Abstract
High frequency of cytidine to thymidine conversions were identified in the genome of several types of cancer cells. In breast cancer cells these mutations are clustered in long DNA regions associated with ssDNA, double-strand DNA breaks (DSBs) and genomic rearrangements. The observed mutational pattern resembles the deamination signature of cytidine to uridine carried out by members of the APOBEC3 family of cellular deaminases. Consistently, APOBEC3B (A3B) was recently identified as the mutational source in breast cancer cells. A3G is another member of the cytidine deaminases family predominantly expressed in lymphoma cells, where it is involved in mutational DSB repair following ionizing radiation treatments. This activity provides us with a new paradigm for cancer cell survival and tumor promotion and a mechanistic link between ssDNA, DSBs and clustered mutations.
The seven human APOBEC3 proteins and their homologs in other mammals are prominent members of a multifunctional family of Zn2+-dependent polynucleotide cytosine deaminases [recently reviewed by (1 and 2–4)]. APOBEC3 proteins elicit DNA cytosine deaminase activity and show broad retrotransposon and retrovirus restriction activities. Two main groups of APOBEC3 proteins can be identified based on their structure: A3A, A3C and A3H have a single zinc-finger domain, whereas A3B, A3D, A3G and A3F bear two zinc-finger domains (5, 6). These structural characteristics may underlie some of the observed differences in protein functionality. In addition to the APOBEC3 deaminases, the APOBEC family includes APOBEC1 and activation-induced cytidine deaminase (AID), as well as the less characterized proteins APOBEC2 and APOBEC4. APOBEC1 is an mRNA editor involved in the metabolism of apolipoprotein B (APOB) in gastrointestinal cells. By editing APOB mRNA residue C666, APOBEC1 generates an early stop codon, resulting in synthesis of a shorter polypeptide. AID is a B-cell-specific DNA deaminase involved in antibody diversification by editing immunoglobulin-gene cytosines to trigger somatic hypermutation and class switch recombination.
In the last decade, APOBEC3 proteins were identified as potent mutators of viral DNA (1). Virtually all APOBEC3 members are involved in hypermutation of viral genomes that replicate via synthesis of ssDNA intermediates, including retroviruses, hepatitis B virus and human papilloma virus. However, the physiological functions of APOBEC3 cytosine deaminases may include roles beyond their involvement in innate immunity. APOBEC3G (A3G) was shown to restrict replication of endogenous retroelements and human endogenous retroviruses (HERV), thus contributing to maintenance of genomic stability (7, 8). In contrast, A3A was implicated in several genome destabilizing activities, including insertion of somatic mutations in human mitochondrial and nuclear DNA (9, 10), demethylation of 5-methylcytosine (11) and induction of DNA breaks in a deaminase-dependent manner (9). In addition A3A is involved in clearance of foreign dsDNA from cells (12). Hence, cellular APOBEC3 deaminases act as potent innate antiviral restriction factors, but may also have diverse effects on genomic regulation and stability (13).
The potentially deleterious mutagenic activity of APOBEC proteins is likely to be highly regulated, as exemplified by ectopic expression of AID by various cytokines leading to carcinogenic mutagenesis of critical cancer-related genes (14–16). However, a similar cancer promoting role for APOBEC3s has only recently emerged. Nik-Zainal and colleagues (17) performed whole genome sequencing of breast cancer cells and non-malignant tissues derived from 21 patients. Analysis of somatic mutation patterns in breast cancer cells revealed that genomes of cancer cells contain regions of hypermutation named “Kataegis”. Base substitutions in these regions were almost exclusively of cytosine at TpC dinucleotides. Remarkably, mutations were closely associated with regions of chromosomal rearrangements and occurred on the same chromosomal strand over long genomic distances, suggesting that they occurred simultaneously or in a progressive manner over a short time span (17). Moreover, short tandem repeats or short stretches of identical sequences at the breakpoints (termed overlapping microhomology) flanked each insertion/deletion (indel) mutation. Repeat-mediated indels were small (1–5 bp), while microhomology-associated indels were mainly larger deletions (up to 50 bp).
These findings are strongly supported by the mutational signature in yeast cells proliferating under chronic DNA-damaging conditions and in other human malignant tumors (18). In both these highly divergent biological systems, mutations were clustered in long ssDNA regions associated with double-strand DNA breaks (DSBs) and genomic rearrangements. In agreement with Nik-Zanihal et al. (17, 18), Roberts and colleagues (18) claimed that “once a mutagen is present (methyl-methane-sulfonate (MMS) in yeast or potentially APOBEC in cancers), the limiting factor in cluster formation appears to be the formation of ssDNA, where the length of ssDNA region and the time it persists are the key parameters determining the cluster’s mutation density and length”. Both reports elicited the signature of the predominant mutations assigned for APOBEC3 deamination activity. A solid support to this assumption was recently provided by Burns and colleagues (19), identifying A3B as a mutational source in breast cancer cells.
Concurrently with the above-mentioned reports, we described a novel role for A3G in promoting DNA double-strand break (DSB) repair in lymphoma cells, which is ssDNA cytidine deamination-dependent (20). The biochemical characteristics of A3G provide the mechanistic platform for the genetic alterations observed in yeast and cancer cells: i. A3G performs cytidine deamination on ssDNA and it is able to interact with the extreme cytosine of the ssDNA 3’-terminus; ii. A3G undergoes intersegmental transfer on ssDNA, a translocation mechanism involving simultaneous binding of two long or short ssDNA segments, directing dispersed hypermutation of cytidine targets. This intersegmental mechanism could be responsible for the appearance of expanded regions of hypermutation, referred to as clusters or “Kataegis”; iii. A3G juxtaposes two 3’ ssDNA termini with minimal terminal microhomology; iv. A3G mixed with HeLa whole cell extracts efficiently supports the end joining of linearized plasmids. Association of A3G with ssDNA ends, together with its ability to mediate interstrand synapsis, led us to speculate that A3G may promote ssDNA end joining. Hence, the formation of end synapses and overlapping microhomology, as well as chromosomal translocations described in yeast cells, breast cancer and lymphomas (17, 18),(21), could be caused by these APOBEC protein activities.
A3G was recently shown to activate ataxia-telangiectasia mutated (ATM) DNA damage checkpoint kinase in HIV-1 infected cells containing deaminated viral DNA (22). High expression of A3G in B cells of patients with diffuse large B-cell lymphoma treated with anthracycline-containing chemotherapy was associated with poor survival (23), suggesting that A3G may promote DNA repair. Several types of cancer cells, such as lymphoma and myeloma cells, display efficient repair of genomic DSBs induced by ionizing radiation (IR) and enhanced cell survival after IR treatment. Interestingly, these cells were found to express high levels of A3G in comparison to their related normal cells. In response to IR, A3G accumulates transiently in the nuclei of these cells and is recruited to DSB repair foci. Consistent with a direct role in DSB repair, inhibition of A3G expression, or its deaminase activity, resulted in reduced DSB repair, whereas reconstitution of A3G expression in A3G-deficient leukemia cells enhanced DSB repair. To confirm the hypothesis that A3G activity is involved in repairing DSBs, an integrated reporter cassette containing an ISceI restriction site was used. Activation of ISceI restriction enzyme in A3G-deficient cells induces a DSB which is repaired mainly by non-homologous end joining (NHEJ), and approximately 10% of DSBs are repaired by homologous recombination (HR) (24). In A3G-expressing cells, the number of DSBs repaired by HR mechanisms was reduced, while mutagenic end-joining increased. These results identified A3G as a component, which promotes DSB repair by NHEJ or microhomology-mediated end joining (MMEJ) (20).
A3G deaminates a CpC target motif (in which the underlined C is preferentially deaminated), whereas the other APOBEC3 enzymes preferentially use TpC ssDNA as a substrate (25, 26). Although the APOBEC3 signature described in breast cancer cells points to APOBEC3 activities distinguished from those of A3G, we speculate that other members of the APOBEC family, predominantly those with two zinc-finger domains (A3B, A3D, A3G and A3F) generally act as cell-specific pro-survival factors. These APOBEC3 proteins may rescue cells from the frequently occurring and highly genotoxic DSB lesions in genomic DNA, via the mechanism we propose for A3G. For instance, A3B is over-expressed in breast cancer cells, and is presumably responsible for many of the C-to-T somatic mutations in these tumor cells (19). Recruitment of APOBEC3 enzymes to the DSB site to participate in DSB repair (see below) could in turn insert mutation via deamination. Indeed, two reports demonstrate association between genomic DNA rearrangements and hypermutation regions of “Kataegis” (17, 18). The fact that the APOBEC3 genes have been selected toward multiplication along evolution (primates/humans encode AID, A1, A2, A4 and seven APOBEC3 proteins, four of which have two zinc fingers, while fish and birds express only AID and APOBEC2 (26)), justifies the multiplication and maintenance of promiscuous and active deaminases in primates despite the potential threat of mutational insertion.
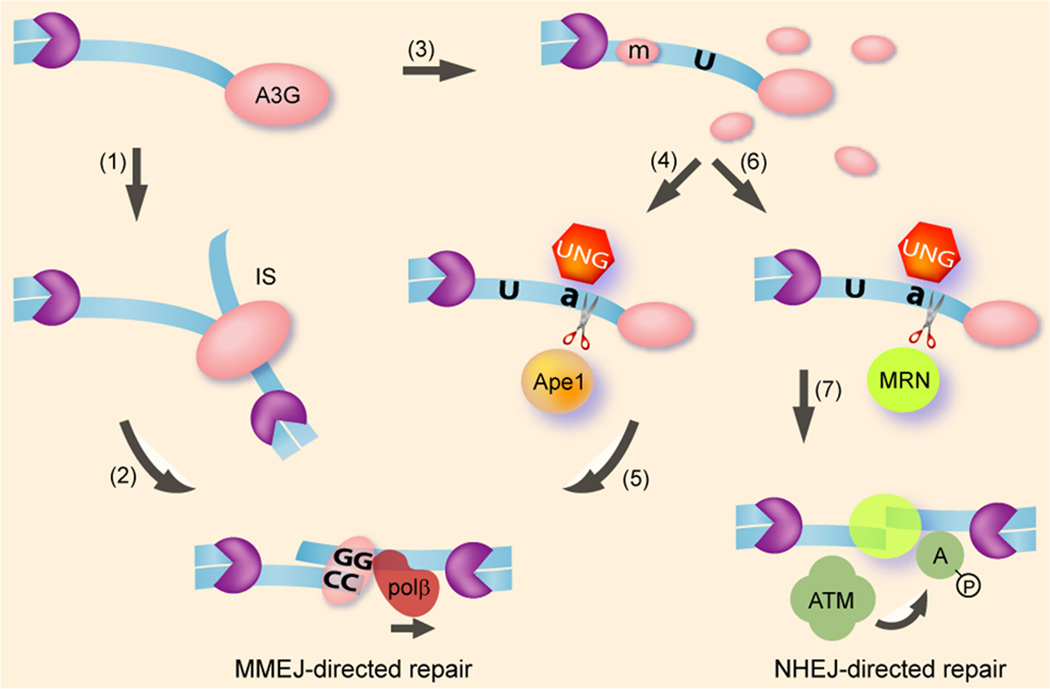
Figure 1 depicts a model which may provide the mechanistic framework for the function of APOBEC3 proteins in the setting of genomic DNA breaks in human cells. Moreover, the results described by Nowarski, Burns and their co-workers (19, 20) and the model described here may shed light on the mechanism of somatic mutations in breast cancer cells (17, 18). The cell-cycle phase and the nature of the DSB lesions determine the DSB repair pathway (27, 28). NHEJ and microhomology-mediated end joining (MMEJ) repair DSBs occurring in all cell-cycle phases and represent the major pathways in G1, while HR functions mainly in G2/S phases. Simple DSBs may be repaired by direct ligation via the NHEJ machinery, while the breaks which often appear in breast cancer cells or are introduced by IR and chemotherapies may require end-processing by nucleolytic end-resection of ssDNA at the DSB, affecting the recruitment of NHEJ, MMEJ and HR factors. C-to-U mutations along the resected ssDNA may lead to formation of a base-excision repair (BER) complex containing RPA and UNG2 that bind directly to dU (29), (30). UNG2 preferentially deglycosylates dU residing on ssDNA. The UNG2 activity forms an abasic site (31, 32), which may lead to ssDNA cleavage by the MRN complex (33). In the case of A3G, BER-mediated cleavage of deaminated ssDNA will generate 3’ ssDNA ending with two or more cytidines. This process can facilitate the search for a homologous micro-polyguanine tract, promoting A3G-mediated MMEJ.
Figure 1.
Deaminase-dependent and -independent models for A3G-mediated DSB repair. DSB end-resection by cellular nucleases/helicases (purple pacman) generates long ssDNA overhangs (blue ribbon). The ssDNA terminus is targeted by A3G multimers that mediate microhomology-based tethering in an interstrand synapse (IS) (1), facilitating MMEJ (2). A3G multimer disassembly generates multiple catalytic dimeric/monomeric mutational units (m), inducing C>U hypermutation of resected ssDNA (3). Uracil DNA glycosylase (UNG) catalyzes removal of the uracil from deoxyuridine, forming an abasic site (a) which is further cleaved by the base excision repair (BER) nuclease ApeI (4). Since A3G acts preferentially on the 3’ cytidine in a polycytidine tract, the truncated ssDNA is likely to end with two or more cytidines, reducing the search for microhomology by an incoming A3G multimer or other MMEJ factors (5). Terminal deamination by A3G may also promote DNA extension by the BER machinery. Alternatively, the MRN complex may associate with an abasic site and mediate cleavage of the ssDNA overhang (6). ssDNA end-bound MRN may then promote tethering of ssDNA termini, ATM activation (A) and recruitment of the NHEJ machinery (7).
DSB end-processing proceeds by a two-stage mechanism initiated by the MRN-CtIP complex that generates short 3’-terminal ssDNA overhangs, which are the substrates for the major NHEJ factor DNA-dependent protein kinase (DNA-PK) complex. These overhangs are then extended by Exo1 and/or BLM helicase to generate extensive ssDNA tracts several kb long, the substrate for HR factors such as Rad51 and Rad52 (34–39). Extensive DSB resection therefore commits DSB repair to HR (40–42). The kinetics of A3G recruitment to DSB foci assigns A3G as a factor associated with persistent DSBs, which are not efficiently repaired by NHEJ (20). Furthermore, the DSB frequency in A3G-depleted cells was comparable to parental cells at early time points following IR, suggesting that A3G is not involved in the classical NHEJ pathway. Taken together with the association of mutation clusters density with ssDNA length and persisting time (18), it is therefore plausible that A3G engages DSBs in the process of repair by HR. Binding of HR mediators occurs following extensive DSB end-resection, estimated to generate an average of 2–4 kb long ssDNA in each side of the break (34). The propensity of A3G to form high order homo-multimers in cells and its ability to simultaneously bind two ssDNA segments both terminally and internally may directly promote HR by juxtaposing resected ssDNA termini to homologous ssDNA regions. It is also plausible that such synapses control the proximity of broken DNA fragments following DSB resection, thus affecting adventitious repair resulting in chromosomal translocation. Alternatively, cytidine deamination may enhance binding of HR factors such as RPA, as recently demonstrated for AID (43). However, we observed a cytidine deamination-dependent shift from HR- to end-joining-mediated repair in irradiated lymphoma cells. This may be the result of resected ssDNA trimming that favors re-association of the NHEJ/MMEJ machinery over HR mediators, as discussed above. Similarly, the observed elevated frequency of mutations and microhomology-mediated indels in BRCA1 or BRCA2 mutant tumors (17) reflects the increased use of MMEJ instead of the impaired HR DSB repair in these breast cancer cells (44, 45). The impaired DSB repair in BRCA1 or BRCA2 mutant tumors is also consistent with the elevated frequency of C>T, G>A mutations induced by A3B (19).
In summary, A3G and probably other APOBEC proteins may promote DSB repair by direct end synapsis and cytidine deamination-dependent cleavage of resected ssDNA, facilitating MMEJ-directed repair and/or recruitment of MRN-ATM. Hence, APOBEC3 proteins play a dual role in promoting survival of cells in vivo, first by enhancing DSB repair following genotoxic treatments or spontaneous breaks, thus preventing cell death, and secondly, by promoting a mutator phenotype that drives tumor progression (46, 47, 48). Inhibition of APOBEC3 expression and/or its catalytic activity would increase sensitivity to genotoxic agents and restrain the progressive accumulation of mutations, which is one of the underlying processes that characterizes the cancer phenotype (49, 50). Hence, anti-APOBEC3 agents could be promising targets for the treatment of cancers in the future. Our preliminary results indicate that peptides aimed at inhibiting A3G activity in cultured tumor cells increase their sensitivity to genotoxic treatments.
Acknowledgment
We thank Drs. R.S. Harris, H. Matsuo, Y.L. Lyubchenko, J. Mueller, C. Schiffer, E. Pikarsky, H. Cedar, M. Goldberg and A. Cohen for critical reading of this manuscript and helpful discussion. This work was carried out in the Peter A. Krueger Laboratory with the generous support of Nancy and Lawrence Glick, and Pat and Marvin Weiss, and supported by grants from the National Institutes of Health (P01 GM091743 to RSH with a subaward to MK). R.N. was a fellow of the Clore Scholars Programme.
References
- 1.Chiu YL, Greene WC. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annual review of immunology. 2008;26:317–353. doi: 10.1146/annurev.immunol.26.021607.090350. [DOI] [PubMed] [Google Scholar]
- 2.Goila-Gaur R, Strebel K. HIV-1 Vif, APOBEC, and intrinsic immunity. Retrovirology. 2008;5:51. doi: 10.1186/1742-4690-5-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malim MH, Emerman M. HIV-1 accessory proteins--ensuring viral survival in a hostile environment. Cell host & microbe. 2008;3:388–398. doi: 10.1016/j.chom.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 4.Conticello SG, Langlois MA, Yang Z, Neuberger MS. DNA deamination in immunity: AID in the context of its APOBEC relatives. Advances in immunology. 2007;94:37–73. doi: 10.1016/S0065-2776(06)94002-4. [DOI] [PubMed] [Google Scholar]
- 5.Jarmuz A, Chester A, Bayliss J, Gisbourne J, Dunham I, Scott J, et al. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics. 2002;79:285–296. doi: 10.1006/geno.2002.6718. [DOI] [PubMed] [Google Scholar]
- 6.LaRue RS, Jonsson SR, Silverstein KA, Lajoie M, Bertrand D, El-Mabrouk N, et al. The artiodactyl APOBEC3 innate immune repertoire shows evidence for a multi-functional domain organization that existed in the ancestor of placental mammals. BMC molecular biology. 2008;9:104. doi: 10.1186/1471-2199-9-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee YN, Malim MH, Bieniasz PD. Hypermutation of an ancient human retrovirus by APOBEC3G. Journal of virology. 2008;82:8762–8770. doi: 10.1128/JVI.00751-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esnault C, Heidmann O, Delebecque F, Dewannieux M, Ribet D, Hance AJ, et al. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature. 2005;433:430–433. doi: 10.1038/nature03238. [DOI] [PubMed] [Google Scholar]
- 9.Landry S, Narvaiza I, Linfesty DC, Weitzman MD. APOBEC3A can activate the DNA damage response and cause cell-cycle arrest. EMBO reports. 2011;12:444–450. doi: 10.1038/embor.2011.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suspene R, Aynaud MM, Guetard D, Henry M, Eckhoff G, Marchio A, et al. Somatic hypermutation of human mitochondrial and nuclear DNA by APOBEC3 cytidine deaminases, a pathway for DNA catabolism. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:4858–4863. doi: 10.1073/pnas.1009687108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carpenter MA, Li M, Rathore A, Lackey L, Law EK, Land AM, et al. Methylcytosine and Normal Cytosine Deamination by the Foreign DNA Restriction Enzyme APOBEC3A. The Journal of biological chemistry. 2012;287:34801–34808. doi: 10.1074/jbc.M112.385161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stenglein MD, Burns MB, Li M, Lengyel J, Harris RS. APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nature structural & molecular biology. 2010;17:222–229. doi: 10.1038/nsmb.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Narvaiza I, Landry S, Weitzman MD. APOBEC3 proteins and genomic stability: the high cost of a good defense. Cell Cycle. 2012;11:33–38. doi: 10.4161/cc.11.1.18706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takai A, Toyoshima T, Uemura M, Kitawaki Y, Marusawa H, Hiai H, et al. A novel mouse model of hepatocarcinogenesis triggered by AID causing deleterious p53 mutations. Oncogene. 2009;28:469–478. doi: 10.1038/onc.2008.415. [DOI] [PubMed] [Google Scholar]
- 15.Okazaki IM, Kotani A, Honjo T. Role of AID in tumorigenesis. Adv Immunol. 2007;94:245–273. doi: 10.1016/S0065-2776(06)94008-5. [DOI] [PubMed] [Google Scholar]
- 16.Endo Y, Marusawa H, Kou T, Nakase H, Fujii S, Fujimori T, et al. Activation-induced cytidine deaminase links between inflammation and the development of colitis-associated colorectal cancers. Gastroenterology. 2008;135:889–898. doi: 10.1053/j.gastro.2008.06.091. 98 e1-3. [DOI] [PubMed] [Google Scholar]
- 17.Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberts SA, Sterling J, Thompson C, Harris S, Mav D, Shah R, et al. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Molecular cell. 2012;46:424–435. doi: 10.1016/j.molcel.2012.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burns MB, Lackey L, Carpenter MA, Rathore A, Land AM, Leonard B, et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013 doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nowarski R, Wilner OI, Cheshin O, Shahar OD, Kenig E, Baraz L, et al. APOBEC3G enhances lymphoma cell radioresistance by promoting cytidine deaminase-dependent DNA repair. Blood. 2012;120:366–375. doi: 10.1182/blood-2012-01-402123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Staudt LM, Wilson WH. Focus on lymphomas. Cancer cell. 2002;2:363–366. doi: 10.1016/s1535-6108(02)00178-2. [DOI] [PubMed] [Google Scholar]
- 22.Norman JM, Mashiba M, McNamara LA, Onafuwa-Nuga A, Chiari-Fort E, Shen W, et al. The antiviral factor APOBEC3G enhances the recognition of HIV-infected primary T cells by natural killer cells. Nat Immunol. 2011 doi: 10.1038/ni.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jais JP, Haioun C, Molina TJ, Rickman DS, de Reynies A, Berger F, et al. The expression of 16 genes related to the cell of origin and immune response predicts survival in elderly patients with diffuse large B-cell lymphoma treated with CHOP and rituximab. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2008;22:1917–1924. doi: 10.1038/leu.2008.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shahar OD, Raghu Ram EV, Shimshoni E, Hareli S, Meshorer E, Goldberg M. Live imaging of induced and controlled DNA double-strand break formation reveals extremely low repair by homologous recombination in human cells. Oncogene. 2012;31:3495–3504. doi: 10.1038/onc.2011.516. [DOI] [PubMed] [Google Scholar]
- 25.Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho SJ, Malim MH. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Current biology : CB. 2004;14:1392–1396. doi: 10.1016/j.cub.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 26.Harris RS, Liddament MT. Retroviral restriction by APOBEC proteins. Nature reviews Immunology. 2004;4:868–877. doi: 10.1038/nri1489. [DOI] [PubMed] [Google Scholar]
- 27.Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell research. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- 28.Sonoda E, Hochegger H, Saberi A, Taniguchi Y, Takeda S. Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA repair. 2006;5:1021–1029. doi: 10.1016/j.dnarep.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 29.Mer G, Bochkarev A, Gupta R, Bochkareva E, Frappier L, Ingles CJ, et al. Structural basis for the recognition of DNA repair proteins UNG2, XPA, RAD52 by replication factor RPA. Cell. 2000;103:449–456. doi: 10.1016/s0092-8674(00)00136-7. [DOI] [PubMed] [Google Scholar]
- 30.Otterlei M, Warbrick E, Nagelhus TA, Haug T, Slupphaug G, Akbari M, et al. Post-replicative base excision repair in replication foci. The EMBO journal. 1999;18:3834–3844. doi: 10.1093/emboj/18.13.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kavli B, Andersen S, Otterlei M, Liabakk NB, Imai K, Fischer A, et al. B cells from hyper-IgM patients carrying UNG mutations lack ability to remove uracil from ssDNA and have elevated genomic uracil. The Journal of experimental medicine. 2005;201:2011–2021. doi: 10.1084/jem.20050042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schrader CE, Linehan EK, Mochegova SN, Woodland RT, Stavnezer J. Inducible DNA breaks in Ig S regions are dependent on AID and UNG. The Journal of experimental medicine. 2005;202:561–568. doi: 10.1084/jem.20050872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Larson ED, Cummings WJ, Bednarski DW, Maizels N. MRE11/RAD50 cleaves DNA in the AID/UNG-dependent pathway of immunoglobulin gene diversification. Molecular cell. 2005;20:367–375. doi: 10.1016/j.molcel.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 34.Chung WH, Zhu Z, Papusha A, Malkova A, Ira G. Defective resection at DNA double-strand breaks leads to de novo telomere formation and enhances gene targeting. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1000948. e1000948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, et al. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- 36.Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, et al. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- 43.Yamane A, Robbiani DF, Resch W, Bothmer A, Nakahashi H, Oliveira T, et al. RPA accumulation during class switch recombination represents 5'-3' DNA-end resection during the S-G2/M phase of the cell cycle. Cell reports. 2013;3:138–147. doi: 10.1016/j.celrep.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Molecular cell. 2001;7:263–272. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 45.Tutt A, Bertwistle D, Valentine J, Gabriel A, Swift S, Ross G, et al. Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. The EMBO journal. 2001;20:4704–4716. doi: 10.1093/emboj/20.17.4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Campbell PJ, Yachida S, Mudie LJ, Stephens PJ, Pleasance ED, Stebbings LA, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–1113. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 48.Loeb LA, Loeb KR, Anderson JP. Multiple mutations and cancer. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:776–781. doi: 10.1073/pnas.0334858100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Loeb LA. Human cancers express mutator phenotypes: origin, consequences and targeting. Nature reviews Cancer. 2011;11:450–457. doi: 10.1038/nrc3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ding Q, Chang CJ, Xie X, Xia W, Yang JY, Wang SC, et al. APOBEC3G promotes liver metastasis in an orthotopic mouse model of colorectal cancer and predicts human hepatic metastasis. The Journal of clinical investigation. 2011;121:4526–4536. doi: 10.1172/JCI45008. [DOI] [PMC free article] [PubMed] [Google Scholar]