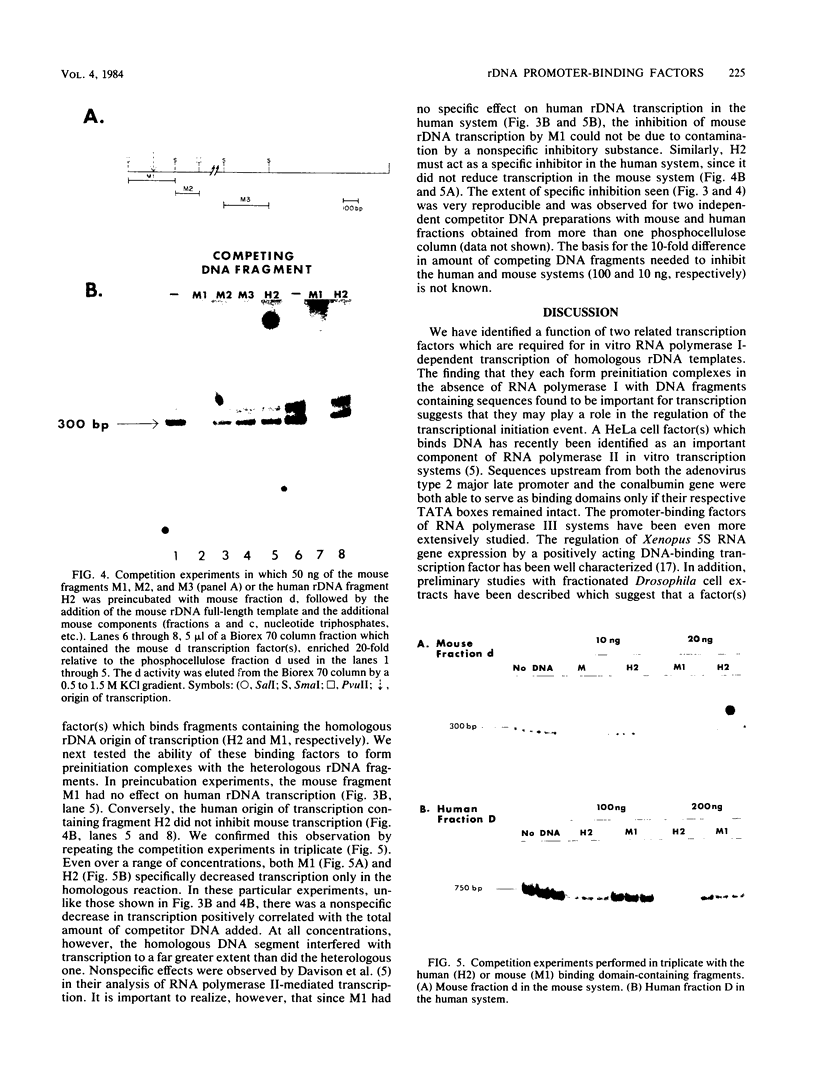
Abstract
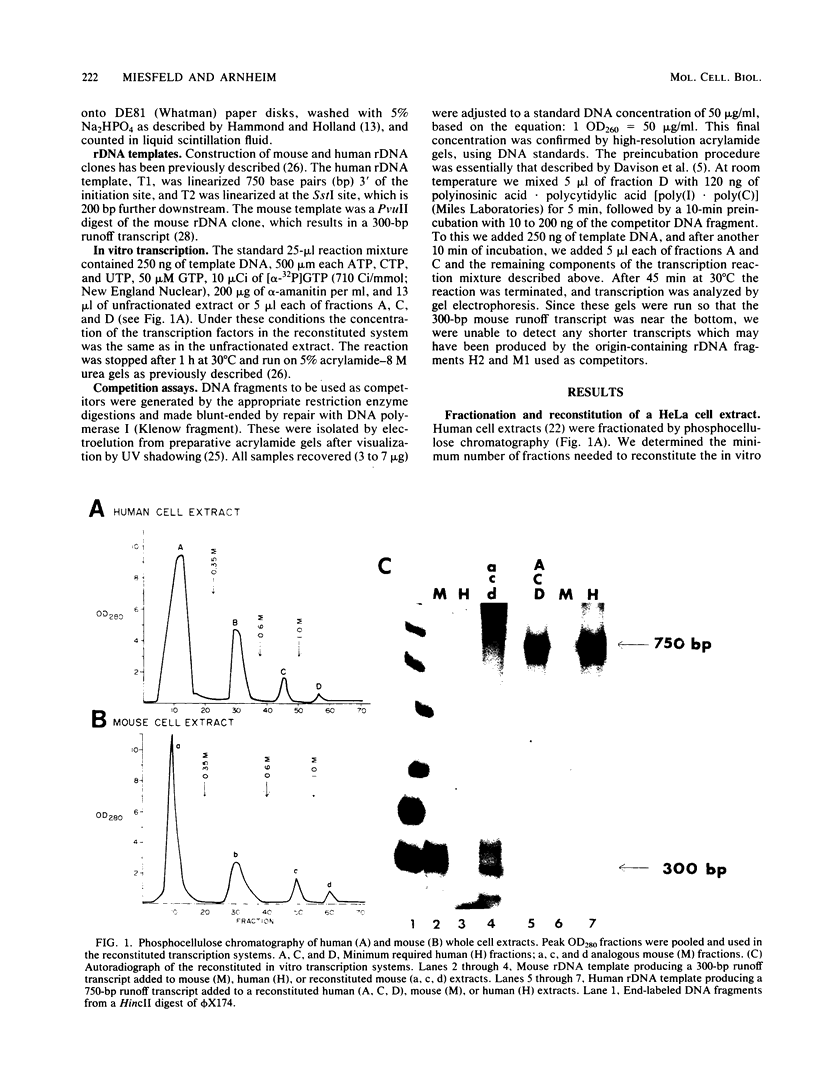
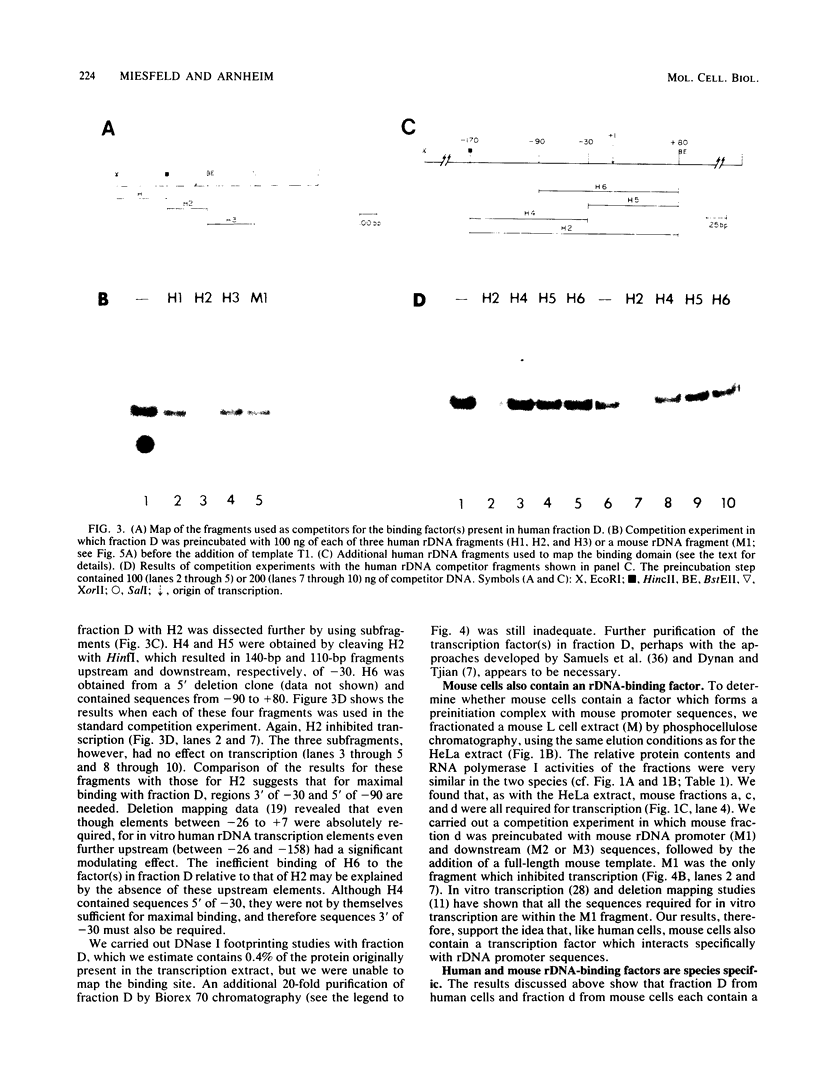
RNA polymerase I transcription factors were purified from HeLa and mouse L cell extracts by phosphocellulose chromatography. Three fractions from each species were found to be required for transcription. One of these fractions, virtually devoid of RNA polymerase I activity, was found to form a stable preinitiation complex with small DNA fragments containing promoter sequences from the homologous but not the heterologous species. These species-specific DNA-binding factors can explain nucleolar dominance in vivo in mouse-human hybrid somatic cells and species specificity in cell-free, RNA polymerase I-dependent transcription systems. The evolution of species-specific transcriptional control signals may be the natural outcome of a special relationship that exists between the RNA polymerase I transcription machinery and the multigene family coding for rRNA.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnheim N., Krystal M., Schmickel R., Wilson G., Ryder O., Zimmer E. Molecular evidence for genetic exchanges among ribosomal genes on nonhomologous chromosomes in man and apes. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7323–7327. doi: 10.1073/pnas.77.12.7323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D. D., Wensink P. C., Jordan E. A comparison of the ribosomal DNA's of Xenopus laevis and Xenopus mulleri: the evolution of tandem genes. J Mol Biol. 1972 Jan 14;63(1):57–73. doi: 10.1016/0022-2836(72)90521-9. [DOI] [PubMed] [Google Scholar]
- Croce C. M., Talavera A., Basilico C., Miller O. J. Suppression of production of mouse 28S ribosomal RNA in mouse-human hybrids segregating mouse chromosomes. Proc Natl Acad Sci U S A. 1977 Feb;74(2):694–697. doi: 10.1073/pnas.74.2.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover G. Molecular drive: a cohesive mode of species evolution. Nature. 1982 Sep 9;299(5879):111–117. doi: 10.1038/299111a0. [DOI] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. Isolation of transcription factors that discriminate between different promoters recognized by RNA polymerase II. Cell. 1983 Mar;32(3):669–680. doi: 10.1016/0092-8674(83)90053-3. [DOI] [PubMed] [Google Scholar]
- Eliceiri G. L., Green H. Ribosomal RNA synthesis in human-mouse hybrid cells. J Mol Biol. 1969 Apr;41(2):253–260. doi: 10.1016/0022-2836(69)90390-8. [DOI] [PubMed] [Google Scholar]
- Eliceiri G. L. The ribosomal RNA of hamster-mouse hybrid cells. J Cell Biol. 1972 Apr;53(1):177–184. doi: 10.1083/jcb.53.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Financsek I., Mizumoto K., Mishima Y., Muramatsu M. Human ribosomal RNA gene: nucleotide sequence of the transcription initiation region and comparison of three mammalian genes. Proc Natl Acad Sci U S A. 1982 May;79(10):3092–3096. doi: 10.1073/pnas.79.10.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I. Nucleotide sequence requirements for specific initiation of transcription by RNA polymerase I. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6908–6911. doi: 10.1073/pnas.79.22.6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I., Roth E., Paule M. R. Ribosomal RNA transcription in vitro is species specific. Nature. 1982 Mar 11;296(5853):173–174. doi: 10.1038/296173a0. [DOI] [PubMed] [Google Scholar]
- Hammond C. I., Holland M. J. Purification of yeast RNA polymerases using heparin agarose affinity chromatography. Transcriptional properties of the purified enzymes on defined templates. J Biol Chem. 1983 Mar 10;258(5):3230–3241. [PubMed] [Google Scholar]
- Hood L., Campbell J. H., Elgin S. C. The organization, expression, and evolution of antibody genes and other multigene families. Annu Rev Genet. 1975;9:305–353. doi: 10.1146/annurev.ge.09.120175.001513. [DOI] [PubMed] [Google Scholar]
- Huet J., Sentenac A., Fromageot P. Spot-immunodetection of conserved determinants in eukaryotic RNA polymerases. Study with antibodies to yeast RNA polymerases subunits. J Biol Chem. 1982 Mar 10;257(5):2613–2618. [PubMed] [Google Scholar]
- Kohorn B. D., Rae P. M. Accurate transcription of truncated ribosomal DNA templates in a Drosophila cell-free system. Proc Natl Acad Sci U S A. 1982 Mar;79(5):1501–1505. doi: 10.1073/pnas.79.5.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuter K. J., Rogers A. The synthesis of ribosomal protein and ribosomal RNA in a rat-mouse hybrid cell line. Exp Cell Res. 1975 Mar 15;91(2):317–325. doi: 10.1016/0014-4827(75)90110-x. [DOI] [PubMed] [Google Scholar]
- Learned R. M., Smale S. T., Haltiner M. M., Tjian R. Regulation of human ribosomal RNA transcription. Proc Natl Acad Sci U S A. 1983 Jun;80(12):3558–3562. doi: 10.1073/pnas.80.12.3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Learned R. M., Tjian R. In vitro transcription of human ribosomal RNA genes by RNA polymerase I. J Mol Appl Genet. 1982;1(6):575–584. [PubMed] [Google Scholar]
- Long E. O., Rebbert M. L., Dawid I. B. Nucleotide sequence of the initiation site for ribosomal RNA transcription in Drosophila melanogaster: comparison of genes with and without insertions. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1513–1517. doi: 10.1073/pnas.78.3.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley J. L., Fire A., Cano A., Sharp P. A., Gefter M. L. DNA-dependent transcription of adenovirus genes in a soluble whole-cell extract. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3855–3859. doi: 10.1073/pnas.77.7.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall C. J. Synthesis of ribosomal RNA in synkaryons and heterokaryons formed between human and rodent cells. J Cell Sci. 1975 Mar;17(3):307–325. doi: 10.1242/jcs.17.3.307. [DOI] [PubMed] [Google Scholar]
- Matsui T., Segall J., Weil P. A., Roeder R. G. Multiple factors required for accurate initiation of transcription by purified RNA polymerase II. J Biol Chem. 1980 Dec 25;255(24):11992–11996. [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Miesfeld R., Arnheim N. Identification of the in vivo and in vitro origin of transcription in human rDNA. Nucleic Acids Res. 1982 Jul 10;10(13):3933–3949. doi: 10.1093/nar/10.13.3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D. A., Dev V. G., Tantravahi R., Miller O. J. Suppression of human nucleolus organizer activity in mouse-human somatic hybrid cells. Exp Cell Res. 1976 Sep;101(2):235–243. doi: 10.1016/0014-4827(76)90373-6. [DOI] [PubMed] [Google Scholar]
- Miller K. G., Sollner-Webb B. Transcription of mouse rRNA genes by RNA polymerase I: in vitro and in vivo initiation and processing sites. Cell. 1981 Nov;27(1 Pt 2):165–174. doi: 10.1016/0092-8674(81)90370-6. [DOI] [PubMed] [Google Scholar]
- Miller O. J., Dev V. G., Miller D. A., Tantravahi R., Eliceiri G. L. Transcription and processing of both mouse and Syrian hamster ribosomal RNA genes in individual somatic hybrid cells. Exp Cell Res. 1978 Sep;115(2):457–460. doi: 10.1016/0014-4827(78)90309-9. [DOI] [PubMed] [Google Scholar]
- Mishima Y., Financsek I., Kominami R., Muramatsu M. Fractionation and reconstitution of factors required for accurate transcription of mammalian ribosomal RNA genes: identification of a species-dependent initiation factor. Nucleic Acids Res. 1982 Nov 11;10(21):6659–6670. doi: 10.1093/nar/10.21.6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagylaki T., Petes T. D. Intrachromosomal gene conversion and the maintenance of sequence homogeneity among repeated genes. Genetics. 1982 Feb;100(2):315–337. doi: 10.1093/genetics/100.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruddle F. H., Creagan R. P. Parasexual approaches to the genetics of man. Annu Rev Genet. 1975;9:407–486. doi: 10.1146/annurev.ge.09.120175.002203. [DOI] [PubMed] [Google Scholar]
- Sakonju S., Bogenhagen D. F., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: I. The 5' border of the region. Cell. 1980 Jan;19(1):13–25. doi: 10.1016/0092-8674(80)90384-0. [DOI] [PubMed] [Google Scholar]
- Samuels M., Fire A., Sharp P. A. Separation and characterization of factors mediating accurate transcription by RNA polymerase II. J Biol Chem. 1982 Dec 10;257(23):14419–14427. [PubMed] [Google Scholar]
- Schaack J., Sharp S., Dingermann T., Söll D. Transcription of eukaryotic tRNA genes in vitro. II. Formation of stable complexes. J Biol Chem. 1983 Feb 25;258(4):2447–2453. [PubMed] [Google Scholar]
- Sharp S., DeFranco D., Dingermann T., Farrell P., Söll D. Internal control regions for transcription of eukaryotic tRNA genes. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6657–6661. doi: 10.1073/pnas.78.11.6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. P. Evolution of repeated DNA sequences by unequal crossover. Science. 1976 Feb 13;191(4227):528–535. doi: 10.1126/science.1251186. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Reeder R. H. The nucleotide sequence of the initiation and termination sites for ribosomal RNA transcription in X. laevis. Cell. 1979 Oct;18(2):485–499. doi: 10.1016/0092-8674(79)90066-7. [DOI] [PubMed] [Google Scholar]
- Swanson M. E., Holland M. J. RNA polymerase I-dependent selective transcription of yeast ribosomal DNA. Identification of a new cellular ribosomal RNA precursor. J Biol Chem. 1983 Mar 10;258(5):3242–3250. [PubMed] [Google Scholar]
- Tantravahi R., Miller D. A., D'Ancona G., Croce C. M., Miller O. J. Location of rRNA genes in three inbred strains of rat and suppression of rat rRNA activity in rat-human somatic cell hybrids. Exp Cell Res. 1979 Mar 15;119(2):387–392. doi: 10.1016/0014-4827(79)90368-9. [DOI] [PubMed] [Google Scholar]
- Tartof K. D. Redundant genes. Annu Rev Genet. 1975;9:355–385. doi: 10.1146/annurev.ge.09.120175.002035. [DOI] [PubMed] [Google Scholar]
- Tsai S. Y., Tsai M. J., Kops L. E., Minghetti P. P., O'Malley B. W. Transcription factors from oviduct and HeLa cells are similar. J Biol Chem. 1981 Dec 25;256(24):13055–13059. [PubMed] [Google Scholar]
- Tsujimoto Y., Hirose S., Tsuda M., Suzuki Y. Promoter sequence of fibroin gene assigned by in vitro transcription system. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4838–4842. doi: 10.1073/pnas.78.8.4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylyk B., Kédinger C., Corden J., Brison O., Chambon P. Specific in vitro initiation of transcription on conalbumin and ovalbumin genes and comparison with adenovirus-2 early and late genes. Nature. 1980 Jun 5;285(5764):367–373. doi: 10.1038/285367a0. [DOI] [PubMed] [Google Scholar]
- Weil P. A., Segall J., Harris B., Ng S. Y., Roeder R. G. Faithful transcription of eukaryotic genes by RNA polymerase III in systems reconstituted with purified DNA templates. J Biol Chem. 1979 Jul 10;254(13):6163–6173. [PubMed] [Google Scholar]
- Wilkinson J. K., Sollner-Webb B. Transcription of Xenopus ribosomal RNA genes by RNA polymerase I in vitro. J Biol Chem. 1982 Dec 10;257(23):14375–14383. [PubMed] [Google Scholar]