Surg Oncol. 2004;11(12):1045–1055. doi: 10.1245/ASO.2004.02.022. [DOI] [PubMed] [Google Scholar]
- Rebbeck TR. Inherited genetic predisposition in breast cancer. A population-based perspective. Cancer. 1999;86(11 Suppl):2493–2501. doi: 10.1002/(sici)1097-0142(19991201)86:11+<2493::aid-cncr6>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Rossner P Jr, Gammon MD, Zhang YJ, Terry MB, Hibshoosh H, Memeo L, Mansukhani M, Long CM, Garbowski G, Agrawal M, Agrawal M, Kalra TS, Gaudet MM, Teitelbaum SL, Neugut AI, Santella RM. Mutations in p53, p53 protein overexpression and breast cancer survival. J Cell Mol Med. 2009;13(9B):3847–3857. doi: 10.1111/j.1582-4934.2008.00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nkhata KJ, Ray A, Schuster TF, Grossmann ME, Cleary MP. Effects of adiponectin and leptin co-treatment on human breast cancer cell growth. Oncol Rep. 2009;21(6):1611–1619. doi: 10.3892/or_00000395. [DOI] [PubMed] [Google Scholar]
- Palanca Suela S, Esteban Cardenosa E, Barragan Gonzalez E, de Juan Jimenez I, Chirivella Gonzalez I, Segura Huerta A, Guillen Ponce C, Montalar Salcedo J, Martinez de Duenas E, Castel Sanchez V, Bolufer Gilabert P. Group for Assessment of Hereditary Cancer of Valencia Community. CASP8 D302H polymorphism delays the age of onset of breast cancer in BRCA1 and BRCA2 carriers. Breast Cancer Res Treat. 2010;119(1):87–93. doi: 10.1007/s10549-009-0316-2. [DOI] [PubMed] [Google Scholar]
- Fanale D, Amodeo V, Corsini LR, Rizzo S, Bazan V, Russo A. Breast cancer genome-wide association studies: there is strength in numbers. Oncogene. 2012;31(17):2121–2128. doi: 10.1038/onc.2011.408. [DOI] [PubMed] [Google Scholar]
- Cherdyntseva NV, Denisov EV, Litviakov NV, Maksimov VN, Malinovskaya EA, Babyshkina NN, Slonimskaya EM, Voevoda MI, Choinzonov EL. Crosstalk between the FGFR2 and TP53 genes in breast cancer: data from an association study and epistatic interaction analysis. DNA Cell Biol. 2012;31(3):306–316. doi: 10.1089/dna.2011.1351. [DOI] [PubMed] [Google Scholar]
- Lo Nigro C, Vivenza D, Monteverde M, Lattanzio L, Gojis O, Garrone O, Comino A, Merlano M, Quinlan PR, Syed N, Purdie CA, Thompson A, Palmieri C, Crook T. High frequency of complex TP53 mutations in CNS metastases from breast cancer. Br J Cancer. 2012;106(2):397–404. doi: 10.1038/bjc.2011.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathoulin-Portier MP, Viens P, Cowen D, Bertucci F, Houvenaeghel G, Geneix J, Puig B, Bardou VJ, Jacquemier J. Prognostic value of simultaneous expression of p21 and mdm2 in breast carcinomas treated by adjuvant chemotherapy with antracyclin. Oncol Rep. 2000;7(3):675–680. doi: 10.3892/or.7.3.675. [DOI] [PubMed] [Google Scholar]
- Blackburn AC, Jerry DJ. Knockout and transgenic mice of Trp53: what have we learned about p53 in breast cancer? Breast Cancer Res. 2002;4(3):101–111. doi: 10.1186/bcr427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett CN, Green JE. Genomic analyses as a guide to target identification and preclinical testing of mouse models of breast cancer. Toxicol Pathol. 2010;38(1):88–95. doi: 10.1177/0192623309357074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardini JE, Napoli M, Piazza S, Rustighi A, Marotta C, Radaelli E, Capaci V, Jordan L, Quinlan P, Thompson A, Mano M, Rosato A, Crook T, Scanziani E, Means AR, Lozano G, Schneider C, Del Sal G. A Pin1/mutant p53 axis promotes aggressiveness in breast cancer. Cancer Cell. 2011;20(1):79–91. doi: 10.1016/j.ccr.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Alsner J, Jensen V, Kyndi M, Offersen BV, Vu P, Borresen-Dale AL, Overgaard J. A comparison between p53 accumulation determined by immunohistochemistry and TP53 mutations as prognostic variables in tumours from breast cancer patients. Acta Oncol. 2008;47(4):600–607. doi: 10.1080/02841860802047411. [DOI] [PubMed] [Google Scholar]
- Bourdon JC, Khoury MP, Diot A, Baker L, Fernandes K, Aoubala M, Quinlan P, Purdie CA, Jordan LB, Prats AC, Lane DP, Thompson AM. p53 mutant breast cancer patients expressing p53gamma have as good a prognosis as wild-type p53 breast cancer patients. Breast Cancer Res. 2011;13(1):R7. doi: 10.1186/bcr2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besaratinia A, Pfeifer GP. Applications of the human p53 knock-in (Hupki) mouse model for human carcinogen testing. FASEB J. 2010;24(8):2612–2619. doi: 10.1096/fj.10-157263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang DK, Ren WH, Yao L, Wang WZ, Peng B, Yu L. Meta-analysis of association between TP53 Arg72Pro polymorphism and bladder cancer risk. Urology. 2010;76(3):765–767. doi: 10.1016/j.urology.2010.04.044. e761. [DOI] [PubMed] [Google Scholar]
- Burger M, Burger SJ, Denzinger S, Wild PJ, Wieland WF, Blaszyk H, Obermann EC, Stoehr R, Hartmann A. Elevated microsatellite instability at selected tetranucleotide repeats does not correlate with clinicopathologic features of bladder cancer. Eur Urol. 2006;50(4):770–775. doi: 10.1016/j.eururo.2006.04.010. discussion 776. [DOI] [PubMed] [Google Scholar]
- Zuiverloon TC, Abas CS, van der Keur KA, Vermeij M, Tjin SS, van Tilborg AG, Busstra M, Zwarthoff EC. In-depth investigation of the molecular pathogenesis of bladder cancer in a unique 26-year old patient with extensive multifocal disease: a case report. BMC Urol. 2010;10:5. doi: 10.1186/1471-2490-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kompier LC, van Tilborg AA, Zwarthoff EC. Bladder cancer: novel molecular characteristics, diagnostic, and therapeutic implications. Urol Oncol. 2010;28(1):91–96. doi: 10.1016/j.urolonc.2009.06.007. [DOI] [PubMed] [Google Scholar]
- Jarmalaite S, Andrekute R, Scesnaite A, Suziedelis K, Husgafvel-Pursiainen K, Jankevicius F. Promoter hypermethylation in tumour suppressor genes and response to interleukin-2 treatment in bladder cancer: a pilot study. J Cancer Res Clin Oncol. 2010;136(6):847–854. doi: 10.1007/s00432-009-0725-y. [DOI] [PubMed] [Google Scholar]
- Lin HY, Huang CH, Yu TJ, Wu WJ, Yang MC, Lung FW. p53 codon 72 polymorphism as a progression index for bladder cancer. Oncol Rep. 2012;27(4):1193–1199. doi: 10.3892/or.2011.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahabreh IJ, Linardou H, Bouzika P, Varvarigou V, Murray S. TP53 Arg72Pro polymorphism and colorectal cancer risk: a systematic review and meta-analysis. Cancer Epidemiol Biomarkers Prev. 2010;19(7):1840–1847. doi: 10.1158/1055-9965.EPI-10-0156. [DOI] [PubMed] [Google Scholar]
- Economopoulos KP, Sergentanis TN, Zagouri F, Zografos GC. Association between p53 Arg72Pro polymorphism and colorectal cancer risk: a meta-analysis. Onkologie. 2010;33(12):666–674. doi: 10.1159/000322210. [DOI] [PubMed] [Google Scholar]
- Wang JJ, Zheng Y, Sun L, Wang L, Yu PB, Dong JH, Zhang L, Xu J, Shi W, Ren YC. TP53 codon 72 polymorphism and colorectal cancer susceptibility: a meta-analysis. Mol Biol Rep. 2011;38(8):4847–4853. doi: 10.1007/s11033-010-0619-8. [DOI] [PubMed] [Google Scholar]
- Goodman JE, Mechanic LE, Luke BT, Ambs S, Chanock S, Harris CC. Exploring SNP-SNP interactions and colon cancer risk using polymorphism interaction analysis. Int J Cancer. 2006;118(7):1790–1797. doi: 10.1002/ijc.21523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu L, Tang Y, Chen C, Wang Q, Xu J, Yang C, Miao X, Wei S, Chen J, Nie S. Polymorphisms in TP53 and MDM2 contribute to higher risk of colorectal cancer in Chinese population: a hospital-based, case–control study. Mol Biol Rep. 2012;39(10):9661–9668. doi: 10.1007/s11033-012-1831-5. [DOI] [PubMed] [Google Scholar]
- Lopez I, PO L, Tucci P, Alvarez-Valin F, AC R, Marin M. Different mutation profiles associated to P53 accumulation in colorectal cancer. Gene. 2012;499(1):81–87. doi: 10.1016/j.gene.2012.02.011. [DOI] [PubMed] [Google Scholar]
- Kanaan Z, Rai SN, Eichenberger MR, Barnes C, Dworkin AM, Weller C, Cohen E, Roberts H, Keskey B, Petras RE, Crawford NP, Galandiuk S. Differential microRNA expression tracks neoplastic progression in inflammatory bowel disease-associated colorectal cancer. Hum Mutat. 2012;33(3):551–560. doi: 10.1002/humu.22021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizat AA, Shahpudin SN, Mustapha MA, Zakaria Z, Sidek AS, Abu Hassan MR, Ankathil R. Association of Arg72Pro of P53 polymorphism with colorectal cancer susceptibility risk in Malaysian population. Asian Pac J Cancer Prev. 2011;12(11):2909–2913. [PubMed] [Google Scholar]
- Wang B, Wang D, Zhang D, Li A, Liu D, Liu H, Jin H. Pro variant of TP53 Arg72Pro contributes to esophageal squamous cell carcinoma risk: evidence from a meta-analysis. Eur J Cancer Prev. 2010;19(4):299–307. doi: 10.1097/CEJ.0b013e32833964bc. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Wang F, Shan S, Qiu X, Li X, Jiao F, Wang J, Du Y. Genetic polymorphism of p53, but not GSTP1, is association with susceptibility to esophageal cancer risk - a meta-analysis. Int J Med Sci. 2010;7(5):300–308. doi: 10.7150/ijms.7.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashash M, Yavari P, Hislop TG, Shah A, Sadjadi A, Babaei M, Le N, Brooks-Wilson A, Malekzadeh R, Bajdik C. Comparison of two diverse populations, British Columbia, Canada, and Ardabil, Iran, indicates several variables associated with gastric and esophageal cancer survival. J Gastrointest Cancer. 2011;42(1):40–45. doi: 10.1007/s12029-010-9228-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duenas M, Santos M, Aranda JF, Bielza C, Martinez-Cruz AB, Lorz C, Taron M, Ciruelos EM, Rodriguez-Peralto JL, Martin M, Larrañaga P, Dahabreh J, Stathopoulos GP, Rosell R, Paramio JM, García-Escudero R. Mouse p53-deficient cancer models as platforms for obtaining genomic predictors of human cancer clinical outcomes. PLoS One. 2012;7(8):e42494. doi: 10.1371/journal.pone.0042494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Morales R, Mendez-Ramirez I, Castro-Hernandez C, Martinez-Ramirez OC, Gonsebatt ME, Rubio J. Polymorphisms associated with the risk of lung cancer in a healthy Mexican Mestizo population: application of the additive model for cancer. Genet Mol Biol. 2011;34(4):546–552. doi: 10.1590/S1415-47572011005000053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeb KK, Michalowska AM, Yoon CY, Krummey SM, Hoenerhoff MJ, Kavanaugh C, Li MC, Demayo FJ, Linnoila I, Deng CX, Lee EY, Medina D, Shih JH, Green JE. Identification of an integrated SV40 T/t-antigen cancer signature in aggressive human breast, prostate, and lung carcinomas with poor prognosis. Cancer Res. 2007;67(17):8065–8080. doi: 10.1158/0008-5472.CAN-07-1515. [DOI] [PubMed] [Google Scholar]
- Campling BG, el-Deiry WS. Clinical implications of p53 mutations in lung cancer. Methods Mol Med. 2003;75:53–77. doi: 10.1385/1-59259-324-0:53. [DOI] [PubMed] [Google Scholar]
- Meylan E, Dooley AL, Feldser DM, Shen L, Turk E, Ouyang C, Jacks T. Requirement for NF-kappaB signalling in a mouse model of lung adenocarcinoma. Nature. 2009;462(7269):104–107. doi: 10.1038/nature08462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Cai DW, Georges RN, Mukhopadhyay T, Grimm EA, Roth JA. Therapeutic effect of a retroviral wild-type p53 expression vector in an orthotopic lung cancer model. J Natl Cancer Inst. 1994;86(19):1458–1462. doi: 10.1093/jnci/86.19.1458. [DOI] [PubMed] [Google Scholar]
- Jiang DK, Yao L, Ren WH, Wang WZ, Peng B, Yu L. TP53 Arg72Pro polymorphism and endometrial cancer risk: a meta-analysis. Med Oncol. 2011;28(4):1129–1135. doi: 10.1007/s12032-010-9597-x. [DOI] [PubMed] [Google Scholar]
- Chen MB, Li C, Shen WX, Guo YJ, Shen W, Lu PH. Association of a LSP1 gene rs3817198T>C polymorphism with breast cancer risk: evidence from 33,920 cases and 35,671 controls. Mol Biol Rep. 2011;38(7):4687–4695. doi: 10.1007/s11033-010-0603-3. [DOI] [PubMed] [Google Scholar]
- Bachelder RE, Crago A, Chung J, Wendt MA, Shaw LM, Robinson G, Mercurio AM. Vascular endothelial growth factor is an autocrine survival factor for neuropilin-expressing breast carcinoma cells. Cancer Res. 2001;61(15):5736–5740. [PubMed] [Google Scholar]
- Eriksson L, Ahluwalia M, Spiewak J, Lee G, Sarma DS, Roomi MJ, Farber E. Distinctive biochemical pattern associated with resistance of hepatocytes in hepatocyte nodules during liver carcinogenesis. Environ Health Perspect. 1983;49:171–174. doi: 10.1289/ehp.8349171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volm M, Koomagi R, Rittgen W. Clinical implications of cyclins, cyclin-dependent kinases, RB and E2F1 in squamous-cell lung carcinoma. Int J Cancer. 1998;79(3):294–299. doi: 10.1002/(sici)1097-0215(19980619)79:3<294::aid-ijc15>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Anayama T, Furihata M, Takeuchi T, Sonobe H, Sasaguri S, Matsumoto M, Ohtsuki Y. Insufficient effect of p27(KIP1) to inhibit cyclin D1 in human esophageal cancer in vitro. Int J Oncol. 2001;18(1):151–155. doi: 10.3892/ijo.18.1.151. [DOI] [PubMed] [Google Scholar]
- Buschges R, Weber RG, Actor B, Lichter P, Collins VP, Reifenberger G. Amplification and expression of cyclin D genes (CCND1, CCND2 and CCND3) in human malignant gliomas. Brain Pathol. 1999;9(3):435–442. doi: 10.1111/j.1750-3639.1999.tb00532.x. discussion 432–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comstock CE, Augello MA, Benito RP, Karch J, Tran TH, Utama FE, Tindall EA, Wang Y, Burd CJ, Groh EM, Hoang HN, Giles GG, Severi G, Hayes VM, Henderson BE, Le Marchand L, Kolonel LN, Haiman CA, Baffa R, Gomella LG, Knudsen ES, Rui H, Henshall SM, Sutherland RL, Knudsen KE. Cyclin D1 splice variants: polymorphism, risk, and isoform-specific regulation in prostate cancer. Clin Cancer Res. 2009;15(17):5338–5349. doi: 10.1158/1078-0432.CCR-08-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata M, Watanabe M, Yamanaka M, Kubota Y, Ito H, Nagao M, Katoh T, Kamataki T, Kawamura J, Yatani R, Shiraishi T. Genetic polymorphisms in cytochrome P450 (CYP) 1A1, CYP1A2, CYP2E1, glutathione S-transferase (GST) M1 and GSTT1 and susceptibility to prostate cancer in the Japanese population. Cancer Lett. 2001;165(2):171–177. doi: 10.1016/s0304-3835(01)00398-6. [DOI] [PubMed] [Google Scholar]
- Uematsu F. Genetic polymorphisms of drug-metabolizing enzymes and susceptibility to lung cancer-relevance to smoking. Nihon Rinsho. 1996;54(2):513–517. [PubMed] [Google Scholar]
- Murray GI, Taylor VE, McKay JA, Weaver RJ, Ewen SW, Melvin WT, Burke MD. Expression of xenobiotic metabolizing enzymes in tumours of the urinary bladder. Int J Exp Pathol. 1995;76(4):271–276. [PMC free article] [PubMed] [Google Scholar]
- Hu YY, Yuan H, Jiang GB, Chen N, Wen L, Leng WD, Zeng XT, Niu YM. Associations between XPD Asp312Asn polymorphism and risk of head and neck cancer: a meta-analysis based on 7,122 subjects. PLoS One. 2012;7(4):e35220. doi: 10.1371/journal.pone.0035220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Niu YM, Wang RX, Li HZ, Chen N. Association between XPD Lys751Gln polymorphism and risk of head and neck cancer: a meta-analysis. Genet Mol Res. 2011;10(4):3356–3364. doi: 10.4238/2011.November.22.6. [DOI] [PubMed] [Google Scholar]
- Kumar A, Pant MC, Singh HS, Khandelwal S. Associated risk of XRCC1 and XPD cross talk and life style factors in progression of head and neck cancer in north Indian population. Mutat Res. 2012;729(1–2):24–34. doi: 10.1016/j.mrfmmm.2011.09.001. [DOI] [PubMed] [Google Scholar]
- Clapper ML. Genetic polymorphism and cancer risk. Curr Oncol Rep. 2000;2(3):251–256. doi: 10.1007/s11912-000-0075-z. [DOI] [PubMed] [Google Scholar]
- Likhin FA, Bartnovskii AE, Vdovichenko KK, Abramov AA, Belokhvostov AS. Characteristics of methyl-specific PCR-test of glutathione-S-transferase P1 gene in plasm DNA and cellular urinary precipitate for differential diagnosis of prostatic adenoma and adenocarcinoma. Urologiia. 2005;4:12–15. [PubMed] [Google Scholar]
- Zhang Z, Liu W, Jia X, Gao Y, Hemminki K, Lindholm B. Use of pyrosequencing to detect clinically relevant polymorphisms of genes in basal cell carcinoma. Clin Chim Acta. 2004;342(1–2):137–143. doi: 10.1016/j.cccn.2003.12.010. [DOI] [PubMed] [Google Scholar]
- Krajinovic M, Lemieux-Blanchard E, Chiasson S, Primeau M, Costea I, Moghrabi A. Role of polymorphisms in MTHFR and MTHFD1 genes in the outcome of childhood acute lymphoblastic leukemia. Pharmacogenomics J. 2004;4(1):66–72. doi: 10.1038/sj.tpj.6500224. [DOI] [PubMed] [Google Scholar]
- Petra BG, Janez J, Vita D. Gene-gene interactions in the folate metabolic pathway influence the risk for acute lymphoblastic leukemia in children. Leuk Lymphoma. 2007;48(4):786–792. doi: 10.1080/10428190601187711. [DOI] [PubMed] [Google Scholar]
- Matakidou A, El Galta R, Rudd MF, Webb EL, Bridle H, Eisen T, Houlston RS. Prognostic significance of folate metabolism polymorphisms for lung cancer. Br J Cancer. 2007;97(2):247–252. doi: 10.1038/sj.bjc.6603830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore LE, Malats N, Rothman N, Real FX, Kogevinas M, Karami S, Garcia-Closas R, Silverman D, Chanock S, Welch R, Tardón A, Serra C, Carrato A, Dosemeci M, García-Closas M. Polymorphisms in one-carbon metabolism and trans-sulfuration pathway genes and susceptibility to bladder cancer. Int J Cancer. 2007;120(11):2452–2458. doi: 10.1002/ijc.22565. [DOI] [PubMed] [Google Scholar]
- Curtin K, Slattery ML, Ulrich CM, Bigler J, Levin TR, Wolff RK, Albertsen H, Potter JD, Samowitz WS. Genetic polymorphisms in one-carbon metabolism: associations with CpG island methylator phenotype (CIMP) in colon cancer and the modifying effects of diet. Carcinogenesis. 2007;28(8):1672–1679. doi: 10.1093/carcin/bgm089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner AS, Beresina OV, Voronina EN, Voropaeva EN, Boyarskih UA, Pospelova TI, Filipenko ML. Polymorphisms in folate-metabolizing genes and risk of non-Hodgkin's lymphoma. Leuk Res. 2011;35(4):508–515. doi: 10.1016/j.leukres.2010.10.004. [DOI] [PubMed] [Google Scholar]
- Lee KM, Lan Q, Kricker A, Purdue MP, Grulich AE, Vajdic CM, Turner J, Whitby D, Kang D, Chanock S, Rothman N, Armstrong BK. One-carbon metabolism gene polymorphisms and risk of non-Hodgkin lymphoma in Australia. Hum Genet. 2007;122(5):525–533. doi: 10.1007/s00439-007-0431-2. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Matsuo K, Hasegawa Y, Hiraki A, Wakai K, Hirose K, Saito T, Sato S, Ueda R, Tajima K. One-carbon metabolism-related gene polymorphisms and risk of head and neck squamous cell carcinoma: case–control study. Cancer Sci. 2007;98(9):1439–1446. doi: 10.1111/j.1349-7006.2007.00533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Lee SH, Choi YL, Wang LH, Park CK, Shin YK. Extensive alteration in the expression profiles of TGFB pathway signaling components and TP53 is observed along the gastric dysplasia-carcinoma sequence. Histol Histopathol. 2008;23(12):1439–1452. doi: 10.14670/HH-23.1439. [DOI] [PubMed] [Google Scholar]
- Gemma A, Uematsu K, Hagiwara K, Takenoshita S, Kudoh S. Mechanism of resistance to growth inhibition by transforming growth factor-beta 1 (TGF-beta 1) in primary lung cancer and new molecular targets in therapy. Gan To Kagaku Ryoho. 2000;27(8):1253–1259. [PubMed] [Google Scholar]
- Jonson T, Albrechtsson E, Axelson J, Heidenblad M, Gorunova L, Johansson B, Hoglund M. Altered expression of TGFB receptors and mitogenic effects of TGFB in pancreatic carcinomas. Int J Oncol. 2001;19(1):71–81. [PubMed] [Google Scholar]
- Franzen P, Ichijo H, Miyazono K. Different signals mediate transforming growth factor-beta 1-induced growth inhibition and extracellular matrix production in prostatic carcinoma cells. Exp Cell Res. 1993;207(1):1–7. doi: 10.1006/excr.1993.1156. [DOI] [PubMed] [Google Scholar]
- Maggio-Price L, Treuting P, Bielefeldt-Ohmann H, Seamons A, Drivdahl R, Zeng W, Lai L, Huycke M, Phelps S, Brabb T, Iritani BM. Bacterial infection of Smad3/Rag2 double-null mice with transforming growth factor-beta dysregulation as a model for studying inflammation-associated colon cancer. Am J Pathol. 2009;174(1):317–329. doi: 10.2353/ajpath.2009.080485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirshfield KM, Rebbeck TR, Levine AJ. Germline mutations and polymorphisms in the origins of cancers in women. J Oncol. 2010;2010:297671. doi: 10.1155/2010/297671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volate SR, Kawasaki BT, Hurt EM, Milner JA, Kim YS, White J, Farrar WL. Gossypol induces apoptosis by activating p53 in prostate cancer cells and prostate tumor-initiating cells. Mol Cancer Ther. 2010;9(2):461–470. doi: 10.1158/1535-7163.MCT-09-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardini MQ, Baba T, Lee PS, Barnett JC, Sfakianos GP, Secord AA, Murphy SK, Iversen E, Marks JR, Berchuck A. Expression signatures of TP53 mutations in serous ovarian cancers. BMC Cancer. 2010;10:237. doi: 10.1186/1471-2407-10-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada S, Mimata A, Sekine M, Mogushi K, Akiyama Y, Fukamachi H, Jonkers J, Tanaka H, Eishi Y, Yuasa Y. Synergistic tumour suppressor activity of E-cadherin and p53 in a conditional mouse model for metastatic diffuse-type gastric cancer. Gut. 2012;61(3):344–353. doi: 10.1136/gutjnl-2011-300050. [DOI] [PubMed] [Google Scholar]
- Sano D, Xie TX, Ow TJ, Zhao M, Pickering CR, Zhou G, Sandulache VC, Wheeler DA, Gibbs RA, Caulin C, Myers JN. Disruptive TP53 mutation is associated with aggressive disease characteristics in an orthotopic murine model of oral tongue cancer. Clin Cancer Res. 2011;17(21):6658–6670. doi: 10.1158/1078-0432.CCR-11-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furth PA, Cabrera MC, Diaz-Cruz ES, Millman S, Nakles RE. Assessing estrogen signaling aberrations in breast cancer risk using genetically engineered mouse models. Ann N Y Acad Sci. 2011;1229:147–155. doi: 10.1111/j.1749-6632.2011.06086.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge R, Hooijberg JH, van Zelst BD, Jansen G, van Zantwijk CH, Kaspers GJ, Peters GJ, Ravindranath Y, Pieters R, Lindemans J. Effect of polymorphisms in folate-related genes on in vitro methotrexate sensitivity in pediatric acute lymphoblastic leukemia. Blood. 2005;106(2):717–720. doi: 10.1182/blood-2004-12-4941. [DOI] [PubMed] [Google Scholar]
- Durfort T, Tkach M, Meschaninova MI, Rivas MA, Elizalde PV, Venyaminova AG, Schillaci R, Francois JC. Small interfering RNA targeted to IGF-IR delays tumor growth and induces proinflammatory cytokines in a mouse breast cancer model. PLoS One. 2012;7(1):e29213. doi: 10.1371/journal.pone.0029213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calogero RA, Cordero F, Forni G, Cavallo F. Inflammation and breast cancer. Inflammatory component of mammary carcinogenesis in ErbB2 transgenic mice. Breast Cancer Res. 2007;9(4):211. doi: 10.1186/bcr1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiff R, Osborne CK. Endocrinology and hormone therapy in breast cancer: new insight into estrogen receptor-alpha function and its implication for endocrine therapy resistance in breast cancer. Breast Cancer Res. 2005;7(5):205–211. doi: 10.1186/bcr1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahm H, Petral-Malec D, Yilmaz-Ceyhan A, Fischer JR, Lorenzoni M, Givel JC, Odartchenko N. Growth stimulation of a human colorectal carcinoma cell line by interleukin-1 and -6 and antagonistic effects of transforming growth factor beta 1. Eur J Cancer. 1992;28A(11):1894–1899. doi: 10.1016/0959-8049(92)90031-v. [DOI] [PubMed] [Google Scholar]
- Yu C, Yao Z, Jiang Y, Keller ET. Prostate cancer stem cell biology. Minerva Urol Nefrol. 2012;64(1):19–33. [PMC free article] [PubMed] [Google Scholar]
- Peng B, Cao L, Ma X, Wang W, Wang D, Yu L. Meta-analysis of association between matrix metalloproteinases 2, 7 and 9 promoter polymorphisms and cancer risk. Mutagenesis. 2010;25(4):371–379. doi: 10.1093/mutage/geq015. [DOI] [PubMed] [Google Scholar]
- Zhang YM, Cao C, Liang K. Genetic polymorphism of epidermal growth factor 61A>G and cancer risk: a meta-analysis. Cancer Epidemiol. 2010;34(2):150–156. doi: 10.1016/j.canep.2010.02.004. [DOI] [PubMed] [Google Scholar]
- Willmarth NE, Ethier SP. Amphiregulin as a novel target for breast cancer therapy. J Mammary Gland Biol Neoplasia. 2008;13(2):171–179. doi: 10.1007/s10911-008-9081-9. [DOI] [PubMed] [Google Scholar]
- Guise TA. Breaking down bone: new insight into site-specific mechanisms of breast cancer osteolysis mediated by metalloproteinases. Genes Dev. 2009;23(18):2117–2123. doi: 10.1101/gad.1854909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix M, Body JJ. Regulation of c-fos and c-jun expression by calcitonin in human breast cancer cells. Calcif Tissue Int. 1997;60(6):513–519. doi: 10.1007/s002239900273. [DOI] [PubMed] [Google Scholar]
- Chen MB, Wu XY, Shen W, Wei MX, Li C, Cai B, Tao GQ, Lu PH. Association between polymorphisms of trinucleotide repeat containing 9 gene and breast cancer risk: evidence from 62,005 subjects. Breast Cancer Res Treat. 2011;126(1):177–183. doi: 10.1007/s10549-010-1114-6. [DOI] [PubMed] [Google Scholar]
- Gu D, Wang M. VEGF 936C>T polymorphism and breast cancer risk: evidence from 5,729 cases and 5,868 controls. Breast Cancer Res Treat. 2011;125(2):489–493. doi: 10.1007/s10549-010-0991-z. [DOI] [PubMed] [Google Scholar]
- Qiu LX, Wang K, Yang S, Mao C, Zhao L, Yao L, Zhang J, Zhang QL, Sun S, Xue K. Current evidences on vascular endothelial growth factor polymorphisms and breast cancer susceptibility. Mol Biol Rep. 2011;38(7):4491–4494. doi: 10.1007/s11033-010-0579-z. [DOI] [PubMed] [Google Scholar]
- Yang DS, Park KH, Woo OH, Woo SU, Kim AR, Lee ES, Lee JB, Kim YH, Kim JS, Seo JH. Association of a vascular endothelial growth factor gene 936 C/T polymorphism with breast cancer risk: a meta-analysis. Breast Cancer Res Treat. 2011;125(3):849–853. doi: 10.1007/s10549-010-1070-1. [DOI] [PubMed] [Google Scholar]
- Xu B, Li JM, Tong N, Tao J, Li PC, Song NH, Zhang W, Wu HF, Feng NH, Hua LX. VEGFA +936C>T polymorphism and cancer risk: a meta-analysis. Cancer Genet Cytogenet. 2010;198(1):7–14. doi: 10.1016/j.cancergencyto.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Liu X, Wang Z, Yu J, Lei G, Wang S. Three polymorphisms in interleukin-1beta gene and risk for breast cancer: a meta-analysis. Breast Cancer Res Treat. 2010;124(3):821–825. doi: 10.1007/s10549-010-0910-3. [DOI] [PubMed] [Google Scholar]
- Oh JS, Kucab JE, Bushel PR, Martin K, Bennett L, Collins J, DiAugustine RP, Barrett JC, Afshari CA, Dunn SE. Insulin-like growth factor-1 inscribes a gene expression profile for angiogenic factors and cancer progression in breast epithelial cells. Neoplasia. 2002;4(3):204–217. doi: 10.1038/sj.neo.7900229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao C, Ying T, Fang JJ, Sun SF, Lv D, Chen ZB, Ma HY, Yu YM, Ding QL, Shu LH, Deng Z-C. Polymorphism of vascular endothelial growth factor -2578C/A with cancer risk: evidence from 11263 subjects. . Med Oncol. 2011;28(4):1169–1175. doi: 10.1007/s12032-010-9613-1. [DOI] [PubMed] [Google Scholar]
- Xue H, Lin B, Ni P, Xu H, Huang G. Interleukin-1B and interleukin-1 RN polymorphisms and gastric carcinoma risk: a meta-analysis. J Gastroenterol Hepatol. 2010;25(10):1604–1617. doi: 10.1111/j.1440-1746.2010.06428.x. [DOI] [PubMed] [Google Scholar]
- Gao LB, Pan XM, Jia J, Liang WB, Rao L, Xue H, Zhu Y, Li SL, Lv ML, Deng W, Chen TY, Wei YG, Zhang L. IL-8–251A/T polymorphism is associated with decreased cancer risk among population-based studies: evidence from a meta-analysis. Eur J Cancer. 2010;46(8):1333–1343. doi: 10.1016/j.ejca.2010.03.011. [DOI] [PubMed] [Google Scholar]
- Liu L, Zhuang W, Wang C, Chen Z, Wu XT, Zhou Y. Interleukin-8–251 A/T gene polymorphism and gastric cancer susceptibility: a meta-analysis of epidemiological studies. Cytokine. 2010;50(3):328–334. doi: 10.1016/j.cyto.2010.03.008. [DOI] [PubMed] [Google Scholar]
- Peng B, Cao L, Wang W, Xian L, Jiang D, Zhao J, Zhang Z, Wang X, Yu L. Polymorphisms in the promoter regions of matrix metalloproteinases 1 and 3 and cancer risk: a meta-analysis of 50 case–control studies. Mutagenesis. 2010;25(1):41–48. doi: 10.1093/mutage/gep041. [DOI] [PubMed] [Google Scholar]
- Wysoczynski M, Ratajczak MZ. Lung cancer secreted microvesicles: underappreciated modulators of microenvironment in expanding tumors. Int J Cancer. 2009;125(7):1595–1603. doi: 10.1002/ijc.24479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Zhang H, Sheng W, Xiang J, Ye Z, Yang J. Adenovirus-mediated ING4 expression suppresses lung carcinoma cell growth via induction of cell cycle alteration and apoptosis and inhibition of tumor invasion and angiogenesis. Cancer Lett. 2008;271(1):105–116. doi: 10.1016/j.canlet.2008.05.050. [DOI] [PubMed] [Google Scholar]
- Gao Y, Cao Y, Tan A, Liao C, Mo Z, Gao F. Glutathione S-transferase M1 polymorphism and sporadic colorectal cancer risk: an updating meta-analysis and HuGE review of 36 case–control studies. Ann Epidemiol. 2010;20(2):108–121. doi: 10.1016/j.annepidem.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Lu S, Wang Z, Cui D, Liu H, Hao X. Glutathione S-transferase P1 Ile105Val polymorphism and breast cancer risk: a meta-analysis involving 34,658 subjects. Breast Cancer Res Treat. 2011;125(1):253–259. doi: 10.1007/s10549-010-0969-x. [DOI] [PubMed] [Google Scholar]
- Qiu LX, Yuan H, Yu KD, Mao C, Chen B, Zhan P, Xue K, Zhang J, Hu XC. Glutathione S-transferase M1 polymorphism and breast cancer susceptibility: a meta-analysis involving 46,281 subjects. Breast Cancer Res Treat. 2010;121(3):703–708. doi: 10.1007/s10549-009-0636-2. [DOI] [PubMed] [Google Scholar]
- Singh V, Parmar D, Singh MP. Do single nucleotide polymorphisms in xenobiotic metabolizing genes determine breast cancer susceptibility and treatment outcomes? Cancer Invest. 2008;26(8):769–783. doi: 10.1080/07357900801953196. [DOI] [PubMed] [Google Scholar]
- Curran JE, Weinstein SR, Griffiths LR. Polymorphisms of glutathione S-transferase genes (GSTM1, GSTP1 and GSTT1) and breast cancer susceptibility. Cancer Lett. 2000;153(1–2):113–120. doi: 10.1016/s0304-3835(00)00361-x. [DOI] [PubMed] [Google Scholar]
- Economopoulos KP, Sergentanis TN, Vlahos NF. Glutathione S-transferase M1, T1, and P1 polymorphisms and ovarian cancer risk: a meta-analysis. Int J Gynecol Cancer. 2010;20(5):732–737. doi: 10.1111/IGC.0b013e3181dedeb5. [DOI] [PubMed] [Google Scholar]
- Zeng FF, Liu SY, Wei W, Yao SP, Zhu S, Li KS, Wan G, Zhang HT, Zhong M, Wang BY. Genetic polymorphisms of glutathione S-transferase T1 and bladder cancer risk: a meta-analysis. Clin Exp Med. 2010;10(1):59–68. doi: 10.1007/s10238-009-0070-0. [DOI] [PubMed] [Google Scholar]
- Moore LE, Baris DR, Figueroa JD, Garcia-Closas M, Karagas MR, Schwenn MR, Johnson AT, Lubin JH, Hein DW, Dagnall CL, Colt JS, Kida M, Jones MA, Schned AR, Cherala SS, Chanock SJ, Cantor KP, Silverman DT, Rothman N. GSTM1 null and NAT2 slow acetylation genotypes, smoking intensity and bladder cancer risk: results from the New England bladder cancer study and NAT2 meta-analysis. Carcinogenesis. 2011;32(2):182–189. doi: 10.1093/carcin/bgq223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukuda M, Nagahara T, Yago T, Matsuda H, Yanoma S. Production of granulocyte colony-stimulating factor by head and neck carcinomas. Biotherapy. 1993;6(3):183–187. doi: 10.1007/BF01878079. [DOI] [PubMed] [Google Scholar]
- Lagadec PF, Saraya KA, Balkwill FR. Human small-cell lung-cancer cells are cytokine-resistant but NK/LAK-sensitive. Int J Cancer. 1991;48(2):311–317. doi: 10.1002/ijc.2910480226. [DOI] [PubMed] [Google Scholar]
- Enzmann V, Faude F, Kohen L, Wiedemann P. Secretion of cytokines by human choroidal melanoma cells and skin melanoma cell lines in vitro. Ophthalmic Res. 1998;30(3):189–194. doi: 10.1159/000055473. [DOI] [PubMed] [Google Scholar]
- Kimura F, Nakamura Y, Sato K, Wakimoto N, Kato T, Tahara T, Yamada M, Nagata N, Motoyoshi K. Cyclic change of cytokines in a patient with cyclic thrombocytopenia. Br J Haematol. 1996;94(1):171–174. doi: 10.1046/j.1365-2141.1996.d01-1783.x. [DOI] [PubMed] [Google Scholar]
- Reinhold D, Bank U, Buhling F, Lendeckel U, Ulmer AJ, Flad HD, Ansorge S. Transforming growth factor-beta 1 (TGF-beta 1) inhibits DNA synthesis of PWM-stimulated PBMC via suppression of IL-2 and IL-6 production. Cytokine. 1994;6(4):382–388. doi: 10.1016/1043-4666(94)90062-0. [DOI] [PubMed] [Google Scholar]
- Nikolova PN, Pawelec GP, Mihailova SM, Ivanova MI, Myhailova AP, Baltadjieva DN, Marinova DI, Ivanova SS, Naumova EJ. Association of cytokine gene polymorphisms with malignant melanoma in Caucasian population. Cancer Immunol Immunother. 2007;56(3):371–379. doi: 10.1007/s00262-006-0193-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman RS, Deavers M, Liu J, Wang E. Peritoneal inflammation - a microenvironment for epithelial ovarian cancer (EOC) J Transl Med. 2004;2(1):23. doi: 10.1186/1479-5876-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarnicki A, Putoczki T, Ernst M. Stat3: linking inflammation to epithelial cancer - more than a “gut” feeling? Cell division. 2010;5:14. doi: 10.1186/1747-1028-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundhaug JE, Fischer SM. Molecular mechanisms of mouse skin tumor promotion. Cancers. 2010;2(2):436–482. doi: 10.3390/cancers2020436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mischek D, Steinborn R, Petznek H, Bichler C, Zatloukal K, Sturzl M, Gunzburg WH, Hohenadl C. Molecularly characterised xenograft tumour mouse models: valuable tools for evaluation of new therapeutic strategies for secondary liver cancers. J Biomed Biotechnol. 2009;2009:437284. doi: 10.1155/2009/437284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiss D. Optimizing molecular-targeted therapies in ovarian cancer: the renewed surge of interest in ovarian cancer biomarkers and cell signaling pathways. Journal of Oncology. 2012;2012:737981. doi: 10.1155/2012/737981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136(5):823–837. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Yan B, Van Waes C. The role of the NF-kappaB transcriptome and proteome as biomarkers in human head and neck squamous cell carcinomas. Biomark Med. 2008;2(4):409–426. doi: 10.2217/17520363.2.4.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada Y, Imamura M. Prognostic factors for esophageal cancer–from the viewpoint of molecular biology. Gan To Kagaku Ryoho. 1996;23(8):972–981. [PubMed] [Google Scholar]
- Ziober BL, Turner MA, Palefsky JM, Banda MJ, Kramer RH. Type I collagen degradation by invasive oral squamous cell carcinoma. Oral Oncol. 2000;36(4):365–372. doi: 10.1016/s1368-8375(00)00019-1. [DOI] [PubMed] [Google Scholar]
- Nuovo GJ. In situ detection of PCR-amplified metalloproteinase cDNAs, their inhibitors and human papillomavirus transcripts in cervical carcinoma cell lines. Int J Cancer. 1997;71(6):1056–1060. doi: 10.1002/(sici)1097-0215(19970611)71:6<1056::aid-ijc23>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Liss C, Fekete MJ, Hasina R, Lam CD, Lingen MW. Paracrine angiogenic loop between head-and-neck squamous-cell carcinomas and macrophages. Int J Cancer. 2001;93(6):781–785. doi: 10.1002/ijc.1407. [DOI] [PubMed] [Google Scholar]
- Stachel D, Albert M, Meilbeck R, Kreutzer B, Haas RJ, Schmid I. Bone marrow Th2 cytokine expression as predictor for relapse in childhood acute lymphoblastic leukemia (ALL) Eur J Med Res. 2006;11(3):102–113. [PubMed] [Google Scholar]
- Melinceanu L, Sarafoleanu C, Lerescu L, Tucureanu C, Caras I, Salageanu A. Impact of smoking on the immunological profile of patients with laryngeal carcinoma. J Med Life. 2009;2(2):211–218. [PMC free article] [PubMed] [Google Scholar]
- Lazar-Molnar E, Hegyesi H, Toth S, Falus A. Autocrine and paracrine regulation by cytokines and growth factors in melanoma. Cytokine. 2000;12(6):547–554. doi: 10.1006/cyto.1999.0614. [DOI] [PubMed] [Google Scholar]
- Inoue K, Wood CG, Slaton JW, Karashima T, Sweeney P, Dinney CP. Adenoviral-mediated gene therapy of human bladder cancer with antisense interleukin-8. Oncol Rep. 2001;8(5):955–964. doi: 10.3892/or.8.5.955. [DOI] [PubMed] [Google Scholar]
- Snarskaia ES, Molochkov VA, Frank GA, Zavalishina LA. Matrix metalloproteinases and their tissue inhibitors in basal cell and metatypical cancer of the skin. Arkhiv Patologii. 2005;67(3):14–16. [PubMed] [Google Scholar]
- Lievre A, Milet J, Carayol J, Le Corre D, Milan C, Pariente A, Nalet B, Lafon J, Faivre J, Bonithon-Kopp C, Olschwang S, Bonaiti-Pellié C, Laurent-Puig P. Members of the ANGH group. Genetic polymorphisms of MMP1, MMP3 and MMP7 gene promoter and risk of colorectal adenoma. BMC Cancer. 2006;6:270. doi: 10.1186/1471-2407-6-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricketts C, Zeegers MP, Lubinski J, Maher ER. Analysis of germline variants in CDH1, IGFBP3, MMP1, MMP3, STK15 and VEGF in familial and sporadic renal cell carcinoma. PLoS One. 2009;4(6):e6037. doi: 10.1371/journal.pone.0006037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M, Garcia A, Martinez-Palones JM, Xercavins J, Reventos J. Susceptibility to endometrial cancer: influence of allelism at p53, glutathione S-transferase (GSTM1 and GSTT1) and cytochrome P-450 (CYP1A1) loci. Br J Cancer. 1997;75(9):1385–1388. doi: 10.1038/bjc.1997.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozhemiakin LA, Bulavin DV, Morozov VI, Osipov EV, Zolotarev DV. Effectiveness of the glutathione s-transferase assay in diagnosing lung cancer. Vopr Onkol. 1995;41(1):33–38. [PubMed] [Google Scholar]
- Schilthuizen C, Broyl A, van der Holt B, de Knegt Y, Lokhorst H, Sonneveld P. Influence of genetic polymorphisms in CYP3A4, CYP3A5, GSTP1, GSTM1, GSTT1 and MDR1 genes on survival and therapy-related toxicity in multiple myeloma. Haematologica. 2007;92(2):277–278. doi: 10.3324/haematol.10618. [DOI] [PubMed] [Google Scholar]
- Lavender NA, Benford ML, VanCleave TT, Brock GN, Kittles RA, Moore JH, Hein DW, Kidd LC. Examination of polymorphic glutathione S-transferase (GST) genes, tobacco smoking and prostate cancer risk among men of African descent: a case–control study. BMC Cancer. 2009;9:397. doi: 10.1186/1471-2407-9-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saadat I, Saadat M. The glutathione S-transferase mu polymorphism and susceptibility to acute lymphocytic leukemia. Cancer Lett. 2000;158(1):43–45. doi: 10.1016/s0304-3835(00)00504-8. [DOI] [PubMed] [Google Scholar]
- Ovsepian VA, Vinogradova E, Sherstneva ES. Cytochrome P4501A1, glutathione S-transferase M1 and T1 gene polymorphisms in chronic myeloid leukemia. Genetika. 2010;46(10):1360–1362. [PubMed] [Google Scholar]
- Vrana D, Pikhart H, Mohelnikova-Duchonova B, Holcatova I, Strnad R, Slamova A, Schejbalova M, Ryska M, Susova S, Soucek P. The association between glutathione S-transferase gene polymorphisms and pancreatic cancer in a central European Slavonic population. Mutat Res. 2009;680(1–2):78–81. doi: 10.1016/j.mrgentox.2009.09.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genes and cancer types included in this meta-analysis.