Abstract
Brassinosteroids (BRs), a group of naturally occurring plant steroidal compounds, are essential for plant growth, development and stress tolerance. Recent studies showed that BRs could induce systemic tolerance to biotic and abiotic stresses; however, the molecular mechanisms by which BRs signals lead to responses in the whole plant are largely unknown. In this study, 24-epibrassinosteroid (EBR)-induced systemic tolerance in Cucumis sativus L. cv. Jinyan No. 4 was analyzed through the assessment of symptoms of photooxidative stress by chlorophyll fluorescence imaging pulse amplitude modulation. Expression of defense/stress related genes were induced in both treated local leaves and untreated systemic leaves by local EBR application. With the suppressive subtractive hybridization (SSH) library using cDNA from the phloem sap of EBR-treated plants as the tester and distilled water (DW)-treated plants as the driver, 14 transcripts out of 260 clones were identified. Quantitative Real Time-Polymerase Chain Reaction (RT-qPCR) validated the specific up-regulation of these transcripts. Of the differentially expressed transcripts with known functions, transcripts for the selected four cDNAs, which encode an auxin-responsive protein (IAA14), a putative ankyrin-repeat protein, an F-box protein (PP2), and a major latex, pathogenesis-related (MLP)-like protein, were induced in local leaves, systemic leaves and roots after foliar application of EBR onto mature leaves. Our results demonstrated that EBR-induced systemic tolerance is accompanied with increased transcript of genes in the defense response in other organs. The potential role of phloem mRNAs as signaling components in mediating BR-regulated systemic resistance is discussed.
Introduction
Brassinosteroids (BRs), a group of naturally occurring plant steroidal compounds, are essential for plant growth and development [1]. They have also been implicated to confer tolerance against a wide spectrum of biotic and abiotic stresses, such as oxidative, thermal, salinity, heavy metal stresses and those caused by pesticides and pathogen attack in many plant species [2]–[5]. Although a large body of evidence has shown that BRs do not undergo long-distance transport between different organs [6], several studies, including ours, showed that they could induce systemic tolerance against both biotic and abiotic stresses [7], [8]. It has been reported that BRs may have an indirect role in long-distance signaling through their effects on other hormones, such as auxins or polyamines [9], [10]. In tobacco, BRs function in an innate immunity system that is distinct from systemic acquired resistance (SAR) and wound-inducible disease resistance [11]. Foliar application of brassinolide reduces the nodule number in a super-nodulating soybean mutant [12]. Further study has indicated that BRs regulate root nodule formation through a change of the polyamine content [10]. Recently, Ding has found that the foliar application of 24-epibrassinosteroid (EBR) enhances the root resistance to Fusarium oxysporum in cucumber plants [8]. Furthermore, Xia and others provided evidence that local EBR treatment activated continuous production of H2O2 by triggering NADPH and thus mediated systemic stress tolerance in cucumber [7]. However the long-distance molecules and related signal transduction cascades involved in the BR-mediated systemic effect remain largely unknown.
Phloem, consisting of companion cell and sieve element, which is also called the sieve-tube system, is the conduit for the transport of photoassimilates, including carbohydrates, amino acids and other nutrients, from source to sink organs [13]. Recently, numerous mRNAs, small RNAs and proteins have been found to be present in the phloem translocation stream and function as components of a long-distance signaling network [14], [15]. These proteins and RNAs have been shown to play important roles in the coordination of different organs in floral induction, tuberization, resource allocation, nutrient signaling and pathogenic defense responses [16]. For example, in tomato, wild-type scion-leaf development was shown to change due to the long distance transport of a homeobox fusion transcript PYROPHOSPHATE-DEPENDENT PHOSPHOFRUCTOKINASE (PFP)-LeT6, after grafting onto the dominant mutant Mouse-ear (Me) [17]. RNA for GAI which is involved in GA signaling has also been demonstrated to regulate leaf development through their long-distance trafficking in transgenic tomato plants [18], and in potato, the movement of BEL1 transcription factor StBEL5 to the stolon tips has been implicated in the enhancement of tuber formation [19]. It is, therefore, highly possible that phloem mRNAs may function as signaling components that mediate BR-regulated systemic resistance.
We were particularly interested in studying the up-regulated mRNAs in phloem sap caused by BRs. Cucumber (Cucumis sativus ), a cucurbit plant, is an ideal species for studying vascular biology because of its distinct vascular bundles and easiness in collecting phloem sap by exudation. To test the above hypothesis, we first analyzed EBR-induced systemic stress tolerance through the assessment of symptoms of photooxidative stress by chlorophyll fluorescence imaging pulse amplitude modulation. Then a suppressive subtractive hybridization (SSH) library with RNA extracted from the phloem sap of distilled water (DW)-treated cucumber as the driver, and EBR-treated as the tester was established to detect the mRNA molecules induced by EBR. mRNAs associated with transcription, protein translation, transport and defense responses were identified. Among them, four identified transcripts involved in defense responses were further studied for their expression in local and distal organs. The results are discussed with the proposal that phloem mRNAs may function as long-distance signaling components involved in the BR-induced systemic resistance in cucumber.
Materials and Methods
Plant Material and Experimental Design
Cucumber (Cucumis sativus L. cv. Jinyan No. 4) seeds were sown in a growth medium containing a mixture of soil and perlite (1∶1, v/v). Seedlings with two cotyledons were transplanted into containers (40 cm×25 cm×15 cm) filled with Hoagland’s nutrient solution. Plants were grown in a greenhouse with a photoperiod of 14 h, average day/night temperatures of 25/18°C, and a maximum photosynthetic photon flux density (PPFD) of 1000 µmol m−2 s−1. Three experiments were carried out to investigate BR-induced changes in stress tolerance and gene transcript in the phloem sap, local and systemic leaves, and roots.
Experiment 1
To determine the systemic effects of BR on tolerance against photooxidative stress and the transcript level of defense-related genes, the 5th leaf from the bottom of a plant at the seven-leaf stage was pretreated with 0.2 µM EBR (Sigma) or DW. Twenty four hours later, the 6th leaf was sprayed with 10 µM paraquat (PQ; Sigma) and stress tolerance was measured on the basis of changes in the maximal photochemical quantum efficiency of photosystem II (Fv/Fm) at 24 h after PQ treatment. Chlorophyll fluorescence was determined with imaging pulse amplitude modulation fluorometer (IMAG-MAXI; Heinz Walz). For the measurement of Fv/Fm, plants were dark-adapted for 30 min. Minimal fluorescence (Fo) was measured during the weak measuring pulses and maximal fluorescence (Fm) was measured during a 0.8-s pulse light exposure to a PPFD of ca. 4000 µmol m−2 s−1. Fv/Fm was determined with the whole leaf as the area of interest [5]. The 5th and 6th leaves were sampled at different time points after EBR treatment for gene expression analysis.
Experiment 2
To investigate EBR-induced transcript profile changes in cucumber phloem sap by SSH library and to confirm the up-regulation of the transcripts detected by SSH library, plants at the seven-leaf stage were used for phloem sap collection. Mature leaves were treated with 0.2 µM EBR (Sigma, USA) or DW at 8 am. The phloem sap was collected at various time intervals (11 am, 2 pm, 5 pm and 8 am in the next morning) from the EBR- and DW- treated plants at the same time. The stem part close to apex and mature leaf petioles were cut, and the phloem sap was collected, as previously described [20], [21]. Briefly, stems or petioles were excised with a sterile razor blade and the cut surface was then blotted with sterile filter paper (3 MM; Whatman, Maidstone, UK) for several times. Phloem sap exuded thereafter was collected using sterile micropipette tips (200 µL) and immediately mixed with 500 µL of TRIzol reagent (Invitrogen, Carlsbad, CA) for phloem sap RNA extraction. Ten plants with ca. seven leaves for each treatment at each time interval were used, and 200 µL of the phloem sap was collected at each time interval. Phloem sap from different time intervals was pooled together for RNA extraction.
Experiment 3
To determine the expression of genes identified from the SSH library in the local and systemic leaves and roots, the 5th leaf at the seven-leaf stage were pretreated with 0.2 µM EBR (Sigma, USA) or DW. The 5th and the 6th leaves were sampled at 0.5, 1, 3 and 6 h later, respectively. Meanwhile, the roots were also sampled at 6, 12, and 24 h after treatment. The samples were stored at −80°C until the RNA extraction.
RNA Isolation from Cucumber Phloem Sap and cDNA Synthesis
Each aliquot (200 µl) of phloem sap was mixed with 500 µl TRIZOL reagent (Invitrogen, Carlsbad, CA, USA). Proteins were then extracted twice with chloroform. The RNA contained in the aqueous phase was precipitated with 2.5 volumes of cold ethanol, centrifuged at 4°C for 45 minutes (min), and then resuspended in diethyl pyrocarbonate-treated water. Total RNA was purified using the RNeasy Mini Kit (Qiagen, Hilden, Germany).
Phloem-sap-derived cDNA from the DW- and EBR-treated plants was amplified by long-distance (LD)-PCR and then used directly for SSH. The SSH library was constructed according to the manual of the Super SMART PCR cDNA Synthesis Kit (Clontech, Palo Alto, CA, USA). cDNA from the control phloem sap in DW-treated plants was used as the driver, while cDNA from the phloem sap in EBR-treated plants was used as the tester. The PCR products from the second amplification were cloned into the pGEM-T easy vector (Promega, Madison, WI, USA). Recombinant plasmids harboring the subtracted cDNA from phloem sap RNA were analyzed by PCR. Independent clones (260) were randomly selected, sequenced (Sangon, Shanghai, China) and then were subjected to BLASTN searches in the Cucurbit Genomics Database (CuGenDB): http://www.icugi.org/cgi-bin/ICuGI/genome/blast.cgi.
Total RNA Extraction and RT-qPCR for Gene Expression Analysis
Total RNA from the phloem sap, leaves and roots of cucumber was extracted using TRIZOL reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s instructions. After extraction, total RNA was dissolved in diethyl pyrocarbonate-treated water. The first-strand cDNA used as the template for RT-qPCR was synthesized using a RevertAid™ First-strand cDNA Synthesis Kit (Fermentas, USA ) from 2 µg of total RNA that was purified by the RNeasy Mini Kit (Qiagen, Hilden, Germany). Gene-specific primers for the RT-qPCR were designed based on EST sequences, which are shown in Table S1. The RT-qPCR was performed with an iCycler iQ Multicolor Real-time PCR Detection System (Bio-Rad, Hercules, CA, USA). Each reaction (20 µl total volume) consisted of 10 µl iQ SYBR Green Supermix, 1 µl of diluted cDNA and 0.1 µM of forward and reserve primers. The PCR cycling conditions were as follows: 95°C for 3 min and 40 cycles of 95°C for 10 seconds (sec) and 58°C for 45 sec. Fluorescence data were collected during the 58°C step. The cucumber actin gene was used as an internal control. The relative gene expression was calculated, as previously described [22].
Statistical Analysis
Plants were arranged in three randomized blocks with three replicates per treatment. The mean values of three replicates were compared using Tukey’s test (P≤0.05).
Results
EBR Induces Systemic Stress Tolerance in Cucumber Plants
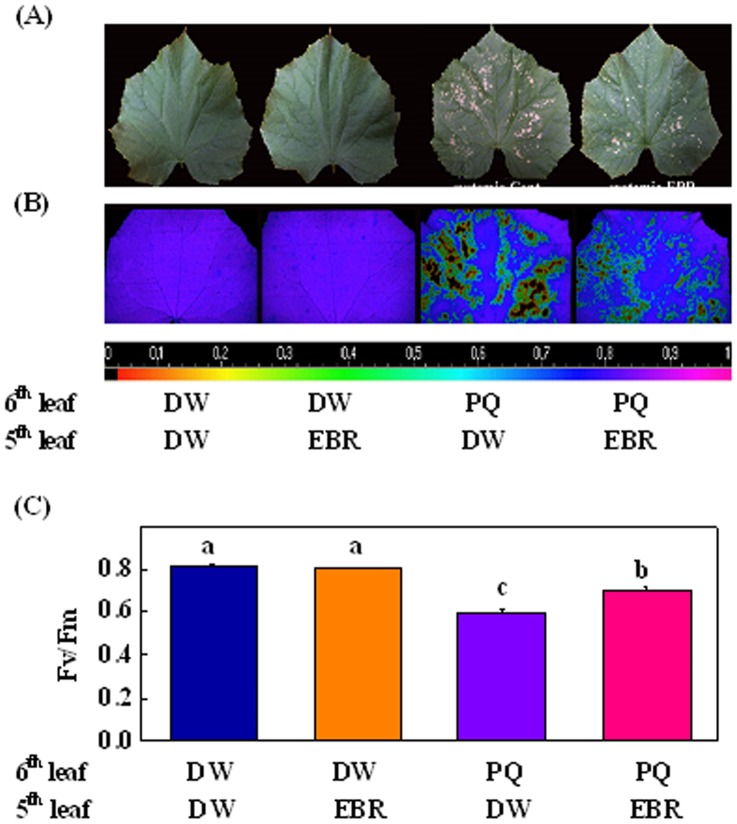
Brassinosteroids are known for their immobility in plants. To determine whether BRs could induce systemic stress tolerance, the 5th leaf (defined as local leaf) of a plant was pretreated with DW or 0.2 µM EBR whilst the 6th leaf (defined as systemic leaf) was sprayed with DW or 10 µM PQ and finally, Fv/Fm was determined at 24 h after the PQ treatment. Exposure of the leaf to PQ resulted in leaf bleaching but bleaching at the 6th leaf was significantly alleviated by the EBR application at the 5th leaf (Figure 1A). Fv/Fm did not change with the DW or EBR treatment in the absence of PQ treatment (Figure 1B, C). Exposure to PQ led to sharp decreases in Fv/Fm in all treatments. The declines in Fv/Fm at 6th leaf, however, were significantly alleviated by the EBR treatment at 5th leaf. Together, these results demonstrated that EBR treatment could induce systemic stress tolerance in cucumber plants.
Figure 1. EBR induced systemic stress tolerance in cucumber plants.
(A) The 5th leaf of cucumber seedlings corresponding to the local treated leaf at the seven-leaf stage was pretreated with distilled water (DW) or 0.2 µM EBR; 24 h later the upper leaf (6th) corresponding to the systemic leaf was exposed to 10 µM paraquat (PQ), photographs were taken at 24 h after PQ treatment. (B) The maximal photochemical quantum efficiency of photosystem II (Fv/Fm) was monitored at 24 h after PQ treatment with imaging pulse amplitude modulation (PAM) to assess the changes in tolerance. (C) Fv/Fm measurement of systemic leaves. Data represent the means of five replicates ± SD. Bars sharing the same letters are not significantly different within the treatment (P≤0.05).
EBR Induces Expression of Defense/stress Related Genes in Local and Systemic Leaves
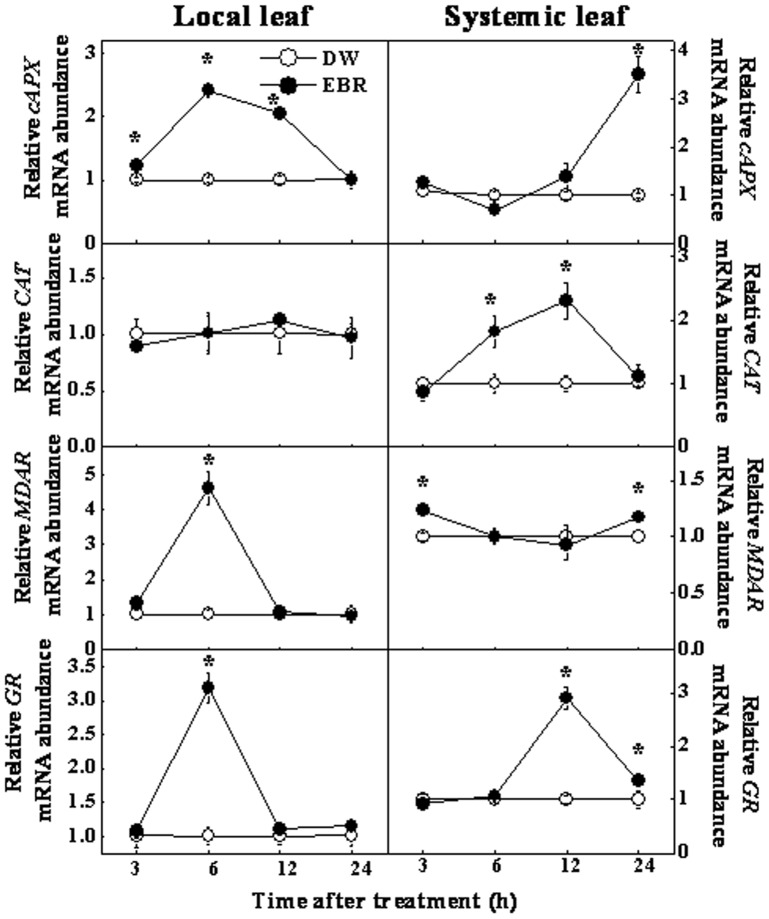
To further study the effects of EBR on the induction of tolerance to oxidative stress in EBR-treated local leaf (5th) and EBR-untreated systemic leaf (6th), we performed a time-course analysis of the expression of a subset of defense/stress related genes involved in biotic and abiotic stresses after EBR application on the 5th leaf (Figure 2). In the local leaf, the transcript level of cytosolic Ascobate peroxidase (cAPX), Monodehydroascorbate reductase (MDAR) and Gutathione reductase (GR) increased significantly at 6 h after EBR treatment, then returned to control levels at 24 h. Interestingly, in the systemic leaf, we observed noticeable changes in the transcript abundance of some of the antioxidant related genes as well. The cAPX, Catalase (CAT) and GR expression was induced to a high level between 12 to 24 h upon EBR treatment, this induction, however, was a little later than that in local leaf. These results suggested that foliar application of EBR not only induced the expression of defense/stress related genes in the local leaf, but also in the systemic leaf in cucumber.
Figure 2. Expression analysis of defense/stress related genes in local and systemic cucumber leaves.
The 5th leaf of cucumber seedlings at the seven-leaf stage was pretreated with distilled water (DW) or 0.2 µM EBR, the 5th leaf corresponding to the local treated leaf and the 6th leaf corresponding to the systemic leaf were harvested at 3, 6, 12, 24 h after EBR treatment. Relative mRNA abundances of cAPX, CAT, MDAR, GR in EBR-treated local and untreated systemic leaves were determined by RT-qPCR analysis. Data represent the means of three replicates ± SD. Asterisks (*) indicate a significant difference from the untreated control at P≤0.05 according to Tukey’s test.
The Identification of EBR-up-regulated Transcripts in Cucumber Phloem Sap by SSH Library Analysis
To determine whether EBR treatment induced changes in the phloem mRNAs, the phloem transcript profile was analyzed by constructing an SSH library. Phloem sap was collected at 3, 6, 9 and 24 h time intervals following the foliar application of DW or EBR on fully expanded, mature cucumber leaves. The DW treatment was set as the driver and the EBR treatment as the tester. A total of 686 clones were obtained, and 260 clones were randomly sequenced, in which there was a high level of redundancy in this population. Bioinformatic analysis revealed that there were only 14 unique clones (Table 1).
Table 1. Differential population of the mRNA transcripts induced by 24-epibrassinosteroid (EBR) in phloem sap of cucumber plants.
| Clone name | Accession Number | Annotation | Best homologs in Cucumis sativus | E-value | GO biological process |
| Cs564 | JG391944 | DNA binding/methylated histone residue binding | Csa009901 | 3.00E-24 | regulation transcription |
| Cs74 | JG391952 | Zinc finger CCCH-type with G patch domain-containing protein | Csa016350 | 5.00E-58 | regulation transcription |
| Cs642 | JG391945 | 40S ribosomal protein S7 | Csa004617 | 1.00E-78 | translation |
| Cs140 | JG391953 | 60S ribosomal protein L38 | Csa007567 | 1.00E-16 | translation |
| Cs418 | JG391947 | tRNA-binding region domain-containing protein | Csa016058 | 1.00E-68 | translation |
| Cs60 | JG391942 | ATG8C (autophagy 8c); microtubule binding | Csa026450 | 4.00E-61 | protein transport |
| Cs623 | JG391955 | MLP-like protein 328 | Csa010594 | 0 | defense response |
| Cs579 | JG391948 | Ankyrin repeat domain-containing protein 2 | Csa013665 | 4.00E-48 | defense response |
| Cs453 | JG391949 | Auxin-responsive protein IAA14 | Csa000125 | 3.00E-57 | response to wounding |
| Cs594 | JG391950 | F-box protein PP2-A14 | Csa012410 | 0 | response to wounding |
| Cs681 | JG391943 | Yippee putative zinc-binding protein | Csa005548 | 1.00E-101 | unknown |
| Cs658 | JG391946 | Unknown | Csa003458 | 1.00E-112 | |
| Cs526 | JG391954 | Unknown | Csa003457 | 1.00E-116 | |
| Cs302 | JG391951 | Unknown | Csa012053 | 6.00E-67 |
EBR-induced changes in the population of mRNAs in the phloem sap of cucumber plants were determined by suppressive subtractive hybridization (SSH) methods. Mature leaves of seven-leaf stage plants were treated with distilled water (DW) or 0.2 µM EBR and phloem sap was collected from stem cut close to apex and at petioles during various time intervals (3, 6, 9 and 24 h) after treatment. RNA was extracted from pooled samples for use in SSH analysis. Data presented reflects the mRNA species up-regulated in EBR-treated vs DW-treated phloem sap.
The bioinformatic analysis was performed to classify these up-regulated RNA species, according to the predicted function of the encoded protein (Table 1). Of the 14 clones obtained, two clones Cs564 and Cs74 encode a putative DNA-binding protein and a zinc finger protein, respectively, which are related with transcription regulation. Three clones Cs642, Cs140 and Cs418 encode a 40S ribosomal protein, a 60S ribosomal protein and a tRNA-binding protein, respectively, which are related with protein translation. Cs60 is predicted to encode a microtubule-binding protein, which is involved in protein transport. Most relevant to our study, four clones were predicted to encode proteins related with defense responses, which included a major latex, pathogenesis-related (MLP)-like protein (Cs623), an ankyrin-repeat protein (Cs579), an F-box protein (Cs594) and an auxin response protein, IAA14 (Cs453). Further sequence alignment of Cs453 with melon phloem sap transcripts by ClustalW2 showed that it has very high similarity (max identities are 97%) with F308 (accession number EB715302) which has been identified as a phloem mobile transcript by grafting experiments in melon plants [23]. In the other clones, Cs681 encoded a putative Yippee-like zinc-binding protein, which has an undescribed biological function. Lastly, three clones, Cs658, Cs526, and Cs302, had no significant similarity to described proteins. By this analysis, we identified four genes that are related with defense responses, which may imply that they are possibly involved in EBR-regulated systemic resistance.
Confirmation of the Up-regulation of EBR-induced Genes in Cucumber Phloem Sap
RT-qPCR was then performed to verify the up-regulation of the transcripts obtained from the SSH library. Nine clones were selected for the additional RT-qPCR analysis (Figure 3). The results indicated that the transcript levels of Cs623 (coding for an MLP-like protein), Cs60 (coding for a microtubule-binding protein), Cs642 (coding for a ribosomal protein) and Cs564 (coding for a DNA-binding protein) increased by 1.5–2-fold in the EBR-treated phloem sap. Additionally, the up-regulated defense-related transcripts, Cs453 (coding for IAA14), Cs579 (coding for a putative ankyrin-repeat protein), and Cs594 (coding for an F-box family protein), were shown to be increased by 2–4-fold. Transcript Cs681 (coding for a putative Yippee-like zinc binding protein) did not increase significantly (p-value≤0.05), this may be caused by the variation between different batches of samples. Above all, these results demonstrated that the SSH library produced reliable results.
Figure 3. Validation of the up-regulated transcripts present in the SSH library by using RT-qPCR.
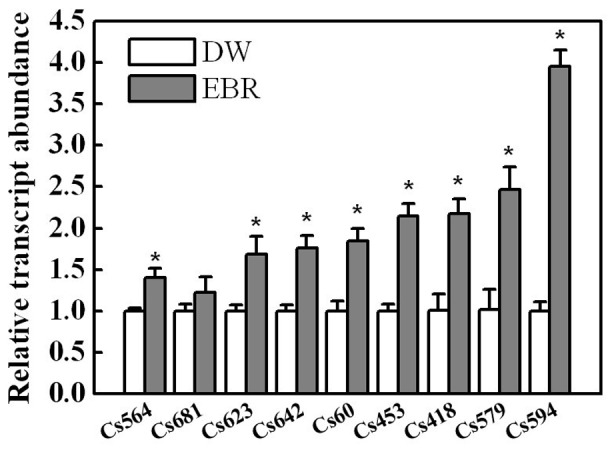
Phloem sap was collected from stem cut close to apex and at petioles during various time intervals (3, 6, 9 and 24 h) after foliar EBR treatment. RNA was extracted from the pooled samples and relative mRNA abundances of transcripts obtained from the SSH library were determined by RT-qPCR. The data were obtained from three separate replicates, each value in the graph shows means ± SD. Asterisks (*) indicate a significant difference from the untreated control at P≤0.05 according to Tukey’s test.
Time-course of Gene Transcripts in Local and Systemic Leaf and Distal Organs after EBR Treatment
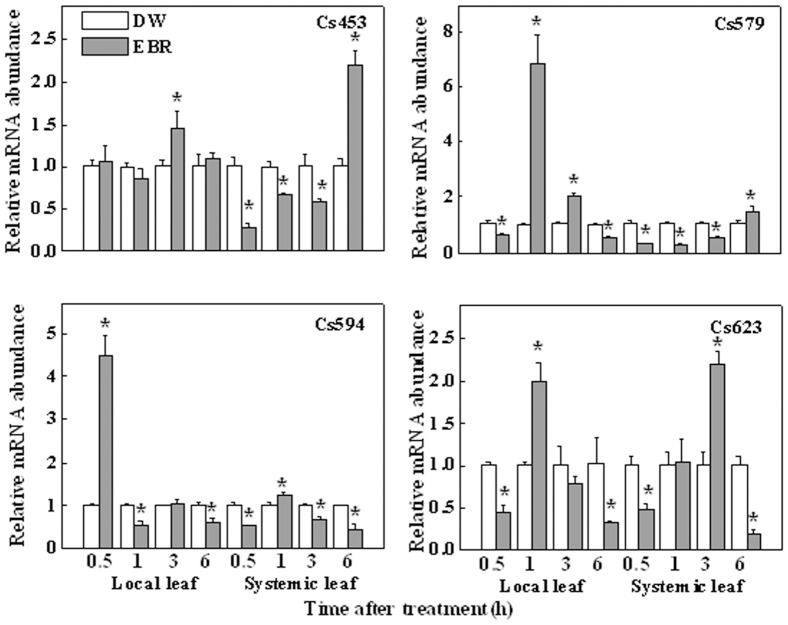
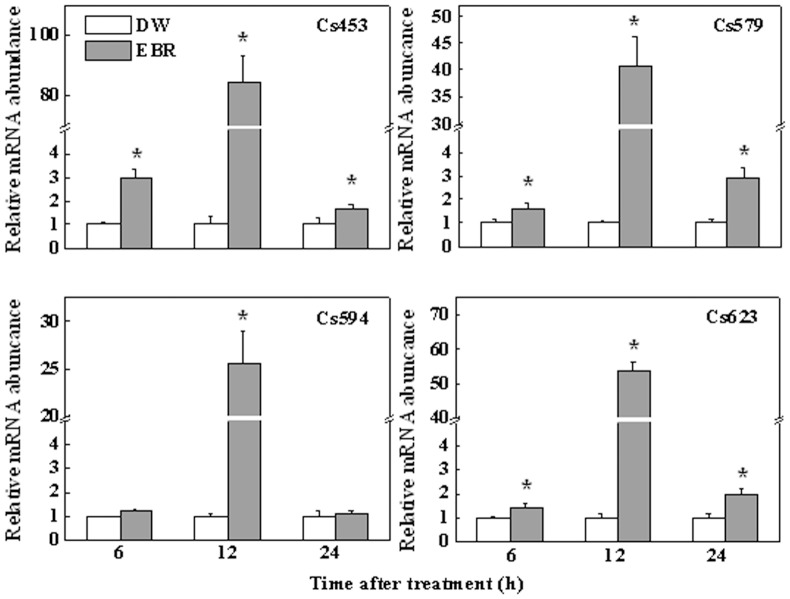
We then examined the EBR-induced changes in the transcripts of genes, Cs453, Cs579, Cs594, and Cs623 identified from the SSH library in the local and systemic leaves (Figure 4) and in the roots (Figure 5). In the local leaves, only a slight increase in the transcript of Cs453 was observed at 3 h after the EBR treatment (Figure 4). However, a 4-fold increase was observed for Cs594 as early as 0.5 h after EBR application, and Cs579 and Cs623 transcripts were upregulated by 7- and 2-fold at 1 h after treatment, respectively. Depending on the genes examined, the transcripts of these genes in the systemic leaves were also altered by the EBR treatment (Figure 4). Transcripts for Cs453 and Cs623 were upregulated by 2-fold in the systemic leaf at 6 and 3 h, respectively. EBR-induced changes in the transcripts for other genes or at other time points were negligible with relative abundance at 0.5∼2-fold. Interestingly, a sharp increase in the abundance of these four genes was observed in the roots. Cs453, Cs579, Cs594 and Cs623 showed a slight elevation at 6 h after the foliar application of EBR; these transcripts were elevated by 80-, 40-, 25-, and 20-fold, respectively, at 12 h, compared with DW treatment, and then declined. All these results suggested that EBR treatment could dynamically increase the transcripts of an array of genes in the phloem.
Figure 4. Expression analysis of defense related transcripts obtained from SSH library in cucumber leaves.
The 5th leaf of cucumber seedlings at the seven-leaf stage was pretreated with distilled water (DW) or 0.2 µM EBR, the 5th leaf corresponding to the local treated leaf and the 6th leaf corresponding to the systemic leaf were harvested at 0.5, 1, 3 and 6 h after EBR treatment. Relative mRNA abundances of Cs453, Cs579, Cs594 and Cs623 obtained from the SSH library were determined in local and systemic leaves by RT-qPCR analysis. The data were obtained from three separate replicates, and each value in the graph shows means ± SD. Asterisks (*) indicate a significant difference from the untreated control at P≤0.05 according to Tukey’s test.
Figure 5. Expression analysis of defense related transcripts obtained from SSH library in cucumber roots.
After EBR (0.2 µM) sprayed on mature leaves, roots were harvested at 6, 12, and 24 h after treatment. Relative mRNA abundances of Cs453, Cs579, Cs594 and Cs623 obtained from the SSH library were determined by RT-qPCR analysis. The data were obtained from three separate replicates, and each value in the graph shows means ± SD. Asterisks (*) indicate a significant difference from the untreated control at P≤0.05 according to Tukey’s test.
Discussion
Over the past several years, genetic and biochemical approaches have yielded significant progress in the understanding of BR signaling. However, the underlying mechanism of the BR regulation of systemic resistance is largely unknown. In this study, we found that EBR induced a systemic tolerance against oxidative stress in cucumber (Figure 1 & 2). This is consistent with our earlier finding that local exposure to EBR led to systemic resistance against oxidative stress in upper and lower leaves and resistance to Fusarium pathogen in the roots of cucumber [7]. Several lines of evidence showed that BRs do not undergo long-distance transport between the shoot and roots [6]. By the transcript analysis of BRs biosynthesis genes, we also confirmed the immobility of exogenously applied EBR in plants of cucumber [7]. Accordingly, the systemic stress tolerance in EBR-untreated leaves and roots could be attributed to signal(s) other than EBR. As the phloem is the major route for the communication between source and sink, mRNAs could function as long-distance information macromolecule to control gene expression in remote tissues [24]. Hence, it is of great interest to investigate the changes in the transcriptional profile of the phloem after BRs applied to the source leaves. To our knowledge, this is the first study to address the effects of BRs on the specific mRNA population in the phloem translocation stream.
By constructing an SSH library using the phloem sap mRNA, we identified 14 transcripts out of 260 clones. It should be noted that this up-regulated group represented a very small change in the total population of phloem transcripts, as compared to the transcripts found in the phloem of other cucurbits [16]. Probably, plants have evolved a highly robust mechanism for the exchange of information macromolecules between the companion cell and the sieve tube system [25]. Furthermore, BR-induced transcription has been described as weak and slow in plants [26]. Compared with the induction by other hormones, such as auxin that has been reported to induce transcripts in excess of 10-fold [27], only a very limited numbers of BR-regulated genes have been shown to be induced by more than 2-fold, as based on studies in Arabidopsis [28]–[31]. Hence, it is not surprising that only a small number of transcripts were identified by our SSH library. The RT-qPCR analysis of the expression of these transcripts also supported this notion (Figure 3). As BR-induced transcripts were time-dependent (Figure 2, 4 & 5), we could not exclude the possibility that other important genes were not identified because the samples were taken at different times after EBR treatment in our study. Finally, it is interesting to note that EBR induced much higher transcripts in the distal roots, suggesting a possible amplifying mechanism in the BR signaling cascades in the long distance communication. Probably, it is attributed to the stronger sink for the roots than the systemic leaf in our study because systemic leaves can also carry out photosynthesis.
The mRNAs represented in the subtractive library were related with different cellular functions, including transcriptional regulation, protein translation, transportation and defense responses. Here, we identified four genes that are related with defense response, which imply that they are possibly involved in EBR-regulated systemic resistance. Further analysis of these four transcripts in the local leaves, systemic leaves and roots showed that they were not only induced in the local leaves, but also induced in the systemic leaves and roots (Figure 4, 5). How these genes were involved in the systemic stress tolerance is of great interest. It is worth noting that EBR-induced transcripts were dynamic and transcripts of these genes were even down-regulated at several time points. Although the magnitude of change is negligible in RT-qPCR and microarray analysis, we could not exclude the interference from other BR-induced signaling such as ROS and the involvement of feedback regulation of the gene transcript. There are evidences that foliar EBR application could induce ROS generation in the phloem cells whilst ROS could transiently decrease auxin signaling [7], [32].
In the four genes identified, Cs623 (an MLP-like protein) is known as a phloem defense-related protein, whereas Cs579 possibly functions as a molecular chaperone [33]. Cs594 was identified as a phloem protein 2 that contains an F-box domain, also known as a phloem defense-related protein [34]. Notably, these three genes were first reported as BR-regulated transcripts. Further study should be carried out to confirm their mobility and analyze the functions of these genes. Cs453 encodes IAA14, which is a member of the auxin/indole-3-acetic acid (Aux/IAA) protein family. In a recent study on the transcript profile of melon phloem-sap, Omid and others [23] identified three auxin signaling-related transcripts, two are Aux/IAA (F-308 and F-571), and the third one is small auxin-up RNA (SAUR, UN-131). An alignment showed that Cs453 is an ortholog of F308. In grafted apple, MpSLR/IAA14, was probed to transport a long distance through the graft union [35]. Much research has indicated that there is crosstalk between the BRs and auxins. A microarray analysis of brassinosteroid-regulated genes in Arabidopsis identified several auxin-related genes, which revealed a marked overlap in the BR- and auxin-signaling pathways [28]. Nakamura and others [36] examined the function of Aux/IAA proteins in auxin- and BR-signaling pathways; they found that IAA proteins function as signaling components that modulate BR sensitivity in an organ-dependent manner. The crosstalk between these two hormones probably occurs through the combinatorial regulation of some common target genes, especially the promoters of the target genes [26]. In this regard, IAA14 functions as a transcriptional activator and may interact with a repressor-type ARF to allow the expression of genes repressed by the ARF [37]. Interestingly, BRs are known for their ability in triggering the generation of ROS which are involved in the regulation in IAA14 [5], [7], [38], and both BRs and IAA14 are involved in the regulation of lateral root development [39]. It is, therefore, plausible that Cs453 is one of the candidates for the mediation of a BR-regulated systemic effect through auxin signaling in the long-distance communication.
Conclusions
By constructing an SSH cDNA library, we identified EBR up-regulated genes in cucumber phloem sap. The further analysis of four cDNAs, which were related with the defense response, suggested that they may be important signaling components functioning in EBR-regulated systemic resistance; in particular, we suggest that Cs453 may be an important candidate. Further study should be carried out to confirm their mobility and analyze the functions of these genes, which will eventually add to our understanding of how BRs regulate the plant systemically.
Supporting Information
Specificity of primers used for RT-qPCR assays.
(DOC)
Acknowledgments
We are grateful to Dr. JH Zhao for excellent technical help with Chl a fluorescence image analysis.
Funding Statement
This work was supported by the National Basic Research Program of China (2009CB119000), National Natural Science Foundation of China (31171999), and the Program for New Century Excellent Talents in University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Sasse JM (2003) Physiological actions of brassinosteroids: An update. J Plant Growth Regul 22: 276–288. [DOI] [PubMed] [Google Scholar]
- 2. Bajguz A, Hayat S (2009) Effects of brassinosteroids on the plant responses to environmental stresses. Plant Physiol Biochem 47: 1–8. [DOI] [PubMed] [Google Scholar]
- 3. Kagale S, Divi UK, Krochko JE, Keller WA, Krishna P (2007) Brassinosteroid confers tolerance in Arabidopsis thaliana and Brassica napus to a range of abiotic stresses. Planta 225: 353–364. [DOI] [PubMed] [Google Scholar]
- 4. Krishna P (2003) Brassinosteroid-mediated stress responses. J Plant Growth Regul 22: 289–297. [DOI] [PubMed] [Google Scholar]
- 5. Xia XJ, Wang YJ, Zhou YH, Tao Y, Mao WH, et al. (2009) Reactive oxygen species are involved in brassinosteroid- induced stress tolerance in cucumber. Plant Physiol 150: 801–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Symons GM, Ross JJ, Jager CE, Reid JB (2008) Brassinosteroid transport. J Exp Bot 59: 17–24. [DOI] [PubMed] [Google Scholar]
- 7. Xia XJ, Zhou YH, Ding J, Shi K, Asami T, et al. (2011) Induction of systemic stress tolerance by brassinosteroid in Cucumis sativus . New Phytol 191: 706–720. [DOI] [PubMed] [Google Scholar]
- 8. Ding J, Shi K, Zhou YH, Yu JQ (2009) Microbial community responses associated with the development of Fusarium oxysporum f. Sp cucumerinum after 24-epibrassinolide applications to shoots and roots in cucumber. Eur J Plant Pathol 124: 141–150. [Google Scholar]
- 9. Nakamura A, Goda H, Shimada Y, Yoshida S (2004) Brassinosteroid selectively regulates PIN gene expression in arabidopsis. Biosci Biotech Biochem 68: 952–954. [DOI] [PubMed] [Google Scholar]
- 10. Terakado J, Yoneyama T, Fujihara S (2006) Shoot-applied polyamines suppress nodule formation in soybean (Glycine max). J Plant Physiol 163: 497–505. [DOI] [PubMed] [Google Scholar]
- 11. Nakashita H, Yasuda M, Nitta T, Asami T, Fujioka S, et al. (2003) Brassinosteroid functions in a broad range of disease resistance in tobacco and rice. Plant J 33: 887–898. [DOI] [PubMed] [Google Scholar]
- 12. Terakado J, Fujihara S, Goto S, Kuratani R, Suzuki Y, et al. (2005) Systemic effect of a brassinosteroid on root nodule formation in soybean as revealed by the application of brassinolide and brassinazole. Soil Sci Plant Nutr 51: 389–395. [Google Scholar]
- 13. van Bel AJE, Ehlers K, Knoblauch M (2002) Sieve elements caught in the act. Trends Plant Sci 7: 126–132. [DOI] [PubMed] [Google Scholar]
- 14. Dinant S, Lemoine R (2010) The phloem pathway: New issues and old debates. C R Biol 333: 307–319. [DOI] [PubMed] [Google Scholar]
- 15. Kehr J (2009) Long-distance transport of macromolecules through the phloem. F1000 Biol Rep 1: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lough TJ, Lucas WJ (2006) Integrative plant biology: Role of phloem long-distance macromolecular trafficking. Annu Rev Plant Biol 57: 203–232. [DOI] [PubMed] [Google Scholar]
- 17. Kim M, Canio W, Kessler S, Sinha N (2001) Developmental changes due to long-distance movement of a homeobox fusion transcript in tomato. Science 293: 287–289. [DOI] [PubMed] [Google Scholar]
- 18. Haywood V, Yu TS, Huang NC, Lucas WJ (2005) Phloem long-distance trafficking of gibberellic acid-insensitive RNA regulates leaf development. Plant J 42: 49–68. [DOI] [PubMed] [Google Scholar]
- 19. Banerjee AK, Chatterjee M, Yu YY, Suh SG, Miller WA, et al. (2006) Dynamics of a mobile RNA of potato involved in a long-distance signaling pathway. Plant Cell 18: 3443–3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ruiz-Medrano R, Xoconostle-Cazares B, Lucas WJ (1999) Phloem long-distance transport of CmNACP mRNA: Implications for supracellular regulation in plants. Development 126: 4405–4419. [DOI] [PubMed] [Google Scholar]
- 21. Yoo BC, Kragler F, Varkonyi-Gasic E, Haywood V, Archer-Evans S, et al. (2004) A systemic small RNA signaling system in plants. Plant Cell 16: 1979–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔCT) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 23. Omid A, Keilin T, Glass A, Leshkowitz D, Wolf S (2007) Characterization of phloem-sap transcription profile in melon plants. J Exp Bot 58: 3645–3656. [DOI] [PubMed] [Google Scholar]
- 24. Lucas WJ, Yoo BC, Kragler F (2001) RNA as a long-distance information macromolecule in plants. Nat Rev Mol Cell Biol 2: 849–857. [DOI] [PubMed] [Google Scholar]
- 25. Ruiz-Medrano R, Moya JH, Xoconostle-Cazares B, Lucas WJ (2007) Influence of cucumber mosaic virus infection on the mRNA population present in the phloem translocation stream of pumpkin plants. Funct Plant Biol 34: 292–301. [DOI] [PubMed] [Google Scholar]
- 26. Hardtke CS, Dorcey E, Osmont KS, Sibout R (2007) Phytohormone collaboration: Zooming in on auxin-brassinosteroid interactions. Trends Cell Biol 17: 485–492. [DOI] [PubMed] [Google Scholar]
- 27. Zhao YD, Dai XH, Blackwell HE, Schreiber SL, Chory J (2003) SIR1, an upstream component in auxin signaling identified by chemical genetics. Science 301: 1107–1110. [DOI] [PubMed] [Google Scholar]
- 28. Goda H, Shimada Y, Asami T, Fujioka S, Yoshida S (2002) Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiol 130: 1319–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mussig C, Fischer S, Altmann T (2002) Brassinosteroid-regulated gene expression. Plant Physiol 129: 1241–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mussig C, Lisso J, Coll-Garcia D, Altmann T (2006) Molecular analysis of brassinosteroid action. Plant Biol 8: 291–296. [DOI] [PubMed] [Google Scholar]
- 31. Yin YH, Vafeados D, Tao Y, Yoshida S, Asami T, et al. (2005) A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120: 249–259. [DOI] [PubMed] [Google Scholar]
- 32. Blomster T, Salojarvi J, Sipari N, Brosche M, Ahlfors R, et al. (2011) Apoplastic reactive oxygen species transiently decrease auxin signaling and cause stress-induced morphogenic response in arabidopsis. Plant Physiol 157: 1866–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shen GX, Kuppu S, Venkataramani S, Wang J, Yan JQ, et al. (2010) ANKYRIN REPEAT-CONTAINING PROTEIN 2A is an essential molecular chaperone for peroxisomal membrane-bound ASCORBATE PEROXIDASE3 in Arabidopsis . Plant Cell 22: 811–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Walz C, Giavalisco P, Schad M, Juenger M, Klose J, et al. (2004) Proteomics of curcurbit phloem exudate reveals a network of defence proteins. Phytochemistry 65: 1795–1804. [DOI] [PubMed] [Google Scholar]
- 35. Kanehira A, Yamada K, Iwaya T, Tsuwamoto R, Kasai A, et al. (2010) Apple phloem cells contain some mRNAs transported over long distances. Tree Genet Genomes 6: 635–642. [Google Scholar]
- 36. Nakamura A, Nakajima N, Goda H, Shimada Y, Hayashi K, et al. (2006) Arabidopsis Aux/IAA genes are involved in brassinosteroid-mediated growth responses in a manner dependent on organ type. Plant J 45: 193–205. [DOI] [PubMed] [Google Scholar]
- 37. Narise T, Kobayashi K, Baba S, Shimojima M, Masuda S, et al. (2010) Involvement of auxin signaling mediated by IAA14 and ARF7/19 in membrane lipid remodeling during phosphate starvation. Plant Mol Biol 72: 533–544. [DOI] [PubMed] [Google Scholar]
- 38. Blomster T, Salojarvi J, Sipari N, Brosche M, Ahlfors R, et al. (2011) Apoplastic reactive oxygen species transiently decrease auxin signaling and cause stress-induced morphogenic response in arabidopsis. Plant Physiol 157: 1866–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fukaki H, Nakao Y, Okushima Y, Theologis A, Tasaka M (2005) Tissue-specific expression of stabilized SOLITARY-ROOT/IAA14 alters lateral root development in Arabidopsis. Plant J 44: 382–395. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Specificity of primers used for RT-qPCR assays.
(DOC)