SUMMARY
Cancer cells undergo a metabolic reprogramming but little is known about metabolic alterations of other cells within tumors. We use mass spectrometry-based profiling and a metabolic pathway-based systems analysis to compare 21 primary human lung tumor cancer-associated fibroblast lines (CAFs) to “normal” fibroblast lines (NFs) generated from adjacent non-neoplastic lung tissue. CAFs are pro-tumorigenic, although the mechanisms by which CAFs support tumors have not been elucidated. We have identified several pathways whose metabolite abundance globally distinguished CAFs from NFs, suggesting that metabolic alterations are not limited to cancer cells. In addition, we found metabolic differences between CAFs from high and low glycolytic tumors that might reflect distinct roles of CAFs related to the tumor’s glycolytic capacity. One such change was an increase of dipeptides in CAFs. Dipeptides primarily arise from the breakdown of proteins. We found in CAFs an increase in basal macroautophagy which likely accounts for the increase in dipeptides. Furthermore, we demonstrate a difference between CAFs and NFs in the induction of autophagy promoted by reduced glucose. In sum, our data suggest increased autophagy may account for metabolic differences between CAFs and NFs and may play additional as yet undetermined roles in lung cancer.
INTRODUCTION
Tumors are comprised of malignant cancer cells and stromal cells that constitute the tumor microenvironment. Research efforts have predominantly focused on understanding the biology of the epithelial component of tumors, leading to the identification of genes and signaling pathways dysregulated in cancer cells. The non-epithelial cellular elements of the tumor mass, although not by themselves malignant, have essential roles in carcinogenesis by supporting the cancer cells (1, 2). Cancer-associated fibroblasts (CAFs) of the tumor microenvironment are pro-tumorigenic, although the exact functions of CAFs have yet to be fully delineated (3, 4). Proposed functions of CAFs include secretion of growth factors, of pro-angiogenic VEGF, and/or of proteases that remodel the extracellular matrix (5-9). The distinct pro-tumorigenic activities of CAFs identified in the different studies might reflect tumor-type specific roles for CAFs.
Proliferation requires growth promoting signaling and an appropriate nutrient milieu. Cancer cells frequently have a predominantly glycolytic metabolism, the “Warburg effect” (10, 11), that is required for the growth of some tumors [e.g., (12)]. The emerging view is that reliance on a predominantly glycolytic metabolism supports the anabolic needs of cancer cells at the expense of efficient production of ATP from glucose (11). Little is known about possible metabolic alterations in the stroma cells of the tumor microenvironment. Alterations in stromal cell metabolism might be advantageous to the cancer cells, as would be the case if stroma cell metabolism was coupled to that of the cancer cells [e.g., (13)].
Here we use mass spectrometry-based profiling of the abundances of 203 biochemicals of 46 metabolic pathways/groups to compare primary human lung tumor CAFs to “normal” fibroblasts (NFs) isolated from non-neoplastic lung tissue located at least 5 cm away from the tumor. We found differences in several metabolic pathways that distinguish CAFs from NFs, suggesting that alterations in cellular metabolism are not limited to epithelial cancer cells of the tumor. Moreover, we found differences between CAFs and NFs that correlate with the glycolytic capacity of the tumor, one of which was a relative increase in CAFs of 17 unrelated dipeptides. The relative difference in dipeptides was most significant between CAFs derived from highly glycolytic tumors and their paired NFs although dipeptides in CAFs from less glycolytic tumors were also increased compared to paired NFs. Unexpectedly, dipeptides were also increased in NFs isolated from lung tissue of individuals with highly glycolytic tumors compared to those from less glycolytic tumors, suggesting either a tumor specific field effect extending beyond 5 cm or metabolic alteration in NFs induced by a widespread inflammatory or other protumorigenic parenchymal milieu. Dipeptides arise from the breakdown of proteins. In CAFs we find evidence for increased autophagy, a lysosome-related catabolic mechanism, which likely explains the increase in dipeptides. We also find that reducing the amount of glucose in the medium from 25 to 5 mM causes an increase of dipeptides and autophagy in NFs but not CAFs, suggesting that CAFs and NFs have different set points for glucose induction of autophagy. There is mounting evidence that increased degradative pathways (e.g., autophagy) in cancer cells, potentially as stress response mechanisms, have roles in tumor maintenance and possibly in resistance to chemotherapy (14-16). Our studies provide evidence that increased autophagy and lysosomal capacity are characteristics of human lung CAFs, alterations that might contribute to the pro-tumorigenic activity of CAFs.
MATERIALS AND METHODS
Human subjects
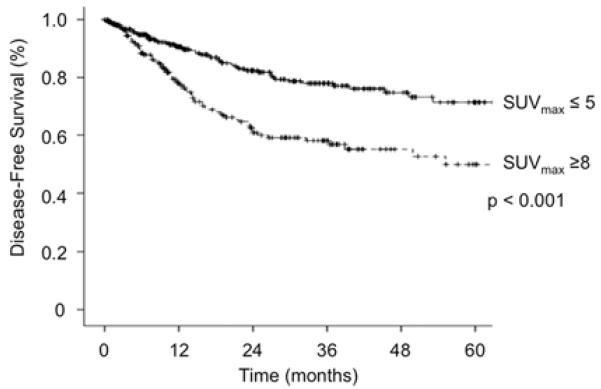
The clinical study (Fig. 3) was an Institutional Review Board approved review of a prospectively assembled thoracic surgery database of patients with non-small cell lung cancer (from February, 2000 to December, 2009). Patient consent was waived. Patients with surgically resected tumors (stage I-III) and documented SUVmax values by positron emission tomography were considered eligible for this review (n=693). Patients were segregated into two cohorts, those with SUVmax≥8 (n=231, 33%) and those with SUVmax≤5 (n=350, 50%). Disease free survival (DFS) for the cohort (from date of surgery to recurrent or death) was evaluated by Kaplan-Meier (KM) survival analysis. Log Rank test was used for comparison of DFS between the two groups. Median follow-up was 23 months.
Figure 3. High tumor SUVmax (SUV ≥8) correlates with decreased disease-free survival.
Disease-free survival of 581 NSCLC patients from February 2000 to December 2009 sorted by SUVmax (SUV ≥8: n=231; SUV≤5: n= 350; median follow-up 23 months).
Cells
Cells were derived from resected tumors and adjacent non-neoplastic tissue in an Institutional Review Board approved study. Patients consented to use of cells derived from their tissue for this study. Tissue, which was processed within 2 hrs of resection, was minced with a razor and incubated in 1 mg/ml collagenase D for 60 min with agitation at 37°C. The material was passed through a 70 μm sieve, red blood cells lysed, and extensively washed in HEPES-buffered DMEM (25 mM glucose) prior to plating in DMEM supplemented with 10% fetal bovine serum (FBS). When the cultures reached ~80% confluence, cells were passaged and expanded further by growth. Cell pellets (≥2.5 × 106 cells) were washed with phosphate-buffered saline, flash frozen in a dry-ice ethanol bath and stored at −80°C. In some cases, cells were expanded in 5 mM glucose-DMEM-10% fetal bovine serum.
Mass spectrometry
Sample Accessioning
Each sample received was accessioned into the Metabolon LIMS system and was assigned by the LIMS a unique identifier that was associated with the original source identifier only. This identifier was used to track all sample handling, tasks, results etc. The samples (and all derived aliquots) were tracked by the LIMS system. All portions of any sample were automatically assigned their own unique identifiers by the LIMS when a new task is created; the relationship of these samples is also tracked. All samples were maintained at −80 °C until processed.
Sample Preparation
Samples were prepared using the automated MicroLab STAR® system from Hamilton Company. A recovery standard was added prior to the first step in the extraction process for QC purposes. Sample preparation was conducted using aqueous methanol extraction process to remove the protein fraction while allowing maximum recovery of small molecules. The resulting extract was divided into four fractions: one for analysis by UPLC/MS/MS (positive mode), one for UPLC/MS/MS (negative mode), one for GC/MS, and one for backup. Samples were placed briefly on a TurboVap® (Zymark) to remove the organic solvent. Each sample was then frozen and dried under vacuum. Samples were then prepared for the appropriate instrument, either UPLC/MS/MS or GC/MS.
Ultrahigh performance liquid chromatography/Mass Spectroscopy (UPLC/MS/MS)
The LC/MS was based on a Waters ACQUITY ultra-performance liquid chromatography (UPLC) and a Thermo-Finnigan linear trap quadrupole (LTQ) mass spectrometer, which consisted of an electrospray ionization (ESI) source and linear ion-trap (LIT) mass analyzer. The sample extract was dried then reconstituted in acidic or basic LC-compatible solvents, each of which contained 8 or more injection standards at fixed concentrations to ensure injection and chromatographic consistency. One aliquot was analyzed using acidic positive ion optimized conditions and the other using basic negative ion optimized conditions in two independent injections using separate dedicated columns. Extracts reconstituted in acidic conditions were gradient eluted using water and methanol containing 0.1% formic acid, while the basic extracts, which also used water/methanol, contained 6.5mM Ammonium Bicarbonate. The MS analysis alternated between MS and data-dependent MS2 scans using dynamic exclusion. Raw data files were archived and extracted.
Gas chromatography/Mass Spectroscopy (GC/MS)
The samples destined for GC/MS analysis were re-dried under vacuum desiccation for a minimum of 24 hours prior to being derivatized under dried nitrogen using bistrimethyl-silyl-triflouroacetamide (BSTFA). The GC column was 5% phenyl and the temperature ramp was from 40° to 300° C in a 16 minute period. Samples were analyzed on a Thermo-Finnigan Trace DSQ fast-scanning single-quadrupole mass spectrometer using electron impact ionization. The instrument was tuned and calibrated for mass resolution and mass accuracy on a daily basis. The information output from the raw data files was automatically extracted as discussed below.
Quality assurance/QC
For QA/QC purposes, additional samples were included with each day’s analysis. These samples included extracts of a pool of well-characterized human plasma, extracts of a pool created from a small aliquot of the experimental samples, and process blanks. QC samples were spaced evenly among the injections and all experimental samples were randomly distributed throughout the run. A selection of QC compounds was added to every sample for chromatographic alignment, including those under test. These compounds were carefully chosen so as not to interfere with the measurement of the endogenous compounds.
Data extraction and compound identification
Raw data was extracted, peak-identified and QC processed using Metabolon’s hardware and software. These systems are built on a web-service platform utilizing Microsoft’s. NET technologies, which run on high-performance application servers and fiber-channel storage arrays in clusters to provide active failover and load-balancing. Compounds were identified by comparison to library entries of purified standards or recurrent unknown entities. More than 2400 commercially available purified standard compounds have been acquired and registered into LIMS for distribution to both the LC and GC platforms for determination of their analytical characteristics.
Data Normalization
Missing values (if any) were assumed to be below the level of detection. However, biochemicals that were detected in all samples from one or more groups but not in samples from other groups were assumed to be near the lower limit of detection in the groups in which they were not detected. In these cases the missing values were replaced with the minimum value obtained for the biochemical within all groups. Each biochemical value was then normalized to cell number and scaled to their median value. The data were also analyzed after normalizing the individual metabolites in each sample to the median of all metabolites in that sample. This scaling accounts for any difference between samples due to a change in the abundance of all metabolites, as would be the case if there were differences in cell size among groups. Similar results were obtained in all comparisons regardless of whether the data for each cell line were scaled to the median value of that cell line. The data sets without and with scaling to the median of all metabolites in that sample are presented in Supplemental table 1. The results presented are the significant differences common to the analyses of the data without and with the scaling.
Heat maps
Heat map data were scaled up for zero mean and unit variance by first subtracting the mean of each row for the matrix needs to be visualized and then dividing each row by its standard deviation. This provides a matrix where each metabolite concentrations have mean zero and standard deviation 1.
Statistical Analysis
An unpaired t-test for differential abundance with a standard two-tailed and two-sample t-test was done using mattes function in MATLAB on each metabolite to calculate the Wilcoxon t-statistic for two groups being compared. An unpaired t-test was then used to perform t-test over the t-scores and fold-change (log2) of metabolites grouped as required for the analysis. Resultant p values were then adjusted for false discovery rate as described by Benjamini-Hochberg correction for multiple testing. The same analysis was repeated with random shuffling (104 times) of the group labels (permutation testing). Results that had a probability of less than 0.05 of appearing in this shuffling exercise were selected as significant. Complete analysis was done in MATLAB R2011b with Bioinformatics (ver 4.0) and Statistics (ver 7.6) toolboxes.
To analyze biochemicals grouped into 7 main groups, we compared the distributions of t-scores of the metabolites of that group (obtained from t-test as described above) to the distribution of the t-scores of the rest of the metabolites. The p-values for this comparison for each group were determined after correcting for multiple testing. To analyze biochemicals grouped into 46 subgroups, we compared the distributions of t-scores (obtained from t-test as described above) and the mean log2-fold change of the metabolites in each group with those of rest of the metabolites combined. Significance was calculated by performing an unpaired t-test assuming normal distributions of unequal variances. P-values corrected for multiple testing were obtained for each sub group as described above. Subgroups having an adjusted p -value <0.05 from t-scores comparisons (which captures the variance of data) and adjusted p-value <0.05 from mean fold change comparison were selected. The data were analyzed in two ways, with and without scaling the metabolites of an individual sample to the median value of all metabolites in that sample. There were only minor differences in the results with and without the scaling. The results presented are the significant differences between groups that were common to the two different methods of analyses.
pH measurements
Lysosomal pH was measured as described previously (17). Cells were incubated for 18 hrs in growth medium supplemented with 2.5 mg/ml fluorescein-rhodamine labeled dextran (Invitrogen, Inc). Following this incubation cells were washed and incubated for an additional 3 hrs in growth medium to chase the dextran into late endosomes and lysosomes. The cells were transferred to a HEPES-balanced salt solution with 10 mM glucose and imaged live by epifluorescence in both fluorescein and rhodamine channels. To construct a pH calibration curve a set of cells similarly labeled were fixed and incubated in monensin containing buffers of pH 3,4,5,6,7,and 8. These cells were imaged with the same settings as the live cells. Metamorph software was used to quantify the rhodamine and fluorescein fluorescence of regions of interest (ROIs) identified in the rhodamine channel. The rhodamine-fluorescein ratios of the ROIs of the fixed cells were used to create the pH calibration curve that was used to assign pH values to the ROIs of the live cells.
LC3, Lamp-2A and Caveolin-1 analyses
Paired CAFs and NFs were grown in culture to 100% confluence in 6 well plates (DMEM-Hams F12 1:1 blend, 10% FBS). Cells were either treated with 30 μM chloroquine for 3 or 5 hours or kept in normal growth media. Protein extracts were prepared in 150 μl of 2× Laemmli buffer, boiled, sonicated, and run on a 15% SDS-PAGE gel. Membranes were incubated overnight with primary antibodies: rabbit anti-LC3B 1:1000 (Invitrogen, Inc); rabbit anti-actin 1:5000 (Cytoskeleton Inc.); rabbit anti-Caveolin-1 antibody (BD Biosciences); and anti-Lamp-2A antibody (Abcam, Inc), and for 1 hour with secondary antibodies (goat anti-rabbit 680 nm, 1:5000). Membranes were read on a LiCor imager and quantified using Odyssey software (version 3.0). Alternatively, LC3 autophagosomes were detected using indirect immunofluorescence in epifluorescence microscopy. Caveolin and LAMP-2A blotting were performed on the non-chloroquine treated extracts only.
RESULTS
Isolation of paired, non-immortalized lung cancer-associated and normal fibroblast cell lines
In an Institution Review Board approved study, we generated by long-term culture 21 CAF lines from surgically resected primary lung cancers and matched NFs from non-neoplastic lung tissue obtained at least 5 cm from the tumor, and we used these samples for mass spectrometry metabolomic profiling of CAFs (Fig.1A). Tumor and non-neoplastic tissues were processed by mechanical and collagenase disruption, and the resultant heterogeneous populations of dispersed cells were plated in growth medium supplemented with 10% FBS. One day after plating, keratin-expressing and vimentin-expressing cells were detected by immunofluorescence (Fig.1B). Overtime in culture without passage, both keratin- and vimentin-expressing cells proliferated, with the keratin-expressing cells growing in clusters characteristic of epithelial cells. However, upon expansion of the cultures by passage, only the vimentin-expressing cells proliferated in samples from both tumor and non-neoplastic tissue (Fig.1C). These vimentin-expressing cells were further expanded and prepared for metabolomic analysis. Numerous previous studies have isolated CAF and NF lines by culture without specific enrichment steps. To be consistent with the terminology in the literature, we refer to the expanded vimentin-expressing cultures as CAFs cell lines and NFs cell lines, although they are not clonal lines. There were no consistent differences in growth rates of the expanded CAF and NF cultures. Pertinent clinical data associated with the 21 CAFs/NFs paired lines are presented in Table I.
Figure 1. Primary non-immortalized human lung CAFs and paired NFs.
A. Flow chart of the protocol to create the CAF and NF lines used in the study.
B. Dispersed cells from a lung tumor (CEM23) maintained in culture for the times noted. Cells were fixed, permeabilized and stained for vimentin (red) and keratin (green). The arrow notes a cluster of keratin-positive cells at 14 days of culture without passage. Images were collected at 20×.
C. Cells from the tumor shown in panel B and corresponding non-neoplastic tissue after repeated passage in culture. Cells were fixed, permeabilized and stained as follows. The same field of cells is shown. In the images on the left, keratin is pseudocolored red, in the middle images vimentin is red and on the right vimentin is red and αSMA green. In all images DNA staining is pseudocolored blue. In the culture conditions used, the vimentin-positive cells proliferate, whereas the keratin-positive cells of the original cultures (from both tumor and non-neoplastic tissue) do not. Therefore only vimentin-positive cells remain after repeated in vitro passages to expand the cultures. In the example shown, the CEM23 CAFs highly expressed αSMA, a marker characteristic of activated fibroblasts, whereas the paired CEM23 NFs did not. Images were collected at 40×. The intensities of the individual colors were identically scaled across all images.
D. Cells from two different CAFs and NFs pairs, after multiple passages in vitro, were stained for vimentin (red), αSMA (green) and DNA (blue). Two different fields for each pair of CAFs and NFs are shown. These data illustrate the heterogeneity of αSMA expression among the different CAFs and NFs. In both cases the CAFs express more αSMA than their paired NFs. CEM248 CAFs are fairly homogenous for αSMA expression compared to CEM254. Some of the CEM254 NFs express significant amounts of αSMA, whereas CEM254 NFs have very low levels of αSMA expression.
E. The expression of caveolin-1 in 6 different pairs of CAFs and corresponding NFs was determined by Western blotting. The antibody detected a doublet at about 20,000 daltons. The actin blot serves as a control for the amount of extracts loaded per lane.
F. Cells from three different CAF and NF pairs, after multiple passages in vitro, were stained for caveolin-1 (green) and DNA (blue). There were no consistent differences in caveolin-1 expression between CAFs and NFs, either in subcellular localization or amount per cell. Images were collected at 40×. The intensities within each fluorescence color are scaled the same across all images.
TABLE I.
| Identifier (CEM#) | SUVmax | Histology | Tumor diameter (cm) |
Nodes |
|---|---|---|---|---|
| 16W | 0.9 | Adenocarcinoma | 1.5 | N |
| 16L | 1.1 | Adenocarcinoma | 1.1 | N |
| 15 | 1.3 | Adenocarcinoma | 2 | N |
| 23 | 1.6 | Adenocarcinoma | 1.5 | N |
| 248 | 1.7 | Adenocarcinoma | 3.2 | N |
| 187 | 3.3 | Squamous | 5 | Y |
| 166 | 4.7 | Adenocarcinoma | 1.7 | N |
| 51 | 5.1 | Adenocarcinoma | 2.5 | Y |
| 232 | 7.5 | Adenocarcinoma | 2.6 | Y |
| 57 | 7.2 | Adenocarcinoma | 6.3 | Y |
| 196 | 8.9 | Squamous | 3.8 | N |
| 217 | 10 | Squamous | 3.2 | N |
| 48 | 11.1 | Adenocarcinoma | 0.8 | N |
| 21 | 11.8 | Adenosquamous | 5 | Y |
| 216 | 13.2 | Squamous | 4 | N |
| 133 | 14.7 | Adenocarcinoma | 8 | Y |
| 160 | 14.8 | Squamous | 4.5 | N |
| 7 | 16.3 | Squamous | 4.5 | N |
| 254 | 16.4 | Squamous | 2.5 | N |
| 212 | 19.7 | Squamous | 6 | Y |
| 172 | 29 | Squamous | 6.5 | N |
Increased expression of α-smooth muscle actin (αSMA) is one commonly reported characteristic of CAFs, presumably reflecting that CAFs are in an “activated state” (3, 4). Consistent with those previous observations, expression of αSMA was generally higher in the CAFs compared to matched NFs, however there was considerable variation in αSMA expression among the different lines (Fig.1D). This heterogeneity was observed in the percentage of cells expressing αSMA within any given CAF and NF pair as well as in the amount of αSMA expression per cell. No correlation between αSMA expression and clinical data were noted. Heterogeneity in αSMA expression in CAFs has been previously reported for CAFs [e.g., (9)].
One group has proposed that reduced caveolin-1 expression drives CAF activation and is responsible for a metabolic reprogramming of CAFs (18). We did not find differences in total caveolin-1 expression between CAFs and paired NFs detected by western blot (Fig.1E). In addition, caveolin-1 was homogeneously expressed at the individual cell level in both CAFs and NFs, labeling both the plasma membranes and internal membranes (Fig.1F). Thus, reduced caveolin-1 expression is not a feature of lung tumor-derived CAFs.
Metabolite groups distinguish cultured primary lung cancer-associated fibroblasts from normal lung fibroblasts
Mass spectrometry was used to profile 203 metabolites in the 21 pairs of cultured primary lung CAFs and their matched NFs as previously described (19, 20). Metabolite levels were corrected for cell numbers and each metabolite was scaled to its median across all samples (Fig.2A & Supplemental Table 1). There was a slight, albeit significant, increase in all 203 metabolites considered as a group in NFs compared to the CAFs (Fig.2B). The data are corrected for cell number and we do not know the reason for this difference. To account for this change, we also analyzed the data after normalizing the metabolites of each individual sample to the median value of all metabolites of each individual sample (Supplemental Table 1). This normalization did not change the results of the CAF to NF comparison. No single metabolite was significantly different between CAFs and NFs in a comparison of the means of the individual metabolites (p ≤ 0.05, two-tailed, unpaired Student’s t test, Benjamini-Hochberg corrected for multiple testing).
Figure 2. Steady-state metabolite profiles in CAFs and paired NFs.
A. Heat map of the abundances of 203 metabolites in 21 pairs of CAFs and NFs. The abundances of each metabolite are scaled to a median value of 1. The color scale is noted below. The CEM numbers are the cell line identifiers. A list of the 203 metabolites is in Supplemental Table I. The heat map panel is shown in a larger format in Supplemental Figure 1A.
B. Distributions of all 203 metabolites in CAFs and NFs.
C. The distributions of “relative” t-scores for the 203 metabolites categorized into 7 biochemical groupings (Supplemental Table I). The numbers of metabolites in each group are shown (n). For display purposes the mean t-score of the (203-n) metabolites not in the group being analyzed was subtracted from the t-scores of the metabolites of the group being analyzed. Thus, the t-score distributions of each of the 7 main pathways are compared to their own reference set. In this way, the further the shift from a relative t-score of 0, the greater the difference from the reference set. The two-tailed unpaired p-values Benjamini-Hochberg corrected for multiple testing of the comparisons of the distributions of the t-scores on the metabolites of the 7 groups to the distributions of the t-scores of the rest of the metabolites are shown on the right.
D & E. The metabolite subgroups whose t-scores distributions (D) and change of abundance (log2(CAF/NF) (E) are both different from their reference data at a p-value ≤ 0.05 (two-tailed unpaired p-values Benjamini-Hochberg corrected for multiple testing) are shown. The numbers of metabolites in the subgroups (n) are shown. The results for all 46 subgroups are shown in Supplemental Fig. 2.
To minimize subject-to-subject variations that are independent of possible CAF to NF differences, we examined the ratio of the individual metabolites in each CAF to the amount of that metabolite in the NF derived from the same individual (Supplemental Fig.1B). In this analysis only the abundance of glucose was significantly different between CAFs and NFs at p ≤ 0.05 (two-tailed unpaired Student’s t test, Benjamini-Hochberg corrected). The biological significance of this change in glucose without corresponding changes in glucose metabolites is not clear.
Cellular metabolism is an interconnected network; therefore, steady state analyses can be confounded because changes in biochemicals of a metabolic pathway will be distributed across individual intermediates of that pathway as well as potentially distributed more broadly over connected pathways. To capture differences in abundance of metabolites of metabolic pathways, and thereby potentially increase the sensitivity of our analysis to detect differences between CAFs and NFs, we assigned the 203 metabolites to 7 groups based on Kyoto Encyclopedia of Genes and Genomes (KEGG) (21) and compared CAFs to NFs based on these groups (Supplemental Table 1). To accomplish this we used the t-test to compare the t-scores of the metabolites of each of the 7 groups to the t-scores of all the other metabolites excluding those of the pathway being examined. In this analysis we determined whether differences in any of the 7 groupings of metabolites between CAFs and NFs were more pronounced than the difference in the abundance of all metabolites between CAFs and NFs. Two of the 7 groups were significantly enriched in CAFs: amino acids and peptides (p ≤ 0.05, unpaired two-tailed Student’s t test, Benjamini-Hochberg corrected) (Fig.2C). These data indicate differences in the cellular metabolisms of CAFs and NFs that were not apparent at an individual metabolite/biochemical level.
To refine the comparison we analyzed the data by further categorizing the metabolites into 46 subgroups based on KEGG designation (21) (Supplemental Table I). We compared both the t-scores and the fold changes (log2) in abundance of the metabolites for each subgroup to the respective t-scores or fold changes of all the other metabolites excluding those of the subgroup being examined (Supplemental Fig.2). Thus, in this analysis we determined whether differences in any of the 46 subgroupings of metabolites between CAFs and NFs were more pronounced than the difference in the abundance of all metabolites between CAFs and NFs. To consider a subgroup significantly different between CAFs and NFs, we imposed the criteria that the distribution of t-scores of the metabolites of a subgroup and the mean log2 fold differences in abundance of the metabolites of a subgroup should both be significantly different (p-values ≤ 0.05, unpaired two-tailed Student’s t-test, Benjamini-Hochberg corrected).
This analysis revealed that 3 subgroups were significantly different between CAFs and NFs (Fig.2D & E). Metabolites of the glycine/serine/threonine subgroup include these amino acids as well as n-acetlylserine and betaine (Supplemental Table I). Creatinine subgroup consists of creatine and creatinine. The dipeptides subgroup consists of 17 unrelated dipeptides (Supplemental Table I). The differences between CAFs and NFs in the glycine/serine/threonine and the creatinine subgroups, which are both part of the KEGG designation amino acid group, likely account for this group distinguishing the CAFs from the NFs (Fig.2C), whereas the change in the dipeptide subgroup accounted for the peptide group distinguishing CAFs from NFs (Fig.2C). The metabolites of the 3 subgroups that were different between CAFs and NFs were all enriched in CAFs.
These metabolite profiling data support the hypothesis that there are steady state metabolic differences between CAFs and NFs.
Different metabolite groups distinguish high and low SUVmax cultured primary lung cancer-associated fibroblasts from paired normal lung fibroblasts
To further interrogate the data, we analyzed the metabolites after subdividing the CAFs and NFs based on the glycolytic activity of the parent primary tumor measured by standardized uptake value (SUVmax) on fluorodeoxyglucose positron emission tomography (Table I). SUVmax, a measure of intracellular accumulation of phosphorylated deoxyglucose, is a surrogate measure of the glycolytic metabolism of tumors (22). To determine whether there were differences in the metabolite profiles of CAFs and NFs related to the SUVmax of the tumor, the CAF/NF pairs were segregated into “low” SUVmax (≤ 5) and “high” SUVmax (≥ 8) groups. Data collected from our clinic show significant differences in disease-free survival for patients based on these SUVmax groupings, in accord with the potential clinical relevance of SUVmax stratification (Fig.3) (23, 24). Based on these criteria, seven CAF/NF pairs were categorized as low SUVmax and eleven as high SUVmax (Table I). The three CAF/NF pairs that fell between SUVmax of 5 and 7 were excluded from this analysis.
No individual metabolites were significantly different between CAFs derived from high SUVmax tumors and their corresponding NFs. Three of the 46 KEGG subgroups were significantly different between high SUVmax CAFs and their matched NFs (Fig. 4A). Both the dipeptides and the glycine/serine/threonine subgroups were significantly enriched in CAFs. These subgroups were also identified in the comparison of CAFs to NFs independent of SUVmax categorization (Fig. 2D). In addition, the mean abundance of metabolites of the lysolipids subgroup (16 different lysolipid species) was reduced in high SUVmax CAFs relative to their paired NFs (Fig. 4A).
Figure 4. Metabolomic profiling reveals influence of tumor SUVmax on CAFs and NFs.
A. Comparison of 11 CAF/NF pairs associated with tumors of SUVmax ≥ 8. The metabolite subgroups whose t-scores distributions (top panel) and change of abundance (log2(CAF/NF) (bottom panel) are both different from their reference data at a p-value ≤ 0.05 (two-tailed unpaired p-values Benjamini-Hochberg corrected for multiple testing) are shown.
B. Comparison of 7 CAF/NF pairs associated with tumors of SUVmax ≤ 5. The subgroups whose t-scores distributions (upper panel) and distributions in change of abundance (log2(CAF/NF) (lower panel) are both different from their reference data at a p-value ≤ 0.05 (two-tailed unpaired p-values Benjamini-Hochberg corrected for multiple testing) are shown. The number of metabolites in the subgroups (n) is shown to the left.
C. Comparison of 11 CAFs from high SUVmax (≥8) to 7 CAFs from low SUVmax (≤5) tumors. The subgroups whose t-scores distributions (upper panel) and distributions in change of abundance (log2(CAF/NF) (lower panel) are both different from their reference data at a p-value ≤ 0.05 (two-tailed unpaired p-values Benjamini-Hochberg corrected for multiple testing) are shown. The number of metabolites in the subgroups (n) is shown to the left.
D. Comparison of 11 NFs from individuals with high SUVmax (≥8) tumors to 7 NFs from individuals with low SUVmax (≤5) tumors. The subgroups whose t-scores distributions (upper panels) and distributions in change of abundance (log2(CAF/NF) (lower panels) are both different from their reference data at a p-value ≤ 0.05 (two-tailed unpaired p-values Benjamini-Hochberg corrected for multiple testing) are shown. The number of metabolites in the subgroups (n) is shown to the left.
No individual metabolites were significantly different between CAFs derived from low SUVmax tumors and their paired NFs. Two of the 46 KEGG subgroups were significantly different between low SUVmax CAFs and their matched NFs (Fig. 4B). As was the case for the comparison of high SUVmax CAFs to NFs, the dipeptide subgroup was significantly enriched in the low SUVmax CAFs relative to their paired NFs, although the relative enrichment in the in low SUVmax CAFs was smaller than in the high SUVmax comparison. There was also a significant decrease in the glycolysis/pyruvate metabolites subgroup in the low SUVmax CAFs relative to their paired NFs (Fig. 4B). The metabolite subgroups that distinguish low SUVmax CAFs from their matched NFs were not the same as those that distinguish high SUVmax CAFs from their paired NFs, suggesting a link between the metabolic alterations in CAFs and the glycolytic activity of the tumor (i.e., SUVmax) from which the CAFs were derived.
Primary lung cancer-associated fibroblasts from high SUVmax tumors have different metabolite profiles than those from low SUVmax tumors
To further explore the possibility of a relationship between CAF metabolism and tumor SUVmax, we compared CAFs from low SUVmax tumors to CAFs from high SUVmax tumors. Two subgroups of metabolites were different between CAFs from low and high SUVmax tumors: lysolipid, and dipeptides (Fig. 4C). The abundance of lysolipids was reduced in the CAFs from high SUVmax tumors, whereas the dipeptides were increased in CAFs from high SUVmax tumors. These two subgroups also distinguished high SUVmax CAFs from their paired NFs (Fig. 4A). Interestingly, in both the comparisons the lysolipids were reduced and dipeptides were increased in the high SUVmax CAFs, suggesting these changes are characteristics of CAFs derived from high SUVmax tumors, reinforcing the hypothesis that metabolic changes in CAFs are in some way linked to the glycolytic capacity of the tumor.
Primary normal lung fibroblasts from individuals with high SUVmax tumors have different metabolite profiles than those derived from individuals with low SUVmax tumors
The above data provide direct support for the hypothesis that there are differences between CAFs from high and low SUVmax tumors. We next compared the metabolite profiles of NFs assigned to either high or low SUVmax groups based on the SUVmax of the tumor from which their corresponding CAFs were derived. Two of the KEGG designated subgroups of metabolites were different between NFs associated with low and high SUVmax tumors (Fig. 4D). Dipeptides were enriched in NFs associated with high SUVmax tumors, whereas the carnitine subgroup (acetylcarnitine, carnitine, deoxycarnitine and hexanoylcarnitine) was enriched in NFs associated with low SUVmax tumors. These data suggest an unexpected link between the glycolytic activity of the tumor (i.e., SUVmax) and the metabolic profile of fibroblasts derived from non-neoplastic tissue of the same lobe as the tumor.
Increased basal autophagy in primary lung cancer-associated fibroblasts relative to normal lung fibroblasts
Dipeptides were increased in each comparison of CAFs to NFs, suggesting an increase in dipeptides is a characteristic of CAFs (Figs. 2 & 4). In addition, dipeptides were also increased in CAFs from high SUVmax tumors relative to those from low SUVmax tumors, and they were increased in NFs associated with high SUVmax tumors relative to those associated with low SUVmax tumors (Fig. 4D). Thus, an increase in dipeptides is also correlated with the higher SUVmax tumors.
Proteolysis is a source of dipeptides and therefore the differences in the dipeptides could reflect differences in proteolysis among the cell groupings. Alterations in autophagy, a lysosome-related proteolytic-degradative mechanism, have been noted in cancer cells, a change that might reflect a response to energy balance alterations in the cancer cells (25). Two autophagic mechanism, macroautophagy and chaperone-mediated autophagy, have been linked to cancer (14-16, 26).
We first determined whether there were differences in macroautophagy between CAFs and NFs. Upon induction of macroautophagy (from here on out referred to as autophagy), microtubule-associated protein 1 light chain 3 (LC3I) is processed by lipidation to form LC3II that associates with membranes during the elongation phase of autophagosome formation. An increase in LC3II, evident by altered electrophoretic migration, is a measure of increased autophagy (Fig. 5A) (27-29). Although there was variability in the total amount of LC3 (LC3I + II) among CAF/NF pairs (reflecting individual-to-individual variation), there were no consistent differences in total LC3 between CAFs and NFs, nor was there a correlation in the total LC3 with the tumor SUVmax (Fig. 5B). There was, however, a significant increase in the basal steady state LC3II as a percent of total LC3 in CAFs, indicating increased autophagy in CAFs (Fig. 5C). To establish that the basal steady state increase in LC3II was due to increased production of LC3II, rather than a decrease in LC3II turnover, we used chloroquine to inhibit degradation of LC3II (29). Chloroquine induced an increase of LC3II in CAFs and NFs lines; however, the relative increase in LC3II as a percent of total LC3 in CAFs was maintained, consistent with the higher basal LC3II in CAFs being due to increased autophagy (Fig. 5C). When we segregated the data for the tumor SUVmax, we found that the increase in LC3II in CAFs relative to their paired NFs was significant for pairs associated with high SUVmax tumors but not for those associated with low SUVmax tumors (Fig. 5C). These results are consistent with the more robust difference in dipeptides between high SUVmax CAF/NF pairs (Fig. 4), and the steady state LC3II measurement may not be of sufficient sensitivity to detect the smaller differences between low SUVmax CAF/NF pairs.
Figure 5. Increased LC3II in CAFs relative to paired NFs.
A. A representative LC3 western blot of a CAF/NF pair (CEM 217) used for quantification of LC3I and LC3II. Data are from cells in growth medium, reflecting the basal steady state levels of LC3 and from cells treated with 10 μM chloroquine for 3 hrs to inhibit degradation of LC3II.
B. The total amount of LC3 (LC3I and LC3II) in each of 8 fibroblasts lines measured by quantitative Western blotting. The values are normalized to total protein (actin). The data are the mean values ± SEM of at least 3 measurements.
C. LC3II as a percent of total LC3 (I + II) in NFs and paired CAFs determined by quantifying Western blots like that shown in panel A. The averages for all 11NF/CAF pairs examined, and for 4 NF/CAF pairs associated with low SUVmax tumors and 7 pairs associated with high SUVmax tumors are shown. * p<0.01.
D. Autophagosomes imaged by LC3 immunofluorescence. Representative fields of control cells (growth medium) or cells following 3 hrs incubation in DMEM medium with no glucose or glutamine stained with LC3 antibody. Autophagosomes containing LC3II are shown in green. Images were collected with a 40× objective. Intensity differences between cells reflect differences in amounts of LC3 staining. DAPI stained nuclei shown in blue.
E. The effect of 3 hrs of incubation in glucose- and glutamine-free medium on the percent of LC3II measured by Western blotting. The data are the average of data from 4 CAF/NF pairs ± SEM. * p< 0.05, paired, one-tailed students t-test.
We also investigated whether there were differences between CAFs and NFs in chaperone-mediated autophagy (CMA), a process in which cytosolic proteins are transported into lysosomes for degradation that has been shown to be increased in some cancer cells (25)(14-16, 26). Upregulation of LAMP-2A is a marker for increased CMA. Although there was individual-to-individual variation in LAMP-2A expression, there were no consistent differences between CAFs and NFs (Supplemental Fig. 3). These data establish that CMA was not upregulated in CAFs.
The above results established an increase in basal autophagy in CAFs. Nutrient limitation is a primary inducer of autophagy, prompting us to investigate if CAFs and NFs differ in the activation of autophagy in response to nutrient reductions. We first determined the effect of acute, stringent nutrient deprivation by incubating cells in medium lacking glucose and glutamine for 3 hrs. Induction of autophagy was investigated by measuring the LC3II in Western blots and by fluorescent microscopy measurement of LC3 stained puncta. Increased basal autophagy in CAFs was evident by an increase in the number and intensity of LC3-positive juxtanuclear structures relative to NFs (Fig. 5D). The LC3 labeling and the concentration in the juxtanuclear region of cells are characteristics of autophagosomes. Acute nutrient deprivation of cells in medium lacking glucose and glutamine resulted in an increase in autophagosomes in CAFs and NFs, although there was some cell-to-cell heterogeneity in both CAFs and NFs (Fig. 5D). Acute nutrient deprivation induced an increase of LC3II, measured by Western blotting, to the same level in CAFs and NFs (Fig. 5E). Thus, despite increased basal autophagy in CAFs, there was no difference between CAFs and NFs in the induction of autophagy in response to acute nutrient deprivation, demonstrating that CAFs further induce autophagy in response to nutrient stress.
We next examined differences in autophagy between CAFs and NFs in response to chronic reduction in glucose availability rather than acute challenge with no glucose. Because of the high glucose demands of cancer cells, glucose available for metabolism by stroma cells within the tumor microenvironment might be chronically limited. Cells are typically grown in culture in media supplemented with 25 mM glucose and our metabolomic profiling was performed in cells expanded in 25 mM glucose. To determine the effect of reduced glucose, cells were grown in media with 25 mM or 5 mM glucose. We used quantitative fluorescence microscopy of LC3 to detect autophagosomes. The amount of LC3 (increased staining intensity) associated with juxtanuclear puncta, characteristic of lysosomes/autophagosome, was increased in NFs grown in 5 mM versus 25 mM glucose, although the increase was not as great as when the cells were incubated in medium lacking glucose and glutamine (Fig. 6A). These data establish that reduced glucose in the growth medium induced autophagy in NFs. However, there was no increase in autophagosomes of CAFs grown in 5 mM glucose relative to growth in 25 mM glucose (Fig. 6A). These data suggest that the set point for induction of autophagy by chronically reduced glucose is different between CAFs and NFs.
Figure 6. Impact of glucose on autophagy.
A. The impact of glucose in the growth media on autophagosomes detected by LC3 fluorescence. Representative images of cells grown in 5 mM, 25 mM glucose or following 3 hr incubation in medium with no glucose and glutamine for 3 hrs are shown. Intensity pseudocolored images are show. The color scale bar is at the bottom of panel A. Images were collected at 40×.
B. Quantification of 17 dipeptides by mass spectrometry from cells grown for at least 7 passages in 25 mM or 5 mM glucose. * p < 0.05, paired, two-tailed student t-test.
C. Representative epifluorescence images of fluorescent dextran endocytosed from the medium during an 18 hr incubation and then chased into lysosomes by an additional 3-hr incubation in medium without dextran. Cells grown in 25 and 5 mM glucose are shown. The images were collected at 40×. DAPI stained nuclei shown in blue.
D. The pH of lysosomes in 7 NF/CAF pairs determined by ratiometric fluorescence microscopy of endocytosed rhodamine-fluorescein (R/F) labeled dextran. The data are the means of the average lysosomal pH of 8 CAF/NF pairs (5 mM glucose) and 2 CAF/NF pairs (25 mM glucose).
To determine if reduced glucose affects dipeptide production, the abundance of the 17 dipeptides measured in the metabolomic profiling were determined for 7 CAFs/NFs grown in 5 mM glucose (Fig. 6B). As was the case for the 21 CAF/NFs pairs (Fig. 2), the abundance of dipeptides was increased in CAFs of this subset of 7 pairs grown in 25 mM glucose. However, there was no difference in the abundance of dipeptides between these CAFs and NFs when the cells were grown in 5 mM glucose (Fig. 6B). The abundance of dipeptides was significantly increased in NFs grown in 5 mM versus 25 mM glucose, whereas the abundance of dipeptides in CAFs was not significantly altered by growth of cells in 5 mM glucose. Thus, the increase in autophagosome formation induced in NFs by growth in reduced glucose was paralleled by an increase of dipeptides.
Growth of CAFs and NFs in media with varying amounts of glucose (25 mM, 5 mM or 2 mM) did not reveal a proliferative advantage of CAFs in response to reduced glucose (Supplemental Fig. 4), although the ability of NFs to up-regulate autophagy upon chronic reduction of glucose might in this in vitro assay mitigate any advantage of the increased basal autophagy in CAFs.
Autophagic degradation ultimately depends on an active lysosomal system. Acidification is essential for the hydrolytic function of lysosomes and regulation of lysosomal pH is one mechanism for controlling lysosomal activity (30-32). We investigated whether there were changes in lysosomal acidification between CAFs and NFs. Cells were incubated with dual labeled fluorescein-rhodamine dextran for 18 hrs and the endocytosed dextran was chased into lysosomes with a 3-hr incubation in medium without dextran. In both CAFs and NFs the label accumulated in puncta distributed throughout the cell but with some degree of concentration in the peri-nuclear region characteristic of lysosomes (Fig.6C). The rhodamine-to-fluorescein fluorescence ratio of the puncta were measured, and converted into a pH values using a pH standard curve (Supplemental Figures 5 & 6). There were no consistent differences in the average lysosomal pH between CAFs and NFs when grown in 25 mM or 5 mM glucose (Fig. 6D). In all conditions lysosomes had a pH value of 4.5, in line with previous measurements of lysosomal pH.
DISCUSSION
Many tumors are metabolically distinct from non-neoplastic tissue (e.g., 18F-deoxyglucose PET activity), reflecting the pronounced glycolytic metabolism of cancer cells. It is not known, however, whether the metabolisms of the non-epithelial cells of the tumor are also altered. To begin to explore that question we used quantitative mass spectrometry profiling of the steady state abundances of 203 metabolites in primary, non-transformed human lung CAFs and their paired NFs. Although we found no differences in individual metabolites distinguishing CAFs from NFs, we report herein the novel observation of significant differences between CAFs and NFs in the steady state abundances of metabolites of select metabolic pathways.
In this initial study, due to some practical limitations of working with primary non-transformed cells, we performed a steady state metabolite profiling. We analyzed the data using a systems pathway approach by identifying changes in KEGG designation groups of metabolites that differed significantly from the overall differences in the abundance of metabolites between comparison samples. In addition to comparing CAFs with NFs, we also analyzed the data after segregating the samples based on the SUVmax of the tumor: “high” SUVmax (≥8) and “low” SUVmax (≤5). The impetus for this classification was that SUVmax is a measure of the glycolytic activity of the tumor and therefore it was of interest to know if this is related to the metabolite profile of CAFs. Furthermore, individuals with high SUVmax tumors have a worse outcome on 5-year disease-free survival, so this classification is of potential clinical significance. We detected changes in groups of metabolites that indicate possible changes in metabolic pathways. We followed up one of these changes, increased dipeptides in CAFs, with additional experiments. Additional future studies are required to investigate other metabolic changes identified in this study. Nonetheless, our results establish for the first time metabolic differences between CAFs and their paired NFs that persist in in vitro culture and that are correlated with the glycolytic activity of the parent tumor.
The relative abundance of the 17 unrelated dipeptides were significantly different in the comparisons of the different fibroblast groups. There was an enrichment of dipeptides, relative to other metabolites, in CAFs versus NFs, and in CAFs from high SUVmax tumors compared to CAFs from low SUVmax tumors. Thus, increase in dipeptide abundance is a general characteristic of CAFs that is correlated with the glycolytic activity of the tumor.
The primary source of cellular dipeptides is protein turnover, suggesting differences in protein degradation among the different fibroblast groups. In parallel with the difference in dipepitde abundance, we found an increase in autophagy in CAFs relative to their paired NFs based on increases of LC3-labeled phagosomes as well as an increase in LC3II/I ratio. Autophagy was originally described as a response to nutrient deprivation in which cellular components are degraded to provide substrates for oxidative energy production. However, autophagy is now recognized to have roles in normal basal cellular physiology by mediating the turnover of long-lived proteins, the clearance of damaged mitochondria and other organelles, and the clearance of potentially toxic protein aggregates as a protection against oxidative damage (14, 25). In addition to these pro-survival activities, autophagy can, under certain conditions, result in cell death (14).
Studies of autophagy in tumors have focused on cancer cells (14). Our data support an increase in autophagy in fibroblasts of the tumor microenvironment, suggesting that alterations in autophagy within tumors are not restricted to the cancer cells. We have also discovered a positive correlation between increased dipeptide abundance/increased basal autophagy in CAFs and the tumor SUVmax, revealing a possible link between the glycolytic activity of the tumor and autophagy.
Increased autophagy is believed to promote survival of cancer cells by providing nutrients (amino acids, lipids and nucleic acids) and/or as a protection against oxidative damage. Increased CAF autophagy might also provide a survival benefit to CAFs in the tumor. Thus, elevated autophagy, by promoting survival of CAFs could indirectly contribute to the pro-tumorigenic effects of CAFs, since CAFs thriving in the tumor would be able to support cancer cells and tumor growth by one or more of the previously described mechanisms (e.g., paracrine support). An alternative and not mutually exclusive view is that increased autophagy is not limited to cell autonomous benefits for CAF survival, but that increased autophagy in CAFs might be of direct benefit to cancer cells. Some possibilities include increased CAF autophagy as a means of sparing nutrients in the tumor microenvironment for use by the cancer cells, or the secretion of nutrients from CAFs generated by autophagy to enrich the nutrient milieu of the tumor microenvironment. The latter is of particular interest with regards to dipeptides since supplementation of media with certain dipeptides enhances in vitro cell viability (33).
CAFs and NFs similarly induced autophagy when acutely challenged with the severe nutrient restriction of removing glucose and glutamine from the medium. However, only the NFs induced autophagy when challenged with a chronic reduction of glucose in the medium, which indicates that NFs have a lower set point for induction of autophagy controlled by alterations in the nutrient milieu. Because of the high glucose consumption of cancer cells, glucose availability for other cells within the tumor is likely limited. Thus, it is tempting to speculate that the basal up-regulation of autophagy in CAFs reflects this difference in set point for induction of autophagy and that it is advantageous for CAF growth/survival in the tumor environment. Although we did not observe consistent differences between growth of CAFs and NFs in normal growth medium (25 mM glucose) or in conditions of “nutrient stress” induced by reduced glucose (5 or 2.5 mM glucose) the in vitro growth assay does not recapitulate the conditions experienced by cells within the tumor microenvironment. Thus, future studies are required to address the functional consequences of increased CAF autophagy.
Regardless, our results establish that autophagy changes in cells of the tumor microenvironment must be taken into account when considering the role of autophagy in solid tumors.
In addition to an increase in the abundance of dipeptides in CAFs from high SUVmax tumors relative to those from low SUVmax tumors, there was an increase, albeit less robust, in the abundance of dipeptides in NFs isolated from individuals with high SUVmax tumors relative to those from individuals with low SUVmax tumors. This link between the SUVmax of the tumor and an increased abundance of dipeptides in fibroblasts outside the tumor microenvironment was unexpected. An explanation for these results is that the possible tumor effects on CAFs are far reaching, and that the less robust difference in the fibroblasts from non-neoplastic lung tissue reflects a diminution of the effect due to distance from the tumor. An alternative possibility is that the increased dipeptide abundance of NFs paired with high SUVmax tumors reflects a pre-existing difference independent of the tumor, and the more robust differences between CAFs from high and low SUVmax tumors is an effect of the tumor re-enforcing this pre-existing difference. Another alternative possibility is that the differences in metabolites between NF derived from high and low SUVmax tumors may reflect differences in host epigenetic alterations in response to exposure to tobacco-related carcinogens.
Here we have focused on the changes in dipeptides, which led us to consider differences in autophagy/proteolysis between CAFs and NFs. However, other groups of metabolites were significantly different in some of the fibroblast comparisons. In a number of comparisons there were changes in metabolites of amino acid pathways, and in each case the change in abundance was in the same direction as that of the dipeptides. These data implicate changes in protein/amino acid metabolism as important differences between CAFs and NFs.
It has been previously suggested that CAFs might undergo alterations in carbohydrate metabolism to support the “Warburg” metabolism of cancer cells. We found a reduction in the steady state abundance of glycolytic intermediates in CAFs in the comparison of CAFs to NFs from low SUVmax tumors, consistent with this hypothesis. However, this difference was not observed in the comparison between high SUVmax CAFs and NFs. A possible explanation for these results is that the cancer cells of the low SUVmax tumors require alterations in CAF carbohydrate metabolism, whereas glycolytic metabolism of the cancer cells of the high SUVmax tumors is sufficiently robust independent of accommodation in the metabolism of CAFs within the tumor. Regardless, our study reveals the importance of considering the glycolytic nature of the tumor in studies of CAFs.
The profiling was performed on cells grown in culture. An advantage of this approach over profiling whole tumor tissue is that we were able to specifically examine the non-epithelial cells of the tumor mass. We focused our analysis on vimentin-positive, keratin-negative cells that grew out of dispersed cells from tumor and adjacent non-neoplastic tissues, which is the standard method for isolating CAFs [e.g., (9)]. Consistent with the literature, the CAF lines as a group expressed more αSMA, a marker for activated fibroblasts, than did paired NFs, but it is important to note that there was considerable heterogeneity among the different lines. However, despite this heterogeneity our analyses revealed differences between CAFs and NFs that were maintained during in vitro culture. Previous studies have shown that CAFs retain differences from NFs after in vitro culture, including differences in gene expression as well as pro-tumorigenic activities measured in vitro and in vivo [e.g.,(9)]. Thus, our findings of metabolic difference between CAFs and NFs maintained after in vitro culture are in agreement with previous studies.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Fred Maxfield, Vivek Mittal, and the members of the McGraw and Altorki group for comments on the manuscript.
FUNDING: Columbia University DIRC P30 DK063608-09 (sub award to NKA & TEM), Neuberger Berman Lung Cancer Research Center (NKA); Seth Sprague Education and Charitable Foundation Award (NKA), WCMC Discovery award (TEM), STARR Cancer Foundation (RS), NSF career award 1054964 (OE), Starr Cancer Foundation (OE) and Clinical and Translational Science Center at Weill Cornell Medical College TL1RR02499 (GS).
Footnotes
The authors have no conflict of interest with regard to the manuscript submitted for review.
REFERENCES
- 1.Albini A, Sporn MB. The tumour microenvironment as a target for chemoprevention. Nature reviews Cancer. 2007;7(2):139–47. doi: 10.1038/nrc2067. Epub 2007/01/16. doi: 10.1038/nrc2067. PubMed PMID: 17218951. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Coussens LM. Accessories to the Crime: Functions of Cells Recruited to the Tumor Microenvironment. Cancer cell. 2012;21(3):309–22. doi: 10.1016/j.ccr.2012.02.022. Epub 2012/03/24. doi: 10.1016/j.ccr.2012.02.022. PubMed PMID: 22439926. [DOI] [PubMed] [Google Scholar]
- 3.Cirri P, Chiarugi P. Cancer-associated-fibroblasts and tumour cells: a diabolic liaison driving cancer progression. Cancer metastasis reviews. 2011 doi: 10.1007/s10555-011-9340-x. Epub 2011/11/22. doi: 10.1007/s10555-011-9340-x. PubMed PMID: 22101652. [DOI] [PubMed] [Google Scholar]
- 4.Ostman A, Augsten M. Cancer-associated fibroblasts and tumor growth--bystanders turning into key players. Current opinion in genetics & development. 2009;19(1):67–73. doi: 10.1016/j.gde.2009.01.003. PubMed PMID: 19211240. [DOI] [PubMed] [Google Scholar]
- 5.Erez N, Truitt M, Olson P, Hanahan D. Cancer-Associated Fibroblasts Are Activated in Incipient Neoplasia to Orchestrate Tumor-Promoting Inflammation in an NF-kappaB-Dependent Manner. Cancer cell. 17(2):135–47. doi: 10.1016/j.ccr.2009.12.041. PubMed PMID: 20138012. [DOI] [PubMed] [Google Scholar]
- 6.Zhang C, Fu L, Fu J, Hu L, Yang H, Rong TH, et al. Fibroblast growth factor receptor 2-positive fibroblasts provide a suitable microenvironment for tumor development and progression in esophageal carcinoma. Clin Cancer Res. 2009;15(12):4017–27. doi: 10.1158/1078-0432.CCR-08-2824. Epub 2009/06/11. doi: 10.1158/1078-0432.CCR-08-2824. PubMed PMID: 19509166. [DOI] [PubMed] [Google Scholar]
- 7.Sadlonova A, Bowe DB, Novak Z, Mukherjee S, Duncan VE, Page GP, et al. Identification of molecular distinctions between normal breast-associated fibroblasts and breast cancer-associated fibroblasts. Cancer Microenviron. 2009;2(1):9–21. doi: 10.1007/s12307-008-0017-0. Epub 2009/03/25. doi: 10.1007/s12307-008-0017-0. PubMed PMID: 19308679; PubMed Central PMCID: PMC2787925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quante M, Tu SP, Tomita H, Gonda T, Wang SS, Takashi S, et al. Bone marrow-derived myofibroblasts contribute to the mesenchymal stem cell niche and promote tumor growth. Cancer cell. 2011;19(2):257–72. doi: 10.1016/j.ccr.2011.01.020. Epub 2011/02/15. doi: 10.1016/j.ccr.2011.01.020. PubMed PMID: 21316604; PubMed Central PMCID: PMC3060401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Navab R, Strumpf D, Bandarchi B, Zhu CQ, Pintilie M, Ramnarine VR, et al. Prognostic gene-expression signature of carcinoma-associated fibroblasts in non-small cell lung cancer. Proc Natl Acad Sci U S A. 2011;108(17):7160–5. doi: 10.1073/pnas.1014506108. Epub 2011/04/09. doi: 10.1073/pnas.1014506108. PubMed PMID: 21474781; PubMed Central PMCID: PMC3084093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134(5):703–7. doi: 10.1016/j.cell.2008.08.021. PubMed PMID: 18775299. [DOI] [PubMed] [Google Scholar]
- 11.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–33. doi: 10.1126/science.1160809. PubMed PMID: 19460998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452(7184):230–3. doi: 10.1038/nature06734. PubMed PMID: 18337823. [DOI] [PubMed] [Google Scholar]
- 13.Koukourakis MI, Giatromanolaki A, Harris AL, Sivridis E. Comparison of metabolic pathways between cancer cells and stromal cells in colorectal carcinomas: a metabolic survival role for tumor-associated stroma. Cancer research. 2006;66(2) doi: 10.1158/0008-5472.CAN-05-3260. PubMed PMID: 16423989. [DOI] [PubMed] [Google Scholar]
- 14.Kimmelman AC. The dynamic nature of autophagy in cancer. Genes & development. 2011;25(19):1999–2010. doi: 10.1101/gad.17558811. Epub 2011/10/08. doi: 10.1101/gad.17558811. PubMed PMID: 21979913; PubMed Central PMCID: PMC3197199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.White EJ, Martin V, Liu JL, Klein SR, Piya S, Gomez-Manzano C, et al. Autophagy regulation in cancer development and therapy. American journal of cancer research. 2011;1(3):362–72. Epub 2011/10/05. PubMed PMID: 21969237; PubMed Central PMCID: PMC3180058. [PMC free article] [PubMed] [Google Scholar]
- 16.Chen HY, White E. Role of autophagy in cancer prevention. Cancer Prev Res (Phila) 2011;4(7):973–83. doi: 10.1158/1940-6207.CAPR-10-0387. Epub 2011/07/08. doi: 10.1158/1940-6207.CAPR-10-0387. PubMed PMID: 21733821; PubMed Central PMCID: PMC3136921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dunn KW, Park J, Semrad CE, Gelman DL, Shevell T, McGraw TE. Regulation of endocytic trafficking and acidification are independent of the cystic fibrosis transmembrane regulator. J Biol Chem. 1994;269(7):5336–45. PubMed PMID: 7508934. [PubMed] [Google Scholar]
- 18.Sotgia F, Martinez-Outschoorn UE, Howell A, Pestell RG, Pavlides S, Lisanti MP. Caveolin-1 and cancer metabolism in the tumor microenvironment: markers, models, and mechanisms. Annual review of pathology. 2012;7 doi: 10.1146/annurev-pathol-011811-120856. [DOI] [PubMed] [Google Scholar]
- 19.Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Analytical chemistry. 2009;81(16):6656–67. doi: 10.1021/ac901536h. Epub 2009/07/25. doi: 10.1021/ac901536h. PubMed PMID: 19624122. [DOI] [PubMed] [Google Scholar]
- 20.Reitman ZJ, Jin G, Karoly ED, Spasojevic I, Yang J, Kinzler KW, et al. Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proc Natl Acad Sci U S A. 2011;108(8):3270–5. doi: 10.1073/pnas.1019393108. Epub 2011/02/04. doi: 10.1073/pnas.1019393108. PubMed PMID: 21289278; PubMed Central PMCID: PMC3044380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanehisa Y, Yoshida M, Taji T, Akagawa Y, Nakamura H. Body weight and serum albumin change after prosthodontic treatment among institutionalized elderly in a long-term care geriatric hospital. Community dentistry and oral epidemiology. 2009;37(6) doi: 10.1111/j.1600-0528.2009.00496.x. Epub 2009/09/29. doi: 10.1111/j.1600-0528.2009.00496.x. PubMed PMID: 19780769. [DOI] [PubMed] [Google Scholar]
- 22.Haberkorn U, Markert A, Mier W, Askoxylakis V, Altmann A. Molecular imaging of tumor metabolism and apoptosis. Oncogene. 2011;30(40):4141–51. doi: 10.1038/onc.2011.169. Epub 2011/05/18. doi: 10.1038/onc.2011.169. PubMed PMID: 21577202. [DOI] [PubMed] [Google Scholar]
- 23.Berghmans T, Dusart M, Paesmans M, Hossein-Foucher C, Buvat I, Castaigne C, et al. Primary tumor standardized uptake value (SUVmax) measured on fluorodeoxyglucose positron emission tomography (FDG-PET) is of prognostic value for survival in non-small cell lung cancer (NSCLC): a systematic review and meta-analysis (MA) by the European Lung Cancer Working Party for the IASLC Lung Cancer Staging Project. J Thorac Oncol. 2008;3(1) doi: 10.1097/JTO.0b013e31815e6d6b. Epub 2008/01/02. doi: 10.1097/JTO.0b013e31815e6d6b. PubMed PMID: 18166834. [DOI] [PubMed] [Google Scholar]
- 24.Stiles BM, Nasar A, Mirza F, Paul S, Lee PC, Port JL, et al. The Annals of thoracic surgery. 2012. Ratio of PET scan maximum standardized uptake value (SUVmax) to tumor size is a better predictor of survival in non-small cell lung cancer than SUVmax. [Google Scholar]
- 25.Singh R, Cuervo AM. Autophagy in the cellular energetic balance. Cell Metab. 2011;13(5) doi: 10.1016/j.cmet.2011.04.004. Epub 2011/05/03. doi: 10.1016/j.cmet.2011.04.004. PubMed PMID: 21531332; PubMed Central PMCID: PMC3099265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kon M, Kiffin R, Koga H, Chapochnick J, Macian F, Varticovski L, et al. Chaperone-mediated autophagy is required for tumor growth. Science translational medicine. 2011;3(109):109ra17. doi: 10.1126/scitranslmed.3003182. Epub 2011/11/18. doi: 10.1126/scitranslmed.3003182. PubMed PMID: 22089453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3(6):542–5. doi: 10.4161/auto.4600. Epub 2007/07/06. PubMed PMID: 17611390. [DOI] [PubMed] [Google Scholar]
- 28.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4(2):151–75. doi: 10.4161/auto.5338. Epub 2008/01/12. PubMed PMID: 18188003; PubMed Central PMCID: PMC2654259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140(3):313–26. doi: 10.1016/j.cell.2010.01.028. Epub 2010/02/11. doi: 10.1016/j.cell.2010.01.028. PubMed PMID: 20144757; PubMed Central PMCID: PMC2852113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trombetta ES, Ebersold M, Garrett W, Pypaert M, Mellman I. Activation of lysosomal function during dendritic cell maturation. Science. 2003;299(5611):1400–3. doi: 10.1126/science.1080106. Epub 2003/03/01. doi: 10.1126/science.1080106. PubMed PMID: 12610307. [DOI] [PubMed] [Google Scholar]
- 31.Majumdar A, Cruz D, Asamoah N, Buxbaum A, Sohar I, Lobel P, et al. Activation of microglia acidifies lysosomes and leads to degradation of Alzheimer amyloid fibrils. Mol Biol Cell. 2007;18(4):1490–6. doi: 10.1091/mbc.E06-10-0975. Epub 2007/02/23. doi: 10.1091/mbc.E06-10-0975. PubMed PMID: 17314396; PubMed Central PMCID: PMC1838985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yogalingam G, Pendergast AM. Abl kinases regulate autophagy by promoting the trafficking and function of lysosomal components. J Biol Chem. 2008;283(51):35941–53. doi: 10.1074/jbc.M804543200. Epub 2008/10/24. doi: 10.1074/jbc.M804543200. PubMed PMID: 18945674; PubMed Central PMCID: PMC2602914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kang S, Mullen J, Miranda LP, Deshpande R. Utilization of tyrosine- and histidine-containing dipeptides to enhance productivity and culture viability. Biotechnology and bioengineering. 2012 doi: 10.1002/bit.24507. Epub 2012/03/27. doi: 10.1002/bit.24507. PubMed PMID: 22447498. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.