Abstract
Despite its importance in central metabolism and bacterial cell signaling, protein histidine phosphorylation has remained elusive with respect to its extent and functional roles in biological systems due to the lack of adequate research tools. We report the development of the first pan-pHis antibody using a stable phosphohistidine (pHis) mimetic as the hapten. This antibody was successfully used in ELISA, Western blot, dot blot, immunoprecipitation, and in detection and identification of histidine-phosphorylated proteins from native cell lysates when coupled with mass spectrometric analysis. We also observed that protein pHis levels in E. coli lysates depend on carbon source and nitrogen availability in the growth media. In particular, we found that pHis levels on PpsA are sensitive to nitrogen availability in vivo and that α-ketoglutarate (α-KG) inhibits phosphotransfer from phosphorylated phosphoenolpyruvate synthase (PpsA) to pyruvate. We expect this antibody to open opportunities for investigating other pHis-proteins and their functions.
INTRODUCTION
Protein phosphorylation is a central player in the regulation of cellular processes.1 Although this class of posttranslational modification (PTM) is known to occur on several amino acids, the available biochemical, pharmacological and proteomic tools for studying the modification are best developed in the context of Ser, Thr and Tyr phosphorylation.2 By contrast, there is a dearth of such tools for studying other protein phosphorylation events, a situation which has greatly hindered our understanding of both the extent and functional roles of these more elusive PTMs.3,4,5 This situation is especially striking for protein histidine phosphorylation, a modification known for 50 years,6 and whose importance is recognized in processes ranging from central metabolism to signaling in bacteria and lower eukaryotes.7 It is currently not possible to monitor global pHis levels within a native proteome, something that is now routine for Ser, Thr and Tyr phosphorylation.8,9 Consequently, we have limited knowledge of how protein histidine phosphorylation changes as a function of cell state or type.
The lack of available tools for studying pHis can be attributed to the evanescent nature of the modification; the chemical instability of the phosphoramidate linkage to acid and certain nucleophiles renders the modification incompatible with standard proteomic workflows and, importantly, has frustrated efforts to raise antibodies against the native modification.3,7,10 Recently, we introduced phosphoryltriazolylalanine (pTza) as a stable mimic of pHis and showed that this unnatural residue could be incorporated into a synthetic peptide that was subsequently used to raise a sequence-specific anti-pHis antibody.11 This triazole-based pHis analog approach has also been pursued by other groups.12,13 While a promising development, it remained unclear whether this pHis mimetic strategy could be adapted to generate a sequence-independent pHis antibody (pan-pHis antibody), which by analogy to the impact of pan-pTyr antibodies14,15 would be a far more useful reagent for studying pHis.
In this report, we address this long-standing analytical deficiency by developing a pan-specific antibody against the pHis modification. We show that this antibody can be used to detect protein histidine phosphorylation in vitro and in vivo. This includes the use of this antibody to perform the first direct detection of pHis levels from native cell lysates. By coupling this new technology to high-resolution nano-flow UPLC-MS, we characterized endogenous pHis sites in E. coli. These investigations led to the finding that histidine phosphorylation of PpsA in E. coli is regulated by nitrogen availability in vivo and that dephosphorylation of this key metabolic enzyme is inhibited by α-KG.
RESULTS
Antibody Generation and In Vitro Characterization
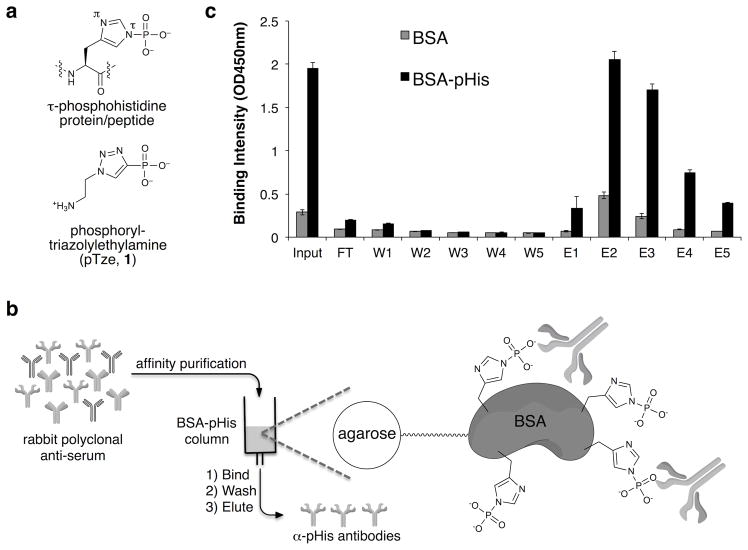
Inspired by the successful use of phosphotyramine as a hapten to develop pan-pTyr antibodies,16 we synthesized the τ-pHis mimic, phosphoryl-triazolylamine (pTze, Fig. 1a). The heterocyclic structure of pTze is expected to closely mimic the size and geometry of pHis. Note, we targeted the more stable τ-pHis isomer, rather than the π-pHis isomer (Fig. 1a and Supplementary Results, Supplementary Fig. 1), since it has been observed more frequently in the small number of pHis-containing proteins that have been rigorously characterized to date.3,7 pTze was conjugated to the carrier protein keyhole limpet hemocyanin (KLH) via its amino group and used to immunize rabbits employing a standard prime-boost protocol. Production of anti-pHis antibodies in the inoculated animals was monitored by ELISA analysis of serum using, as an analyte, bovine serum albumin (BSA) that had been chemically phosphorylated on its native histidine residues (BSA-pHis) (Supplementary Fig. 2). Following antigen boosts, the crude rabbit anti-serum was affinity purified over an agarose column containing immobilized BSA-pHis (Fig. 1b). ELISA analysis showed enrichment of the desired pHis binding antibodies over serum proteins and other nonspecific antibodies (Fig. 1c and Supplemental Fig. 3).
Figure 1. Affinity purification of antibodies raised against pTze.
a) Structures of τ-pHis and its stabile mimic, pTze (1). b) Schematic depicting the strategy to affinity purify pHis polyclonal antibody. c) ELISA analysis of input, flow through (FT), wash fractions (W1, W2, W3, W4, W5), and elution fractions (E1, E2, E3, E4, E5) from the affinity purification for pHis binding affinity. Fractions E2 and E3 displayed the highest affinity for BSA-pHis compared to the BSA control (n = 4, mean ± s.d.).
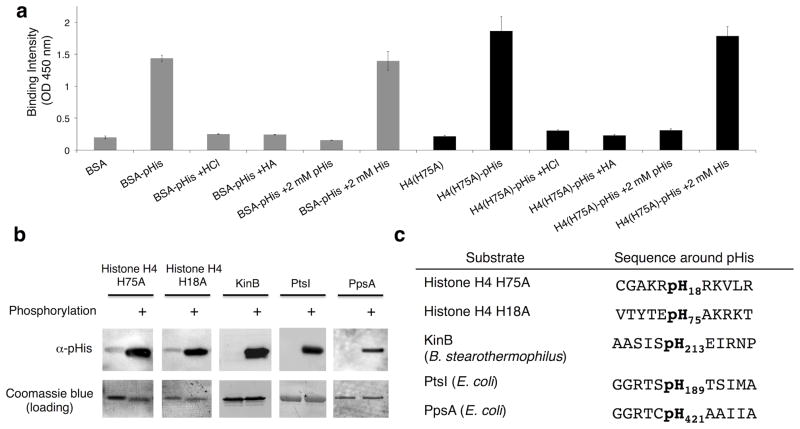
The purified antibodies were found to bind robustly to both BSA-pHis and chemically phosphorylated histone H4 (H4(H75A)-pHis) by ELISA (Fig. 2a). Pre-treatment of these pHis containing proteins with acid or hydroxylamine (HA), both of which result in dephosphorylation of pHis,17 reduced binding to levels comparable with the non-phosphorylated control proteins. Moreover, binding to BSA-pHis and H4(H75A)-pHis was inhibited by the presence of free pHis amino acid but not histidine, providing further evidence that the antibodies selectively recognized pHis within the proteins (Fig. 2a).
Figure 2. Affinity purified antibodies raised against pTze are cross-reactive to phosphohistidine residues and are sequence independent.
a) ELISA data of affinity-purified antibodies measured against BSA-pHis (gray bars) and histone H4(H75A)-pHis (H4(H75A)-pHis) (black bars), as well as the indicated controls (n = 4, mean ± s.d.). b) Western blots of pHis-containing proteins phosphorylated in vitro. The Coomassie Blue stain was included as the loading control. See Supplementary Fig. 18 for full Western blots. c) Sequences of pHis proteins recognized by the anti-pHis antibody (α-pHis) in panel b.
Encouraged by the ELISA results, we next tested the purified antibody in Western blotting experiments, initially using a series of purified recombinant proteins. Gratifyingly, the antibody could be used to detect histone H4 chemically phosphorylated on either His18 or His75, as well as a bacterial histidine kinase KinB, and the E. coli metabolic proteins enzyme I (PtsI) and phosphoenolpyruvate synthetase (PpsA) which had been enzymatically autophosphorylated in vitro (Fig. 2b). In all cases, the signals were abolished upon treatment with acid or hydroxylamine, confirming that detection is mediated through pHis (Supplementary Fig. 4). Importantly, the flanking sequences around the pHis sites in these proteins vary widely (Fig. 2c), suggesting the antibody will be quite generally applicable and context-independent.
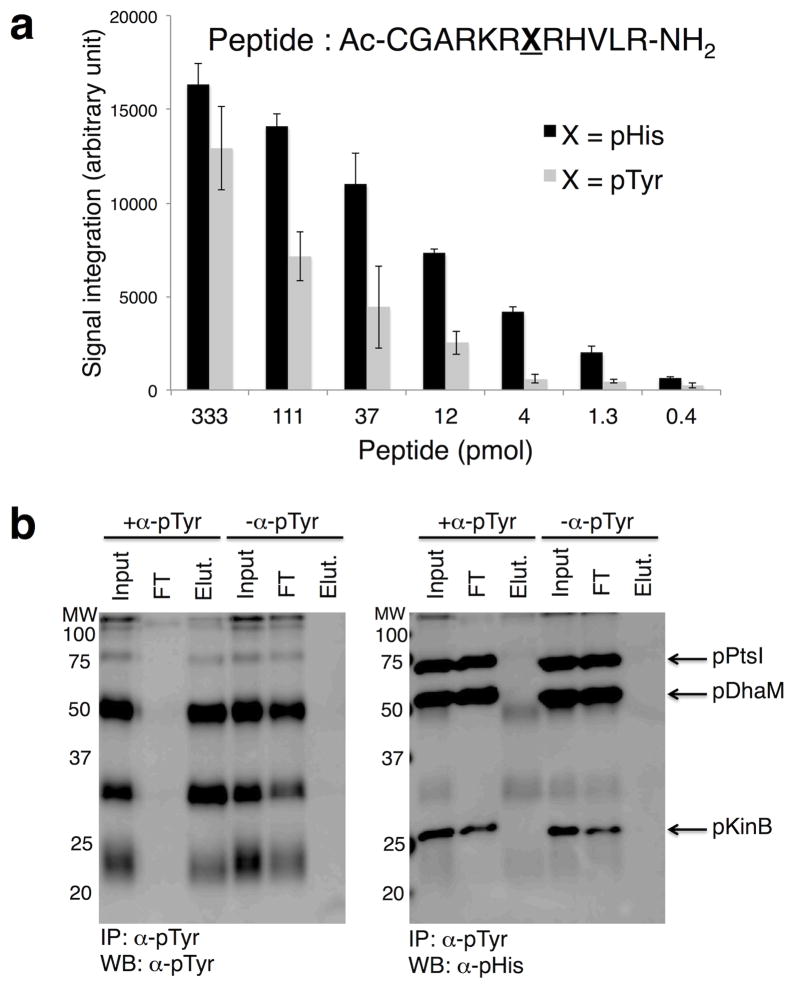
In addition, we performed a series of control experiments to measure the cross-reactivity of the pHis antibody to other phosphoamino acids. The antibody did not recognize pSer or pThr conjugated to BSA, however, cross-reactivity with BSA-pTyr was observed by ELISA analysis (Supplementary Fig. 5). Peptide dot-blots indicated that the pHis antibody exhibits ~10 fold higher sensitivity towards pHis peptide over pTyr (Fig. 3a and Supplementary Fig. 6). This cross-reactivity represents a potential complication for the analysis of native proteomes, particularly those from higher eukaryotes. A solution to this problem was found through the observation that a series of commercially available α-pTyr antibodies show minimal cross-reactivity to pHis-containing proteins (Supplementary Fig. 7). This discrimination permitted the immuno-depletion of pTyr-containing proteins from a mixture without removing pHis proteins (Fig. 3b), thereby filtering out any potential unwanted pTyr signals, however low abundance they may be.
Figure 3. Cross-reactivity of pHis antibody to pTyr.
a) Dot blot analysis of pHis and pTyr peptides derived from histone H4 sequence. The signal intensities are quantified using ImageJ program and plotted against the peptide amount (n = 3, mean ± s.d.). b) Western blot analysis of proteins immunoprecipitated with a monoclonal pTyr antibody. Proteins modified with pTyr (pTyr MW markers, Calbiochem) were mixed with pHis-containing KinB (pKinB), DhaM (pDhaM), and PtsI (pPtsI). In the left panel, pTyr proteins were successfully immunoprecipitated with α-pTyr (+α-pTyr) but not in the mock IP control (−α-pTyr). pHis proteins (pKinB, pDhaM, pPtsI) were not immunoprecipitated by α-pTyr and were found only in the flow through (FT) (right panel). Input refers to the sample prior to immunoprecipitation, FT is the supernatant after immunoprecipitation, Elut is the elution sample. See Supplementary Fig. 19 for full Western blot and Coomassie Blue stain of membranes.
Detection of pHis in Cellular Proteins
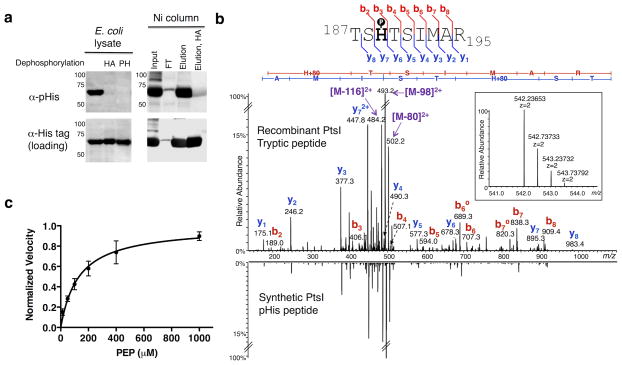
We next explored whether our pHis antibody could detect endogenously phosphorylated pHis containing proteins following overexpression in cells. As a test case we selected PtsI, the first enzyme in the phosphoenolpyruvate:sugar phosphotransferase system (PTS), the main route for bacterial glucose uptake (Supplementary Fig. 8).18 Western blot analysis of crude lysate from E. coli overexpressing a His6-tagged version of PtsI showed robust pHis signal with our antibody (Fig. 4a). Treatment of this lysate with hydroxylamine or a mammalian phosphohistidine phosphatase (PHPT1) both led to complete loss of the pHis signal (Fig. 4a). As further confirmation that our antibody recognized authentic endogenous histidine phosphorylation on PtsI, we purified the protein using Nickel-NTA resin and observed phosphorylation on His-189 directly by LC-MS/MS following tryptic digestion of the protein (Fig. 4a and 4b). Moreover, the LC retention time and the MS/MS fragmentation pattern of this tryptic peptide were essentially identical to that of a pHis-bearing synthetic version of this PtsI peptide (Fig. 4b and Supplementary Fig. 9a). We also prepared and analyzed an isoform of the PtsI peptide in which the phosphoryl group is on a serine instead of the histidine residue (TpSHTSIMAR) and observed that both the LC retention time and MS/MS fragmentation patterns, particularly the absence of a prominent neutral loss of 80 amu, were distinct from the pHis peptide (Supplementary Fig. 9b).
Figure 4. Analysis of histidine phosphorylation on PtsI.
a) Western blots of E. coli lysates expressing His6-tagged PtsI. Left: α-pHis blot of crude lysates and those treated with hydroxylamine (HA) or phosphohistidine phosphatase (PH). Right: Crude lysates were purified over Ni-NTA beads and the indicated fractions probed with the α-pHis antibody. As loading controls, the membranes were stripped and re-blotted with an α-His-tag antibody. See Supplementary Fig. 20 for full Western blots. b) Overexpressed PtsI was digested with trypsin and analyzed by high-resolution nano-UPLC-MS. Shown is the MS/MS spectrum from the tryptic peptide ion bearing pHis at the canonical His-189 site, with the matched b- and y- ions indicated in the spectrum and in the sequence flag diagram above (inset: MS spectrum of the precursor ion species, including its accurate mass measurement). For comparison, the MS/MS spectrum of a synthetic version of the pHis-bearing peptide is shown in mirror image below. c) A dot blot assay was developed to measure the kinetics of autophosphorylation of PtsI by PEP. A plot of the reaction velocity as a function of PEP concentration was used to determine an apparent Km value of 135 ± 30 μM (n = 3, mean ± s.d.).
We used recombinant PtsI as a test case for the development of an α-pHis dot-blot assay for use in biochemical studies. Using this high-throughput method, we measured the Km for phosphorylation of PtsI with phosphoenolpyruvate (PEP) as the phosphoryl donor (Fig. 4c and Supplementary Fig. 10). From the plot of reaction velocity versus PEP concentration, we obtained an apparent Km of 135 ± 30 μM, which is in agreement with previously reported Km values.19,20 We also used this assay to investigate recent reports that millimolar concentrations of α-ketoglutarate (α-KG) inhibit PtsI.20,21 These studies provide evidence that α-KG inhibits PtsI; however, direct analyses of pHis levels were not made. With our pHis dot blot assay, we confirmed that millimolar concentrations of α-KG led to decreased histidine phosphorylation on PtsI by PEP (Supplementary Fig. 11). We stress that it should be possible to adapt this α-pHis dot-blot assay for use in the kinetic analysis of other pHis-containing enzymes, obviating the use of onerous radio-assays involving labeled cofactors.
Metabolic Changes Affect pHis Protein Levels
We next asked whether the antibody could be used to detect endogenous pHis-containing cellular proteins. This seemed likely given our results with overexpressed PtsI (Fig. 4a), as well as analogous results we obtained with another pHis protein in the PTS pathway, PTS-dependent dihydroxyacetone kinase, phosphotransferase subunit (DhaM, Supplementary Fig. 12). Moreover, the antibody could efficiently immunoprecipitate both of these overexpressed pHis proteins from lysates (Supplementary Fig. 13), enabling crucial applications in proteomics-type experiments (vide infra).
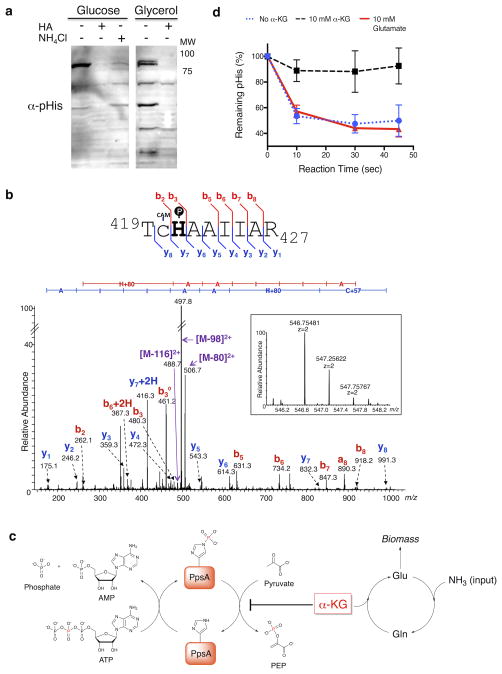
Since several proteins in the PTS pathway contain pHis as the enzymatic intermediate,18 we decided to survey the pHis-proteome of E. coli as a function of nitrogen availability or carbon source, both of which can influence metabolic flux through this pathway.21 Prototrophic E. coli NCM 3722 cells were grown on minimal media containing glucose and arginine as the limiting nitrogen source. Western blotting revealed a strong band at 85 kDa that was sensitive to hydroxylamine treatment (Fig. 5a). Switching the carbon source to glycerol, led to a marked increase in the number of pHis proteins detected using our antibody (Fig. 5a). This is likely explained by a lack of phosphorylation flux through PTS proteins in cells grown in the absence of glucose, resulting in a buildup of pHis signal on PTS proteins.
Figure 5. pHis levels in PpsA are sensitive to nitrogen availability and are regulated by α-KG.
a) Western blotting of NCM 3722 cells grown on minimal media containing glucose or glycerol as the carbon source and arginine as the nitrogen source. α-pHis signals were sensitive to hydroxylamine (HA) treatment of the lysates and nitrogen upshift in the growth media (NH4Cl). As a loading control, the membranes were imaged with colloidal gold stain (Supplementary Fig. 21). See Supplementary Fig. 22 for full Western blot. b) MS/MS of an endogenous PpsA tryptic pHis peptide identified from fractionated glucose-fed E. coli lysate (Supplementary Fig. 16). The gel band at 85 kDa was analyzed by high-resolution nano-UPLC-MS after trypsin digestion. The spectrum indicates pHis at the canonical His421 site, with the matched b- and y- ions indicated in the spectrum and in the sequence flag diagram above (CAM = S-carboxyamidomethyl) (inset: MS spectrum of the precursor ion species, including its accurate mass measurement). c) Model for regulation of PpsA catalytic cycle. Intracellular levels of α-KG can be significantly increased by nitrogen limitation. Inhibition of the PpsA dephosphorylation by the increased α-KG will lead to the buildup of phosphorylated enzyme. d) Dephosphorylation assay of autophosphorylated PpsA. The dephosphorylation was inhibited by α-KG, but not by glutamate (n = 3, mean ± s.d.).
We were struck by the rapid loss of the pHis signal in the 85 kDa region upon nitrogen upshift (NH4Cl for 30 mins.) in the glucose fed cells and, thus, sought to identify the protein(s) involved (Fig. 5a). Accordingly, we used our antibody to immunoprecipitate the pHis-containing proteome of E. coli NCM 3722 cells grown under glucose and arginine conditions. Trypsin digestion followed by LC-MS/MS analysis of proteins in the 85 kDa region led to the identification of three proteins known to be histidine phosphorylated: PEP synthetase (PpsA), phosphoenolpyruvate-protein phosphotransferase (PtsP), and polyphosphate kinase (Ppk) (see Supplementary Table 1 for full protein list). We overexpressed a His6-tagged variant of each candidate protein in NCM 3722 cells and found that the pHis levels in only PpsA-overexpressing cells were sensitive to ammonia, suggesting that PpsA is the 85 kDa protein detected in the native lysate (Supplementary Fig. 14 and 15). To confirm this, we fractionated lysate from glucose-fed E. coli by ammonium sulfate precipitation, and the 85 kDa gel band was digested and analyzed by LC-MS/MS (Supplementary Fig. 16). A pHis-containing peptide was observed in the digest and, gratifyingly, corresponded to that of the expected PpsA tryptic peptide containing pHis at the canonical phosphorylation site (His421, Fig. 5b).
We were intrigued by the buildup of phospho-PpsA under nitrogen limitation conditions. PpsA catalyzes the generation of PEP by employing the β-phosphoryl group of ATP as the phosphoryl donor for its autophosphorylation on a histidine residue and subsequent phosphoryl transfer to pyruvate (Fig. 5c).22 While the intracellular levels of ATP, PEP, and pyruvate do not show significant change upon varying nitrogen levels in NCM 3722 cells, α-KG accumulates dramatically under limited nitrogen, but quickly decreases upon addition of ammonia.21,23 α-KG is known to inhibit PpsA activity in vitro but it is not clear at which step: the ATP-catalyzed autophosphorylation on histidine or the phosphoryl transfer from the pHis to pyruvate.22 In vitro PpsA phosphorylation and dephosphorylation assays involve the use of [β-32P]ATP, which is not commercially available. Using the much more convenient dot-blot assay enabled by our pHis antibody, we did not observe a difference in the autophosphorylation step in the presence or absence of α-KG (Supplementary Fig. 17). However, we found the second, phosphoryl transfer step to pyruvate to be much slower in the presence of α-KG compared to the absence of this metabolite or to a glutamate control (Fig. 5d). This shows that α-KG can act as a regulator of pHis levels in PpsA. Thus, our observation of PpsA phosphorylation build-up can be explained by the fact that α-KG accumulates under limiting nitrogen conditions and, at these higher concentrations, can inhibit phosphoryl transfer from phosphorylated PpsA to pyruvate. Subsequent replenishment of ammonia reduces α-KG levels and restores phosphoryl transfer to pyruvate causing a decrease in pHis levels of PpsA (Fig. 5c).
DISCUSSION
Protein phosphorylation is central to many cellular processes.1 Consequently, tools that permit phospho-protein levels to be monitored as a function of cell type or state have proven invaluable in biomedical research.2,8 While strategies are now well established for cataloguing pSer, pThr and pTyr levels in native proteomes, little progress has been made in the analogous study of protein histidine phosphorylation. The latter represents a very serious analytical blind-spot, given that pHis plays a critical role in signaling cascades in bacteria and lower eukaryotes and occurs commonly in metabolic enzymes in all cell types.3,7 At the heart of this problem lies the chemical nature of the pHis modification. The intrinsic instability of the phosphoramidate linkage within pHis has frustrated the development of even the most basic biochemical tools, in particular a modification specific antibody. Thus, our ability to study the extent and dynamics of the pHis modification in native proteomes has lagged far behind that of other phospho-protein modifications.3 In this study, we employed a pHis mimetic approach to develop the first pan-specific antibody against the modification. This research tool was used to directly detect pHis-containing proteins through ELISA, dot blot, Western blot and immunoprecipitation experiments. Our pan antibody, utilized in conjunction with mass spectrometry, also enabled the detection and identification of endogenously phosphorylated pHis proteins from cell lysates.
The potential utility of anti-pHis antibodies has long been appreciated and efforts to develop such reagents date back over ten years.4 The short half-life of native phosphohistidine in serum makes it of little value as a hapten for raising modification-specific antibodies.4,7 Faced with this situation, alternative strategies based on the use of more stable pHis analogs or mimetics have been considered.3 These include the use of the more stable thiophosphorylated analog of pHis, which can be generated chemically or enzymatically, the latter using ATP-γ-S.10,24 While pHis-specific antibodies have yet to be reported using this approach, the unique nucleophilicity of the thiophosphoryl group can be used to install a secondary immunogenic site via attachment of a suitable hapten.10 Replacement of the labile phosphoramidate linkage with a more inert chemical bond has also been explored. Stable phosphonate linkages can be incorporated into various five-membered heterocyclic frameworks, leading to a range of presumptive pHis mimetics.11,12,13,25 Previously, we described one such mimetic based on a triazole framework, phosphoryltriazolylalanine.11 Antibodies raised against this unnatural residue in the context of a specific peptide sequence, cross-react with native pHis in the same sequence context. This study is important since it establishes that pHis mimetics can be used in antibody development. Still, it remained unclear whether sequence-independent pHis antibodies, which are far more valuable for proteomics studies, could be generated using this mimetic approach. Indeed, it has been questioned whether phosphoryltriazole-based molecules can be used to generate sequence-independent pHis antibodies since such antibodies should rely solely on the recognition of the pHis sidechain, which modeling indicates differs, albeit subtly, in its electrostatic surface potentials from the phosphoryltriazole.11,12 In this report, we showed that it is indeed possible to develop pan pHis antibodies using phosphoryltriazole as the hapten. We speculate that electrostatic differences in the imidazole and triazole ring systems in pHis and the mimetic, respectively, are outweighed, at least in terms of antibody recognition, by the anionic phosphoryl group common to both.
One limitation of the current pan-pHis antibody is its mild cross-reactivity with pTyr residues. Fortunately, pHis and pTyr can be biochemically distinguished from each other based on the stability towards acid or hydroxylamine (Supplementary Figs. 4 and 19). In addition, we observed that commercial pan pTyr antibodies show little affinity towards pHis proteins. Taking advantage of the affinity differences, we were able to selectively immunodeplete pTyr-containing proteins in the presence of pHis proteins. Parenthetically, this result argues against the idea of using a pTyr antibody as a surrogate for the pHis antibody.26,27 A practical drawback of our antibody is its finite supply, being a polyclonal antibody. Generation of monoclonal antibodies, potentially carried out with next-generation pHis analogs to minimize pTyr cross-reactivity, will address those issues in the future.
The availability of pan-pHis antibodies opens the way to a number of investigations both biochemical and cell-based. Monitoring protein histidine phosphorylation in a test tube currently involves the use of radiolabeling, with for example 32P-ATP, which has its attendant safety and cost (i.e. disposal) issues.28 As demonstrated herein, the availability of a robust immuno-detection platform overcomes many of these problems, potentially allowing higher throughput studies of biochemical systems where pHis is involved, for example two-component signaling.29 Complementing the in vitro applications, we imagine that pan-pHis antibodies will also find broad use in cell-based studies by allowing global pHis levels to be monitored in different contexts, for instance various cell types or as a function of some cellular perturbation. As an illustration of the latter, we were able to probe global protein histidine phosphorylation in E. coli under different metabolic states. This led to the discovery that nitrogen availability and carbon source dramatically affects the pHis levels in a number of proteins. Among those proteins, PEP synthase, PpsA, was found to undergo rapid dephosphorylation in vivo upon the relief of nitrogen limitation. Subsequent in vitro PpsA phosphorylation and dephosphorylation assays showed that the dephosphorylation to form PEP is inhibited by α-KG, a key metabolite tightly regulated by the nitrogen availability. These observations clarified a long-standing question about the mechanism of PpsA inhibition by α-KG. PpsA is a key enzyme in E. coli metabolic engineering efforts geared towards channeling carbon flux for production of shikimic acid and downstream aromatic metabolites.30 Thus, we believe our findings on the regulation of PEP synthase activity will provide further insights on the bioengineering of the bacterial central metabolic pathway.
In conclusion, we have successfully developed the first pan-pHis antibody and used this reagent in variety of assays that commonly employ modification-specific antibodies. These applications of our pan-pHis antibody help address the long-standing dearth of adequate tools in histidine phosphorylation research, which has been the major roadblock to progress in the field. Further studies, including the investigation of histidine phosphorylation in mammalian systems and antibody development for the other pHis isomer, are currently underway.
ONLINE METHODS
Preparation and characterization of pTze, phosphorylated peptides and proteins are described in detail in the Supplementary Notes 2 and 3.
Affinity purification of anti-pHis polyclonal antiserum
Affinity resin consisting of phosphorylated BSA immobilized onto agarose was prepared by covalently attaching BSA to agarose beads using SulfoLink® Resin (Thermo Scientific) according to the manufacturers instructions. Chemical phosphorylation of the immobilized BSA-agarose resin was performed by pre-equilibrating 1 mL of resin in TBS (25 mM Tris pH 8.5, 137 mM NaCl, 2.7 mM KCl) followed by incubation of the resin in 500 mM potassium phosphoramidate in Tris-Buffered Saline pH 8.5 (TBS) overnight at RT with mixing on a nutator. The resin was then washed with 4 column volumes of TBS. 600 μL of crude polyclonal pHis antiserum diluted 5-fold into TBS was then added to the resin and incubated for 1 h at RT on a nutator. The flow through was collected and the resin was washed with 6 column volumes of TBS. Antibodies were eluted from the column in 1 mL fractions with elution buffer (100 mM glycine pH 2.5). After elution off of the column, the elution fractions were immediately neutralized by adding 100 μL of 1 M Tris (pH 8.0). Fractions containing pHis antibody were determined by SDS-PAGE and ELISA. Further details can be found in Supplementary Note 4.
ELISAs of pHis proteins
pHis protein samples were diluted in coating buffer (0.032 M Na2CO3, 0.068 M NaHCO3, pH 9.6) and added to a Nunc-Immuno Maxisorp 96-well plate (Thermo Scientific). The plate was incubated for 2 h at RT on a nutator. The wells were then washed three times with wash buffer (25 mM Tris, pH 8.5, 137 mM NaCl, 2.7 mM KCl, 0.1% v/v Tween 20). The wells were then blocked by adding 1% BSA in wash buffer at 50 μL/well and incubating for 45 min at RT on a nutator. The blocking solution was then removed from each well. pHis antibody was diluted in wash buffer and then added to the wells at 50 μL/well and incubated for 45 min at RT on a nutator. The wells were then emptied and washed three times with wash buffer. Goat-anti rabbit-HRP secondary antibody was diluted 5000-fold into wash buffer and added to the wells at 50 μL/well and incubated for 45 min at RT on a nutator. The wells were emptied and washed four times with wash buffer. 50 μL of Ultra TMB ELISA substrate (Thermo Scientific) was added to each well and incubated for 5 min at RT followed by addition of 50 μL of 2 N H2SO4 to quench the reaction. The absorbance at 450 nm was measured on a Spectramax M3 plate reader. Further details for ELISA experiments can be found in Supplementary Note 4.
Western blotting using pHis antibody
Samples were diluted into 4x basic loading buffer (160 mM Tris, pH 8.5, 40% (v/v) glycerol, 4% (w/v) SDS, 0.08% (w/v) bromophenol blue, and 8% (v/v) BME) and resolved by SDS-PAGE. The resolved proteins were electroblotted onto a PVDF membrane in Towbin buffer (25 mM Tris base, 192 mM glycine, pH 8.3, 10 % (v/v) methanol) at 100 V for 60 min. The membrane was blocked with 3% BSA in wash buffer (25 mM Tris, 137 mM NaCl, 2.7 mM KCl, 0.1% (v/v) Tween-20, pH 8.5) for 1 h at RT. The membrane was then incubated with affinity-purified anti-pHis antibody diluted 1:100 in wash buffer with 3% BSA for 1 h at RT. The membrane was washed with wash buffer (3 × 5 min) and then incubated with goat anti-rabbit IgG-HRP conjugate (diluted 1:5000 in wash buffer with 3% BSA) or Protein A-HRP (diluted 1:8000 in wash buffer with 3% BSA) for 1 h at RT. The membrane was washed with wash buffer (3 × 5 min), drained, and was incubated with ECL chemiluminescence substrate solution (Thermo Scientific) for 1 min at RT. The chemiluminescence from the membrane was imaged using ImageQuant LAS 4000. The membrane was washed with water and stained with Coomassie blue or colloidal gold (Bio-Rad) for the visualization and quantification of total protein. Further details for Western blot experiments can be found in Supplementary Note 4.
Peptide dot blots comparing pHis and pTyr affinity
The H4-His18 peptide (1 mM solution) was chemically phosphorylated at the histidine residues with potassium phosphoramidate (final 100 mM) overnight at RT. 1 mM of the H4-pHis18 and H4-pTyr18 peptides were serially diluted 3-fold with ddH2O to give solutions of 333, 111, 37, 12, 4, 1.3, 0.4 μM. Each solution was spotted onto two nitrocellulose membranes, and the membranes were air-dried. One membrane was stained with colloidal gold, dried, and scanned. The other membrane was analyzed by Western blot with affinity-purified pHis antibody following the general Western blot procedure above. The signal intensity from the blot was measured using the ImageJ software.
Selective immunoprecipitation of pTyr proteins using α-pTyr mAb
2.0 μg of phosphorylated PtsI (pPtsI), DhaM (pDhaM) and KinB (pKinB) were combined and diluted into 250 μL TBST (25 mM Tris pH 8.5, 137 mM NaCl, 2.7 mM KCl, 0.1% Tween 20) containing 3 μL pTyr protein ladder (Calbiochem). The sample was then transferred to 250 μL TBST containing 30 μL protein G resin with or without 20 μg anti-pTyr mAb antibody prebound to the resin. After adding the protein mixture to the protein G resin, the samples were incubated for 1 h at 4 °C on a nutator. The samples were centrifuged to pellet the resin and the supernatant was discarded. The pelleted resin was then washed with 5 × 200 μL of TBST buffer. Proteins were eluted from the resin by treatment with 30 μL 50 mM phosphotyrosine for 15 min at RT. Input, flow through, and elution samples were analyzed by Western blot. Full details can be found in Supplementary Note 6.
Western blot of NCM 3722 E. coli lysate under varying carbon and nitrogen sources
NCM 3722 cells were grown in nitrogen-limited minimal media (Gutnick minimal salts, 2.5 mM arginine, 0.4% w/v glucose or glycerol) at 37 °C to an OD600 of 0.6. The glucose-fed cells were then split into 2 × 10 mL aliquots in sterile 50 mL conical plastic tubes, and one aliquot was treated with NH4Cl (final concentration: 10 mM). Both aliquots were then grown at 37 °C for 30 min. 10 mL of the glucose- and glycerol-fed cells were then pelleted by centrifugation (4500 xg for 5 min at 4 °C). The pellets were then resuspended in 400 μL loading buffer (160 mM Tris pH 8.5, 40% v/v glycerol, 4% w/v SDS, 0.08% w/v Bromophenol Blue, 8% v/v 2-mercaptoethanol) and sonicated (3 × 5 s at 45% power) on ice. 50 μL of this lysate was incubated with 500 mM hydroxylamine for 1 h at 37 °C. 15 μL of each sample was analyzed by Western blot. Full experimental details can be found in Supplementary Note 7.
Immunoprecipitation of NCM 3722 E. coli lysate
NCM 3722 cells were grown in 40 mL of nitrogen-limited culture medium (Gutnick minimal salts, 2.5 mM arginine, 0.4% w/v glucose) overnight at 37 °C to an OD600 of 1.0. The cells were then pelleted by centrifugation (4500 xg for 5 min at 4 °C). The pellets were then resuspended in 1.2 mL lysis buffer (25 mM Tris pH 8.5, 137 mM NaCl, 2.7 mM KCl supplemented with Complete® protease inhibitor (Roche) and phosphatase inhibitor cocktail 2 and 3 (Sigma-Aldrich, diluted 200-fold from stock solution) and sonicated (3 × 5 s at 45% power) on ice. The lysate was then centrifuged at 17,000 xg for 10 min at 4 °C to remove cell debris.
50 μL of the lysate was diluted into 500 μL TBST (25 mM Tris pH 8.5, 137 mM NaCl, 2.7 mM KCl, 0.1% Tween 20). This protein mixture was then added to 30 μL of protein G agarose resin pre-bound with 30 μg anti-pHis antibody (IP sample) or protein G agarose with no antibody (mock IP sample). The antibody was pre-bound to the resin by incubating in 1 mL of TBS (25 mM Tris pH 8.5, 137 mM NaCl, 2.7 mM KCl) with 30 μL of protein G agarose resin for 2 h at 4 °C. After adding the lysate to the resin, it was incubated for 45 min at 4 °C on a nutator and then centrifuged (10,000 xg at RT for 10 s) to pellet the resin. The supernatant was removed and the resin was then washed with 4 x 200 μL TBST. Bound proteins were eluted from the resin by adding 25 μL of 20 mM pTze in wash buffer and incubating for 15 min at RT. The eluted proteins were then treated with 5 μL 4x loading buffer (160 mM Tris pH 8.5, 40% v/v glycerol, 4% w/v SDS, 0.08% w/v Bromophenol Blue, 8% v/v 2-mercaptoethanol). 20 μL of the IP and mock IP elution samples were then boiled at 100 °C and loaded onto a 12% Bis-Tris Gel and resolved by electrophoresis for 60 min at 165 V. The gel region between 70 kDa to 100 kDa was excised from the gel and submitted for proteomic analysis by the Proteomics Resource Center at the Rockefeller University. (Note: Boiling of these samples precluded the detection and analysis of pHis-containing peptides by mass spectrometry. Thus, this analysis primarily focused on the identification of immunoprecipitated proteins, not necessarily on the detection of actual pHis sites on these proteins.)
Dot blot assays of PpsA phosphorylation and dephosphorylation
Phosphorylation of PpsA
PpsA (~25 μM) was diluted in assay buffer (100 mM Tris (pH 8.0), 5 mM MgCl2) to a final concentration of 500 nM. 170 μL of the PpsA sample was then placed into a siliconized Eppendorf tube followed by addition of 10 μL α-KG (final concentration of 10 or 15 mM) or 10 μL of assay buffer. The mixture was cooled to 15 °C and the reaction was initiated by adding 20 μL of 100 μM ATP (final 10 μM). At time points (t = 0, 10, 30, and 45 s), 20 μL of the reaction mix was taken out and added into 100 μL of quenching buffer (6 M guanidine hydrochloride in 100 mM Tris buffer, pH 8). The quenched reaction mixtures were loaded into the wells of a dot-blot apparatus (Bethesda Research Laboratories) assembled with a nitrocellulose membrane (Thermo scientific). The reaction mixtures were blotted onto the membrane using vacuum and the wells were washed with 200 μL of TBS (25 mM Tris pH 8.5, 137 mM NaCl, 2.7 mM KCl). The apparatus was disassembled and the membrane was blocked with 3% BSA in TBST (25 mM Tris pH 8.5, 137 mM NaCl, 2.7 mM KCl, 0.1% Tween 20) for 1 h at RT. The membrane was then incubated with anti-pHis antibody (1:100 dilution in TBST with 3% BSA) for 1 h at RT, washed with TBST (3 × 5 min), and incubated with anti-Rabbit fluorescent antibody (Li-Cor, 800 nm, 1:15000 dilution in TBST with 3% BSA) for 1 h at RT. The membrane was washed (3 × 5 min) with TBST, rinsed with deionized water, and imaged on Li-Cor Odyssey Infrared Imager at 800 nm. The signal intensity of each dot was quantified using the Odyssey software.
Dephosphorylation Assay of PpsA
50 μL of PpsA stock (~25 μM) was diluted with 50 μL of assay buffer (final concentration ~12 μM) followed by addition of 3 μL of 1 mM ATP (final concentration 33 μM). The reaction mixture was incubated for 10 min at RT and then diluted with 500 μL of assay buffer. The mixture was then concentrated back to 100 μL using a Vivaspin® 500 microcolumn (MWCO 30,000) (Sartorius) to remove excess ATP. The protein concentration was determined by measuring the absorbance at 280 nm. The phosphorylated PpsA sample was then diluted with assay buffer to a final concentration 500 nM. 170 μL of this phosphorylated PpsA stock solution was transferred to a siliconized Eppendorf tube followed by addition of 10 μL of 200 mM α-KG stock (final 10 mM), 10 μL of 200 mM glutamate stock (final 10 mM) or 10 μL of assay buffer. The sample was then incubated at RT for 10 min. The reaction was initiated by adding 20 μL of 100 μM sodium pyruvate (final 10 μM). At time points (t=0, 10, 30, and 45 s), 20 μL of the reaction mix was taken out and added into 100 μL of Quenching Buffer. The quenched reaction mixtures were loaded into the wells of a dot-blot apparatus (Bethesda Research Laboratories) assembled with a nitrocellulose membrane and analyzed by Western blot as described above. Full details can be found in Supplementary Note 8.
High-resolution nano-UPLC-MS analysis of pHis proteins and peptides
LC-MS and MS/MS analyses of pHis samples were performed on high-resolution, high-mass-accuracy, reversed-phase nano-UPLC-MS platforms, consisting of either a nano-flow capillary ultra high performance LC system (Nano Ultra 2D Plus, Eksigent) coupled to an LTQ-Orbitrap XL hybrid mass spectrometer (ThermoFisher Scientific) outfitted with a Triversa NanoMate ion source robot (Advion), or an Easy nLC Ultra 1000 nano-UPLC system (ThermoFisher Scientific) coupled to a Velos Pro-Orbi Elite hybrid mass spectrometer (ThermoFisher Scientific) equipped with a Flex Ion source (Proxeon Biosystems). Sample concentration and washing was accomplished online using a trapping capillary column (150 μm x ca. 40 mm, packed with 3 μm, 100 Å Magic AQ C18 resin (Michrom)) at a flow rate of 4 μL/min for 4 min, while separation was achieved using an analytical capillary column (75 μm x ca. 15 cm, packed with 1.7 μm 100 Å BEH C18 resin (Waters)), under a linear gradient of A and B solutions (solution A: 3% acetonitrile/0.1% formic acid/0.1% acetic acid; solution B: 97% acetonitrile/0.1% formic acid/0.1% acetic acid) over 90 min at a flow rate of 250–300 nL/min. On the Orbi XL platform, nanospray ionization was carried out using the NanoMate ion source at 1.74 kV, with the LTQ heated capillary set to 200 °C, while on the Orbi Elite platform, nanospray was achieved using sprayer tips made from PicoFRIT capillaries (New Objective) using a voltage of 2.1 kV, with the Velos heated capillary at 200 °C. Full-scan mass spectra were acquired in the Orbitrap in positive-ion mode over the m/z range of 335–1800 at a resolution of 100,000 (Orbi XL) or 120,000 (Orbi Elite). MS/MS spectra were simultaneously acquired using CID in the LTQ for the seven (Orbi XL) or fifteen (Orbi Elite) most abundant multiply charged species in the full-scan spectrum having signal intensities of >1000 NL. For protein identification experiments, dynamic exclusion was set such that MS/MS was acquired only once for each species over a period of 120 s. All spectra were acquired in profile mode. See Supplementary Note 9 for full description of MS Data Analysis.
Supplementary Material
Acknowledgments
We would like to acknowledge S. Darst and M. Bick for providing KinB, and J. Rabinowitz and M. Reaves for providing NCM 3722 cells. We thank J. Rabinowitz, M. Reaves, and B. Wang for stimulating discussions, S. Kyin and H. Shwe for their assistance in MS. This work was funded by the U. S. National Institutes of Health (5R01GM095880). J-M.K. and R.C.O. were supported by a postdoctoral fellowship from Damon Runyon Cancer Research Foundation (DRG-2005-09) and a National Institutes of Health Research Service Award (1F32CA167901), respectively.
Footnotes
AUTHOR CONTRIBUTIONS
J-M.K. and R.C.O. and T.W.M. designed the experiments. J-M.K., R.C.O and D.H.P. performed the experiments. J-M.K. and R.C.O. prepared new reagents. Experimental results were analyzed and the manuscript written by J-M.K., R.C.O., D.H.P., and T.W.M.
COMPETING FINANCIAL INTERESTS
J.-M.K. and T.W.M. are co-inventors of a patent application on stable phosphohistidine analogs.
References
- 1.Walsh CT. posttranslational modification of proteins: expanding nature’s inventory. Roberts and Company Publishers; Greenwood Village: 2006. Protein phosphorylation by protein kinases; pp. 35–80. [Google Scholar]
- 2.Grimsrud PA, Swaney DL, Wenger CD, Beauchene NA, Coon JJ. Phosphoproteomics for the masses. ACS Chem Biol. 2010;5:105–119. doi: 10.1021/cb900277e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kee JM, Muir TW. Chasing Phosphohistidine, an elusive sibling in the phosphoamino acid family. ACS Chem Biol. 2012;7:44–51. doi: 10.1021/cb200445w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matthews HR. Protein kinases and phosphatases that act on histidine, lysine, or arginine residues in eukaryotic proteins: a possible regulator of the mitogen-activated protein kinase cascade. Pharmacol Therapeut. 1995;67:323–350. doi: 10.1016/0163-7258(95)00020-8. [DOI] [PubMed] [Google Scholar]
- 5.Attwood PV, Besant PG, Piggott MJ. Focus on phosphoaspartate and phosphoglutamate. Amino Acids. 2010;40:1035–1051. doi: 10.1007/s00726-010-0738-5. [DOI] [PubMed] [Google Scholar]
- 6.Boyer PD, Peter JB, Ebner KE, Deluca M, Hultquist DE. Identification of phosphohistidine in digests from a probable intermediate of oxidative phosphorylation. J Biol Chem. 1962;237:3306–3308. [PubMed] [Google Scholar]
- 7.Attwood PV, Piggott MJ, Zu XL, Besant PG. Focus on phosphohistidine. Amino Acids. 2007;32:145–156. doi: 10.1007/s00726-006-0443-6. [DOI] [PubMed] [Google Scholar]
- 8.Rikova K, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 9.Olsen JV, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 10.Carlson HK, et al. Use of a semisynthetic epitope to probe histidine kinase activity and regulation. Anal Biochem. 2010;397:139–143. doi: 10.1016/j.ab.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kee JM, Villani B, Carpenter LR, Muir TW. Development of stable phosphohistidine analogues. J Am Chem Soc. 2010;132:14327–14329. doi: 10.1021/ja104393t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mcallister TE, Nix MG, Webb ME. Fmoc-chemistry of a stable phosphohistidine analogue. Chem Commun (Camb) 2011;47:1297–1299. doi: 10.1039/c0cc04238b. [DOI] [PubMed] [Google Scholar]
- 13.Mukai S, et al. Stable triazolylphosphonate analogues of phosphohistidine. Amino Acids. 2011;43:857–874. doi: 10.1007/s00726-011-1145-2. [DOI] [PubMed] [Google Scholar]
- 14.Hunter T. Tyrosine phosphorylation: thirty years and counting. Curr Opin Cell Biol. 2009;21:140–146. doi: 10.1016/j.ceb.2009.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rush J, et al. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol. 2004;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 16.Frackelton AR, Posner M, Kannan B, Mermelstein F. Generation of monoclonal antibodies against phosphotyrosine and their use for affinity purification of phosphotyrosine-containing proteins. Meth Enzymol. 1991;201:79–92. doi: 10.1016/0076-6879(91)01010-y. [DOI] [PubMed] [Google Scholar]
- 17.Duclos B, Marcandier S, Cozzone AJ. Chemical-properties and separation of phosphoamino acids by thin-layer chromatography and or electrophoresis. Meth Enzymol. 1991;201:10–21. doi: 10.1016/0076-6879(91)01004-l. [DOI] [PubMed] [Google Scholar]
- 18.Meadow ND, Fox DK, Roseman S. The bacterial phosphoenol-pyruvate: glycose phosphotransferase system. Annu Rev Biochem. 1990;59:497–542. doi: 10.1146/annurev.bi.59.070190.002433. [DOI] [PubMed] [Google Scholar]
- 19.Waygood EB, Steeves T. Enzyme-I of the phosphoenolpyruvate-sugar phosphotransferase system of Escherichia-coli - purification to homogeneity and some properties. Can J Biochem. 1980;58:40–48. doi: 10.1139/o80-006. [DOI] [PubMed] [Google Scholar]
- 20.Venditti V, Ghirlando R, Clore GM. Structural basis for enzyme I inhibition by α-Ketoglutarate. ACS Chem Biol. 2013 doi: 10.1021/cb400027q. 130329145716008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doucette CD, Schwab DJ, Wingreen NS, Rabinowitz JD. α-ketoglutarate coordinates carbon and nitrogen utilization via enzyme I inhibition. Nat Chem Biol. 2011;7:894–901. doi: 10.1038/nchembio.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chulavatnatol M, Atkinson DE. Phosphoenolpyruvate synthetase from Escherichia coli Effects of adenylate energy charge and modifier concentrations. J Biol Chem. 1973;248:2712–2715. [PubMed] [Google Scholar]
- 23.Yuan J, et al. Metabolomics-driven quantitative analysis of ammonia assimilation in E. coli. Mol Syst Biol. 2009;5:302. doi: 10.1038/msb.2009.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lasker M, et al. Protein histidine phosphorylation: increased stability of thiophosphohistidine. Protein Sci. 1999;8:2177–2185. doi: 10.1110/ps.8.10.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schenkels C, Erni B, Reymond J. Phosphofurylalanine, a stable analog of phosphohistidine. Bioorg Med Chem Lett. 1999;9:1443–1446. doi: 10.1016/s0960-894x(99)00209-7. [DOI] [PubMed] [Google Scholar]
- 26.Klumpp S, Krieglstein J. Reversible phosphorylation of histidine residues in proteins from vertebrates. Sci Signal. 2009;2:pe13. doi: 10.1126/scisignal.261pe13. [DOI] [PubMed] [Google Scholar]
- 27.Frackelton AR, Ross AH, Eisen HN. Characterization and use of monoclonal antibodies for isolation of phosphotyrosyl proteins from retrovirus-transformed cells and growth factor-stimulated cells. Mol Cell Biol. 1983;3:1343–1352. doi: 10.1128/mcb.3.8.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paul R, et al. Allosteric regulation of histidine kinases by their cognate response regulator determines cell fate. Cell. 2008;133:452–461. doi: 10.1016/j.cell.2008.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Ann Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 30.Patnaik R, Liao JC. Engineering of Escherichia coli central metabolism for aromatic metabolite production with near theoretical yield. Appl Environ Microbiol. 1994;60:3903–3908. doi: 10.1128/aem.60.11.3903-3908.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.