Abstract
In recent years there has been significant growth in the use of patient-specific models to predict the effects of neuromodulation therapies such as deep brain stimulation (DBS). However, translating these models from a research environment to the everyday clinical workflow has been a challenge, primarily due to the complexity of the models and the expertise required in specialized visualization software. In this paper, we deploy the interactive visualization system ImageVis3D Mobile, which has been designed for mobile computing devices such as the iPhone or iPad, in an evaluation environment to visualize models of Parkinson’s disease patients who received DBS therapy. Selection of DBS settings is a significant clinical challenge that requires repeated revisions to achieve optimal therapeutic response, and is often performed without any visual representation of the stimulation system in the patient. We used ImageVis3D Mobile to provide models to movement disorders clinicians and asked them to use the software to determine: 1) which of the four DBS electrode contacts they would select for therapy; and 2) what stimulation settings they would choose. We compared the stimulation protocol chosen from the software versus the stimulation protocol that was chosen via clinical practice (independently of the study). Lastly, we compared the amount of time required to reach these settings using the software versus the time required through standard practice. We found that the stimulation settings chosen using ImageVis3D Mobile were similar to those used in standard of care, but were selected in drastically less time. We show how our visualization system, available directly at the point of care on a device familiar to the clinician, can be used to guide clinical decision making for selection of DBS settings. In our view, the positive impact of the system could also translate to areas other than DBS.
Index Terms: Biomedical and Medical Visualization, Mobile and Ubiquitous Visualization, Computational Model, Clinical Decision Making, Parkinson’s Disease
1 Introduction
Neuromodulation is the alteration of neural activity by means of implanted devices. Most neuromodulation systems consist of a multi-electrode lead that is surgically implanted in the brain, and is connected to a subcutaneous implantable pulse generator (IPG) in the torso. The basic concept behind neuromodulation is that stimulation-induced current flows from electrode(s) through surrounding brain tissue, which in turn causes a therapeutic functional response. One important example of this approach is deep brain stimulation (DBS), which is an established therapy for treating the motor symptoms of Parkinson’s disease (PD) [18], [36], as well as a variety of other disorders [35]. The Medtronic DBS system consists of an electrode lead with four cylindrical contacts (each 1.27 mm diameter, 1.5 mm height, 1.5 mm or 0.5 mm spacing between contacts) and an IPG that delivers continuous stimulation (see Figure 2 ). The electrode location is chosen during surgery based on brain anatomy, imaging data and intra-operative electrophysiology. The stimulation parameters are selected post-operatively, and are titrated to provide good therapeutic benefit with minimal side effects. A persistent problem with neuromodulation techniques such as DBS has been the selection of stimulation settings for optimal response. To achieve this, patients must often undergo lengthy and repeated clinic visits to determine the best settings. A study by Hunka et al. [24] found that the total time spent programming the stimulator and assessing DBS patients ranged from 18–36 hours per patient. Part of the reason for this length of time is the amount of trial and error involved in choosing the best stimulation protocol without any visual guidance on the location of the electrode or the effects of stimulation on nearby brain tissue.
Fig. 2.
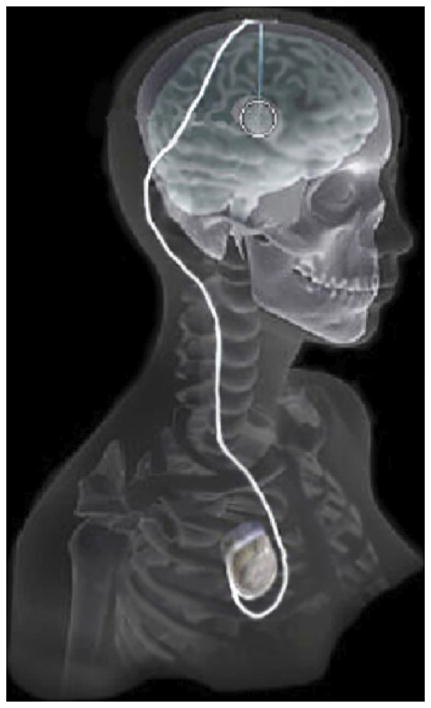
Overview of the DBS system. The DBS electrode is implanted in the brain during stereotactic surgery. The electrode is attached via an extension wire to the IPG, which is implanted in the torso. The entire system is subcutaneous and is designed to deliver continuous stimulation for several years at a time.
Neurologists and neurosurgeons are experts at solving the classic “black box” problem where the practitioner can study the inputs and outputs to a system but cannot see within it. In the application to neuromodulation, the input is the stimulation protocol, the output is the patient response and the “black box” is the patient. This approach has persisted for decades, primarily because the computational tools necessary to visualize the effects of stimulation were not available. While significant progress has been made over the last few years in the sophistication of computational models available for neuromodulation, very few of these have been introduced into clinical practice for several possible reasons: complex software that lacks a simple interface; complex visualizations that are difficult to interpret; and new software is perceived as increasing the demands on clinicians who are often under intense time pressure. As a result, the approaches that tend to persist in the clinical application of neuromodulation are reductionist methods where empirical data are used to select stimulation protocols from the tens of thousands that are available. However, evidence-based practices for this approach have not yet emerged, which is one of the main reasons why selection of stimulation parameters is more of an art than a science.
In this paper we set out to demonstrate how patient-specific models of DBS can be combined into a decision support system that can be easily used at the point of care. We hypothesized that ImageVis3D Mobile would enable clinicians to choose DBS parameters that were comparable to standard of care but in much less time. Furthermore, simplified interfaces common to such platforms would lower the barrier to entry and be more readily accepted. To test this we have chosen to use devices that clinicians are accustomed to in their daily routines: their smartphones. In the current setting we use iPhone class hardware (this includes iPod touch and iPad devices), but the concepts presented here are in no way restricted to this platform. We have implemented a mobile visualization environment called ImageVis3D Mobile. This environment consists not only of interactive volume and geometry rendering implementations, optimized for OpenGL ES 1.1 and OpenGL ES 2.0 mobile devices, but in particular it also contains means of receiving and exploring data, as well as sharing it between devices. Both renderers can coexist, enabling seamless interleaving of volumes and opaque or transparent geometry. To transfer data to a device we utilize techniques from instant messaging systems that allow the end user to view a new dataset with literally a single touch.
We tested our hypothesis by asking five clinicians who have extensive experience with DBS programming to choose stimulation settings using patient-specific computational models of DBS on ImageVis3D Mobile. The feasibility and accuracy of the computational models has been reported in previous studies (see Section 2.1 ). The DBS settings and time required were compared to retrospective data that was gathered during standard clinical care (independent of the study). We show that mobile visualization of patient-specific DBS models have compelling features for clinical decision making.
The remainder of this paper is structured as follows: The next section presents related work. Section 3 then gives an overview of the evaluation and the motivation behind it. In Section 4, we briefly outline the functional details of our visualization environment, especially focusing on the subsystems relevant for the evaluation. Section 5 and Section 6 describe the evaluation in detail and discuss the results achieved. We conclude with predictions based on our results and possible future directions.
2 Related Work
Over the last few years a body of work has emerged on computational methods to predict the effects of neuromodulation therapy. However, it has proven very difficult to create a visualization system that can be integrated into clinical care.
2.1 Computational Models of DBS
Recently, computational models have been developed to predict and visualize the effects of DBS on an individual patient basis [3], [8], [9], [10], [11], [28], [30]. Briefly, finite element models that are derived from patient medical image volumes are used to determine the location of the electrode in the brain, calculate the bioelectric fields produced during stimulation, and predict the neural response to the applied electric field. The primary outcome of this approach is a model-predicted volume of tissue activated (VTA) (Figure 1 ), which is the region of neural tissue that is affected by DBS. These models have been validated by comparing model-predicted outcomes to clinically measured responses in PD patients [4], [5], have been used retrospectively and prospectively to determine how activation of certain anatomical regions is correlated with motor [6], [27] and neuropsychological [7] outcomes in PD, and have been shown to guide clinicians to select stimulation parameters that improve cognitive and motor outcomes [21]. However, two problems persist:
Fig. 1.
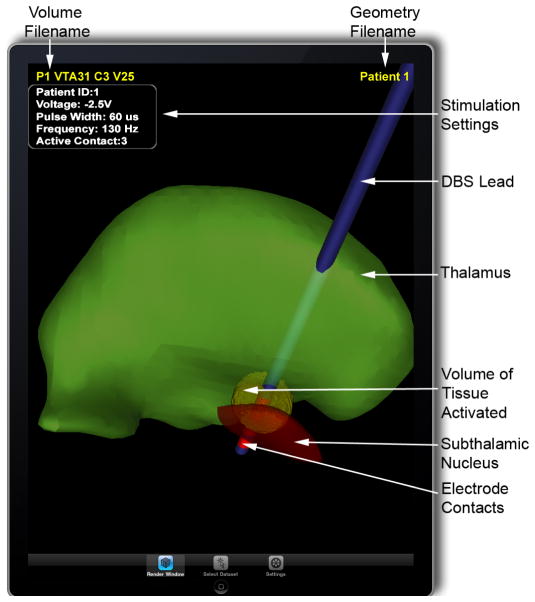
A patient-specific model of deep brain stimulation (DBS) is used to show the location of the electrode lead relative to the surrounding nuclei in a Parkinson’s disease patient. A model-predicted volume of tissue activated (VTA) during DBS (yellow part) is shown surrounding the distal electrode contact. With this model it is possible to view the overlap between the VTA and nearby anatomical structures, which is a key feature in clinical decision making when choosing stimulation settings.
DBS programming is often performed without any visual guidance on the location of the DBS electrodes or the effects of stimulation on surrounding structures.
The software required to perform such visualization can require significant training and is not widely available in a clinical setting.
Hence, there is a need for a simple, intuitive application that can visualize the effects of DBS on an individual basis to facilitate clinical decision making.
2.2 Visualization on Mobile Devices
In the past two decades texture-based volume rendering on graphics hardware has positioned itself as a powerful tool for interactive visual analysis of volumetric datasets. While early volume rendering systems required supercomputers and expensive graphics subsystems, over the years hardware requirements have become more and more relaxed. Nowadays, commodity PCs and even notebooks are sufficient to visualize even extremely large datasets interactively.
In parallel with commodity hardware, mobile devices have caught the attention of the visualization community as another viable and interesting platform. Even before today’s powerful mobile devices were available Encarnação et al. [19] discussed the general issues in using mobile devices to obtain and access data. Later Paelke et al. [33] discussed user interface design aspects for mobile devices. More recently, Chittaro [15] focused on the general issues of visualizing content on mobile devices.
When capable mobile devices became available, Chang and Ger [13] implemented a ray-caster for opaque geometry on PocketPC devices. They argued that the performance of ray-casting and ray-tracing approaches is dominated by the number of pixels, and therefore mobile devices, where hardware capabilities are expected to grow but screen sizes will remain relatively stagnant, are a perfect fit for these approaches. Their system realized a client-server model, whereby desktop systems could be utilized to accelerate rendering on the mobile device. Burigat and Chittaro [2] described a VRML-based system for visualizing what a user sees as they roam a city. Lluch et al. [26] presented a client/server surface rendering system. A server holds a scene graph and uses it, along with client view information, to select an appropriate resolution from a multi-resolution representation on disk. Scene access is done in an out-of-core fashion, allowing very large models to be visualized. Even when rendering is done on the server, for large data a single machine may not be able to provide updates to the mobile device quickly enough for mobile users. For this reason Lamberti and Sanna [25] introduce a Chromium-based [23] rendering system which encodes the data as MPEG and streams it to be decoded on the mobile device. With motion estimation being the most expensive process of MPEG4 encoding, Cheng et al. [14] are able to significantly improve this step by directly retrieving motion vectors from information available in the rendering pipeline. Their algorithm can be implemented on a GPU, further increasing encoding speed.
Most closely related to our rendering subsystem is a contribution by Moser and Weiskopf [31]. In particular our OpenGL ES 1.1 volume renderer is based on their findings. Park et al. [34] developed a system for collaborative medical visualization, using parallel server-based volume rendering techniques. Very recently Meir and Rubinsky [29] investigate the use of mobile devices as a cost-effective component of a distributed system for performing ultrasounds. Their system combines simple-to-use, inexpensive ultrasound devices at the client site, which generate ultrasound data. The data is sent to a server which performs volume rendering at pre-defined camera angles, and sends the images back down to mobile devices for analysis in the field.
3 Overview
In this paper, we evaluate the deployment of our mobile visualization system ImageVis3D Mobile in a real-world environment to support clinical decision making in DBS for PD. Using our system, clinicians were able to make decisions similar to current standard practice but in substantially less time. This improvement was attributable to the use of patient-specific models in the interactive visualization system, but the system also conferred indirect benefits. As we will show, the system provided a framework for comparison of alternate programming strategies employed by different users, which is a significant ongoing subject of debate in the clinical application of DBS. In addition, the system has several attributes which are attractive for clinical workflow.
Under the current standard of care for DBS, patients return to the clinic a few weeks after implantation of the system for their initial programming, which is performed an outpatient setting such as an exam room. Each center and practitioner performs this process slightly differently, but the general trend is to first perform a monopolar review with each electrode contact as the cathode and the IPG case as the anode. Common initial stimulation settings are 60 μsec and 130 Hz, while a range of voltage amplitudes are tested. If satisfactory results are not achieved then more complex stimulation protocols are considered. This process can include substantial trial and error, which is partly attributable to the lack of visualization of the patient anatomy or the effects of stimulation.
In this study we evaluated the accuracy and speed of DBS programming using ImageVis3D Mobile compared to standard of care. To do so we identified four Parkinson’s disease patients who previously received DBS leads implanted in the subthalamic nucleus (STN) and who were good responders to the therapy. We then constructed patient-specific models of DBS and provided them to the clinicians in ImageVis3D Mobile. The clinicians were blinded to the actual identity of the patients, and were asked to use ImageVis3D Mobile to determine the best electrode contact to use for monopolar stimulation, as well as the stimulation amplitude that would provide the best therapeutic benefit with minimal side effects. The values chosen in the study were compared to those used for each patient’s clinical DBS settings, which were determined through standard medical care outside of this study. Lastly, we determined the amount of time necessary to program patients using ImageVis3D Mobile compared to the time required for standard clinical practice, which was estimated using data gathered from the patients’ medical records.
4 Visualization System
ImageVis3D Mobile is a mobile, interactive visualization system for volume and geometry data, implemented for Apple’s iOS software platform. iOS runs on a large number of devices and is the platform of choice for our target users in the evaluation, who are familiar with the user interface and interaction metaphors the platform provides. As the hardware specifications of iPhone, iPod and iPad reflect the design of many other mobile devices, our findings in this evaluation should be applicable to a wide range of mobile hardware.
In the following subsections we outline the main components of ImageVis3D Mobile, the rendering system and the data transfer. The focus is on functionality relevant for the evaluation rather than on technical details. We conclude with a description of the evaluation datasets rendered in ImageVis3D Mobile as seen by the clinicians.
4.1 Rendering
ImageVis3D Mobile provides volume and geometry rendering capabilities, which have been implemented to support both OpenGL ES 1.1 and OpenGL ES 2.0. Due to the lack of support for 3D textures in OpenGL ES, three axis aligned stacks of 2D textures are used to access volumetric data on the GPU as described by Hadwiger et al. [22]. The volume renderer implements manual trilinear filtering and volumetric lighting in OpenGL ES 2.0.
A key feature of ImageVis3D Mobile is furthermore to interleave multiple datasets and render them together. To interleave volumes and geometry, semi-transparent geometry is sorted in back-to-front order in each frame and inserted in-between the volume slices for correct compositing. This feature is required for our evaluation where geometric data of a patient’s nuclei including the placed electrode shaft needs to be overlapped with VTAs which indicate the effects of DBS.
It is worth noting that on touch enabled screens such as our target platform, better frame rates are required than on traditional mouse operated systems. This is due to the fact that the user expects the data to move in sync with their finger, otherwise the fingers and the dataset feel decoupled. Therefore, we provide a number of methods to increase speed during periods of interaction, such as a reduction of the render target resolution, the texture sampling quality, the volume quality, and an option to disable lighting on interaction. In addition to these means in the volume renderer the precision of visibility sorting can be reduced to speed up the geometry rendering as well.
4.2 Data Transfer
ImageVis3D Mobile is the mobile counterpart of the desktop visualization system ImageVis3D, which builds on the out-of-core volume rendering system library Tuvok as described by Fogal and Krüger [20]. To prepare data for rendering on the mobile device we extended Tuvok’s modular IO subsystem with the capability to write out ImageVis3D Mobile data files. This allows our pipeline to accept a number of volume and geometry formats and convert those automatically into ImageVis3D Mobile data. Amongst those formats are SCIRun [17] volumes and geometry in which the input data for this survey is stored.
Transfer of these datasets to the device can be done in several ways. In our particular use case, simple data transfer not requiring any technical expertise is a key requirement. Therefore, transfer via a direct cable link from a data server is not desirable for several reasons. First, it requires direct access to the server, which is most likely located in a computer laboratory away from the point of care. Second, this solution requires expertise on how to connect a device to the server, and then how to use additional software to select and transfer datasets. To avoid these issues, ImageVis3D Mobile uses instant messaging technology coupled with a wireless connection to access data on a server.
Our data distribution system thereby builds on a particular technology of the iOS called Push Notifications. Push Notifications are Apple’s means of being able to send messages to devices without the need to constantly run custom receiver software on the device. Instead, a single daemon runs and distributes instant messages to any application that supports the feature. While Push Notifications are only available on iOS devices, our general concept can be applied to other mobile devices as well, it would just require ImageVis3D Mobile to be run in daemon mode, listening on a network port.
To distribute data, we keep track of all devices and datasets with a central management application, which can be used to select datasets and notify registered devices about their availability. Notifications are initiated by an operator at the server, and the notification process can be automated so certain users automatically receive new datasets attributed to them. Notifications are passed from the server to the Apple Push Notification Service, which delivers them via an accredited and encrypted IP connection to a device directly if it is available or as soon as it comes online [1]. When the notification is delivered, a dialog window appears on the mobile device allowing the user to accept or decline the download of the data. Accepting a download will automatically start ImageVis3D Mobile and initiates the download and display of the data. Note that the whole process requires just a single tap from the end user who is only interested in reviewing the datasets. Should a user decline the download of a new dataset in the notification dialog he can still access it later. For this purpose, ImageVis3D Mobile provides a list menu where all datasets available for this client are listed, tapping one or multiple of those list entries initiates the download of the datasets to the mobile client similar to accepting them in the notification dialog. This way, a user can access all datasets at a later point without requiring another push notification. Also, datasets can be exchanged between devices with a bluetooth connection through a similar list menu. In all cases, the user’s single point of interaction is ImageVis3D Mobile.
4.3 Evaluation Datasets
Once the datasets have been transferred to a device, they are ready for inspection. For our evaluation, data for each patient was divided into a geometric component (see Figure 3a) and a volumetric component (see Figure 3b). The geometric component consists of surface representations of nearby anatomical nuclei (thalamus and subthalamic nucleus, see labels in Figure 4 ), as well as the DBS lead and electrode contacts. We deliberately chose geometrically simple surfaces of common anatomical structures that mimicked the types of atlas representations that physicians are likely to be familiar with. We constructed the anatomical surfaces by coregistering each patient MRI to an atlas brain using a 3D nonlinear warping algorithm [16]. Surfaces for the DBS lead and electrode contacts were constructed using SCIRun [17]. The volumetric component is the VTA. In total, 36 VTAs were provided for each patient (9 for each electrode contact, representing a range of voltages from −1V to −5V, all at 130 Hz, 60 μsec pulse width). While indeed special desktop software is required to produce ImageVis3D Mobile-compatible visualization data from raw input, this process can be automated and is independant from ImageVis3D Mobile’s simplified interface. Figure 4 shows both components interleaved in ImageVis3D Mobile. Annotations were provided to distinguish between patients (the geometric component, top right), as well as to convey the VTA (top left). DBS stimulation settings for the VTA were also provided. Text size can be adapted by the user.
Fig. 3.
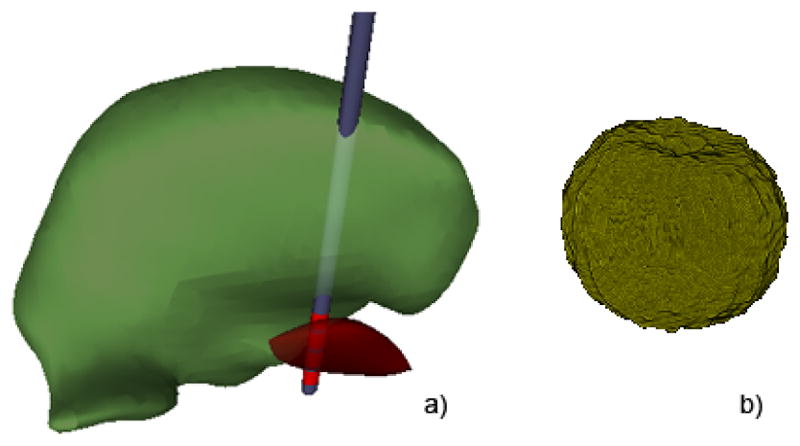
Patient thalamus (green), subthalamic nucleus (red), and DBS lead with four electrode contacts (a). Volume of tissue activated (VTA) (b).
Fig. 4.
Interleaved view in ImageVis3D Mobile. The VTA for −2.5V at contact 3 is shown for a specific patient.
This combination of visualization components was ideally suited to our evaluation for several reasons. First, the use of geometric and volume components allowed us to visualize each in their native format as generated in SCIRun. Second, the text annotations provided details necessary for the users to know which patient and stimulation settings are being evaluated. Third, the overlay of volume and geometry data allowed the user to quickly determine the amount of overlap between the VTA and nearby anatomical structures, which is the feature that most strongly guides their decision making. Lastly, the volumetric format is highly extensible, and should allow us to easily incorporate additional dimensions of information (see Section 7 ).
5 Evaluation
In order to evaluate the utility of ImageVis3D Mobile for clinical decision making, we constructed patient-specific models of four PD patients who were good responders to DBS. Models were created in SCIRun [17] using previously described methods [5] and subsequently transferred to ImageVis3D Mobile. We provided these models to five clinicians (three movement disorders neurologists, one neurosurgeon and one nurse) who have extensive experience with programming DBS systems for PD patients. Each clinician was asked to select DBS parameters using ImageVis3D Mobile on an iPad without knowing the identity of the patients. We then compared their selections to data collected via standard of care, along with the amount of time required. Institutional Review Board (IRB) approval was obtained prior to conducting the study.
5.1 Standard of Care
PD patients who were evaluated in this study received DBS via standard care, independent of our experimental protocol, as follows:
Pre-operative motor and neuropsychological evaluation: based on these evaluations the physicians a) determine if the subject is a good DBS candidate, b) choose an anatomical target for DBS implantation, and c) decide whether the patient should receive unilateral or bilateral DBS leads (one hemisphere or both hemispheres). In this study we examined unilateral PD DBS patients with DBS leads implanted in the subthalamic nucleus (STN).
Pre-operative imaging (magnetic resonance imaging (MRI) and computed tomography (CT)): whole-brain images are acquired to identify anatomical targets (MRI) and to determine the position of the stereotactic frame on the head (CT). The neurosurgeon loads the MRI and CT into a surgical planning system to determine the entry location, trajectory and depth of the DBS lead.
DBS surgery: patients undergo stereotactic surgery to implant the DBS lead. Briefly, a burr hole is drilled in the head and an incision is made in the dura. Microelectrode recordings are acquired to confirm the target location prior to implanting the DBS lead. Both microelectrodes and DBS leads are precisely positioned using the stereotactic frame and the surgical planning system. Once the lead is implanted the patient is stimulated briefly in the operating room to confirm therapeutic response (note that the patient is awake for some portions of the surgery). Following implantation of the DBS lead, the IPG is implanted in the torso and connected to the lead using an extension wire.
Selection of stimulation parameters: Four to six weeks after surgery the IPG is turned on for the first time. The clinician works with the patient to determine the stimulation parameters that provide the best therapeutic response with minimal side effects. This is done through a process of activating each of the four individual electrode contacts and testing a range of stimulation parameters (voltage, pulse width and frequency). This process is usually performed over several visits to the clinic. The patients examined in this study had an average of three to four visits requiring over four hours of time with a clinician to perform DBS programming.
In our study we compared the amount of time required to perform initial programming (step 4 above) to the amount of time required to choose stimulation parameters using IV3D. The time required via standard care was estimated from retrospective chart review. In addition, we compared the stimulation settings chosen using each approach.
5.2 Training
Prior to the experimental protocol, each clinician was trained as follows:
The clinician was informed of the objectives of the study.
They were shown an example dataset in ImageVis3D Mobile on an iPad and the following interactions were demonstrated: rotating, translating and scaling models in the viewer; loading individual patient models of anatomical nuclei and electrode location; overlaying and selecting VTAs.
After the demonstration, the clinician was given the opportunity to have hands-on experience with ImageVis3D Mobile.
The total training time was approximately 10 minutes for each clinician.
5.3 Experimental Protocol
After training, the following experimental protocol was conducted to evaluate ImageVis3D Mobile for DBS parameter selection:
Patient DBS models were announced via push notification and transferred automatically to an iPad running ImageVis3D Mobile.
An individual patient model was loaded in ImageVis3D Mobile. While the patients in our study were previously treated by the clinicians involved in the ImageVis3D Mobile evaluation, patients were anonymized and clinicians were blinded to their identity. No identifying information was provided other than a patient number, and it was not possible to determine the patient identity from the ImageVis3D Mobile visualization.
The clinician was asked to select the most appropriate electrode contact for stimulation based on the location of the DBS electrode relative to nearby anatomical nuclei (thalamus and subthalamic nucleus).
VTAs were loaded for the chosen electrode contact, starting with −1V amplitude. On-screen annotations provided verification of stimulation settings.
The clinician stepped through a range of VTAs from −1V to −5V in 0.5V increments for the chosen electrode contact. From these, the most appropriate voltage value was chosen.
The clinician was allowed to choose a different electrode contact and repeat the previous step if none of the VTAs seemed appropriate.
These steps were timed on a per-patient basis.
6 Results and Discussion
We tested our experimental protocol among five clinicians who examined models for four PD DBS patients. We found that the amount of time required to choose stimulation settings was significantly faster using ImageVis3D Mobile compared to standard clinical care. Selection of stimulation settings required an average of 1.7±0.8 minutes per patient across all clinicians, compared to an average of 4±1.4 hours required for programming via standard of care to reach stable settings with good therapeutic response (usually within three to four clinic visits). In addition, we found that the stimulation settings chosen using ImageVis3D Mobile were very similar to those selected via standard of care. The voltages selected using ImageVis3D Mobile were generally equal to or smaller than the voltages selected using standard care (Table 1 ), and in fact this is a trend that has been observed previously [21]. The active electrode contacts chosen using ImageVis3D Mobile were either the same as or adjacent to the contact chosen using standard care (Table 2 ). Prior studies have noted comparable therapeutic benefit from more than one electrode contact [32]. Hence, we consider this degree of variability to be within the range that is observed clinically.
TABLE 1.
DBS voltages chosen with ImageVis3D Mobile versus standard care.
| Patient ID | Standard Care | IV3DM (Average) |
|---|---|---|
| 1 | 4.1V | 2.35±0.34V |
| 2 | 2.3V | 2.4±0.74V |
| 3 | 2.5V | 2.05±0.76V |
| 4 | 2.2V | 2.0±0.71V |
TABLE 2.
Electrode contact (C) chosen with ImageVis3D Mobile versus standard care.
| Patient ID | Standard Care | Clinician Number | ||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | ||
| 1 | C2 | C2 | C3 | C3 | C3 | C3 |
| 2 | C2 | C2 | C2 | C1 | C1 | C1 |
| 3 | C2 | C1 | C2 | C1 | C1 | C2 |
| 4 | C1 | C2 | C2 | C2 | C1 | C2 |
In addition, feedback on this system from clinicians has been very positive. The user interface is intuitive, especially for existing iPhone users. The ability to interactively visualize patient models provides a level of understanding that is not currently available. This is perceived as a welcome alternative to the current process, and clinicians who have used this system are optimistic about its long-range potential to provide the optimal DBS therapy more rapidly than previously possible. Hence, the salient features of this for clinical decision making are the abilities to: easily retrieve data; view the DBS electrode location relative to surrounding anatomy on an individual patient basis at the point of care; view how the DBS-induced VTAs overlap with nearby anatomical structures; interact with the visualization using an intuitive touch screen interface.
In this study the clinicians were not provided with any information on how VTAs should be selected relative to their overlap with surrounding anatomical structures. In fact, the verbal feedback they provided during the experiment indicated slightly different approaches to parameter selection: three of them tried to maximize VTA overlap with the STN; one chose VTAs that were superior to the STN; two tried to avoid VTA overlap with thalamus as much as possible. This reflects ongoing discussion in the DBS community about optimal target locations for stimulation, and we feel that this accounts for some of the variance in our results. Hence, even with detailed visualization of patient-specific data, there is not currently a consensus on the best stimulation target for PD patients.
6.1 Interpretation and Potential Influence on Clinical Workflow
Our results showed a dramatic decrease in time required to select stimulation setting using ImageVis3D Mobile compared to standard care. However, an important question remains: what is responsible for the observed time difference? There are many possible explanations that may not be attributable to ImageVis3D Mobile. First, during standard care patients often receive a brief motor exam after each change in DBS parameters. This was not possible in our study design because the clinicians were blinded to the patients’ identities, and because one purpose of our study was to evaluate the utility of the software without motor evaluation. Second, while our study focused on selection of DBS voltage alone (pulse width and frequency were fixed), clinicians will sometimes explore these variables during initial programming. Current guidelines suggest that good response to DBS can be achieved with pulse widths ranging from 60 μsec to 210 μsec and frequencies from 130 to 185 Hz. Hence, the parameter space that is explored during standard care is somewhat larger than the range that we tested. Third, each patient model required between 30 and 60 minutes of preparation by a trained technician prior to transferring the data to the mobile device, though we anticipate that this amount of time could be reduced in the future by creating a semi-automated system for model generation. Despite the differences in these two approaches and the difficulties of making direct comparisons, we believe that these factors cannot completely account for the large effect size we observed (a 99.5% reduction in the amount of time required to choose DBS parameters). In addition, it is important to recognize that our approach could facilitate a fundamentally different clinical workflow. Specifically, the use of ImageVis3D Mobile and patient-specific DBS models could allow the clinicians to quickly converge on a small range of parameters that are likely to provide good therapeutic response. From these initial settings we anticipate that the clinician will evaluate motor outcomes while exploring nearby settings. Hence, instead of performing a comprehensive review of motor outcomes at a wide range of stimulation settings for all DBS contacts, clinicians could focus their effort on a much smaller parameter space prior to beginning motor exams.
The ability to conduct these experiments on mobile computing devices is a subtle but important feature. First, as indicated earlier, clinicians became proficient at using ImageVis3D Mobile for DBS parameter selection in very little time. We believe this is a reflection of the simplified interface to transfer, select and interact with datasets and the representation of information such as electrodes, anatomical nuclei and VTAs in a familiar manner. While we did not compare ImageVis3D Mobile to an equivalent desktop-based system, we anticipate that the latter would require clinicians to spend substantially more time to become used to the interface and access their datasets for review. Second the use of mobile devices with wireless data delivery is far more convenient for clinical workflow, and does not require clinicians to rely on desktop computers which might not be available directly at the point of care. In particular, significant attention has been paid recently to the role of mobile computing devices in a clinical environment for this very reason. Hence, we believe that our implementation could be a welcome addition to a healthcare delivery system that is attempting to reduce reliance on desktop based architectures. However, our current system is in a prototype state and requires further testing before introducing it into a clinical environment. This is especially true for the data distribution sub-systems when considering issues issues authentication and protected health information.
6.2 Insights into Visualization Applications
We believe that some of the developments we made in this study are of much broader interest. In particular:
Use of Instant Messaging for Data Distribution Most of the proposed systems focus mainly on the renderer and present efficient means of visualizing data as fast as possible at maximum quality. While this is a very important characteristic of a visualization environment, the system becomes useless if the intended users are unable to transfer their data onto the device. We hide the hardware and software details of the data transfer process. This is achieved through the combination of wireless networks and instant messaging technology.
Natural Multi-Touch Interfaces While multi-touch technology dates back to the early eighties [12], only very recently, with the introduction of the iPhone, have such devices become popular. In a short period multi-touch hardware has become available for almost any type of hardware (e.g. large display systems, workstations, mobile devices). While we are certainly not the first to point out this fact, we believe that in particular visualization applications can benefit significantly from the integration of these interaction metaphors.
User Familiarity We believe that it is a widely underestimated fact that users are more effective with devices they know well. As most people spend quite a decent amount of time per day using their smartphones, it seems only natural to use them for as many tasks as possible, even if this means working with suboptimal hardware environments. Interestingly, in this work we found that clinicians are more than willing to ignore the disadvantages of the small display in favor of working with their own well-known handheld devices.
6.3 Limitations
One goal of this study was to evaluate the potential of patient-specific DBS models and ImageVis3D Mobile to reduce the burden for both patients and clinicians in the selection of DBS parameters, and in this regard the results are very promising. However, our approach has a few limitations that should be taken into consideration. First, this was a retrospective study and therefore we did not test the stimulation settings chosen using ImageVis3D Mobile in each patient. Hence, it is possible that the chosen settings could be better than, equal to or worse than the settings chosen via standard of care. Lastly, the VTAs available within ImageVis3D Mobile did not allow exploration of all parameters available to the clinician. In particular the clinicians could change stimulation amplitude (voltage) while pulse width and frequency were fixed at common values. However, this is not a limitation of our approach but was rather a deliberate simplification of the study design to focus on its primary advantage over existing methods: it could drastically decrease the amount of time required for DBS programming by providing a good starting point. Hence, rather than testing a range of stimulation settings at all electrode contacts, the settings chosen using ImageVis3D Mobile could be used for initial clinical evaluation and then titrated as needed. In a future study we plan to assess the efficacy of model-selected parameters, and benefits are conferred to the patient as a result.
7 Conclusions & Future Directions
We anticipate that this system could provide a significant step forward in clinical practice for several reasons: mobile computing platforms such as the iPhone are widely used by physicians, and new hardware devices such as the iPad have generated significant interest in the clinical community; computational models are gaining acceptance by practitioners, and are being used more often for clinical decision making; the system described here has a simple, intuitive interface and can be used at the patient bedside. A final advantage of this approach is more subtle. In the course of the experiment we realized that the interactive visualization provided a structure for comparison of different approaches to DBS programming. One persistent problem in neuromodulation is that the vocabulary for describing target locations is somewhat imprecise, and alternate programming approaches are employed by different practitioners. To put this in perspective, consider the fact STN DBS does not refer strictly to the STN but rather to the region around the STN encompassing a volume of approximately 1000mm3. In comparison, a DBS electrode contact is approximately 6mm3, and a VTA at typical DBS settings is approximately 250mm3. Hence, there is the possibility for substantial variation in the exact position of the VTA in the STN region, and the interactive visualization provided using this approach allows for a structured comparison despite imprecise vocabulary. We feel that this is a significant indirect benefit of the system.
Even though feedback has been positive, one area of future work is to assess whether use of this system improves patient outcomes. We plan to test this by prospectively comparing neuropsychological and motor outcomes from DBS using this system versus the current standard of care. In doing so, we will take advantage of the extensibility of the volumetric visualization in ImageVis3D Mobile to add new dimensions of information for decision making. In a future study we will include volumetric target locations from a probabilistic atlas to determine whether the inclusion of evidence gathered from other patients results in further improvements in the selection of DBS parameters. Recent work has begun to develop methods to define optimal stimulation targets from retrospective and prospective multi-patient clinical studies [6], [27]. By including both classes of information the clinician can first see the interaction of the stimulation system with surrounding anatomical nuclei, and secondly see the interaction with target regions where stimulation has therapeutic effects or side effects as defined by probabilistic atlases that are compiled from multi-patient studies.
While previous attempts have been made to provide interactive visualization of patient-specific DBS models, these require significant amounts of training and domain knowledge to become proficient. An advantage of the system described here is the minimal amount of training required and its attractive features for clinical workflow. We predict that this approach could have significant impact not only in DBS for PD but also in other neuromodulation methods where interactive patient-specific models could provide useful insights into the best way to prescribe the therapy. We conclude that the use of patient-specific models of DBS in a mobile computing device running ImageVis3D Mobile has strong potential to improve patient outcomes by facilitating clinical decision making.
Acknowledgments
This research was made possible in part by an Advancing a Healthier Wisconsin grant at the Medical College of Wisconsin, by the Intel Visual Computing Institute, by the Cluster of Excellence “Multimodal Computing and Interaction” at the Saarland University, by the NIH/NCRR Center for Integrative Biomedical Computing, P41-RR12553-10 and by Award Number R01EB007688 from the National Institute Of Biomedical Imaging And Bioengineering, and by the Office of Advanced Scientific Computing Research, Office of Science, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231 through the Scientific Discovery through Advanced Computing (SciDAC) program’s Visualization and Analytics Center for Enabling Technology. The content is under sole responsibility of the authors.
Biographies

Christopher R. Butson Christopher Butson received a B.S. in Mechanical Engineering from the University of Maryland, M.S. in Electrical Engineering from George Washington University and Ph.D. in Biomedical Engineering from the University of Utah. He completed post-doctoral training at the Cleveland Clinic and is now an Associate Professor in the Departments of Neurology & Neurosurgery at the Medical College of Wisconsin, as well as an Adjunct Professor in the Department of Biomedical Engineering at Marquette University. He is an active member of the Society for Neuroscience (SFN), the Institute of Electrical & Electronics Engineers (IEEE) and the Engineering in Medicine & Biology Society (EMBS). He is currently leading a research laboratory which focuses on neuromodulation, specifically therapeutic and diagnostic brain stimulation.

Georg Tamm Georg Tamm studied media and communication computer science at Reutlingen University and graduated with a masters degree in 2009. In addition, he studied games software development at Sheffield Hallam University in England and finished with a masters degree in 2008. Current research topics at the Interactive Visualization and Data Analysis group in Saarbrücken focus on scalable, interactive rendering of large data sets.

Sanket Jain Sanket Jain studied biomedical engineering at Thadomal Shahani Engineering College (Mumbai), earning his bachelors in 2006. Since completing his masters degree in Biomedical Engineering at Marquette University in 2009 he has been working at Medical College of Wisconsin with Dr. Christopher Butson in the areas of Deep Brain Stimulation and Cortical Stimulation.

Thomas Fogal studied computer science at the University of New Hampshire, earning his bachelors in 2006 and his masters in 2011. In 2008, he joined the Scientific Computing and Imaging Institute, where he continues to work on volume rendering tools and parallel visualization and analysis platforms.

Jens Krüger studied computer science at the Rheinisch-Westfälische Technische Hochschule Aachen where he received his diploma in 2002. In 2006 he finished his PhD in 2006 at the Technische Universität München and after Post Doc positions in Munich and at the Scientific Computing and Imaging (SCI) Institute he joined the Cluster of Excellence in 2009 to head the Interactive Visualization and Data Analysis group. In addition to his position within the Cluster, Jens Krüger also holds an adjunct faculty title of the University of Utah and is a principal investigator of multiple projects in the Intel Visual Computing Institute.
Contributor Information
Christopher R. Butson, Email: cbutson@mcw.edu, Departments of Neurology and Neurosurgery at the Medical College of Wisconsin.
Georg Tamm, IVDA and DFKI.
Sanket Jain, Department of Neurology at the Medical College of Wisconsin.
Thomas Fogal, SCI.
Jens Krüger, IVDA, DFKI, Intel VCI, and SCI.
References
- 1.Apple. Apple Push Notification Service; 2010. http://tinyurl.com/ApplePushNotification. [Google Scholar]
- 2.Burigat S, Chittaro L. Location-aware visualization of vrml models in gps-based mobile guides. Web3D ’05: Proceedings of the tenth international conference on 3D Web technology; New York, NY, USA. ACM; 2005. pp. 57–64. http://doi.acm.org/10.1145/1050491.1050499. [Google Scholar]
- 3.Butson C, McIntyre CC. Role of electrode design on the volume of tissue activated during deep brain stimulation. J Neural Eng. 2006;3(1):1–8. doi: 10.1088/1741-2560/3/1/001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butson CR, Cooper SE, Henderson JM, McIntyre CC. Predicting the effects of deep brain stimulation with diffusion tensor based electric field models. Med Image Comput Comput Assist Interv. 2006;9(Pt 2):29–437. doi: 10.1007/11866763_53. [DOI] [PubMed] [Google Scholar]
- 5.Butson CR, Cooper SE, Henderson JM, McIntyre CC. Patient-specific analysis of the volume of tissue activated during deep brain stimulation. NeuroImage. 2007;34(2):661–70. doi: 10.1016/j.neuroimage.2006.09.034. http://linkinghub.elsevier.com/retrieve/pii/S1053811906009669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butson CR, Cooper SE, Henderson JM, Wolgamuth B, McIntyre CC. Probabilistic analysis of activation volumes generated during deep brain stimulation. NeuroImage. 2011;54:2096–2104. doi: 10.1016/j.neuroimage.2010.10.059. http://www.ncbi.nlm.nih.gov/pubmed/20974269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butson CR, Driesslein K, Hung S, Matthews M, Sheridan CJ, Kopell BH, Bobholz J. Probabilistic atlas of neuropsychological outcomes from subthalamic nucleus deep brain stimulation. Movement Disorders Society Congress; 2010. [Google Scholar]
- 8.Butson CR, Maks CB, McIntyre CC. Sources and effects electrode impedance during deep brain stimulation. Clin Neurophysiol. 2006;117(2):447–54. doi: 10.1016/j.clinph.2005.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Butson CR, McIntyre CC. Tissue and electrode capacitance reduce neural activation volumes during deep brain stimulation. Clin Neurophysiol. 2005;116(10):2490–500. 1388–2457. doi: 10.1016/j.clinph.2005.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butson CR, McIntyre CC. Differences among implanted pulse generator waveforms cause variations in the neural response to deep brain stimulation. Clin Neurophysiol. 2007;118(8):1889–94. doi: 10.1016/j.clinph.2007.05.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butson CR, McIntyre CC. Current steering to control the volume of tissue activated during deep brain stimulation. Brain Stimulation. 2008;1(1):7–15. doi: 10.1016/j.brs.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buxton B. Multi-Touch Systems that I Have Known and Loved. 2007 Jan; http://www.billbuxton.com/multitouchOverview.html.
- 13.Chang C, Ger S. Enhancing 3D graphics on mobile devices by image-based rendering. PCM í02: Proceedings of the Third IEEE Pacific Rim Conference on Multimedia; Springer-Verlag; 2002. pp. 1105–1111. http://www.cs.nthu.edu.tw/~chunfa/pcm2002.pdf. [Google Scholar]
- 14.Cheng L, Bhushan A, Pajarola R, El Zarki M. Real-time 3d graphics streaming using mpeg-4. Proceedings IEEE/ACM Workshop on Broadband Wireless Services and Applications; 2004. http://www.broadnets.org/2004/workshop-papers/Broadwise/ChengL.pdf. [Google Scholar]
- 15.Chittaro L. Visualizing information on mobile devices. Computer. 2006;39(3):40–45. http://dx.doi.org/10.1109/MC.2006.109. [Google Scholar]
- 16.Christensen GE, Joshi SC, Miller MI. Volumetric transformation of brain anatomy. IEEE Trans Med Imaging. 1997;16(6):864–77. doi: 10.1109/42.650882. [DOI] [PubMed] [Google Scholar]
- 17.S. Computing and I. I. (SCI) SCIRun: A scientific computing problem solving environment. http://www.scirun.org.
- 18.Deuschl G, Schade-Brittinger C, Krack P, Volkmann J, Schafer H, Botzel K, Daniels C, Deutschlander A, Dillmann U, Eisner W, Gruber D, Hamel W, Herzog J, Hilker R, Klebe S, Kloss M, Koy J, Krause M, Kupsch A, Lorenz D, Lorenzl S, Mehdorn HM, Moringlane JR, Oertel W, Pinsker MO, Reichmann H, Reuss A, Schneider GH, Schnitzler A, Steude U, Sturm V, Timmermann L, Tronnier V, Trottenberg T, Wojtecki L, Wolf E, Poewe W, Voges J. A randomized trial of deep-brain stimulation for parkinson’s disease. N Engl J Med. 2006;355(9):896–908. doi: 10.1056/NEJMoa060281. http://www.ncbi.nlm.nih.gov/pubmed/16943402. [DOI] [PubMed] [Google Scholar]
- 19.Encarnação JL, Frühauf M, Kirste T. Mobile visualization: Challenges and solution concepts. Proc. CAPE’95; 1995. http://citeseer.ist.psu.edu/159692.html. [Google Scholar]
- 20.Fogal T, Krüger J. Tuvok - an architecture for large scale volume rendering. Proceedings of the 15th Vision, Modeling and Visualization Workshop; 2010; 2010. http://ivda.cs.uni-saarland.de/fileadmin/publications/2010/VMV2010a.pdf. [Google Scholar]
- 21.Frankemolle AM, Wu J, Noecker AM, Voelcker-Rehage C, Ho JC, Vitek JL, McIntyre CC, Alberts JL. Reversing cognitive-motor impairments in parkinson’s disease patients using a computational modelling approach to deep brain stimulation programming. Brain. 2010;133(Pt 3):746–61. doi: 10.1093/brain/awp315. http://brain.oxfordjournals.org/cgi/content/abstract/awp315v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hadwiger M, Kniss JM, Rezk-Salama C, Weiskopf D, Engel K. Real-time Volume Graphics. A. K. Peters, Ltd; 2006. http://www.real-time-volume-graphics.org/ [Google Scholar]
- 23.Humphreys G, Houston M, Ng R, Frank R, Ahern S, Kirchner PD, Klosowski JT. Chromium: a stream-processing framework for interactive rendering on clusters. ACM Trans Graph. 2002;21(3):693–702. http://doi.acm.org/10.1145/566654.566639. [Google Scholar]
- 24.Hunka K, Suchowersky O, Wood S, Derwent L, Kiss ZH. Nursing time to program and assess deep brain stimulators in movement disorder patients. Journal of Neuroscience Nursing. 2005;37(4):204–10. doi: 10.1097/01376517-200508000-00006. http://journals.lww.com/jnnonline/Abstract/2005/08000/NursingTimetoProgramandAssessDeepBrain.6.aspx. [DOI] [PubMed] [Google Scholar]
- 25.Lamberti F, Sanna A. A Streaming-Based Solution for Remote Visualization of 3D Graphics on Mobile Devices. IEEE Transactions on Visualization and Computer Graphics. 2007;13(2):247–260. doi: 10.1109/TVCG.2007.29. http://dx.doi.org/10.1109/TVCG.2007.29. [DOI] [PubMed] [Google Scholar]
- 26.Lluch J, Gaitán R, Camahort E, Vivó R. Interactive three-dimensional rendering on mobile computer devices. ACE ’05: Proceedings of the 2005 ACM SIGCHI International Conference on Advances in computer entertainment technology; New York, NY, USA. ACM; 2005. pp. 254–257. http://doi.acm.org/10.1145/1178477.1178520. [Google Scholar]
- 27.Maks CB, Butson CR, Walter BL, Vitek JL, McIntyre CC. Deep brain stimulation activation volumes and their association with neurophysiological mapping and therapeutic outcomes. J Neurol Neurosurg Psychiatry. 2009 doi: 10.1136/jnnp.2007.126219. (In Press) http://www.ncbi.nlm.nih.gov/pubmed/18403440. [DOI] [PMC free article] [PubMed]
- 28.McIntyre CC, Mori S, Sherman DL, Thakor NV, Vitek JL. Electric field and stimulating influence generated by deep brain stimulation of the subthalamic nucleus. Clin Neurophysiol. 2004;115(3):589–95. doi: 10.1016/j.clinph.2003.10.033. [DOI] [PubMed] [Google Scholar]
- 29.Meir A, Rubinsky B. Distributed network, wireless and cloud computing enabled 3-D ultrasound; a new medical technology paradigm. PLoS ONE. 2009;4(11):e7974. doi: 10.1371/journal.pone.0007974. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2775631/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miocinovic S, Maks CB, Noecker AM, Butson CR, McIntyre CC. Cicerone: Deep brain stimulation neurosurgical navigation software system. Acta Neurochir Suppl (Wien) 2007;97:561–567. doi: 10.1007/978-3-211-33081-4_65. [DOI] [PubMed] [Google Scholar]
- 31.Moser M, Weiskopf D. Interactive Volume Rendering on Mobile Devices. Workshop on Vision, Modelling, and Visualization VMV ’08; 2008. pp. 217–226. http://www.vis.uni-stuttgart.de/~weiskopf/publications/vmv08.pdf. [Google Scholar]
- 32.Okun MS, Fernandez HH, Wu SS, Kirsch-Darrow, BL, BD, Suelter FM, Jacobson CE, Wang X, Gordon CW, Zeilman P, Romrell J, Martin P, Ward H, Rodriguez RL, Foote KD. Cognition and mood in parkinson’s disease in subthalamic nucleus versus globus pallidus interna deep brain stimulation: the COMPARE trial. Annals of Neurology. 2009;65:586–95. doi: 10.1002/ana.21596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paelke V, Reimann C, Rosenbach W. A visualization design repository for mobile devices. AFRIGRAPH ’03: Proceedings of the 2nd international conference on Computer graphics, virtual Reality, visualisation and interaction in Africa; New York, NY, USA. ACM; 2003. pp. 57–62. http://doi.acm.org/10.1145/602330.602341. [Google Scholar]
- 34.Park S, Kim W, Ihm I. Mobile collaborative medical display system. Computer Methods and Programs in Biomedicine. 2008;89(3):248–260. doi: 10.1016/j.cmpb.2007.11.012. http://dx.doi.org/10.1016/j.cmpb.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 35.Schwalb JM, Hamani C. The history and future of deep brain stimulation. Neurotherapeutics. 2008;5(1):3–13. doi: 10.1016/j.nurt.2007.11.003. http://www.journals.elsevierhealth.com/periodicals/nurt/article/S1933-7213(07)00263-2/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weaver FM, Follett K, Stern M, Hur K, Harris C, Marks J, WJ, Rothlind J, Sagher O, Reda D, Moy CS, Pahwa R, Burchiel K, Hogarth P, Lai EC, Duda JE, Holloway K, Samii A, Horn S, Bronstein J, Stoner G, Heemskerk J, Huang GD. Bilateral deep brain stimulation vs best medical therapy for patients with advanced parkinson disease: a randomized controlled trial. JAMA. 2009;301(1):63–73. doi: 10.1001/jama.2008.929. http://www.ncbi.nlm.nih.gov/pubmed/19126811. [DOI] [PMC free article] [PubMed] [Google Scholar]