Abstract
Cells are constantly exposed to various oxidants, either generated endogenously due to metabolic activity or exogenously. One way that cells respond to oxidants is through the action of redox-regulated proteins. These proteins also play important roles in oxidant signaling and protein biogenesis events. The key sensors built into redox-regulated proteins are cysteines, which undergo reversible thiol oxidation in response to changes in the oxidation status of the cellular environment. In this review, we discuss three examples of redox-regulated proteins found in bacteria, mitochondria, and chloroplasts. These proteins use oxidation of their redox-sensitive cysteines to reversibly convert large structural domains into more disordered regions or vice versa. These massive structural rearrangements are directly implicated in the functions of these proteins.
Introduction
Cellular processes rely heavily on and are regulated by reversible post-translational modifications, including cysteine oxidation, phosphorylation, and acetylation. These modifications provide the proteome with enormous diversity, flexibility, and the ability to undergo rapid changes in cellular pathways and cell fate in response to changes in environment and metabolic state [1–3]. Post-translational modifications result in reversible covalent changes in specific amino acids, affecting charge and hydrophobicity and leading to alterations in recognition sites and protein shapes. These changes can be tightly regulated and are generally faster and energetically less expensive than transcription or translational means of regulation [2]. Recent studies suggest that the functional diversity and plasticity of proteins may be increased further by the introduction of post-translational modifications at residues located within disordered regions[ 4–9]. These post-translational modifications have the potential to promote reversible large-scale order ⇔ disorder transitions, converting structured domains into disordered regions and vice versa. One type of post-translational folding switch is constituted by redox-sensitive cysteine thiols, whose capacity to form reversible disulfide bonds promotes order ⇔ disorder transitions that reversibly alter the function of the protein depending on the redox environment [10–14].
A redox-controlled order-to-disorder transition: activation of the stress-specific chaperone Hsp33
Exposure of bacteria to hypochlorous acid (HOCl), the active ingredient of bleach and a potent antimicrobial produced by cells of the innate immune system, causes oxidative protein unfolding and aggregation [15]. As a defense mechanism, bacterial Hsp33, a chaperone specialized to rapidly sense oxidative stress conditions that lead to protein unfolding is activated[ 15–17]. Once activated, Hsp33 protects proteins against toxic protein aggregation and thereby shields bacteria against oxidative stress-induced cell death. How do stress conditions that oxidize, unfold, and inactivate countless other proteins lead to the activation of this chaperone? Oxidative stress conditions elicit the same response in Hsp33 as they do in many other oxidation sensitive proteins—they oxidize and unfold this protein. However, instead of inactivating this chaperone, unfolding serves as the trigger for Hsp33’s activation. Thus, Hsp33 not only represents a new paradigm in chaperone action but belongs to a growing number of proteins, which gain activity upon protein unfolding [18]. Since it has been recognized that many if not most proteins that have been categorized as intrinsically disordered, do under at least some physiologically relevant conditions gain structure, we will refer to them as conditionally rather than intrinsically disordered proteins [19].
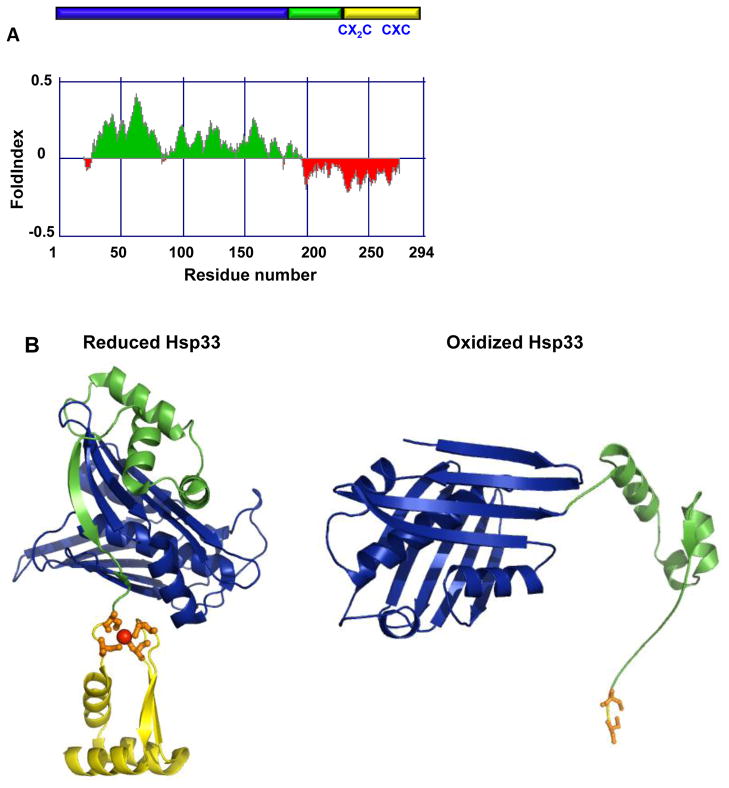
How is Hsp33’s unfolding regulated and, even more importantly, how does unfolding lead to its functional activation? Under non-stress conditions, Hsp33 is a compactly folded zinc-binding protein with negligible chaperone activity (Figure 1). Coordinated by four absolutely-conserved cysteine thiolates, zinc confers stability to the zinc-binding domain and keeps Hsp33 inactive as long as reducing, non-stress conditions prevail. Exposure to fast-acting oxidants, such as HOCl, causes the rapid formation of two intramolecular disulfide bonds. Though disulfide bonds are normally stabilizing and structure forming, in a surprising twist, their formation in Hsp33 causes the release of zinc and with that the unfolding of the zinc-binding domain [11,20,21]. This oxidation-induced unfolding is facilitated by the near absence of stabilizing hydrophobic core amino acids in the zinc-binding domain. Even more surprising and by a yet to be identified mechanism, disulfide bond formation in the C-terminal zinc-binding domain of Hsp33 extends its destabilizing effect almost 50 residues upstream of the zinc binding domain. It affects a flexible linker region, which is folded in reduced Hsp33 but adopts a disordered conformation once Hsp33 is oxidized. In the reduced form of Hsp33, a close network of inter-atomic interactions connects certain residues in Hsp33’s N-terminal domain (aa 1–178) with those in the C-terminal redox switch domain. One possible model is that an oxidation-mediated shift in the position of these few highly interactive residues works in a zipper-like fashion, decreasing the tight inter-domain packing one residue at a time and eventually causing the observed destabilization of the linker region. Hsp33’s zinc-coordinating cysteines thus serve as oxidant sensors that control the thermodynamic stability of the adjacent linker region via their oxidation state. It appears that the folding status of the linker region helps to determine Hsp33’s affinity for client proteins. This idea is consistent with the finding that introduction of a single linker-destabilizing mutation causes the constitutive activation of reduced, zinc-coordinated Hsp33 [22].
Figure 1.
The redox-regulated chaperone Hsp33. (A) Schematic representation of the Hsp33 structure. The N-terminal domain is depicted in blue, the linker region is depicted in green and the Zn-binding domain is shown in yellow. Disorder prediction of E. coli Hsp33 was made using the FoldIndex software[ 69]. Predicted disordered regions are shown in red whereas predicted ordered regions are shown in green. (B) Model of reduced E. coli Hsp33 based on the structure of reduced B. subtilis Hsp33 (pdb:1VZY) [22] and structure of a truncated form of oxidized E. coli Hsp33 (pdb:1HW7). Cysteines involved in Hsp33’s activation process are shown on the linear presentation in (A) and on the structure (B) by stick presentation.
Hsp33 is not the only molecular chaperone that uses conditionally disordered regions to bind to unfolding client proteins. The acid-activated chaperone HdeA, for instance, undergoes pH-induced unfolding. This unfolding allows HdeA to bind other acid-denatured proteins and prevent their aggregation at low pH conditions [23,24]. Also, in the small heat shock proteins, widely conserved among bacteria and eukaryotes, disordered regions appear to be involved in client binding [25,26]. At first glance, the concept that chaperones use conditionally disordered regions to interact with unfolding proteins is very appealing as the plasticity of binding inherent to these regions could provide a long-sought explanation as to how individual chaperones can bind multiple different client proteins. This idea is also consistent with the fact that disordered regions are often found in proteins that have several different partner proteins, acting as flexible “hubs “in protein-protein interactions [27–30]. Moreover, the highly hydrophilic nature of the interactions between disordered regions and unfolding client proteins will certainly increase the solubility of the client proteins and counteract protein aggregation. However, one of the hallmarks of many chaperone client proteins is that they have hydrophobic surfaces, which are transiently exposed, prone to aggregation, and in need of protection[ 31]. So how do conditionally disordered chaperones recognize and bind their clients? Moreover, why do conditionally disordered proteins not become client proteins for other chaperones? Answers to these questions may help to change how we think about chaperones, as well as conditionally and intrinsically disordered proteins.
An enhanced understanding of the role of conditionally disordered regions in client binding has resulted from H/D exchange experiments with Hsp33 and Hsp33-client protein complexes. These experiments showed that Hsp33’s linker region selectively binds to partially structured substrates, using them as a scaffold to refold the linker region and increasing complex stability [32]. A similar mechanism where disordered domains are utilized to recognize misfolded substrates was recently reported for another biological system involved in protein quality control, namely the yeast nuclear PQC ubiquitin ligase San1 [33]. San1 specifically recognizes misfolded ubiquitinated proteins via disordered C- and N-terminal regions [33]. In the case of San1, computational analysis predicts the presence of ordered stretches of ~20 aa sequences, interspersed at regular intervals with disordered regions. The authors suggest that this combination of motifs might be responsible for binding misfolded clients [33]. Whether this is also the case for Hsp33 and other conditionally disordered chaperones remains to be elucidated.
A redox-controlled disorder-to-order transition: activation of the copper chaperone COX17
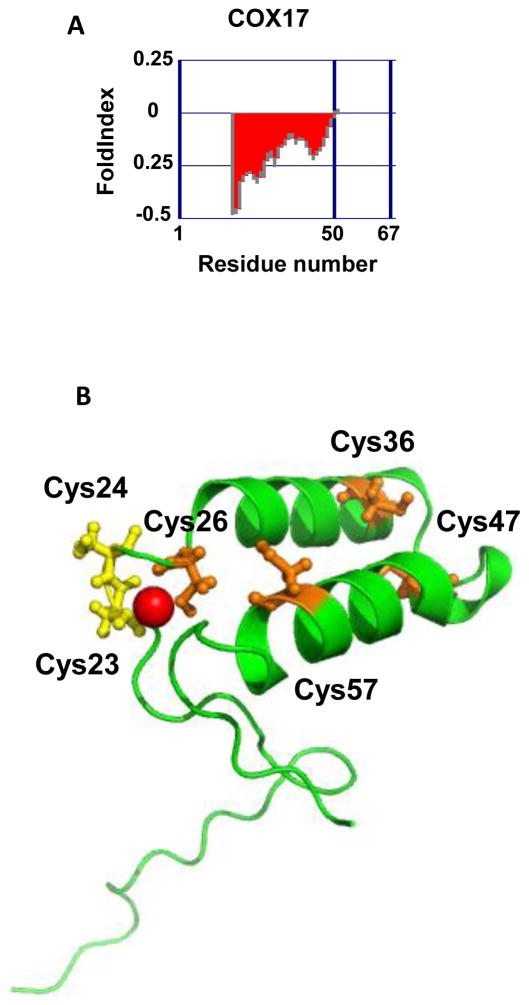
Mammalian cytochrome c oxidase is a 13-subunit complex located in the mitochondrial inner membrane. The loading of copper into this complex is a finely tuned process that involves several mitochondrial proteins, of which one of the most important is the small ~60 aa copper chaperone called COX17 [34–36]. This cysteine-rich protein undergoes a redox-mediated disorder-to-order transition upon its entry into the mitochondria. This transition impacts copper binding and the ability of COX17 to transfer copper to cytochrome c oxidase. Fully reduced and largely disordered when present within the reducing environment of the cytosol [12,37,38], COX17 interacts with the oxidoreductase/chaperone Mia40 upon entering the mitochondrial inter-membrane space [38]. Hydrophobic interactions combined with intermolecular disulfide bond formation between Mia40 and COX17 lead to the formation of the first helix in COX17. Formation of the first disulfide bond stabilizes this helix, which then serves as a scaffold to form the second helix in COX17, whose formation is concomitant with the second disulfide bond formation [38]. Thus, the introduction of two disulfide bonds converts the cytosolically unstructured COX17 into a structured coiled coil-helix-coiled coil-helix (CHCH) protein. The cysteines involved in this redox-controlled disorder-to-order transition are located within a conserved twin C-X9-C motif (Figure 2). In addition, COX17 contains a C-C motif (residues 23 and 24), which binds one Cu(I) ion [12]. Mass spectrometric analysis of the porcine homolog of COX17 revealed that copper binding strongly depends on the redox status of the protein. Whereas the fully reduced Cox17 cooperatively binds four Cu(I) ions in the form of a solvent-shielded cluster, COX17 with two disulfide bonds binds one Cu(I) ion. At this point, it is not entirely clear which oxidation state is the physiologically relevant form of COX17 in mitochondria, and how many Cu(I) ions are transferred to the COX complex.
Figure 2.
The copper chaperone Cox17. (A) Disorder prediction of S. cerevisiae Cox17 was made using the FoldIndex software [69]. Predicted disordered regions are shown in red whereas predicted ordered regions are shown in green. (B) Representative NMR structure of S. cerevisiae Cox17 with Cu(I) ion (pdb:1U96). Cysteines involved in disulfide bonds that stabilize the helical structure of the oxidized COX17 are shown in orange. The copper-coordinating cysteines are shown in yellow.
Recent NMR studies by the Pierattelli group characterized yeast COX17 and revealed that the unfolded, reduced form of COX17 consists of an ensemble of partially folded helices, which likely contributes to the rapid formation of the first helix in COX17 and disulfide bond formation within this protein, supportive of the “conformation selection” model [37]. This result emphasizes that the characterization of the nature of the structural ensembles present in intrinsically and conditionally disordered proteins should be of use in helping to elucidate the mechanism of order-to-disorder transitions and should particularly be helpful in discriminating between the “folding upon binding” and “conformational selection” models of protein-protein interaction. The “folding upon binding” model suggests that proteins fold upon binding their ligand or partner protein, whereas the “conformation selection” model suggests that proteins harbor several conformations that differ in their thermodynamic stabilities [39–43]. In the conformational selection model, ligand binding shifts the structural equilibrium towards a more stable conformation [41,42]. It was recently proposed by Espinoza-Fonseca[ 44] that both mechanistic models should be merged into a single one, “the synergistic model”, which could provide a useful basis for understanding how intrinsically disordered proteins bind their partners.
Another redox-controlled disorder-to-order transition: the role of CP12 as regulator of the Calvin Cycle
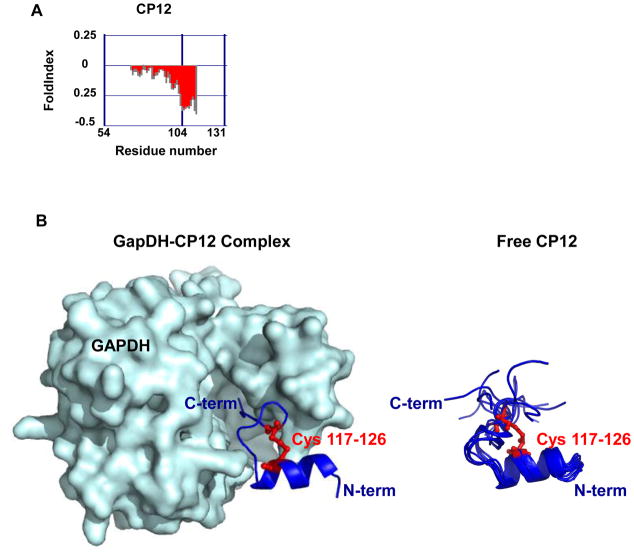
The redox-regulated protein CP12 serves as the “on-off” switch of the Calvin cycle in plants; it is responsible for turning CO2 fixation on under light conditions and off in the dark [45–47]. CP12 is an 80 amino acid intrinsically disordered protein[ 13,14,48,49] (Figure 3); it is widespread in oxygenic photosynthetic organisms and has recently been reported in viruses that infect marine cyanobacteria [50]. In the presence of light, when sufficient reducing equivalents and ATP are produced by photosynthesis to fuel conversion of CO2 into triosephosphates, all four cysteines in CP12 are reduced. In this redox state, CP12 is disordered and inactive [48,49]. In the dark, however, when reducing equivalents become scarce, CP12 forms two disulfide bonds and adopts a partially structured conformation. This makes CP12, like Hsp33 and HdeA, examples of conditionally disordered rather than “intrinsically” disordered proteins. Once oxidized and partially folded, CP12 forms a ternary complex with glyceraldehyde-3-phosphate dehydrogenase (GapDH) (Figure 3) and phosphoribonuclease (PRK), two enzymes critical for CO2 assimilation. This complex formation effectively blocks their enzymatic activities, thus causing the inactivation of the Calvin cycle in the dark. Structural studies revealed that conformational changes in CP12 occur upon oxidation and disulfide bond formation [13,14] and showed that the structural formation of CP12 upon binding GapDH stabilizes the complex [13,14,51,52]. Once photosynthesis restarts and reducing conditions are re-established, thioredoxin reduces CP12’s disulfide bonds, causing the dissociation of the complex and the reactivation of GAPDH and PRK [53,54].
Figure 3.
CP12 is a redox-regulator of the Calvin Cycle. (A) Disorder prediction of A. Thaliana CP12 was made using the FoldIndex software [69]. Predicted disordered regions are shown in red whereas predicted ordered regions are shown in green. (B, left) Crystal structure of the GAPDH - CP12 complex (pdb:3RVD). GapDH is shown in light blue surface representation while CP12 is shown in dark blue cartoon representation. Due to the disordered features of CP12, only a small fragment (aa 110-131) of the protein is structured. (B, right) Solution structures of the C-terminal fragment of CP12 (aa110-131) (pdb:2LJ9). Cysteines that form disulfide bond in oxidized CP12 are shown in red.
Conclusions
This review summarizes recent advances in our understanding of how redox-sensitive cysteines function as reversible protein folding state switches, controlling the activity of redox proteins through disulfide-mediated folding or unfolding. The thiol-disulfide switch can be tightly regulated by either oxidants or the prevailing redox conditions of the environment, making this mechanism an excellent strategy to rapidly and reversibly affect protein structure and function under specific stress conditions. The example proteins illustrated in this review have in common the propensity to interact with other proteins. They also refold their disordered regions upon binding their partners or clients. This makes them good examples of the recently recognized class of conditionally disordered proteins. The involvement of intrinsic plasticity in molecular recognition is often explained in terms of promiscuity and flexibility [5,27,55,56]. Beyond these notions, however, little is known as to how conditionally disordered proteins are selected to recognize aggregation-prone, partially unfolded intermediates in the case of chaperones like Hsp33, or fully functional enzymes, such as GapDH in the case of CP12 or Mia40 in the case of COX17. Current studies fall short in providing high-resolution structures of these complexes, making any mechanistic assessment challenging. It is clear that “disordered” or “unstructured” proteins exist as highly dynamic structural ensembles, and considerable effort has been made to characterize individual ensembles [57,58]. Yet it is not trivial to characterize how one ensemble differs from another, and a complex issue to relate ensemble differences to their respective functional differences. The situation is even more complex if we attempt to characterize the structural outcome of post-translational modifications, which provide the rapid “on-off” mechanism. In the most likely scenario, only one endpoint of this process, usually the most thermodynamically stable, is structurally defined. On a positive note, studies of protein disorders have prompted the development and adaptation of modern technologies, especially in the field of nuclear magnetic resonance (NMR), small angle X-ray scattering (SAXS), and mass spectrometry [37,59–68]. By using these technologies, we have obtained initial descriptive snapshots of conditionally disordered regions and structural ensemble selections. However, we still have a long way to go before we fully understand the structural mechanisms that promote highly regulated unfolding-folding transitions under specific physiological conditions and how specific residue modifications control these transitions.
Highlights.
Disulfide bond formation promotes large-scale order ⇔ disorder transitions
Redox-mediated unfolding of Hsp33 leads to its activation as chaperone
Redox-mediated folding of COX17 impacts its function as copper chaperone
Redox-mediated folding of CP12 makes it the “on-off” switch of the Calvin Cycle
Developments in structural techniques show promise in revealing protein disorder
Acknowledgments
We thank James Bardwell for critically reading this manuscript. This work was supported by a National Institutes of Health RO1 GM065318 award to U.J.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
- 1.Markiv A, Rambaruth ND, Dwek MV. Beyond the genome and proteome: targeting protein modifications in cancer. Curr Opin Pharmacol. 2012;12:408–413. doi: 10.1016/j.coph.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 2.Walsh CT, Garneau-Tsodikova S, Gatto GJ., Jr Protein posttranslational modifications: the chemistry of proteome diversifications. Angew Chem Int Ed Engl. 2005;44:7342–7372. doi: 10.1002/anie.200501023. [DOI] [PubMed] [Google Scholar]
- 3.Foster MW, Hess DT, Stamler JS. Protein S-nitrosylation in health and disease: a current perspective. Trends Mol Med. 2009;15:391–404. doi: 10.1016/j.molmed.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marin M, Ott T. Phosphorylation of intrinsically disordered regions in remorin proteins. Front Plant Sci. 2012;3:86. doi: 10.3389/fpls.2012.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitrea DM, Yoon MK, Ou L, Kriwacki RW. Disorder-function relationships for the cell cycle regulatory proteins p21 and p27. Biol Chem. 2012;393:259–274. doi: 10.1515/hsz-2011-0254. [DOI] [PubMed] [Google Scholar]
- **6.Mittag T, Orlicky S, Choy WY, Tang X, Lin H, Sicheri F, Kay LE, Tyers M, Forman-Kay JD. Dynamic equilibrium engagement of a polyvalent ligand with a single-site receptor. Proc Natl Acad Sci U S A. 2008;105:17772–17777. doi: 10.1073/pnas.0809222105. Analysis of the interaction between the intrinsically disordered protein Sic1 and its partner Cdc4 shows that protein complexes cannot be viewed as static structures. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hagai T, Azia A, Toth-Petroczy A, Levy Y. Intrinsic disorder in ubiquitination substrates. J Mol Biol. 2011;412:319–324. doi: 10.1016/j.jmb.2011.07.024. [DOI] [PubMed] [Google Scholar]
- *8.Shental-Bechor D, Smith MT, Mackenzie D, Broom A, Marcovitz A, Ghashut F, Go C, Bralha F, Meiering EM, Levy Y. Nonnative interactions regulate folding and switching of myristoylated protein. Proc Natl Acad Sci U S A. 2012;109:17839–17844. doi: 10.1073/pnas.1201803109. Interdisciplinary study, which discusses how myristoylation affects protein folding and binding. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iakoucheva LM, Radivojac P, Brown CJ, O’Connor TR, Sikes JG, Obradovic Z, Dunker AK. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wouters MA, Fan SW, Haworth NL. Disulfides as redox switches: from molecular mechanisms to functional significance. Antioxid Redox Signal. 2010;12:53–91. doi: 10.1089/ars.2009.2510. [DOI] [PubMed] [Google Scholar]
- 11.Ilbert M, Horst J, Ahrens S, Winter J, Graf PC, Lilie H, Jakob U. The redox-switch domain of Hsp33 functions as dual stress sensor. Nat Struct Mol Biol. 2007;14:556–563. doi: 10.1038/nsmb1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arnesano F, Balatri E, Banci L, Bertini I, Winge DR. Folding studies of Cox17 reveal an important interplay of cysteine oxidation and copper binding. Structure. 2005;13:713–722. doi: 10.1016/j.str.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 13.Fermani S, Trivelli X, Sparla F, Thumiger A, Calvaresi M, Marri L, Falini G, Zerbetto F, Trost P. Conformational selection and folding-upon-binding of intrinsically disordered protein CP12 regulate photosynthetic enzymes assembly. J Biol Chem. 2012;287:21372–21383. doi: 10.1074/jbc.M112.350355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gontero B, Maberly SC. An intrinsically disordered protein, CP12: jack of all trades and master of the Calvin cycle. Biochem Soc Trans. 2012;40:995–999. doi: 10.1042/BST20120097. [DOI] [PubMed] [Google Scholar]
- 15.Winter J, Ilbert M, Graf PC, Ozcelik D, Jakob U. Bleach activates a redox-regulated chaperone by oxidative protein unfolding. Cell. 2008;135:691–701. doi: 10.1016/j.cell.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jakob U, Muse W, Eser M, Bardwell JC. Chaperone activity with a redox switch. Cell. 1999;96:341–352. doi: 10.1016/s0092-8674(00)80547-4. [DOI] [PubMed] [Google Scholar]
- 17.Winter J, Linke K, Jatzek A, Jakob U. Severe oxidative stress causes inactivation of DnaK and activation of the redox-regulated chaperone Hsp33. Mol Cell. 2005;17:381–392. doi: 10.1016/j.molcel.2004.12.027. [DOI] [PubMed] [Google Scholar]
- 18.Bardwell JC, Jakob U. Conditional disorder in chaperone action. Trends Biochem Sci. 2012;37:517–525. doi: 10.1016/j.tibs.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Gall T, Romero PR, Cortese MS, Uversky VN, Dunker AK. Intrinsic disorder in the Protein Data Bank. J Biomol Struct Dyn. 2007;24:325–342. doi: 10.1080/07391102.2007.10507123. [DOI] [PubMed] [Google Scholar]
- 20.Vijayalakshmi J, Mukhergee MK, Graumann J, Jakob U, Saper MA. The 2. 2 A crystal structure of Hsp33: a heat shock protein with redox-regulated chaperone activity. Structure. 2001;9:367–375. doi: 10.1016/s0969-2126(01)00597-4. [DOI] [PubMed] [Google Scholar]
- 21.Graf PC, Martinez-Yamout M, VanHaerents S, Lilie H, Dyson HJ, Jakob U. Activation of the redox-regulated chaperone Hsp33 by domain unfolding. J Biol Chem. 2004;279:20529–20538. doi: 10.1074/jbc.M401764200. [DOI] [PubMed] [Google Scholar]
- 22.Cremers CM, Reichmann D, Hausmann J, Ilbert M, Jakob U. Unfolding of metastable linker region is at the core of Hsp33 activation as a redox-regulated chaperone. J Biol Chem. 2010;285:11243–11251. doi: 10.1074/jbc.M109.084350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malki A, Le HT, Milles S, Kern R, Caldas T, Abdallah J, Richarme G. Solubilization of protein aggregates by the acid stress chaperones HdeA and HdeB. J Biol Chem. 2008;283:13679–13687. doi: 10.1074/jbc.M800869200. [DOI] [PubMed] [Google Scholar]
- **24.Tapley TL, Korner JL, Barge MT, Hupfeld J, Schauerte JA, Gafni A, Jakob U, Bardwell JC. Structural plasticity of an acid-activated chaperone allows promiscuous substrate binding. Proc Natl Acad Sci U S A. 2009;106:5557–5562. doi: 10.1073/pnas.0811811106. One of the first detailed studies of conditionally disordered chaperones. The authors show how plasticity of the acid-activated chaperone HdeA promotes its binding to a variety of different folding intermediates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sudnitsyna MV, Mymrikov EV, Seit-Nebi AS, Gusev NB. The role of intrinsically disordered regions in the structure and functioning of small heat shock proteins. Curr Protein Pept Sci. 2012;13:76–85. doi: 10.2174/138920312799277875. [DOI] [PubMed] [Google Scholar]
- *26.Jaya N, Garcia V, Vierling E. Substrate binding site flexibility of the small heat shock protein molecular chaperones. Proc Natl Acad Sci U S A. 2009;106:15604–15609. doi: 10.1073/pnas.0902177106. This study characterizes the interaction between the small heat shock protein PsHsp18.1 and two different client proteins by using photo-activatable crosslinkers. The authors demonstrate that the intrinsically disordered N-terminal part of PsHsp18.1 is involved in client interaction, demonstrating the importance of flexibility for substrate binding. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uversky VN. Multitude of binding modes attainable by intrinsically disordered proteins: a portrait gallery of disorder-based complexes. Chem Soc Rev. 2011;40:1623–1634. doi: 10.1039/c0cs00057d. [DOI] [PubMed] [Google Scholar]
- 28.Meszaros B, Simon I, Dosztanyi Z. The expanding view of protein-protein interactions: complexes involving intrinsically disordered proteins. Phys Biol. 2011;8:035003. doi: 10.1088/1478-3975/8/3/035003. [DOI] [PubMed] [Google Scholar]
- 29.Stein A, Pache RA, Bernado P, Pons M, Aloy P. Dynamic interactions of proteins in complex networks: a more structured view. FEBS J. 2009;276:5390–5405. doi: 10.1111/j.1742-4658.2009.07251.x. [DOI] [PubMed] [Google Scholar]
- 30.Shimizu K, Toh H. Interaction between intrinsically disordered proteins frequently occurs in a human protein-protein interaction network. J Mol Biol. 2009;392:1253–1265. doi: 10.1016/j.jmb.2009.07.088. [DOI] [PubMed] [Google Scholar]
- 31.Frydman J. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu Rev Biochem. 2001;70:603–647. doi: 10.1146/annurev.biochem.70.1.603. [DOI] [PubMed] [Google Scholar]
- *32.Reichmann D, Xu Y, Cremers CM, Ilbert M, Mittelman R, Fitzgerald MC, Jakob U. Order out of disorder: working cycle of an intrinsically unfolded chaperone. Cell. 2012;148:947–957. doi: 10.1016/j.cell.2012.01.045. This study reveals how the redox-regulated chaperone Hsp33 uses its own order-to-disorder transition to promote disorder-to-order transitions in its bound client proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *33.Rosenbaum JC, Fredrickson EK, Oeser ML, Garrett-Engele CM, Locke MN, Richardson LA, Nelson ZW, Hetrick ED, Milac TI, Gottschling DE, et al. Disorder targets misorder in nuclear quality control degradation: a disordered ubiquitin ligase directly recognizes its misfolded substrates. Mol Cell. 2011;41:93–106. doi: 10.1016/j.molcel.2010.12.004. A comprehensive study that shows how the ubiquitin ligase San1 interacts with mis-folded substrates via intrinsically disordered N- and C-terminal domains. The authors characterize the architecture of these binding regions, which are built as modules of ordered and disordered regions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abajian C, Yatsunyk LA, Ramirez BE, Rosenzweig AC. Yeast cox17 solution structure and Copper(I) binding. J Biol Chem. 2004;279:53584–53592. doi: 10.1074/jbc.M408099200. [DOI] [PubMed] [Google Scholar]
- 35.Horng YC, Cobine PA, Maxfield AB, Carr HS, Winge DR. Specific copper transfer from the Cox17 metallochaperone to both Sco1 and Cox11 in the assembly of yeast cytochrome C oxidase. J Biol Chem. 2004;279:35334–35340. doi: 10.1074/jbc.M404747200. [DOI] [PubMed] [Google Scholar]
- 36.Leary SC. Redox regulation of SCO protein function: controlling copper at a mitochondrial crossroad. Antioxid Redox Signal. 2010;13:1403–1416. doi: 10.1089/ars.2010.3116. [DOI] [PubMed] [Google Scholar]
- 37.Bertini I, Felli IC, Gonnelli L, Vasantha Kumar MV, Pierattelli R. High-resolution characterization of intrinsic disorder in proteins: expanding the suite of (13)C-detected NMR spectroscopy experiments to determine key observables. Chembiochem. 2011;12:2347–2352. doi: 10.1002/cbic.201100406. [DOI] [PubMed] [Google Scholar]
- 38.Banci L, Bertini I, Cefaro C, Cenacchi L, Ciofi-Baffoni S, Felli IC, Gallo A, Gonnelli L, Luchinat E, Sideris D, et al. Molecular chaperone function of Mia40 triggers consecutive induced folding steps of the substrate in mitochondrial protein import. Proc Natl Acad Sci U S A. 2010;107:20190–20195. doi: 10.1073/pnas.1010095107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koshland DE., Jr Enzyme flexibility and enzyme action. J Cell Comp Physiol. 1959;54:245–258. doi: 10.1002/jcp.1030540420. [DOI] [PubMed] [Google Scholar]
- 40.Changeux JP, Edelstein S. Conformational selection or induced fit? 50 years of debate resolved. F1000 Biol Rep. 2011;3:19. doi: 10.3410/B3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frauenfelder H, Sligar SG, Wolynes PG. The energy landscapes and motions of proteins. Science. 1991;254:1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 42.Csermely P, Palotai R, Nussinov R. Induced fit, conformational selection and independent dynamic segments: an extended view of binding events. Trends Biochem Sci. 2010;35:539–546. doi: 10.1016/j.tibs.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hammes GG, Chang YC, Oas TG. Conformational selection or induced fit: a flux description of reaction mechanism. Proc Natl Acad Sci U S A. 2009;106:13737–13741. doi: 10.1073/pnas.0907195106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Espinoza-Fonseca LM. Reconciling binding mechanisms of intrinsically disordered proteins. Biochem Biophys Res Commun. 2009;382:479–482. doi: 10.1016/j.bbrc.2009.02.151. [DOI] [PubMed] [Google Scholar]
- 45.Wedel N, Soll J, Paap BK. CP12 provides a new mode of light regulation of Calvin cycle activity in higher plants. Proc Natl Acad Sci U S A. 1997;94:10479–10484. doi: 10.1073/pnas.94.19.10479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pohlmeyer K, Paap BK, Soll J, Wedel N. CP12: a small nuclear-encoded chloroplast protein provides novel insights into higher-plant GAPDH evolution. Plant Mol Biol. 1996;32:969–978. doi: 10.1007/BF00020493. [DOI] [PubMed] [Google Scholar]
- 47.Tamoi M, Miyazaki T, Fukamizo T, Shigeoka S. The Calvin cycle in cyanobacteria is regulated by CP12 via the NAD(H)/NADP(H) ratio under light/dark conditions. Plant J. 2005;42:504–513. doi: 10.1111/j.1365-313X.2005.02391.x. [DOI] [PubMed] [Google Scholar]
- 48.Graciet E, Gans P, Wedel N, Lebreton S, Camadro JM, Gontero B. The small protein CP12: a protein linker for supramolecular complex assembly. Biochemistry. 2003;42:8163–8170. doi: 10.1021/bi034474x. [DOI] [PubMed] [Google Scholar]
- 49.Marri L, Trost P, Trivelli X, Gonnelli L, Pupillo P, Sparla F. Spontaneous assembly of photosynthetic supramolecular complexes as mediated by the intrinsically unstructured protein CP12. J Biol Chem. 2008;283:1831–1838. doi: 10.1074/jbc.M705650200. [DOI] [PubMed] [Google Scholar]
- 50.Thompson LR, Zeng Q, Kelly L, Huang KH, Singer AU, Stubbe J, Chisholm SW. Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism. Proc Natl Acad Sci U S A. 2011;108:E757–764. doi: 10.1073/pnas.1102164108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Avilan L, Puppo C, Erales J, Woudstra M, Lebrun R, Gontero B. CP12 residues involved in the formation and regulation of the glyceraldehyde-3-phosphate dehydrogenase-CP12-phosphoribulokinase complex in Chlamydomonas reinhardtii. Mol Biosyst. 2012;8:2994–3002. doi: 10.1039/c2mb25244a. [DOI] [PubMed] [Google Scholar]
- 52.Matsumura H, Kai A, Maeda T, Tamoi M, Satoh A, Tamura H, Hirose M, Ogawa T, Kizu N, Wadano A, et al. Structure basis for the regulation of glyceraldehyde-3-phosphate dehydrogenase activity via the intrinsically disordered protein CP12. Structure. 2011;19:1846–1854. doi: 10.1016/j.str.2011.08.016. [DOI] [PubMed] [Google Scholar]
- 53.Howard TP, Metodiev M, Lloyd JC, Raines CA. Thioredoxin-mediated reversible dissociation of a stromal multiprotein complex in response to changes in light availability. Proc Natl Acad Sci U S A. 2008;105:4056–4061. doi: 10.1073/pnas.0710518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marri L, Zaffagnini M, Collin V, Issakidis-Bourguet E, Lemaire SD, Pupillo P, Sparla F, Miginiac-Maslow M, Trost P. Prompt and easy activation by specific thioredoxins of calvin cycle enzymes of Arabidopsis thaliana associated in the GAPDH/CP12/PRK supramolecular complex. Mol Plant. 2009;2:259–269. doi: 10.1093/mp/ssn061. [DOI] [PubMed] [Google Scholar]
- 55.Oldfield CJ, Meng J, Yang JY, Yang MQ, Uversky VN, Dunker AK. Flexible nets: disorder and induced fit in the associations of p53 and 14-3-3 with their partners. BMC Genomics. 2008;9 (Suppl 1):S1. doi: 10.1186/1471-2164-9-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tompa P, Csermely P. The role of structural disorder in the function of RNA and protein chaperones. FASEB J. 2004;18:1169–1175. doi: 10.1096/fj.04-1584rev. [DOI] [PubMed] [Google Scholar]
- 57.Fisher CK, Stultz CM. Constructing ensembles for intrinsically disordered proteins. Curr Opin Struct Biol. 2011;21:426–431. doi: 10.1016/j.sbi.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mittag T, Forman-Kay JD. Atomic-level characterization of disordered protein ensembles. Curr Opin Struct Biol. 2007;17:3–14. doi: 10.1016/j.sbi.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 59.Choy WY, Forman-Kay JD. Calculation of ensembles of structures representing the unfolded state of an SH3 domain. J Mol Biol. 2001;308:1011–1032. doi: 10.1006/jmbi.2001.4750. [DOI] [PubMed] [Google Scholar]
- 60.Schneider R, Huang JR, Yao M, Communie G, Ozenne V, Mollica L, Salmon L, Jensen MR, Blackledge M. Towards a robust description of intrinsic protein disorder using nuclear magnetic resonance spectroscopy. Mol Biosyst. 2012;8:58–68. doi: 10.1039/c1mb05291h. [DOI] [PubMed] [Google Scholar]
- 61.Wright PE, Dyson HJ. Linking folding and binding. Curr Opin Struct Biol. 2009;19:31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Salmon L, Nodet G, Ozenne V, Yin G, Jensen MR, Zweckstetter M, Blackledge M. NMR characterization of long-range order in intrinsically disordered proteins. J Am Chem Soc. 2010;132:8407–8418. doi: 10.1021/ja101645g. [DOI] [PubMed] [Google Scholar]
- 63.Kaaki W, Woudstra M, Gontero B, Halgand F. Exploration of CP12 conformational changes and of quaternary structural properties using electrospray ionization traveling wave ion mobility mass spectrometry. Rapid Commun Mass Spectrom. 2013;27:179–186. doi: 10.1002/rcm.6442. [DOI] [PubMed] [Google Scholar]
- 64.Smith DP, Giles K, Bateman RH, Radford SE, Ashcroft AE. Monitoring copopulated conformational states during protein folding events using electrospray ionization-ion mobility spectrometry-mass spectrometry. J Am Soc Mass Spectrom. 2007;18:2180–2190. doi: 10.1016/j.jasms.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bernado P, Svergun DI. Structural analysis of intrinsically disordered proteins by small-angle X-ray scattering. Mol Biosyst. 2012;8:151–167. doi: 10.1039/c1mb05275f. [DOI] [PubMed] [Google Scholar]
- *66.Devarakonda S, Gupta K, Chalmers MJ, Hunt JF, Griffin PR, Van Duyne GD, Spiegelman BM. Disorder-to-order transition underlies the structural basis for the assembly of a transcriptionally active PGC-1alpha/ERRgamma complex. Proc Natl Acad Sci U S A. 2011;108:18678–18683. doi: 10.1073/pnas.1113813108. This work examines the interaction between peroxisome proliferator activated receptor (PPAR), γ coactivator-1α (PGC-1α) and estrogen-related receptor (ERR). The authors show that PGC-1α undergoes a large disorder-to-order transition upon binding the ER receptor, providing a solution model of the complex. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Balasubramaniam D, Komives EA. Hydrogen-exchange mass spectrometry for the study of intrinsic disorder in proteins. Biochim Biophys Acta. 2012 doi: 10.1016/j.bbapap.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rob T, Liuni P, Gill PK, Zhu S, Balachandran N, Berti PJ, Wilson DJ. Measuring dynamics in weakly structured regions of proteins using microfluidics-enabled subsecond H/D exchange mass spectrometry. Anal Chem. 2012;84:3771–3779. doi: 10.1021/ac300365u. [DOI] [PubMed] [Google Scholar]
- 69.Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, Silman I, Sussman JL. FoldIndex: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics. 2005;21:3435–3438. doi: 10.1093/bioinformatics/bti537. [DOI] [PubMed] [Google Scholar]