Abstract
This study investigated the effects of pregnane X receptor (PXR/NR1I2) and CYP2B6 genetic variants on sodium ferulate (SF)-mediated induction of bupropion hydroxylation. The pharmacokinetics of bupropion and hydroxybupropion were evaluated after an oral dose of bupropion (150 mg) administered with and without SF pretreatment for 14 days in 33 healthy subjects. The area under the time-concentration curve (AUC) ratio of AUC_hyd (AUC(0-∞) of hydroxybupropion)/AUC_bup (AUC(0-∞) of bupropion) represents the CYP2B6 hydroxylation activity, which was significantly lower in CYP2B6*6 carriers (NR1I2 TGT noncarriers or carriers) than in noncarriers in both the basal and SF-induced states (p-value<0.05). AUC ratio and AUC_hyd of NR1I2 -24113AA variant were markedly lower than GA and GG genotypes (7.5±2.1 versus 14.5±3.3 and 20.6±1.1, and 8873±1431 versus 14,504±2218 and 17,586±1046) in the induced states. However, -24020(-)/(-) variant didn't show significant difference in the induction of CYP2B6 hydroxylation activity by SF compared with other -24020[GAGAAG]/(-) genotypes. NR1I2 TGT haplotype (-25385T+g.7635G+g.8055T) carriers exhibited a significantly decreased AUC ratio, compared with TGT noncarriers, in the basal states (7.6±1.0 versus 9.7±1.0), while this result wasn't observed in CYP2B6*6 noncarriers. Moreover, individuals with complete mutation-type [CYP2B6*6/*6+NR1I2 TGT+ -24113AA+ -24020 (-)/(-)] showed even lower percent difference of AUC ratio (8.7±1.2 versus 39.5±8.2) than those with complete wild-type. In conclusion, it is suggested that NR1I2 variants decrease the bupropion hydroxylation induced by SF treatment, particularly in CYP2B6*6 carriers.
Trial Registration
ChiCTR.org ChiCTR-TRC-11001285
Introduction
Human pregnane X receptor (hPXR) may regulate metabolic pathways in response to changes in the environment by appropriate alterations in gene expression of key metabolic enzymes [1]. hPXR is a member of the nuclear receptor superfamily, and encoded by the NR1I2 gene, which is located in the chromosome 13q11-13 and composed of nine exons. It consists of 434 amino acids with a molecular weight of 49.7 kDa [2]. It is abundantly expressed in liver and intestine, and plays an important role in ligand-activated transcription and regulation of genes involved in xenobiotic and endobiotic metabolism [3]–[5]. The transcriptional activity of NR1I2 is mediated through the ligand binding pathway. Many chemical drugs, including rifampin [4], [5], phenobarbital [6], [7], dexamethasone [7], [8], and herbs such as Kava [7], St. John's wort (hyperforin) [9], [10], and Ginkgo biloba extract (ginkgolide A) [11] have been verified as regulators of NR1I2 transcriptional activity. Meanwhile, hPXR binds to DNA response elements as a heterodimer with the 9-cis retinoid X receptor (RXRα) [12] and acts as a transcriptional regulator of many important genes that are encoding drug-metabolizing enzymes, such as phase I enzymes (e.g., CYP3A4, 3A5, 2A6, 2B6, 2C9, 2C19, and 1A1), phase II enzymes (e.g., UGT1A1, 1A3, 1A4, and 1A6), and phase III enzymes (e.g., MDR1, MRP2, and OATP1B1) [13]–[15].
The genetic variants of NR1I2 affect the disposition and interaction of various drugs through an induction pathway of CYPs, which may explain the interindividual difference of PXR activity [16], [17]. Previous studies showed that variant alleles of NR1I2 single nucleotide polymorphisms (SNPs) -25385C>T (rs3814055), -24113G>A (rs2276706), g.7635A>G (rs6785049), or g.8055C>T (rs2276707) were associated with increased NR1I2 transcriptional activity [18], and -24020[GAGAAG]>(-) (rs3842689) completed loss of NR1I2 promoter activity in HepG2 cells [19]. NR1I2 TGT haplotype (-25385T+g.7635G+g.8055T) and -25385C>T variants were also reported to be associated with reduced induction of bupropion hydroxybupropion by rifampin [20]. However, these controversial results of NR1I2 variants functions in vitro and in vivo have not been consistently validated.
Sodium ferulate (3-methoxy-4-hydroxy-cinnamate sodium, C10H9NaO4, SF) is the sodium salt of ferulic acid (FA), which is widely distributed in herbs and Chinese formulas such as Ligusticum, Chuanxiong and Chaihu–Sugan–San [21], [22]. It is usually used as food supplements or herbal medicine in countries or areas accepting the theory of Traditional Chinese Medicine (TCM). With the anti-oxidant, anti-atherogenic, anti-platelet clotting, anti-inflammatory, lipid-lowering, cholesterol biosynthesis inhibitory and analgesic effects [23]–[26], FA has the potential to be developed into an effective pure compound for prevention and treatment of cardiovascular diseases. Presently, SF has been approved by State Food and Drugs Administration of China (SFDA) as a clinical therapy for cardiovascular and cerebrovascular diseases [27], [28].
Combination of SF and CYP2B6 substrate drugs has the potential possibility of drug-drug interactions and may lead to undesirable and harmful clinical consequences. Although human CYP2B6 represents approximately 1% of total hepatic CYP content, it shows a relative contribution of 2% to 10% in total hepatic CYP activity, and participates in the metabolism of a variety of substances including bupropion, selegiline, valproic acid, cyclophosphamide, ifosfamide, nevirapine, efavirenz, propofol, ketamine, and synthetic opioid methadone [29]. In particular, the metabolic pathway of hydroxybupropion is almost exclusively catalyzed by CYP2B6 that is a standard model for studies of drug-drug interactions of CYP2B6 substrate drugs [30]–[33]. Recently, we verified that bupropion hydroxylation metabolism (represents CYP2B6 metabolism activity) was induced by a 14 days pre-treatment of 150 mg SF in healthy volunteers [34]. Our previous investigations in HepG2 cells suggested that FA may increase 67% transcriptional expression of CYP2B6 through PXR activation compared with control group (unpublished data). Due to frequently combinational use, especially in China, of SF and other drugs during the treatment of clinical diseases, it is important to be aware of the possibility of interactions of combination of SF and CYP2B6 substrate drugs, and to prevent harmful clinical toxicity.
Effects of CYP2B6 variations on protein expression levels and enzyme activities may cause up to hundreds fold of interindividual difference in exposure to drugs [35]. Early studies indicated CYP2B6*6 carriers contributed to the interindividual difference in CYP2B6 substrate drugs disposition [36], [37]. The low-activity and high-frequency of allele CYP2B6*6 was very prevalent in African Americans (32.8%), Papua New Guineans (62%) and Asians (21%) [35]. It can be simply speculated that the number of abnormal population of CYP2B6 variants will be marked, when other CYP2B6 functional variants are added. Therefore, it is very critical to investigate whether CYP2B6 variants effect the induction of bupropion hydroxylation by SF.
Based on above studies, SF-induced CYP2B6 activity is suggested to be associated with both CYP2B6*6 polymorphisms and other factors such as NR1I2 genetic variants. Furthermore, both NR1I2 and CYP2B6 variants may be associated with the clinical pharmacokinetics and/or interactions of SF and bupropion. However, clinical pharmacogenetics study data of NR1I2 and CYP2B6 variants is still scarce. Further studies are still necessary to investigate the functions of NR1I2 and CYP2B6 genetic variants in clinic. The purpose of this paper is to evaluate the effect of NR1I2 and CYP2B6 genetic variants, and to demonstrate the relationship between these genetic variants and metabolic induction of bupropion hydroxylation by SF administration in Chinese individuals.
Materials and Methods
The protocol for this trial and supporting CONSORT checklist are available as supporting information; see Checklist S1 and Protocol S1.
Genotyping
Genomic DNA was isolated from peripheral blood samples using SQ Blood DNA KitII(Omega Bio-Tic, Georgia, USA).
NR1I2. NR1I2 -25385C>T (rs3814055), -24113G>A (rs2276706), -24020 [GAGAAG]>(-) (rs3842689), g.7635A>G (rs6785049) and g.8055C>T (rs2276707) were amplified by polymerase chain reaction (PCR) and genotyped. The sequences of forward and reverse primers were 5′-CAAGGCAAGCATCCACTTGA -3′, 5′- GTTGAT- TCTGTTCACTTGGG-3′ for -25385C>T, 5′- AGGCAGCGGCTCCTTGGTAA -3′, 5′-AGGACAGCAGCATGA- CAGTC-3′ for -24113G>A and -24020[GAGAAG]>(-), 5′- CAAGCTCAGTGGGTG- GAGTT -3′, 5′- TTCTCCCCAGGTGAGGATCT-3′ for g.7635A>G and g.8055C>T, respectively. The PCRs used a thermocycling profile of initial denaturation at 95°C for 5 min, followed by 30 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s and extension at 72°C for 1 min. The final volume of the PCR was 50 µl , consisting of 20 ng of DNA, 10 µM of each primer pair, 2.5 µM of dNTPs, 5 µl of 10× reaction buffer, and 2.5 U of Taq DNA polymerase (MBI Fermentas, Ontario, Canada). SNPs were identified by DNA sequencing according to a standard protocol with ABI PRISM Big Dye Terminator Cycle Sequencing Ready Reaction Kit and ABI 3700 DNA Analyzer (Applied Biosystems, Foster City, California, USA).
CYP2B6. The wild-type allele CYP2B6*1 was defined as 516G/785A, and CYP2B6*6 was defined as 516T/785G. CYP2B6*6 was detected in a haplotype assay using a two-step allele-specific PCR as described previously [38]. The validity of the method was confirmed by sequencing.
Subjects
To detect NR1I2 and CYP2B6 genetic polymorphisms in the Chinese population, a total of 152 individual samples (from the DNA bank, Hunan Key laboratory of Pharmacogenetics, Central South University) were genotyped. Thirty-four healthy male volunteers (eighteen CYP2B6*1/*1, nine CYP2B6*1/*6 and seven CYP2B6*6/*6) were enrolled in the clinical trial with informed consent form signed (aged 20 to 24 years; weight range: 52–77 kg; body mass index range: 18–25 kg/m2), and divided into CYP2B6*6 noncarriers (CYP2B6*1/*1, wild-type) and CYP2B6*6 (CYP2B6*1/*6+CYP2B6*6/*6) carriers. The health status of subjects were ascertained by checking medical history and taking a full clinical examination, drug screening, and standard hematologic and blood chemical laboratory tests. Standardized protein-rich diets with no vegetables, fruits or cereals were provided for subjects for 2 weeks prior to study and during the whole study, in order to exclude the influence of food-originated FA. Drugs, alcohol, soft drinks, tobaccos, vitamins and caffeine-containing beverages, any nutritional supplements were refrained for 2 weeks before study commencement and throughout the study. Regular heavy drinkers, smokers, users of glucocorticoids and those with body weight exceeded their ideal weight by 20% were excluded. Finally, 33 of recruited subjects finished the trial (the date range of subject enrollment was from April 24, 2011 to July 25, 2011).
This study was approved by the ethics committee of Central South University, Changsha, Hunan, P. R. China (approved number: CTXY- 110003) and registered in the Chinese Clinical Trial Registry (registration number: ChiCTR-TRC -11001285, name: The effect of ferulic acid on metabolism of bupropion by CYP2B6). Overall clinical trial procedures abided by the Good Clinical Practice of the International Conference on Harmonization (ICH-GCP).
Study Design
This study was carried out in a two-phase, randomized, crossover manner with a 2-week washout period between phases. In each phase, after an overnight fast, subjects were given pretreatment with or without three 50-mg SF tablets (one tablet, three times a day) of the same batch (Lot No.: 100810; HengDa ShengKang Pharmaceutical Co., Sichuan, China) for fourteen days. The signature of subjects, supervision of investigators and detection of plasma concentrations of FA were carried out to assure subject compliance to treatment. On day 15, after an overnight fast, a single dose of 150 mg bupropion (two tablets of 75 mg Zyban SR; WanTe, Hainan, China) was given to each subject by oral administration with 200 ml water at 8:00 a.m. Subjects fasted for another 4 h after drug administration, except water drinking 2 h after dosing. Standard meals were provided for all of the participants. Serial blood samples for PK analysis (5 ml) were collected using a forearm in-dwelling venous catheter (anticoagulation with sodium heparin) before dosing and at 0.5, 1, 2, 3, 4, 5, 6, 8, 10, 12, 24, 36, 48, 60 and 72 h after bupropion ingestion.
Drug Concentration Analysis
The plasma concentrations of bupropion and hydroxybupropion were examined by liquid chromatography–mass spectrometry using Waters Micromass Quattro Micro API LC/MS/MS instrument (Milford, MA, USA). An Angel kromasil C18 (5 µm, 150×2.1 mm) and a mobile phase (acetonitrile∶0.1% formic acid∶20 mM ammonium formate = 4∶3∶3) at a flow rate of 0.2 ml/min were applied. Propranolol was used as the internal standard. The ion transitions monitored were as follows: m/z 240 to 184 for bupropion,m/z 256 to 238 for hydroxybupropion and m/z 260 to 183 for propranolol. These transitions represent the product ions of the [M+H]+ ions. The lower limits of detection for bupropion and hydroxybupropion were 0.25 ng/ml and 1.168 ng/ml, and the assay ranges used were 0.42–430.1 ng/ml and 2.344–1200 ng/ml, respectively. The linear correlation coefficient for bupropion calibration curves was 0.998, and for hydroxybupropion was 0.996. The highest bupropion and hydroxybupropion plasma concentration measured were 323.70 ng/ml and 615.66 ng/ml. The mean extraction recovery and precision of bupropion and hydroxybupropion was assessed by determining quality control (QC) plasma samples at three concentration levels (concentrations of bupropion and hydroxybupropion were 2.344, 37.5, 600 ng/ml and 0.84, 13.438, 430 ng/ml, respectively). The recovery of bupropion and hydroxybupropion, determined at each concentration level was 92.1±7.1, 93±5.3, 91.8±3.6 and 90.6±4.7, 91.5±4.8, 92.4±6.3 (%, n = 5), respectively, and did not exceed 10% of the relative standard deviation (R.S.D.). The precision (R.S.D.) of bupropion at three concentration levels in intra-day and inter-day was 8.67, 5.67, 7.32 (%) and 4.19, 6.57, 8.92 (%), respectively. Similarly, the precision (R.S.D.) of hydroxybupropion in intra-day and inter-day was 7.34, 3.58, 8.49 (%) and 4.25, 7.13, 9.12 (%), respectively. The precision of both bupropion and hydroxybupropion was less than 10% of R.S.D.
Pharmacokinetic Analysis
The maximum plasma concentration (C max) and the time to C max (T max) were obtained by inspection of the concentration-time data. The AUC to the last quantifiable concentration AUC0-t was determined by use of the linear trapezoidal rule. ke is the elimination rate constant determined from the terminal slope of the log concentration-time plot. The elimination half-life (t 1/2) was calculated as 0.693/ke. The area under the concentration-time curve extrapolated to infinity AUC0-∞ was calculated as AUC0-∞ = AUC0–72+C72/ke, where C72 is the plasma concentration measured 72 h after drug administration. The oral clearance (CL/F) of bupropion was calculated by dividing the bupropion dose by the AUC of bupropion and the subject's weight.
Statistical Analysis
Study sample sizes were estimated based on prior bupropion PK data. The planned sample size, statistical power, and alpha level were performed using the NCSSV2007 program. For instance, if three subjects in TGT carriers for CYP2B6*6 noncarriers group were enrolled, the power (AUC ratio) is up to 0.92371 (α = 0.05, β = 0.2) according to prior bupropion PK data. Likewise, in TGT carriers for CYP2B6*6 carriers group, if five subjects participated in this study, the power will add to 0.93165 (α = 0.05, β = 0.2) (calculation methods as described NCSS2007 instruction). The bioequivalence approach was used to determine clinically relevant interactions [39]. AUC ratio, namely AUC_hyd (AUC(0-∞) of hydroxybupropion) was divided by AUC_bup (AUC(0-∞) of bupropion) for each period, representing CYP2B6 activity. The percent differences in the PK parameters between the basal and SF-treated states were calculated as an absolute of 100×(induced-basal)/basal. WinNonlin (version 5.2; Pharsight, Mountain View, CA) was used for the PK analysis. The paired two-tailed t-tests were used to determine the difference between basal and induced states, and logarithmic transformation was used for the non–normally distributed data before analysis. The differences in PK parameters between noncarriers and carriers groups of NR1I2 TGT haplotype and CYP2B6*6 genotypes were obtained using the Wilcoxon rank-sum test. The difference among NR1I2 -25385C >T (CC, CT and TT) and -24113G>A (GG, GA and AA) genotype groups was obtained by use of the Kruskal-Wallis test. The Fisher's exact test was used to detect difference of genotype distributions between CYP2B6*1/*1 and CYP2B6*1/*6+CYP2B6*6/*6. Results were expressed as mean ± standard deviation in the text and tables, and as mean ± standard error in the figures. Linkage disequilibrium (LD) analysis and haplotype construction were performed using the Haploview 4.2 program (Broad Institute of Harvard and MIT, Cambridge, MA) and Phase 2.0 (UW Center for Commercialization; University of Washington), Data were analyzed with SPSS software (IBM SPSS; version 13.0). The chosen statistical significance level was p<0.05.
Results
Genetic Polymorphisms of NR1I2 in the Chinese Population
The frequencies of NR1I2 alleles -25385T, -24113A, -24020(-), g.7635G and g.8055T were 0.283, 0.181, 0.208, 0.522, and 0.625, respectively. The distribution of those genotypes was consistent with Hardy-Weinberg equilibrium (p-value >0.05, χ2 test). NR1I2 -25385C>T, -24113G>A, and -24020[GAGAAG]>(-) displayed a slight LD (r 2≥0.56). Also, NR1I2 g.7635A>G and g.8055C>T showed a slight LD (r 2≥0.62). In view that compared with previous NR1I2 haplotypes investigation, we selected three SNPs -25385C>T, g.7635A>G, and g.8055C>T to perform haplotype analysis, and eight haplotypes were inferred based on these SNPs in Chinese: CAC, CGT, TGT, TAC, CGC, TGC, CAT, and TAT. The population frequencies of these haplotypes were 0.221, 0.491, 0.136, 0.035, 0.073, 0.043, 0.001 and 0.000, respectively. CAC haplotype was regarded as the wild-type allele, and TGT haplotype was the mutation-type allele. The subjects were divided into TGT carrier (n = 10) and noncarrier (n = 23) groups, based on the existence of NR1I2 TGT haplotype or not.
Results of the Clinical Study
According to the ICH-GCP guideline [40], no serious drug-related adverse event was observed from the 33 subjects during the course of this study. No clinically significant alterations were observed in heart rate, blood pressure or body temperature. No significant difference was shown in the demographic characteristics of the volunteers or in the distributions of CYP2B6 genotypes among NR1I2 haplotypes (see Table 1).
Table 1. Demographic data and genotypes of the subjects.
| TGT Noncarriers (n = 23) | TGT Carriers (n = 10) | p | |
| Age (yr) | 22±0.6 | 21±0.3 | 0.105a |
| Weight (kg) | 63±1.7 | 62±2.3 | 0.384a |
| High (cm) | 1.70±0.01 | 1.71±0.02 | 0.603a |
| BMI (kg/m2) | 22.0±0.4 | 21.7±0.5 | 0.124a |
| 2B6*1/*1 (18) | 14 | 4 | 0.448b |
| 2B6*1/*6 (9)+*6/*6 (6) | 9 | 6 |
Wilcoxon rank-sum test between TGT Noncarriers and TGT Carriers groups.
Fisher's exact test between CYP2B6*1/*1 and CYP2B6*1/*6+CYP2B6*6/*6, X2 = 1.224, p = 0.448. BMI: body mass index.
After SF treatment, in complete wild-type groups, AUC_bup was significantly lower (657±73 versus 853±102), while AUC_hyd was significantly higher (15,594±2799 versus 12,732±2448). The AUC ratio (AUC_hyd/AUC_bup), which represents the metabolic activity of bupropion into hydroxybupropion, markedly increased (17.7±2.1 versus 13.1±1.7, see Table 2). However, No significant difference was shown in -24113AA, -24020(-)/(-), and TGT carriers (CYP2B6*6 carriers), AUC_hyd and AUC ratio after SF treatment (p-value>0.05, paired t test, see Table 3 and 4). Furthermore, in -24113AA variant, AUC ratio and AUC_hyd was significantly lower (7.5±2.1 versus 14.5±3.3 and 20.6±1.1, and 8873±1431 versus 14,504±2218 and 17,586±1046) than -24113GA and GG genotypes in the induced states. Moreover, -24113 AA variant also showed significantly lower AUC ratio (6.8±0.7 versus 11.3±2.5 and 15.5±0.8) than -24113GA and GG genotypes in the basal states (see Table 3). Further research found that -24020(-)/(-) variant only showed slight difference (8895±1328 versus 15,116±1797 and 15,518±1168, p-value>0.05) in AUC_hyd in the induced states compared to other -24020[GAGAAG]/(-) genotypes, and no significant impact was shown on the induction of bupropion hydroxybupropion by SF compared with other -24020[GAGAAG]/(-) genotypes.
Table 2. Effects of complete wild-type and mutation-type individuals on SF-mediated metabolic induction of bupropion hydroxylation.
| Complete wild-types (n = 6) | Complete mutation-types (n = 2) | |
| AUC (0–∞)_bup (ng·h/ml) | ||
| Basal | 853±102 | 1262±344a |
| Induced | 657±73 | 1191±254a |
| % differenceb | 26.9±0.7* | 5.4±0.6a |
| AU C(0–∞)_hyd (ng·h/ml) | ||
| Basal | 12,732±2448 | 6523±384a |
| Induced | 15,594±2799 | 7562±890a |
| % differenceb | 25.2±5.9* | 15.5±6.8a |
| AUC_hyd/AUC_bup | ||
| Basal | 13.1±1.7 | 7.0±1.4a |
| Induced | 17.7±2.1 | 7.6±1.6a |
| % differenceb | 39.5±8.2* | 8.7±1.2a |
p<0.01, paired t test between the basal and induced states.
p<0.05, Wilcoxon rank-sum test between [CYP2B6*1/*1+NR1I2 CAC + -24113GG+-24020[GAGAAG]/[GAGAAG]] (complete wild-type) and [CYP2B6*6/*6+NR1I2 TGT+ -24113AA+-24020(-)/(-)] (complete mutation-type) groups.
% difference represents the percent difference between basal and induced state, calculated as an absolute of 100×(induced-basal)/basal.
Table 3. Effects of NR1I2 SNP polymorphisms on SF-mediated metabolic induction of bupropion hydroxylation.
| -24113GG (n = 23) | -24113GA (n = 7) | -24113AA (n = 3) | -24020[GAGAAG]/[GAGAAG] (n = 22) | -24020[GAGAAG]/(-) (n = 8) | -24020(-)/(-) (n = 3) | |
| AUC(0–∞)_bup (ng·h/ml) | ||||||
| Basal | 977±64 | 1062±136 | 1138±207 | 1011±64 | 1114±131 | 1125±186 |
| Induced | 960±55 | 1031±169 | 1083±206 | 995±53 | 1000±157 | 1086±214 |
| % differenceb | 8.9±1.5 | 0.6±0.4 | 0.5±0.3 | 6.8±1.3 | 4.3±0.5 | 1.2±0.4 |
| AU C(0–∞)_hyd (ng·h/ml) | ||||||
| Basal | 13,296±881 | 11,601±1367 | 8126±1573 | 12,063±952 | 11,814±1101 | 8185±1436 |
| Induced | 17,586±1046 | 14,504±2218 | 8873±1431a | 15,518±1168 | 15,116±1797 | 8895±1328 |
| % differenceb | 32.0±5.1* | 27.9±6.9* | 12.5±6.3 | 31.2±5.3* | 28.0±6.0* | 17.5±4.7 |
| AUC_hyd/AUC_bup | ||||||
| Basal | 15.5±0.8 | 11.3±2.5 | 6.8±0.7a | 12.4±1.0 | 11.9±1.9 | 7.3±0.8 |
| Induced | 20.6±1.1 | 14.5±3.3 | 7.5±2.1a | 16.8±1.2 | 15.6±3,1 | 9.5±2.0 |
| % differenceb | 34.4±5.0* | 30.6±6.3* | 11.3±7.9 | 39.4±5.2* | 28.6±4.9* | 20.6±8.5 |
p<0.01, paired t test between the basal and induced states.
p<0.05, Kruskal-Wallis test for NR1I2 -24113G>A and -24020[GAGAAG]>(-) groups.
% difference represents the percent difference between basal and induced state, calculated as an absolute of 100×(induced-basal)/basal.
Table 4. Effects of NR1I2 haplotype and CYP2B6 genotypes on SF -mediated metabolic induction of bupropion hydroxylation.
| CYP2B6 * 6 noncarriers (wild-type) (n = 18) | CYP2B6 * 6 carriers (n = 15) | pd | |||
| TGT Noncarriers (n = 14) | TGT Carriers (n = 4) | TGT Noncarriers (n = 9) | TGT Carriers (n = 6) | ||
| AUC(0–∞)_bup (ng·h/ml) | |||||
| Basal | 869±63 | 924±73 | 1186±100b | 1247±161 | 0.004 |
| Induced | 800±99 | 898±51 | 1175±81b | 1194±90 | 0.002 |
| % differencee | 7.9±0.8 | 2.8±0.5 | 2.2±0.8 | 1.0±0.8 | 0.229 |
| AU C(0–∞)_hyd (ng·h/ml) | |||||
| Basal | 12,484±1239 | 12,057±1100 | 11,485±1499 | 8844±822c | 0.044 |
| Induced | 16,759±1624 | 14,078±676 | 14,030±1891 | 11,029±1225c | 0.021 |
| % differencee | 32.5±6.5* | 18.1±5.2a | 24.9±9.1* | 24.4±7.5 | 0.048 |
| AUC_hyd/AUC_bup | |||||
| Basal | 14.5±1.5 | 13.9±1.1 | 9.7±1.0b | 7.6±1.0a , c | 0.000 |
| Induced | 19.1±1.9 | 18.6±2.7 | 11.6±1.1b | 9.1±1.5c | 0.000 |
| % differencee | 34.8±7.1* | 30.1±6.7* | 23.2±7.0 | 17.1±9.5a | 0.012 |
p<0.01, paired t test between the basal and induced states.
p<0.05, Wilcoxon rank-sum test for the NR1I2 groups within each CYP2B6 genotype group.
p<0.05, Wilcoxon rank-sum test for the CYP2B6 groups with TGT noncarriers.
p<0.05, Wilcoxon rank-sum test for the CYP2B6 groups with TGT carriers.
Wilcoxon rank-sum test for the CYP2B6 groups; p<0.05 is indicated in bold.
% difference represents the percent difference between basal and induced state, calculated as an absolute of 100×(induced-basal)/basal.
The effects of CYP2B6 genotypes and NR1I2 TGT haplotype were considered as a whole and are showed by subgroups (see Table 4). CYP2B6*6 carriers always showed significantly lower AUC ratio than that of noncarrier group in both the basal and induced states, and also in TGT noncarrier or carrier groups. NR1I2 TGT carriers (CYP2B6*6 carriers) had a significantly lower AUC ratio and percent difference of AUC ratio (7.6±1.0 versus 9.7±1.0, and 17.1±9.5 versus 23.2±7.0) than noncarriers. However, there was no significant difference observed in AUC ratio and percent difference of AUC ratio between NR1I2 TGT carrier and noncarrier groups for CYP2B6*6 noncarriers. Furthermore, TGT carriers (CYP2B6*6 noncarriers) had significantly lower percent difference of AUC_hyd (18.1±5.2 versus 32.5±6.5) than noncarriers, while there was no significant difference in CYP2B6*6 carriers (see Table 4). In addition, the complete mutation-type [CYP2B6*6/*6+NR1I2 TGT+ -24113AA+-24020(-)/(-)] individuals exhibited even lower percent difference of AUC ratio (8.7±1.2 versus 39.5±8.2) than those of complete wild-types (p-value<0.05, see Table 2).
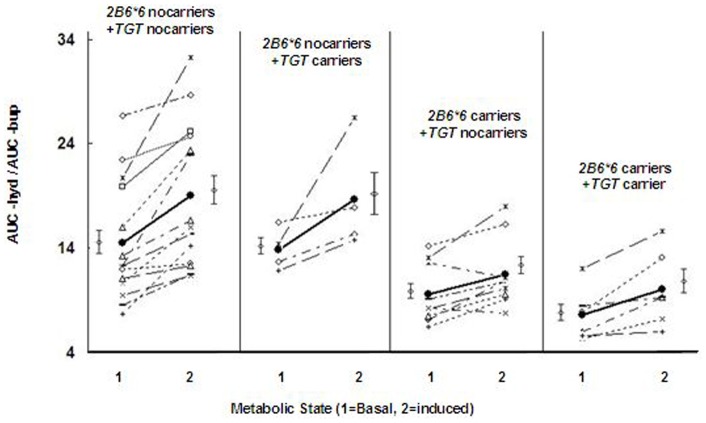
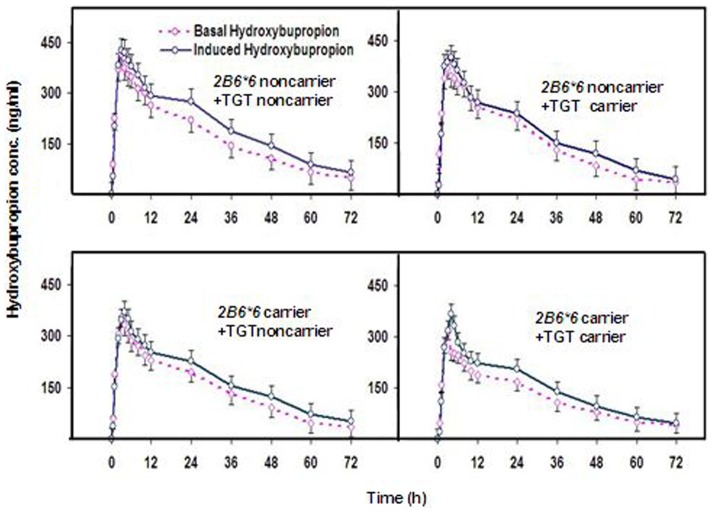
As shown in Fig. 1, individual plots of CYP2B6 and NR1I2 variants indicated that the combination of NR1I2 TGT haplotype and CYP2B6*6 affected the AUC ratio in both the basal and induced states. The concentration-time profiles of hydroxybupropion were very different for CYP2B6*6+NR1I2 TGT carriers from other groups, with the lowest values in both the basal and induced states (see Fig. 2). The Cmax values of hydroxybupropion in the basal and induced states showed no significant difference between NR1I2 TGT carriers and noncarriers (unpublished data). However, the hydroxybupropion Cmax in CYP2B6*6+NR1I2 TGT carriers was significantly lower than CYP2B6*6+NR1I2 TGT noncarriers in both the basal and induced states (284.3±40.8 versus 395.2±40.5, and 332.9±27.6 versus 424.5±32.8, p-value<0.05).
Figure 1. Effects of NR1I2 TGT and CYP2B6*6 on induction of bupropion hydroxylation by SF.
Individual profiles showing AUC_hyd/AUC_bup ratios for the basal and SF-induced states in the NR1I2 and CYP2B6 genotype groups (n = 33).
Figure 2. Concentration (conc.)–time profiles of hydroxybupropion.
Mean plasma concentration-time profiles of hydroxybupropion after oral administration of 150 mg of bupropion or bupropion+SF treatment in subjects after 14 days in the NR1I2 and CYP2B6 genotype groups.
Discussion
In the present study, we investigated the NR1I2 genetic polymorphism in Chinese population and found that the allelic frequencies of -25385T and -24113A in Chinese (0.283, 0.181) were lower than the reported frequencies of Korean (0.32, 0.32), European descent (0.39, 0.32), and Africa American (0.39, 0.32) [18], [20]. The allelic frequency of -24020(-) was significantly lower (0.218) than that of Japanese (0.274) and Korean (0.319) [19], [20], and the allelic frequency of g.7635G was an intermediate value (0.522) between European descent (0.35) and Africa American (0.77). However, g.8055T has markedly higher (0.625) allele frequency compared with that in Brazilian (0.125), European descent (0.15), African (0.18), Indian (0.24), Korean (0.41), and Malay (0.43) [18], [20], [41], [42]. The NR1I2 allelic frequency observed in this study was very similar to the previous report in Chinese [43].
Further haplotypes analysis showed that compared with the Korean, the composition and frequencies of NR1I2 TGT haplotypes in Chinese people were completely different. Eight haplotypes were inferred based on SNPs in positions -25385C>T, g.7635A>G, and g.8055, and the frequency distribution of TGT haplotype was slightly lower (0.136) than that of Korean (0.199). Furthermore, linkage manners of NR1I2 SNPs were inconsistent with the Korean. A slight but not complete LD among -25385C>T, -24113G>A, and -24020[GAGAAG]>(-) and between g.7635A>G and g.8055C>T was observed [20]. These results suggest that the NR1I2 gene has unique characteristics of high polymorphism and significant interethnic variants.
As shown in Table 4, we found in the clinical investigation that in TGT noncarriers (TGT noncarriers, n = 14), the overall pharmacokinetic parameters of bupropion and hydroxybupropion (AUC_bup, AUC_hyd, and AUC ratio) indicated the strongest effects including basal activities, induced activities and their percent differences. However, with the emergence of TGT and CYP2B6*6 variants (TGT carriers, n = 4 and TGT noncarriers, n = 9, respectively), the strongest effects of the overall pharmacokinetic parameters of bupropion and hydroxybupropion became weaker and smaller, until TGT variant existed in CYP2B6*6 carriers (TGT carriers, n = 6). Moreover, in each step of the attenuation effects of the pharmacokinetic parameters, TGT and CYP2B6*6 carriers always showed smaller effects than noncarriers. Therefore, this result suggests that the decreased metabolism of bupropion with SF treatment is affected by both NR1I2 TGT and CYP2B6*6 variants. Also, similar findings were obtained from -24113G>A, -24020 [GAGAAG]> (-), and complete wild/mutation-type individuals (see Table 2 and 3). In short, our data strongly support the hypothesis that NR1I2 TGT haplotype, -24113AA, and CYP2B6*6 variants play very important roles in bupropion disposition.
Interestingly, in previous study, NR1I2 TGT carriers slightly manifested stronger effects on some pharmacokinetics parameters of bupropion and hydroxybupropion than the corresponding noncarrier groups (p-value>0.05). Conversely, CYP2B6*6 carriers showed smaller effects on AUC_hyd, and AUC ratio than the noncarriers (p-value<0.05) [20]. Reports also showed that CYP2B6 expression increased in the basal state while decreased in the induced state when treated with rifampin in PXR.2 cells [44]. However, these results were not observed in our study. Rifampin is a known selective human PXR activator with little cross-interaction with other receptors, such as small heterodimer partner (SHP) and hepatocyte nuclear factor-4α(HNF-4α) [45], [46]. Early study indicated that interaction of PXR with HNF-4αand its coactivators, peroxisome proliferator-activated receptor-γ-coactivator-1α(PGC-1α) contributed to the strong induction of CYP3A4 by rifampin, whereas gene expression of SHP was simultaneously inhibited by PXR, which weakened inhibitory effect of SHP on CYP3A4 expression and strengthened the HNF-4αinducibility of CYP3A4 [45], [47]. Therefore, we may assume that under the situation that some interfering factors of SHP gene expression exceeded the effects of PXR variants (PXR function variant led to increased SHP gene expression), the activity of CYP did not decline but increased. More attention should be paid to the study of SHP gene expression and regulation in the near future. Moreover, Owen's group reported that genetic variability in constitutive androstane receptor (CAR) was involved in the metabolism and disposition of CYP2B6 substrate drugs recently [48].
In our study, further analysis showed that TGT carriers, only in the basal states, had significantly lower AUC ratio and percent differences (7.6±1.0 versus 9.7±1.0, and 17.1±9.5 versus 23.2±7.0) than TGT noncarriers. However, CYP2B6*6 carriers exhibited significant differences in the most of pharmacokinetic parameter values of bupropion and hydroxybupropion (AUC_bup, AUC_hyd, and AUC ratio) compared with CYP2B6*6 noncarriers in both the basal and induced states (see Table 4). In addition, from Fig. 1 and Fig. 2, a tenuous distinction existed in the concentration (conc.) – time curves of hydroxybupropion and AUC ratio when TGT carriers appeared; while the curves and AUC ratio values quickly dropped when CYP2B6*6 carriers came. This result suggests that CYP2B6*6 variants had stronger reduced metabolic capacity than NR1I2 TGT haplotype. Interestingly, the complete mutation-type [CYP2B6*6/*6+NR1I2 TGT+ -24113AA+-24020(-)/(-)] individuals indicated even lower metabolism activities (8.7±1.2 versus 39.5±8.2) than the complete wild-types (see Table 2). Therefore, we tentatively conclude that reduced metabolic capacity is more significant in individuals including CYP2B6*6 mutations, NR1I2 TGT haplotype, and other NR1I2 variants with reduced functions.
To date, the verified sites and positions of the SNPs (or haplotypes) of NR1I2 functional variants were as follows: -25385C>T (5′-UTR), -24622A>T (5′-UTR), -24446C>A (Exon 1), -24113G>A (Intron 1), -24020[GAGAAG]>(-) (Intron 1), 106G>A (*2, Exon 3), 2904C>T (*5, Exon 3), 4321G>A (*4, Exon 4), 4374G>A (*10, Exon 4), 4444A>G (*11, Exon 4), 7635A>G (Intron 5), 8055C>T (Intron 6), 8528G>A (*12, Exon 8), 8561C>T (*7, Exon 8), 9863A>G (*8, Exon 9), 10620C>T (3′-UTR), 10799G>A (3′-UTR), 11156A>C (3′-UTR), 11193T>C (3′-UTR), and NR1I2*1B (8055C>T, Intron 6+2654T>C, 3′-UTR) [18], [20], [42], [43], [49], [50]. However, some investigations of clinical pharmacogenetics of the NR1I2 functional variants were not consistent with their findings in vitro. For instance, NR1I2 -25385C>T, -24113G>A, 7635A>G, or 8055C>T was reported to be associated with higher magnitude of induction of intestinal CYP3A by rifampin in vitro [18], but recently, the subjects with -25385C>T or TGT (-25385T+g.7635G+g.8055T) carriers were verified to have decreased CYP2B6 activity (AUC ratio) induced by rifampin in Korean, and NR1I2*1B (8055C>T+2654T>C) haplotype was strongly associated with its downstream target genes of MDR1 in Asian breast cancer patients [42]. Moreover, the result for our subjects with -24113AA showed the lowest percent differences of AUC ratio (11.3±7.9) after SF induction compared with wild genotypes (see Table 3). The specific mechanism of this difference in vitro and in vivo is yet unknown. However, we must admit that the result in vitro was easily interfered by diverse uncontrollable factors. These results highlighted the important role of NR1I2 pharmacogenetics in the disposition of putative drug substrates. It can be assumed that NR1I2 genetic polymorphisms will play an essential role in affecting interethnic variations in drug disposition.
Previous researches reported that oxysterol, 24(S), 25- epoxycholesterol (LXR agonists), glucocorticoid (GR agonists), and vitamin D (VDR agonists) could induce expression of CYP2B6 through the binding co-activators of the corresponding ligands and PXR [51]–[53]. Moreover, gender factor also affected the results of clinical trials [54], [55]. It is worth mentioning that these interference factors were well balanced through our strict subject exclusion criteria and good clinical trial control. However, there are also some deficiencies in our study. For instance, we paid more attention to the NR1I2 variants, which had the functions reported in vitro or in vivo such as -25385C>T, -24113G>A, -24020[GAGAAG]/(-), 7635A>G, and 8055C>T [18], [19], [20], [42], [49], [50], [56]–[58]. The rarely reported or less concerned NR1I2 variants were not included in this research. Future clinical pharmacogenetics research of NR1I2 variants should be focused on the reported functional variants with lower distribution frequencies, which have potential possibility to play more important roles than the star variants. In addition, lower concomitance mutation frequencies and strict subject exclusion criteria also affected our subjects enrollment. Relatively small and uneven numbers of individuals with various genotypes were investigated in our study, which were the limitations of drawing of conclusions based upon this sample sizes. Further clinical studies of the NR1I2 variants pharmacogenetics should be operated in larger groups, and even different ethnic populations [59].
Conclusions
As the data indicated, NR1I2 TGT haplotype, -24113AA, CYP2B6*6, and the complete mutation-type [CYP2B6*6/*6+NR1I2 TGT+ -24113AA+-24020 (-)/(-)] individuals have strong evidence to show the ability to reduce the metabolic capacity of CYP2B6 after SF administration in Chinese individuals. Individuals/ethnic populations with different genetic backgrounds may show significant differences in drug metabolism and efficacy, sometimes even manifested as severe adverse drug reactions or no efficacy. Our findings provide an important reference for carrying out the gene oriented individual/interethnic therapy of CYP2B6 substrate drugs, and avoiding the adverse effects of SF and CYP2B6 substrate drugs combination. Whether other NR1I2 and regulator variants also have impact on the disposition of CYP2B6 substrate drugs by SF requires further exploration. Large-scale population pharmacokinetic, pharmacodynamic, and nosazontology analysis using pharmacogenomics method are needed to clarify the role of NR1I2 and CYP2B6*6 variants in the efficacy, safety, and drug interactions of CYP2B6 substrate drugs, even disease susceptibility between individuals.
Supporting Information
CONSORT checklist.
(DOC)
Trial protocol.
(DOC)
Acknowledgments
We'd like to take this opportunity to thank distinguished Prof. Howard L. McLeod of Institute of Pharmacogenomics and Individualized Therapy, University of North Carolina at Chapel Hill, North Carolina, for his important comments on our paper and Dr. Yuli Liu, Dr. Yichen Liu of Pharmacogenetics Research Institute, Central South University, for their substantial contributions to prepare this manuscript, and use of the WinNonlin Software in the PK Analysis.
Funding Statement
This work was supported by the National Scientific Foundation of China (No. 81001476), 863 Project (No. 2012AA02A518), and Program for Changjiang Scholars and Innovative Research Team in University (IRT0946). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bailey I, Gibson GG, Plant K, Graham M, Plant N (2011) A PXR-mediated negative feedback loop attenuates the expression of CYP3A in response to the PXR agonist pregnenalone-16alpha-carbonitrile. PLoS One 6: e16703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Saradhi M, Krishna B, Mukhopadhyay G, Tyagi RK (2005) Purification of full-length human pregnane and Xenobiotic receptor: polyclonal antibody preparation for immunological characterization. Cell Research 15: 785–795. [DOI] [PubMed] [Google Scholar]
- 3. Kliewer SA, Moore JT, Wade L, Staudinger JL, Watson MA, et al. (1998) An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell 92: 73–82. [DOI] [PubMed] [Google Scholar]
- 4. Blumberg B, Sabbagh WJ, Juguilon H, Bolado JJ, van Meter CM, et al. (1998) SXR, a novel steroid and xenobiotic-sensing nuclear receptor. Genes Dev 12: 3195–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bertilsson G, Heidrich J, Svensson K, Asman M, Jendeberg L, et al. (1998) Identification of a human nuclear receptor defines a new signaling pathway for CYP3A induction. Proc Natl Acad Sci USA 95: 12208–12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jones SA, Moore LB, Shenk JL, Wisely GB, Hamilton GA, et al. (2000) The pregnane X receptor: a promiscuous xenobiotic receptor that has diverged during evolution. Mol Endocrinol 14: 27–39. [DOI] [PubMed] [Google Scholar]
- 7. Yueh MF, Kawahara M, Raucy J (2005) High volume bioassays to assess CYP3A4-mediated drug interactions: induction and inhibition in a single cell line. Drug Metab Dispos 33: 38–48. [DOI] [PubMed] [Google Scholar]
- 8. Pascussi JM, Drocourt L, Gerbal-Chaloin S, Fabre JM, Maurel P, et al. (2001) Dual effect of dexamethasone on CYP3A4 gene expression in human hepatocytes. Sequential role of glucocorticoid receptor and pregnane X receptor. Eur J Biochem 268: 6346–6358. [DOI] [PubMed] [Google Scholar]
- 9. Dresser GK, Schwarz UI, Wilkinson GR, Kim RB (2003) Coordinate induction of both cytochrome P4503A and MDR1 by St John's wort in healthy subjects. Clin Pharmacol Ther 73: 41–50. [DOI] [PubMed] [Google Scholar]
- 10. Moore LB, Goodwin B, Jones SA, Wisely GB, Serabjit-Singh CJ, et al. (2000) St. John's wort induces hepatic drug metabolism through activation of the pregnane X receptor. Proc Natl Acad Sci U S A 97: 7500–7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lau AJ, Yang G, Rajaraman G, Baucom CC, Chang TK (2010) Human pregnane X receptor agonism by Ginkgo biloba extract: assessment of the role of individual ginkgolides. J Pharmacol Exp Ther 335: 771–780. [DOI] [PubMed] [Google Scholar]
- 12. Lehmann JM, McKee DD, Watson MA, Willson TM, Moore JT, et al. (1998) The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. J Clin Invest 102: 1016–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang HB, Faucette S, Sueyoshi T, Moore R, Ferguson S, et al. (2003) A novel distal enhancer module regulated by pregnane X receptor/constitutive androstane receptor is essential for the maximal induction of CYP2B6 gene expression. J Biol Chem 278: 14146–14152. [DOI] [PubMed] [Google Scholar]
- 14. Frank C, Makkonen H, Dunlop TW, Matilainen M, Väisänen S, et al. (2005) Identification of pregnane X receptor binding sites in the regulatory regions of genes involved in bile acid homeostasis. J Mol Biol 346: 505–519. [DOI] [PubMed] [Google Scholar]
- 15. Olinga P, Elferink MG, Draaisma AL, Merema MT, Castell JV, et al. (2008) Coordinated induction of drug transporters and phase I and II metabolism in human liver slices. Eur J Pharm Sci 33: 380–389. [DOI] [PubMed] [Google Scholar]
- 16. Hustert E, Zibat A, Presecan-Siedel E, Eiselt R, Mueller R, et al. (2001) Natural protein variants of pregnane X receptor with altered transactivation activity toward CYP3A4. Drug Metab Dispos 29: 1454–1459. [PubMed] [Google Scholar]
- 17. Wang XD, Li JL, Su QB, Guan S, Chen J, et al. (2009) Impact of the haplotypes of the human pregnane X receptor gene on the basal and St John's wort-induced activity of cytochrome P450 3A4 enzyme. Br J Clin Pharmacol 67: 255–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang J, Kuehl P, Green ED, Touchman JW, Watkins PB, et al. (2001) The human pregnane X receptor: genomic structure and identification and functional characterization of natural allelic variants. Pharmacogenetics 11: 555–572. [DOI] [PubMed] [Google Scholar]
- 19. Uno Y, Sakamoto Y, Yoshida K, Hasegawa T, Hasegawa Y, et al. (2003) Characterization of six base pair deletion in the putative HNF1-binding site of human PXR promoter. J Hum Genet 48: 594–597. [DOI] [PubMed] [Google Scholar]
- 20. Chung JY, Cho JY, Lim HS, Kim JR, Yu KS, et al. (2011) Effects of pregnane X receptor (NR1I2) and CYP2B6 genetic polymorphisms on the induction of bupropion hydroxylation by rifampin. Drug Metab Dispos 39: 92–97. [DOI] [PubMed] [Google Scholar]
- 21. Chen LL, Qi J, Chang YX, Zhu DN, Yu BY (2009) Identification and determination of the major constituents in Traditional Chinese Medicinal formula Danggui-Shaoyao-San by HPLC–DAD–ESI-MS/MS. Journal of Pharmaceutical and Biomedical Analysis 50: 127–137. [DOI] [PubMed] [Google Scholar]
- 22. Zhang YJ, Huang X, Wang Y, Xie Y, Qiu XJ, et al. (2011) Ferulic acid-induced anti-depression and prokinetics similar to Chaihu-Shugan-San via polypharmacology. Brain Res Bull 86: 222–228. [DOI] [PubMed] [Google Scholar]
- 23. Wang BH, Ouyang JP, Liu YM, Yang JW, Wei L, et al. (2004) Sodium ferulate inhibits atherosclerogenesis in hyperlipidemia rabbits. J Cardiovasc Pharmacol 43: 549–554. [DOI] [PubMed] [Google Scholar]
- 24. Traore M, Hong JL, Xu CL (2003) Influence of Angelicae sinensis extract on lipid accumulation in rabbit aortic smooth muscle cells induced by oxidized LDL. Biorheology 40: 389–394. [PubMed] [Google Scholar]
- 25. Barone E, Calabrese V, Mancuso C (2009) Ferulic acid and its therapeutic potential as a hormetin for age-related diseases. Biogerontology 10: 97–108. [DOI] [PubMed] [Google Scholar]
- 26. Liu HQ, Sun Y, Gao Y, Chen FF, Xu MB, et al. (2010) The analgesic effect and mechanism of the combination of sodium ferulate and oxymatrine. Neurochem Res 35: 1368–1375. [DOI] [PubMed] [Google Scholar]
- 27. Srinivasan M, Sudheer AR, Menon VP (2007) Ferulic Acid: therapeutic potential through its antioxidant property. J Clin Biochem Nutr 40: 92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang BH, Ou-Yang JP (2005) Pharmacological actions of sodium ferulate in cardiovascular system. Cardiovasc Drug Rev 23: 161–172. [DOI] [PubMed] [Google Scholar]
- 29. Wang HB, Tompkins LM (2008) CYP2B6: new insights into a historically overlooked cytochrome P450 isozyme. Curr Drug Metab 9: 598–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Faucette SR, Hawke RL, Lecluyse EL, Shord SS, Yan B, et al. (2000) Validation of bupropion hydroxylation as a selective marker of human cytochrome P450 2B6 catalytic activity. Drug Metab Dispos 28: 1222–1230. [PubMed] [Google Scholar]
- 31. Hesse LM, Venkatakrishnan K, Court MH, von Moltke LL, Duan SX, et al. (2000) CYP2B6 mediates the in vitro hydroxylation of bupropion: potential drug interactions with other antidepressants. Drug Metab Dispos 28: 1176–1183. [PubMed] [Google Scholar]
- 32. Fan L, Wang JC, Jiang F, Tan ZR, Chen Y, et al. (2009) Induction of cytochrome P450 2B6 activity by the herbal medicine baicalin as measured by bupropion hydroxylation. Eur J Clin Pharmacol 65: 403–409. [DOI] [PubMed] [Google Scholar]
- 33. Hogeland GW, Swindells S, McNabb JC, Kashuba ADM, Yee GC, et al. (2007) Lopinavir/ritonavir reduces bupropion plasma concentrations in healthy subjects. Clin Pharmacol Ther 81: 69–75. [DOI] [PubMed] [Google Scholar]
- 34. Gao LC, Huang X, Tan ZR, Fan L, Zhou HH (2012) The effects of sodium ferulate on the pharmacokinetics of bupropion and its active metabolite in healthy men. European Review for Medical and Pharmacological Sciences (in press).. [PubMed] [Google Scholar]
- 35. Zhang H, Sridar C, Kenaan C, Amunugama H, Ballou DP, et al. (2011) Polymorphic variants of cytochrome P450 2B6 (CYP2B6.4–CYP2B6.9) exhibit altered rates of metabolism for bupropion and efavirenz: a charge-reversal mutation in the K139E variant (CYP2B6.8) impairs formation of a functional cytochrome p450-reductase complex. J Pharmacol Exp Ther 338: 803–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Saitoh A, Sarles E, Capparelli E, Aweeka F, Kovacs A, et al. (2007) CYP2B6 genetic variants are associated with nevirapine pharmacokinetics and clinical response in HIV-1-infected children. AIDS 21: 2191–2199. [DOI] [PubMed] [Google Scholar]
- 37. Nyakutira C, Röshammar D, Chigutsa E, Chonzi P, Ashton M, et al. (2008) High prevalence of the CYP2B6 516G>T(*6) variant and effect on the population pharmacokinetics of efavirenz in HIV/AIDS outpatients in Zimbabwe. Eur J Clin Pharmacol 64: 357–365. [DOI] [PubMed] [Google Scholar]
- 38. Lee AM, Jepson C, Hoffmann E, Epstein L, Hawk LW, et al. (2007) CYP2B6 genotype alters abstinence rates in a bupropion smoking cessation trial. Biol Psychiatry 62: 635–641. [DOI] [PubMed] [Google Scholar]
- 39. Chen J, Jiang WM, Gao XL, Jiang X, Zhang QZ, et al. (2004) Bioequivalence evaluation of two rabeprazole enteric coated formulations in healthy Chinese volunteers. Eur J Drug Metab Pharmacokinet 29: 103–106. [DOI] [PubMed] [Google Scholar]
- 40.ICH Guideline for Good Clinical Practice CPMP/ICH/135/95. Available: http://www.emea.europa.eu/pdfs/human/ich/013595en.pdf. Accessed 25 May 2012.
- 41. Moreira RPP, Jorge AAL, Mendonca BB, Bachega TASS (2011) Frequency of genetic polymorphisms of PXR gene in the Brazilian population. Clinics (Sao Paulo) 66: 1041–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sandanaraj E, Lal S, Selvarajan V, Ooi LL, Wong ZW, et al. (2008) PXR pharmacogenetics: association of haplotypes with hepatic CYP3A4 and ABCB1 messenger RNA expression and doxorubicin clearance in Asian breast cancer patients. Clin Cancer Res 14: 7116–7126. [DOI] [PubMed] [Google Scholar]
- 43. Wang XD, Li JL, Su QB, Deng XY, Lu Y, et al. (2007) A pharmacogenetic study of pregnane X receptor (NR1I2) in Han Chinese. Curr Drug Metab 8: 778–786. [DOI] [PubMed] [Google Scholar]
- 44. Lin YS, Yasuda K, Assem M, Cline C, Barber J, et al. (2009) The major human pregnane X receptor (PXR) splice variant, PXR.2, exhibits significantly diminished ligand-activated transcriptional regulation. Drug Metab Dispos 37: 1295–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lim YP, Huang JD (2008) Interplay of pregnane X receptor with other nuclear receptors on gene regulation. Drug Metab Pharmacokinet 23: 14–21. [DOI] [PubMed] [Google Scholar]
- 46. Ourlin JC, Lasserre F, Pineau T, Fabre JM, Sa-Cunha A, et al. (2003) The small heterodimer partner interacts with the pregnane X receptor and represses its transcriptional activity. Mol Endocrinol 17: 1693–1703. [DOI] [PubMed] [Google Scholar]
- 47. Li TG, Chiang JYL (2003) Rifampicin induction of CYP3A4 requires pregnane X receptor cross talk with hepatocyte nuclear factor 4αand coactivators, and suppression of small heterodimer partner gene expression. Drug Metab Dispos 34: 756–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wyen C, Hendra H, Siccardi M, Platten M, Jaeger H, et al. (2011) Cytochrome P450 2B6 ( CYP2B6) and constitutive androstane receptor ( CAR ) polymorphisms are associated with early discontinuation of efavirenz-containing regimens. J Antimicrob Chemother 6: 1–7. [DOI] [PubMed] [Google Scholar]
- 49. Lamba J, Lamba V, Schuetz E (2005) Genetic variants of PXR (NR1I2) and CAR (NR1I3) and their implications in drug metabolism and pharmacogenetics. Curr Drug Metab 6: 369–383. [DOI] [PubMed] [Google Scholar]
- 50. Svärd J, Spiers JP, Mulcahy F, Hennessy M (2010) Nuclear receptor-mediated induction of CYP450 by antiretrovirals: functional consequences of NR1I2 (PXR) polymorphisms and differential prevalence in whites and sub-Saharan Africans. J Acquir Immune Defic Syndr 55: 536–549. [DOI] [PubMed] [Google Scholar]
- 51. Duniec-Dmuchowski Z, Ellis E, Strom SC, Kocarek TA (2007) Regulation of CYP3A4 and CYP2B6 expression by liver X receptor agonists. Biochem Pharmacol 74: 1535–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Drocourt L, Ourlin JC, Pascussi JM, Maurel P, Vilarem MJ (2002) Expression of CYP3A4, CYP2B6, and CYP2C9 is regulated by the vitamin D receptor pathway in primary human hepatocytes. J Biol Chem 277: 25125–25132. [DOI] [PubMed] [Google Scholar]
- 53. Wang H, Faucette SR, Gilbert D, Jolley SL, Sueyoshi T, et al. (2003) Glucocorticoid receptor enhancement of pregnane X receptor mediated CYP2B6 regulation in primary human hepatocytes. Drug Metab Dispos 31: 620–630. [DOI] [PubMed] [Google Scholar]
- 54. Mukonzo JK, Röshammar D, Waako P, Andersson M, Fukasawa T, et al. (2009) A novel polymorphism in ABCB1 gene, CYP2B6*6 and sex predict single-dose efavirenz population pharmacokinetics in Ugandans. Br J Clin Pharmacol 68: 690–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lamba V, Lamba J, Yasuda K, Strom S, Davila J, et al. (2003) Hepatic CYP2B6 expression: gender and ethnic differences and relationship to CYP2B6 genotype and CAR (constitutive androstane receptor) expression. J Pharmacol Exp Ther 307: 906–922. [DOI] [PubMed] [Google Scholar]
- 56. King CR, Xiao M, Yu J, Minton MR, Addleman NJ, et al. (2007) Identification of NR1I2 genetic variation using resequencing. Eur J Clin Pharmacol 63: 547–554. [DOI] [PubMed] [Google Scholar]
- 57. Lamba J, Lamba V, Strom S, Venkataramanan R, Schuetz E (2008) Novel single nucleotide polymorphisms in the promoter and intron 1 of human pregnane X receptor/NR1I2 and their association with CYP3A4 expression. Drug Metab Dispos 36: 169–181. [DOI] [PubMed] [Google Scholar]
- 58. Folwaczny M, Tengler B, Glas J (2012) Variants of the human NR1I2 (PXR) locus in chronic periodontitis. J Periodontal Res 47: 174–179. [DOI] [PubMed] [Google Scholar]
- 59. Ni WJ, Ji J, Dai ZY, Papp A, Johnson AJ, et al. (2010) Flavopiridol pharmacogenetics: clinical and functional evidence for the role of SLCO1B1/OATP1B1 in flavopiridol disposition. PLoS One 5: e13792. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
CONSORT checklist.
(DOC)
Trial protocol.
(DOC)