Abstract
Physical exercise is associated with positive neural functioning. Here we examined the gene expression consequences of 1 week of voluntary wheel running in adolescent male mice. We assayed expression levels of genes associated with synaptic plasticity, signaling pathways, and epigenetic modifying enzymes. Two regions were examined: the hippocampus, which is typically examined in exercise studies, and the cerebellum, an area directly involved in motor control and learning. After 1 week of exercise, global acetylation of histone 3 was increased in both brain regions. Interestingly this was correlated with increased brain derived neural growth factor in the hippocampus, as noted in many other studies, but only a trend was found in cerebellum. Differences and similarities between the two areas were noted for genes encoding functional proteins.
In contrast, the expression pattern of DNA methyltransferases (Dnmts) and histone deacetylases (Hdacs), genes that influence DNA methylation and histone modifications in general, decreased in both regions with exercise. We hypothesize that epigenetic mechanisms, involving many of the genes assessed here, are essential for the positive affects of exercise on behavior and suspect these data have relevance for adolescent boys.
Keywords: Epigenetic, Exercise, Puberty, Adolescent, Cerebellum, BDNF
1. Introduction
Over the past 20 years, research conducted in humans and rodents has demonstrated beneficial effects of physical exercise on the brain including enhanced learning and memory, structural plasticity and neuroprotection against neurodegenerative disorders (as reviewed by Cotman and Berchtold, 2002; Thomas et al., 2012; van Praag, 2009). Due to the urgent need to ameliorate symptoms associated with Alzheimer’s disease, a significant portion of this research focuses on the aging brain and the ability of exercise to reverse cognitive decline via increased neurogenesis and neural plasticity in the hippocampus (Cotman and Berchtold, 2002; Kramer et al., 2006). Markedly less attention has been devoted to the effects of exercise on the young brain (i.e. children and adolescents). Yet, exercise not only plays an important role in maintaining a healthy brain during aging, but also seems essential in promoting normal brain growth and maturation during development (Tomporowski et al., 2008). For example, studies reviewed by Chaddock et al. (2011), show that low levels of physical activity in children and adolescents correlates with poor cognitive abilities sometimes associated with reduced volume in hippocampal and basal ganglia brain regions. In contrast, acute exercise improves executive function and alleviates symptoms in children with attention deficit hyperactivity disorder (ADHD) (Archer and Kostrzewa, 2012; Chang et al., 2012). Moreover, recent behavioral studies, conducted in children and adolescents, show long term beneficial effects of exercise on spatial memory, visual discrimination and the consolidation of information into long-term memory (Aberg et al., 2009; Coles and Tomporowski, 2008; Fedewa and Ahn, 2011; Herting and Nagel, 2012; Sibley and Etnier, 2003).
Identified mechanisms that presumably underlie improved cognition as a consequence of exercise include neurogenesis, synaptogenesis, synaptic plasticity and mediation by neurotrophic factors (Farmer et al., 2004; Ferreira et al., 2010; Garcia et al., 2012; Lou et al., 2008; Molteni et al., 2002; Uysal et al., 2005; Vaynman et al., 2006). Brain-derived neurotrophic factor (BDNF), a molecule implicated in learning and memory, has shown consistent up-regulation in the hippocampus, dentate gyrus and perirhinial cortex in response to wheel or treadmill running (Adlard et al., 2005; Gomes da Silva et al., 2012; Griffin et al., 2009; Hopkins et al., 2011; Zoladz and Pilc, 2010). Other genes assessed, including those associated with synaptic trafficking and plasticity, signal transduction and transcription regulation, show significant up-regulation in response to exercise (Farmer et al., 2004; Molteni et al., 2002; Tong et al., 2001). However, these studies are confined to adult rat hippocampus. Moreover, the regulatory basis of exercise-induced gene expression is currently under exploration. The powerful and immediate effect of exercise on gene expression changes in the brain likely engages epigenetic mechanisms. In fact, a recent study shows that epigenetic factors regulate hippocampal Bdnf expression in response to exercise (Gomez-Pinilla et al., 2011).
In the present study, we used juvenile mice to examine gene expression changes in the brain produced by exercise. We examined genes associated with neuronal function and epigenetic modifications. Along with evaluation of genes that regulate histone acetylation and DNA methylation we asked if total acetylation on histone 3 (H3) was altered by exercise. Finally, we selected to examine gene expression in two very distinct regions of the brain that differ on both a structural and functional level, the cerebellum and hippocampus. This focus allowed us a preliminary look at genes that might be involved in the motoric versus the cognitive affects of exercise. Here we show a complex and regionally specific effect of exercise on a network of genes. Moreover, our results suggest that epigenetic modifications, including histone acetylation and differential histone deacetylase and DNA methyltransferase activity, may underlie many of these gene changes.
2. Methods
2.1. Animals
C57BL/6J mice were born, reared and housed at the University of Virginia School of Medicine Animal Facility in Jordan Hall on a 12:12 h light:dark cycle (lights on at 0600 h). Food (Harlan Teklad Mouse/Rat Sterilizable Diet #7012) and water was provided ad libitum.
2.2. Exercise
We used a voluntary running-wheel model as previously described (Waters et al., 2004). Briefly, 46-day-old male mice were randomized to modified cages equipped with a running wheel. For the exercised group, 10 mice were placed individually in cages with a freely moving wheel attached to a magnetic sensing mechanism which allowed tracking of running activity, as a function of time and distance, by a computer. Mice were given free access to the wheel over a period of 7 days and ran predominately at night. Mice ran on average 8.7 km on Day 1 and steadily increased their distance to an average of 12.2 km by the end of the week. For the sedentary control group, 10 mice were placed individually in cages equipped with the same type of wheel secured to the cage to prevent rotational running activity and were similarly housed in the same room and handled the same way as the exercised mice.
2.3. Brain tissue extraction
Sedentary and exercised mice were randomly removed from cages following 1 week of activity, anesthetized using euthanasol and the brains were rapidly removed. The hippocampus and cerebellum were carefully dissected on ice, frozen on dry ice and pulverized into a fine powder under liquid nitrogen. The powder was then divided into two aliquots and stored at −80 °C until further processing for either histone or RNA extraction.
2.4. Histone extraction and immunoblotting
Histones were extracted and immunoblotting performed as previously described (Tsai et al., 2009). Briefly, mouse brain pulverized tissues were homogenized in RIPA buffer, centrifuged and the pellets containing nuclei were resuspended with sulfuric acid and continuously mixed for 1 h at 4 °C. The samples were again centrifuged and the separated supernatant was precipitated overnight to generate a nuclear pellet, which was resuspended in RIPA buffer. The lysate protein concentrations for each sample were determined by BCA (bicinchoninic acid) protein assays. For immunoblotting, samples (5 µg protein each) were subjected to electrophoresis on 16% polyacrylamide-SDS gels, transferred to nitrocellulose, blocked overnight, rinsed and incubated with the primary antibodies against acetylated K9/14 H3 (Millipore-Upstate in Temecula, CA #06-599; 1:5000) and total H4 (Millipore- Upstate, #07-108). Blots were either incubated for 1 h in a horseradish peroxidase (HRP)-conjugated donkey anti-rabbit IgG secondary antibody and detected on X-ray film with chemiluminescence or IRDye goat-anti-mouse secondary antibody (Licor, Lincoln, NE) and detected with Odyssey® near infrared imager (Licor). For each data point, the amount of acetylated H3 was normalized to total histone H4 to control for differences in the amount of nuclear extract protein loaded on the gel across samples.
2.5. Quantitative real-time PCR (qRT-PCR)
An RNeasy® Lipid Tissue Mini Kit (Qiagen, Valencia, CA) was used to isolate total RNA according to the manufacturer’s protocol. The quantity and quality of the RNA were determined using a NanoVue™ Spectrophotometer. cDNA templates were prepared using an AffinityScript qPCR cDNA Synthesis Kit (Agilent Technologies, Santa Clara, CA) according to the manufacturer’s protocol. The ABI StepOnePlus real-time PCR system was used to perform qRT-PCR. Either TaqMan® Probe- or SYBR® Green-Based Detection (Applied Biosystems, Carlsbad, CA) were used to detect PCR products of interest. The following TaqMan® Gene Expression Assays were used: estrogen receptor α (Esr1, Mm00433149_m1), estrogen receptor β (Esr2, Mm00599821_m1), cerebellin 1 precursor protein (Cbln1, Mm01247195_m1), c-src tyrosine kinase (Csk, Mm00432751_m1), methyl CpG binding protein 2 (Mecp2, Mm01193537_g1), fragile X mental retardation syndrome 1 homolog (Fmr1, Mm00484415_m1), brain derived neurotrophic factor (Bdnf, Mm01334043_m1), calbindin (Calb1, Mm00486645_m1), reelin (Reln, Mm00465200_m1), calcium/calmodulin-dependent protein kinase IV (Camk4, Mm01135329_m1), glutamate receptor, ionotropic, delta 2 (Grid2, Mm00515053_m1), synaptophysin (Syp, Mm00436850_m1), synapsin I (Syn1, Mm00449772_m1*), neuronal calcium sensor 1 (Ncs1, Mm00490552_m1). All samples were normalized to either beta-2 microglobulin (B2m, Mm00437762_m1) or mouse beta-actin (Actb, #4352933E). Oligonucleotide primers were designed for SYBR-Green based analysis using consensus sequences and Blast from the NCBI genomic alignment database and subsequently synthesized by Invitrogen (Carlsbad, CA) as detailed by Table 1. Validation experiments were conducted to test for equally efficient target and endogenous control gene amplification and primers were between 90 and 110% efficient for all amplifications. For TaqMan and SYBR Green based detection, target and endogenous control genes were measured in triplicate for each cDNA sample during each real-time run to avoid inter-sample variance. For SYBR Green based qRT-PCR, a no-reverse transcriptase reaction was run in parallel to cDNA synthesis samples to control for contamination and genomic amplification. Each of these qRT-PCR reactions was verified for a single PCR product of expected size with the disassociation melting curve stage. All genes of interest were analyzed with StepOne™ software using the comparative cycle thresholds method (CT) method.
Table 1.
Oligonucleotide primers designed for SYBR-Green based quantitative real-time PCR (qRT-PCR).
| Gene | Forward primer nucleotide sequence | Reverse primer nucleotide sequence |
|---|---|---|
| Dnmt1 | 5′-CCGCAGGCGGCTCAAGACTT-3′ | 5′-GTCCCGGTTGGCGGGACAAC-3′ |
| Dnmt3a | 5′-GAGGGAACTGAGACCCCAC-3′ | 5′-CTGGAAGGTGAGTCTTGGCA-3′ |
| Dnmt3b | 5′-AGCGGGTATGAGGAGTGCAT-3′ | 5′-GGGAGCATCCTTCGTGTCTG-3′ |
| Hdac1 | 5′-AGGCTCTGTCGCAAGTGCTGTG-3′ | 5′-TCTTTGCATGGTGCAGGCCC-3′ |
| Hdac2 | 5′-TGGTTCAGTTGCTGGGGCTGTG-3′ | 5′-TCCTCCAGCCCAATTGACAGCCAT-3′ |
| Hdac3 | 5′-ATGACAGGACTGACGAGGCCGA-3′ | 5′-TGGGTGCTTCTGGCCTGCTGTA-3′ |
| Hdac4 | 5′-AACATGGCTTTCACGGGTGGCC-3′ | 5′-AACCACCGTTCTGAAGGCTGCC-3′ |
| Hdac5 | 5′-GACATCACAGCAGCTCCGCCC-3′ | 5′-CCATCTGCCGACTCGTTGGGA-3′ |
| Hdac7 | 5′-GCCCTTGAGAGAACAGTCCAT-3′ | 5′-AAGCTGCTAAGCACGGAGC-3′ |
| Hdac8 | 5′-CCGCCAACAGTGGGCATTCACT-3′ | 5′-TACTGGCCCGTTTGGGGACCTT-3′ |
| B2m | 5′-GGCTCACACTGAATTCACCCCCAC-3′ | 5′-ACATGTCTCGATCCCAGTAGACGGT-3′ |
| Actb | 5′-GCCACCAGTTCGCCATGGAT-3′ | 5′-TCTGGGCCTCGTCACCCACATA-3′ |
| Bdnf | 5′-TCATACTTCGGTTGCATGAAGG-3′ | 5′-AGACCTCTCGAACCTGCCC-3′ |
Dnmt1 = DNA methyltransferase 1, Dnmt3a = DNA methyltransferase 3a, Dnmt3b = DNA methyltransferase 3b, Hdac1 = histone deacetylase 1, Hdac2 = histone deacetylase 2, Hdac3 = histone deacetylase 3, Hdac4 = histone deacetylase 4, Hdac5 = histone deacetylase 5, Hdac7 = histone deacetylase 7, Hdac8 = histone deacetylase 8, B2m = beta-2 microglobulin, Actb = beta actin, Bdnf = brain derived neurotrophic factor.
2.6. Statistical analyses
Data collected comparing sedentary to exercised mice, was analyzed by Student’s t-tests. Pearson product–moment or Spearman rank order correlation tests were used to analyze the relationship between two variables. Correlation matrices representing all variables were generated with Sigma Plot and corrected for multiple comparisons.
3. Results
3.1. Voluntary exercise affects expression of neuroplasticity-related genes in the cerebellum and hippocampus
Animals engaged in voluntary exercise through free access to a running wheel for 1 week, a length of time that produces changes in mouse hippocampal Bdnf expression (Adlard et al., 2005). Table 2 shows the expression profile of genes related to synaptic plasticity and cell signaling in the cerebellum and hippocampus of exercising animals. This table gives a summary of percent increase or decrease in gene expression, which occurred in exercising, as compared to sedentary, animals. The most dramatic change was a 128% increase in Bdnf hippocampal expression (p < .0001). A much smaller, (21%) trend (p < .07) was noted in the cerebellum of running animals. Fmr1, which regulates RNA trafficking to dendrites (Zukin et al., 2009), was increased in both the cerebellum (9%, p < .03) and the hippocampus (14%, p < .001). Surprisingly, Ncs, which encodes a protein that colocalizes with synaptophysin and is involved in synaptic transmission (Noh et al., 2005), was decreased in both brain regions. Interestingly, the expression profiles for a number of genes were not the same in hippocampus and cerebellum in response to exercise. For example, Syn1 and Syp, genes encoding proteins important in synaptic transmission via trafficking and retrieval of synaptic vesicles (Cesca et al., 2010; Kwon and Chapman, 2011) showed increased expression in only the hippocampus. However, Cbln1, which encodes a synaptogenic and maintenance protein that predominates in the cerebellum (Yuzaki, 2011), was significantly upregulated in the cerebellum (up 21%, p < .00) compared to the hippocampus (down 36%, p < .05). Moreover, Reln, which modulates presynaptic neurotransmitter release (Hellwig et al., 2011), showed a marked increase in the cerebellum of exercising animals (27%, p < .05) compared to no change in the hippocampus. Genes associated with cell signaling also differed in the two brain regions examined in response to exercise. CamkIV, which activates new protein synthesis required for synaptic potentiation (Sinagra et al., 2008), was up 13% in the cerebellum (p < .05) but unchanged in the hippocampus. In contrast, Calb1, which encodes a protein that can act as both a calcium binding and sensor protein underlying synaptic plasticity (Schmidt, 2012), was unchanged in the cerebellum as compared to a small increase in the hippocampus (p < .05).
Table 2.
Percent change in relative gene expression, in the cerebellum and hippocampus of exercised compared to sedentary mice assessed by qRT-PCR in brains of adolescent male mice that were sedentary (n = 10) or exercised with voluntary wheel running (n = 9) for 1 week prior to sacrifice. Gene expression changes that were significant (p < .05) are presented in the table. Positive values indicate percent increased expression and negative values indicate percent decreased expression.
| Gene | Gene function | % change in runners relative to sedentary group |
|
|---|---|---|---|
| Cerebellum | Hippocampus | ||
| Bdnf | Mediates activity-dependent synaptic modulation (Lu, 2003) | 21* | 128 |
| Calb1 | Synaptic plasticity (Schmidt, 2012) | NS | 16 |
| CamK4 | Activates new protein synthesis required for synaptic potentiation (Toyoda et al., 2010) | 13 | NS |
| Cbln1 | Synaptogenesis and maintenance; predominates in cerebellum (Yuzaki, 2011) | 21 | 36 |
| Csk | Regulates synaptic vesicle trafficking (Messa et al., 2010) | NS | NS |
| Fmr1 | Regulates RNA trafficking to dendrites (Zukin et al., 2009) | 9 | 14 |
| Grid2 | Primarily localized to Purkinje cells and regulates synaptic plasticity (Yuzaki, 2003, 2004) | NS | NS |
| Ncs1 | Synaptic transmission, colocalizes to synaptophysin in cerebellum and hippocampus (Jinno et al., 2004; Noh et al., 2005) | −9 | −23 |
| Reln | Modulates presynaptic neurotransmitter release and stimulates dendritic spine mRNA translation (Dong et al., 2003; Hellwig et al., 2011) | 27 | NS |
| Syn1 | Fine tuning of neuronal plasticity via directing trafficking of synaptic vesicles in presynaptic terminals (Cesca et al., 2010) | NS | 20 |
| Syp | Regulates synaptic vesicle retrieval following neuronal stimulation (Kwon and Chapman, 2011) | NS | 20 |
| Esr1 | Mediates spinogenesis at localized synaptic sites and regulates Bdnf expression (Mukai et al., 2010; Solum and Handa, 2002) | NS | −31 |
| Esr2 | Regulates hippocampal synaptic plasticity by increasing presynaptic proteins (Liu et al., 2008) | NS | NS |
Bdnf = brain derived growth factor, Calb = calbindin d28k, Camk4 = calcium/calmodulin-dependent kinase 4, Cbln1 = cerebellin 1 precursor protein, Csk = c-src, Fmr1 = fragile X mental retardation protein, Grid2 = glutamate receptor, Ncs = neuronal calcium sensor 1 Reln = reelin, Syn = synapsin, Syp = synaptophysin, Esr1 = estrogen receptor α, Esr2 = estrogen receptor β, NS = not significant.
p < .07.
3.2. The relationship between synaptic plasticity and cell signaling genes changes as a function of running
Given that a number of genes related to synaptic plasticity and cell signaling responded to exercise, we examined the relationship between these genes as a function of exercise. Table 3 shows correlation matrices of the genes examined in both the hippocampus and the cerebellum of sedentary and exercised animals. Only genes with significant correlations (p < .05) are represented in each matrix. In both the hippocampus and the cerebellum, the correlation matrices generated from exercising animals (Table 3c and d) are markedly different than the matrices of sedentary animals (Table 3a and b) suggesting that the relationship between synaptic plasticity and cell signaling genes changed as a function of exercise. Of particular interest is the strong positive relationship that exists between Bdnf and Esr1 in the hippocampus (r = .89, p < .03) and cerebellum (r = .81, p < .03) of exercising animals compared to no relationship in sedentary animals. Moreover, in the hippocampus of sedentary animals, we did not find a correlation between the cell signaling genes CamkIV and Csk and other synaptic plasticity genes (Table 3a). However, strong correlations between CamKIV and SYP and Csk and Grid2 where revealed in exercising animals (Table 3c). Similarly, in the cerebellum, a significant CamkIV-Syn correlation (r = .71, p < .05) is present in exercising animals (Table 3d) compared to sedentary animals (Table 3b). The correlation present in sedentary mouse hippocampus between Reln and Esr2 (r = .98, p < .001) (Table 3a) actually increased dramatically in response to exercise (Table 3c). Likewise, in the cerebellum, the correlations between the cell-signaling gene, Calb1, and two genes associated with synaptic plasticity, Grid2 and Syp both increased in exercising animals (Table 3d) by approximately 17% compared to sedentary animals (Table 3b).
Table 3.
Correlations between gene expression levels of all structural and signaling genes assessed by qRT-PCR in brains of adolescent male mice that were sedentary (n = 10) or exercised with voluntary wheel running (n = 9) for 1 week prior to sacrifice. Two brain regions were analyzed, the hippocampus and cerebellum. Correlations that were significant (p < 0.05) are presented in the table. Correlation values in red were negative and in black were positive.
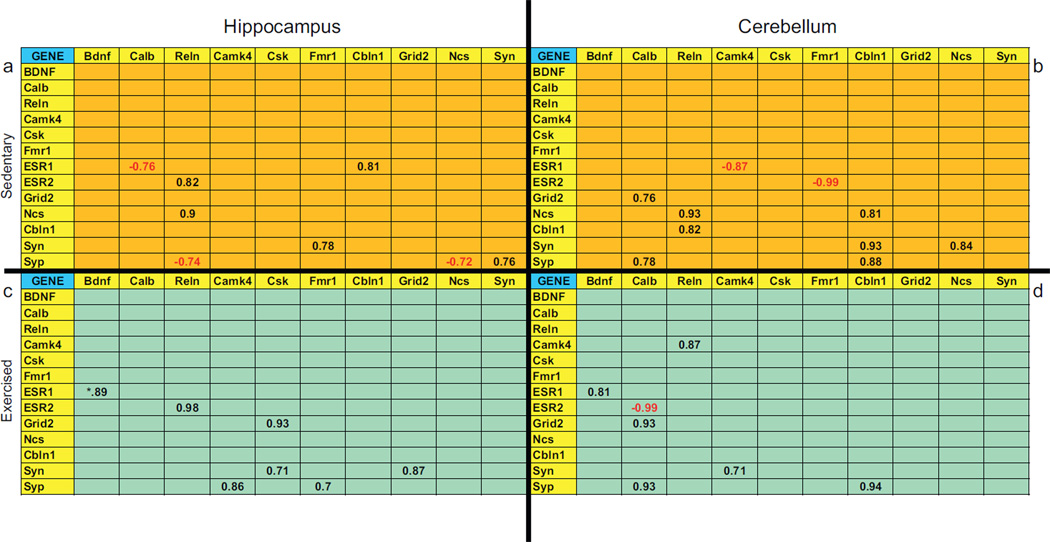 |
Bdnf = brain derived growth factor, Calb = calbindin d28k, Camk4 = calcium/calmodulin-dependent kinase 4, Cbln1 = cerebellin 1 precursor protein, Csk = c-src tyrosine kinase, Esr1 = estrogen receptor α, Esr2 = estrogen receptor β, Fmr1 = fragile X mental retardation protein, Grid2 = glutamate receptor, Ncs = neuronal calcium sensor1, Reln = reelin, Syn = synapsin, Syp = synaptophysin.
Finally, although the relationship between synaptic plasticity and cell signaling genes changed as a function of exercise, the changes observed are generally region dependent, with the exception of Bdnf and Esr1 in exercising animals. Moreover, the relationship between genes in the sedentary animals is also region dependent with the exception of a significant correlation between Ncs and Relin that occurred in both regions of sedentary mouse brains. This correlation was not present in exercising mice.
3.3. Changes in histone acetylation and epigenetic genes occur in response to exercise
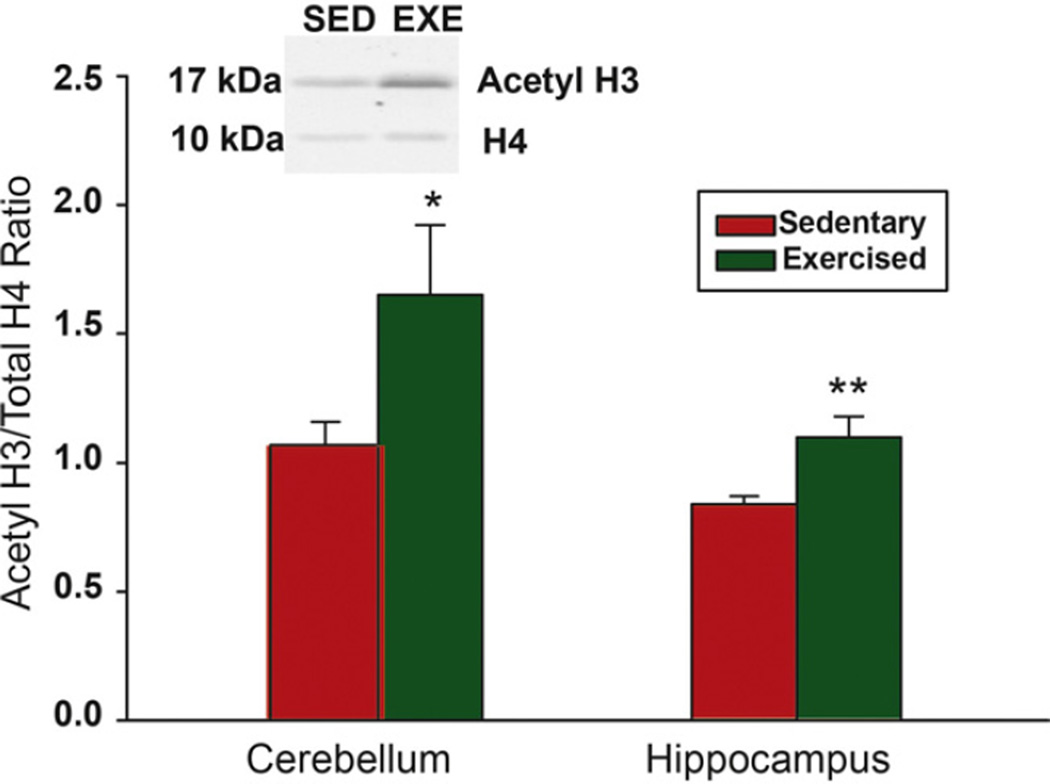
Since a number of genes increase and decrease in expression in both the cerebellum and the hippocampus in response to exercise, we examined epigenetic changes that occurred in response to exercise. We examined H3 acetylation, a mark of gene transcription because this epigenetic mark associates with increased levels of Bdnf expression in the hippocampus of exercising adult rats (Gomez-Pinilla et al., 2011). Fig. 1 shows that H3 acetylation was significantly increased in both the cerebellum and the hippocampus of exercising compared to sedentary animals. Moreover, several of the histone deacetylase (Hdac) genes and genes for DNA methyl transferases (Dnmt’s) were also affected by exercise. Table 4 shows the expression profile of genes related to histone deacetylase and DNA methyl transferase activity in the cerebellum and hippocampus of exercising animals. In the hippocampus, all genes encoding the DNA methyltransferases (Dnmt1, Dnmt3a and Dnmt3b) were significantly down regulated in exercising animals from 14% to 19% (p < .01). Moreover, Hdac5, Hdac7 and Hdac8 showed a similar decrease in expression of 22% (p < .001), 20% (p < .05) and 10% (p < .02), respectively, in exercising animals. Interestingly, Mecp2, which encodes a protein that acts as a transcriptional repressor involved in neuronal maintenance (Guy et al., 2011), did not change in expression in response to exercise. However, this gene showed a 23% decrease (p < .03) in the cerebellum of exercising compared to control animals. Moreover, Dnmt1 and Hdac8 were significantly depressed by 19% (p < .02) and 17% (p < .03) in the cerebellum of exercising animals. Surprisingly, in this region, Hdac2, showed a significant up regulation (28%, p < .002) in exercising animals.
Fig. 1.
Voluntary exercise increases histone H3 acetylation in the cerebellum and hippocampus of juvenile mice. Mice were sacrificed on postnatal Day 53 after 1 week in a cage with a locked running wheel (sedentary red histograms) or with an unlocked running wheel (exercised green histograms). Levels of acetylated histone H3 were assessed by Western Blotting. Representative immunoblot from the cerebellum of a sedentary (SED) compared to exercised (EXE) mouse. For each mouse, the amount of acetylated histone H3 in the cerebellum (n = 8 per group) and hippocampus (n = 5 per group) was divided by the total amount of histone 4 in each area to normalize the amount of protein loaded across samples. Each bar represents the mean ± SEM. Significantly different from sedentary *p = 0.05, **p < 0.01. Acetyl = acetylation, H3 = histone 3, H4 = histone 4.
Table 4.
Percent change in relative gene expression, associated with epigenetic changes, in the cerebellum and hippocampus of exercised compared to sedentary mice assessed by qRT-PCR in brains of adolescent male mice that were sedentary (n = 10) or exercised with voluntary wheel running (n = 9) for 1 week prior to sacrifice. Gene expression changes that were significant (p < .05) are presented in the table. Positive values indicate percent increased expression and negative values indicate percent decreased expression.
| % change in runners relative to sedentary group | ||
|---|---|---|
| Gene | Cerebellum | Hippocampus |
| Mecp2 | −23 | NS |
| Hdac1 | NS | NS |
| Hdac2 | 28 | NS |
| Hdac3 | NS | NS |
| Hdac4 | NS | NS |
| Hdac5 | NS | −22 |
| Hdac7 | NS | −20 |
| Hdac8 | −17 | −10 |
| Dnmt1 | −19 | −14 |
| Dnmt3a | NS | −17 |
| Dnmt3b | NS | −19 |
Mecp2 = methyl CpG binding protein, Hdac1 = histone deacetylase 1, Hdac2 = histone deacetylase 2, Hdac3 = histone deacetylase 3, Hdac4 = histone deacetylase 4, Hdac5 = histone deacetylase 5, Hdac7 = histone deacetylase 7, Hdac8 = histone deacetylase 8, Dnmt1 = DNA methyltransferase 1, Dnmt3a = DNA methyltransferase 3a, Dnmt3b = DNA methyltransferase 3b, NS = not significant.
3.4. The relationship between synaptic plasticity and repressive mark genes change in response to exercise
Given that a number of genes related to synaptic plasticity and cell signaling changed expression in response to exercise, we wanted to determine how these genes and genes associated with repressive marks relate to each other. Table 5 shows correlation matrices of synaptic plasticity crossed with repressive mark genes examined in both the hippocampus and the cerebellum of sedentary and exercised animals. In both the hippocampus and the cerebellum, the correlation matrices generated from exercising animals (Table 5c and d) are markedly different than the matrices of sedentary animals (Table 5a and b) suggesting that the relationship between synaptic plasticity and repressive mark genes change as a function of exercise. Most notable was an increased number of negative correlations in the hippocampus of sedentary compared to exercising animals. For example, Esr1 shows significant negative correlations with Hdac1 (r = −.83, p < .01), Hdac2 (r = −.87, p < .005), Hdac3 (r = −.72, p < .05) and Hdac5 (r = −.70, p < .05) in sedentary hippocampus (Table 5a), which are all absent in the hippocampus of exercising animals (Table 5c). These negative correlations suggest that these Hdac genes may reduce expression of Esr1 or vice versa. Interestingly, the cerebellum did not show a similar shift in response to exercise. In fact, negative correlations were generally absent from the cerebellum of sedentary animals, yet markedly present in exercising animals. Two correlations of significance in this area (Table 5d) were the high negative correlations between Bdnf and Hdac1 (r = −.92, p < .003) and Hdac2 (r = −.90, p < .006) suggesting the possibility that higher levels of Bdnf expression associate with low levels of these typically repressive genes.
Table 5.
Correlations between gene expression levels of all structural and signaling genes versus epigenetic genes and distances ran (in the exercised group only) assessed by qRT-PCR in brains of adolescent male mice that were sedentary (n = 10) or exercised with voluntary wheel running (n = 9) for 1 week prior to sacrifice. Two brain regions were analyzed, the hippocampus and cerebellum. Correlations that were significant (p < 0.05) are presented in the table. Correlation values in red were negative and in black were positive.
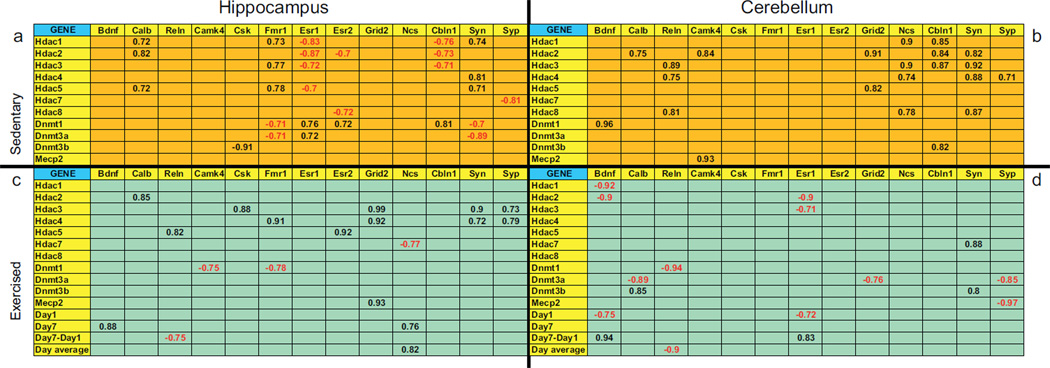 |
Bdnf = brain derived growth factor, Calb = calbindin d28k, Camk4 = calcium/calmodulin-dependent kinase 4, Cbln1 = cerebellin 1 precursor protein, Csk = csrc tyrosine kinase, Dnmt1 = DNA methyltransferase 1, Dnmt3a = DNA methyltransferase 3a, Dnmt3b = DNA methyltransferase 3b, Esr1 = estrogen receptor α, Esr2 = estrogen receptor β, Fmr1 = fragile X mental retardation protein, Grid2 = glutamate receptor, Hdac1 = histone deacetylase 1, Hdac2 = histone deacetylase 2, Hdac3 = histone deacetylase 3, Hdac4 = histone deacetylase 4, Hdac5 = histone deacetylase 5, Hdac7 = histone deacetylase 7, Hdac8 = histone deacetylase 8, Mecp2 = methyl CpG binding protein 2, Ncs = neuronal calcium sensor 1 Reln = reelin, Syn = synapsin, Syp = synaptophysin.
For exercised mice only Day 1 = average distance run on the first day of wheel exposure, Day 7 = average distance run on the seventh and final day of wheel exposure, Day 7–Day 1 = average difference in distance run on the first versus last day of wheel exposure, Day average = average distance run for all 7 days of wheel exposure.
3.5. Distance run positively correlates with Bdnf expression in the cerebellum and hippocampus
In a final analysis, we asked if individual variation in the amount of running subjects engaged in, yielded any novel correlations with the genes we measured. Also included in the exercise matrices (Table 5c and d) are the running distance for Day 1, Day 7, the difference between Day 1 and Day 7 and the running distance averaged across the 7 days. Surprisingly, few genes correlated with any of the variables that reflect distance run by the mice. However, Bdnf mRNA was positively correlated with the amount of running on Day 7 in the hippocampus (r = .88, p < .008) and between Day 7 and Day 1 in the cerebellum (r = .94, p < .001). This suggests that increased running stimulates higher expression levels of Bdnf in both the cerebellum and the hippocampus.
4. Discussion
Exercise in adolescent male mice produced significant changes in the expression levels of genes associated with synaptic plasticity and signaling pathways. Furthermore, in the two regions examined we noted that expression profiles were region dependent. Some genes showed similar changes in expression in both areas while other changes were unique to either the cerebellum or hippocampus. Moreover, H3 acetylation, identified with gene transcription, increased in both brain regions in response to wheel running. In contrast, the expression pattern of DNA methyltransferases (Dnmts) and histone deacetylases (Hdacs), genes which influence DNA methylation and histone modifications, respectively, displayed subtle differences in the two brain regions but in general decreased with exercise. These findings suggest that epigenetic changes may regulate the unique patterns of gene expression, associated with synaptic plasticity and signaling pathways, which occur in adolescent male brain in response to exercise.
4.1. Exercise affects Bdnf and other genes associated with synaptic plasticity and cell signaling
The most dramatic change in gene expression was a strong increase in Bdnf mRNA in the hippocampus of exercising mice. Historically, Bdnf has been the primary target gene examined in animal studies aimed at understanding the molecular basis underlying the positive effects of exercise on the brain (for review see Zoladz and Pilc, 2010). The positive relationship between Bdnf and wheel running, which correlated with distance run per night in the hippocampus and caudal cortex, was demonstrated first by Neeper et al. (1996). In fact, Bdnf levels remained elevated several days after exercise was stopped and exercise facilitated Bdnf induction following re-exposure (Berchtold et al., 2005). Other work confirmed a link between increased hippocampal Bdnf levels and improved object recognition and spatial memory in response to exercise (Cassilhas et al., 2012; Griffin et al., 2009; Vaynman et al., 2004). Moreover, blocking hippocampal BDNF in rats, prevented exercise-induced cognitive enhancement (Vaynman et al., 2004). Exercise-induced BDNF mediates synaptic plasticity by engaging its downstream targets, cAMP-response element binding (CREB) protein, synapsin I and synapthophysin, while simultaneously increasing its own mRNA and its receptor, tyrosine kinase B (TrkB) (Vaynman et al., 2003, 2006). Clearly, in the hippocampus, increased Bdnf expression in response to exercise directs the expression of downstream targets related to synaptic plasticity, which in turn orchestrate improved learning. Our data replicate previous work in adult rats as we report increased Bdnf, Syn1 and Syp mRNA expression in mouse hippocampus in response to exercise, suggesting that exercise may prime the brain for improved learning via a Bdnf mediated mechanism which enhances synaptic plasticity gene expression.
In contrast, neither Bdnf, Syn1 nor Syp mRNA were significantly increased in the cerebellum following exercise. This confirms a recent study which showed no changes in Syn1 and Syp protein levels in the cerebellum of adolescent rats in response to treadmill running (Garcia et al., 2012). Because these genes were not up regulated in the cerebellum, it is likely that changes in genes involved in synaptic plasticity occurred independent of Bdnf regulation. Cbln1, a gene primarily associated with the cerebellum and involved in synaptogenesis and maintenance of parallel fiber-Purkinje cell synapses (Yuzaki, 2011) and Relin, associated with presynaptic neurotransmitter release (Hellwig et al., 2011) were both significantly increased in the cerebellum in response to exercise compared to a down-regulation and no change, respectively, in the hippocampus. In fact, since increased Bdnf expression coincidently suppresses Cbln1 expression in the cerebellum (Iijima et al., 2009), we were not surprised to see an increase in Cbln1 mRNA in light of the lack of a significant increase in Bdnf levels. Moreover, we found high Bdnf levels coincident with low Cbln1 gene expression in the hippocampi of exercising mice. Finally, CamkIV, which activates new protein synthesis, required for synaptic potentiation (Sinagra et al., 2008), showed increased expression in the cerebellum in response to running while the hippocampus showed no change. CamkIV plays a role in cerebellar long-term depression which underlies the long term memory necessary for acquiring the conditioned eye-blink response (Lee et al., 2009). Since exercising rats show significantly more conditioned eye-blink responses than sedentary rats (Green et al., 2011), this raises the possibility that increased levels of CamkIV account for this effect.
Two genes associated with synaptic plasticity and function, Fmr1 and Ncs1, changed in opposite directions and did so similarly in the hippocampus and cerebellum of exercised compared to sedentary mice. In both regions, Fmr1, which regulates RNA trafficking to dendrites (Zukin et al., 2009), was significantly increased. In contrast, Ncs1, which colocalizes to synaptophysin (Noh et al., 2005), was down-regulated in exercised mice. Interestingly, although Calb1, which encodes a calcium binding and sensor protein associated with synaptic plasticity (Schmidt, 2012), did not change expression in the cerebellum in response to exercise, it increased significantly in the hippocampus. Knock down of this protein in the hippocampus impairs spatial learning suggesting a role in this form of learning (Molinari et al., 1996). This implies that the increase in hippocampal Calb1 expression we observed may in part underlie exercised induced improved spatial memory described by others (Cassilhas et al., 2012; van Praag et al., 1999; Vaynman et al., 2004).
4.2. Exercise induces epigenetic changes associated with gene transcription and repression
Gene expression changes can occur in the absence of altering the underlying DNA sequence. When this happens it is due to epigenetic regulation, which can either activate or repress gene transcription through chromatin modifications including posttranslational histone modifications and covalent modifications of genomic DNA (Takizawa and Meshorer, 2008). In the brain, epigenetic mechanisms regulate synaptic plasticity (Sultan and Day, 2011). Therefore, we predicted that any changes we observed in gene expression related to synaptic plasticity and signal transduction in response to exercise would occur coincident with epigenetic gene expression changes. Our results demonstrate a number of gene expression changes for genes that regulate chromatin modifications. First, global H3 acetylation, associated with enhanced gene transcription, significantly increased in both the hippocampus and the cerebellum of adolescent mice in response to exercise. This finding confirms an earlier study showing that voluntary exercise increases acetylated H3 in adult rat hippocampus (Gomez-Pinilla et al., 2011). Moreover, using an acetyl-histone H3 chromatin immunoprecipitation (ChIP) assay, these authors further showed that Bdnf was one of the genes up regulated directly in response to increased H3 acetylation. Similarly, in adult rat medial frontal cortex, long-term potentiation (LTP) increased H3 acetylation of both Bdnf and Reln genes thereby increasing expression (Sui et al., 2012). Histone acetylation regulates memory processing in the adult hippocampus and other brain areas and can promote memory via increased transcription of genes related to synaptic plasticity and signaling (Sultan and Day, 2011). Our results suggest that, similar to an adult, the hippocampus and cerebellum of adolescent brain may also be primed for facilitated learning and memory by increased H3 acetylation in response to exercise. However, in the case of the cerebellum, Bdnf is likely not the target of H3 acetylation.
We also examined expression of several HDAC genes which regulate histone acetylation by removing acetyl groups from histones thereby creating a compacted chromatin structure closed to transcription factors (Delcuve et al., 2012). In the hippocampus, the genes for theses enzymes either showed no change in expression or a significant down-regulation (Hdac5, Hdac7 and Hdac8) in response to exercise. These down-regulated HDACs may in part be responsible for the increased acetylation we observe in the hippocampus in response to running. Interestingly, although H3 acetylation also increased in the cerebellum in response to exercise, the gene expression levels of the HDACs did not change with the exception of Hdac2, which surprisingly increased in gene expression in response to exercise. However, the protein product of Hdac2 requires phosphorylation in order to be incorporated into a corepressor complex (Segre and Chiocca, 2011). Since we only examined gene expression changes in response to exercise, it is quite possible that changes in these genes do not directly correspond to changes in their enzymatic products. Alternatively, the Hdac2 protein product may decrease the expression of genes that are repressive in nature. For example, the gene encoding protein phosphatase 1 (PP1), which suppresses memory, is down regulated in response to fear conditioning via increased methylation of its promoter site (Miller and Sweatt, 2007). Finally, a recent study showed that Bdnf mediated spine density and morphology and quantal synaptic neurotransmitter release was actually reduced in CA1 hippocampal neurons in response to reduced HDAC activity suggesting that a delicate balance exists between histone acetylation and deacetylation in regulating hippocampal synaptic activity (Calfa et al., 2012).
In addition to the covalent histone modifications discussed above, DNA methylation is another form of epigenetic control of gene expression associated with gene repression. DNA methylation is carried out by DNA methyltransferases (DNMT) converting cytosines at CpG dinucleotide sites to 5-methylcytosine in order to directly or indirectly repress gene transcription in neurons (Sultan and Day, 2011). In the cerebellum, Dnmt1 decreases in response to running compared to no change in Dnmt3a and Dnmt3b gene expression levels. This finding was of particular interest when compared to the gene expression increase in Reln we observed in the cerebellum of exercised mice. We found a strong negative correlation (−.94) between Dnmt1 and Reln in the cerebellum of exercised mice. This correlation is absent in sedentary mice. Dnmt1 regulates Reln expression in mouse GABAergic neurons such that reduced levels of this methyltransferase result in an increase in reelin protein (Noh et al., 2005). In the cerebellum, granule cells synthesize reelin and express Dnmt mRNA (Sinagra et al., 2008; Veldic et al., 2004). Reelin has been implicated in both presynaptic neurotransmitter release and dendritic mRNA translation and promotes synaptic plasticity in the brain (Dong et al., 2003; Hellwig et al., 2011; Weeber et al., 2002). Therefore, our findings suggest that decreased Dnmt1 expression, induced by exercise, promotes cerebellar synaptic plasticity by possibly increasing Reln expression in granule cells.
In contrast, we noted a significant decrease in Dnmt1, Dnmt3a and Dnmt3b in the hippocampus of exercised, compared to sedentary mice. Depolarization of cortical neurons reduces both mRNA and protein expression of Dnmt1 and Dnmt3a while elevating in parallel Bdnf exon-1 and exon-4 (Sharma et al., 2008). Likewise, we show a highly significant increase in Bdnf in the hippocampus of mice in response to exercise, which coincides with depressed DNA methyltransferase gene expression. This suggests that repressed Dnmt expression in response to adolescent exercise may be responsible for up-regulated Bdnf activity. In contrast, fear conditioning leads to an up regulation of Dnmt3a and Dnmt3b in the CA1 area of adult rat hippocampus (Miller and Sweatt, 2007). In fact, DNA methylation plays a positive functional role in behavior related to the formation and perseverance of hippocampal memory in response to fear conditioning by inhibiting the expression of memory repressive genes, such as PP1 (Sultan and Day, 2011). In this sense, Dnmt’s may be repressing genes that normally inhibit or interfere with synaptic plasticity genes (Levenson et al., 2006). These findings demonstrate that the type of environmental intervention or sensory stimulation governs the direction of epigenetic and downstream gene changes that occur in the hippocampus.
4.3. Exercise influences the expression of genes associated with synaptic plasticity and epigenetic regulation differently in adolescent cerebellum versus hippocampus
Our findings show that gene expression profiles reflecting synaptic plasticity and signal transduction and generated in response to exercise in adolescent brain, differ remarkably in the cerebellum and hippocampus. Moreover, epigenetic changes, occurring under the influence of exercise, similarly differ in these two brain regions. These findings are not surprising given that the cerebellum and hippocampus are two brain regions that are fundamentally different in both function and structure. For example, motor dependent learning and memory predominate in the cerebellum with neural plasticity distributed between cerebellar cortex and deep cerebellar nuclei (Raymond et al., 1996). In contrast, cognitive dependent learning and memory underlie hippocampal function driven primarily by long-term potentiation (LTP) and long-term depression (LTD) in areas CA1, CA2 and CA3 (Caruana et al., 2012; Shapiro, 2001). Moreover, the cellular mechanisms underlying postsynaptic LTP and LTD in the cerebellum are in marked contrast to their counterparts in the hippocampus (Jorntell and Hansel, 2006). Finally, the nature of the environmental variable we used, exercise, has direct effects on the cerebellum but likely second order effects on the hippocampus. However, despite these differences, the cerebellum and hippocampus are functionally related and recent work has suggested they act in concert (Baillieux et al., 2008; Stoodley and Schmahmann, 2010). We hypothesize that epigenetic regulation may be one mechanism maintaining coordination between these brain regions.
4.4. Summary
A meta-analysis of adult human studies conducted over the past 45 years, found a positive impact of exercise on neurocognitive performance including improved attention and processing speed, executive function and memory (Smith et al., 2010). Recent human studies targeted at juvenile and adolescent age groups show they can also benefit cognitively from the effects of exercise (Voss et al., 2011). In fact, acute exercise improves executive function and alleviates symptoms in children with attention deficit hyperactivity disorder (ADHD) (Archer and Kostrzewa, 2012; Chang et al., 2012; Pontifex et al., 2012). In rats, exposure to exercise in adolescence increases Bdnf levels and improves hippocampal plasticity and spatial memory as adults (Gomes da Silva et al., 2012). Our study shows that exercise is a powerful environmental intervention capable of inducing gene expression changes associated with synaptic plasticity, signaling pathways and epigenetic regulation in both the hippocampus and the cerebellum of adolescent brain. Future studies should be directed at identifying cells in the cerebellum and hippocampus responsible for these changes. Since these brain regions can be associated with autism, schizophrenia and ADHD, all disorders affecting juveniles and adolescents with a higher incidence, exercise may prove to be a valuable tool in ameliorating symptoms associated with these diseases in young people.
Acknowledgements
We thank Drs. Vitor Lira and Zhen Yan for allowing us to use their exercise set up and for their expert advice. We thank Jackson Ball for technical assistance. This work was funded by NIH R01 MH057759.
References
- Aberg MA, Pedersen NL, Toren K, Svartengren M, Backstrand B, Johnsson T, Cooper-Kuhn CM, Aberg ND, Nilsson M, Kuhn HG. Cardiovascular fitness is associated with cognition in young adulthood. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:20906–20911. doi: 10.1073/pnas.0905307106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adlard PA, Perreau VM, Cotman CW. The exercise-induced expression of BDNF within the hippocampus varies across life-span. Neurobiology of Aging. 2005;26:511–520. doi: 10.1016/j.neurobiolaging.2004.05.006. [DOI] [PubMed] [Google Scholar]
- Archer T, Kostrzewa RM. Physical exercise alleviates ADHD symptoms: regional deficits and development trajectory. Neurotoxicity Research. 2012;21:195–209. doi: 10.1007/s12640-011-9260-0. [DOI] [PubMed] [Google Scholar]
- Baillieux H, De Smet HJ, Paquier PF, De Deyn PP, Marien P. Cerebellar neurocognition: insights into the bottom of the brain. Clinical Neurology and Neurosurgery. 2008;110:763–773. doi: 10.1016/j.clineuro.2008.05.013. [DOI] [PubMed] [Google Scholar]
- Berchtold NC, Chinn G, Chou M, Kesslak JP, Cotman CW. Exercise primes a molecular memory for brain-derived neurotrophic factor protein induction in the rat hippocampus. Neuroscience. 2005;133:853–861. doi: 10.1016/j.neuroscience.2005.03.026. [DOI] [PubMed] [Google Scholar]
- Calfa G, Chapleau CA, Campbell S, Inoue T, Morse SJ, Lubin FD, Pozzo-Miller L. HDAC activity is required for BDNF to increase quantal neurotransmitter release and dendritic spine density in CA1 pyramidal neurons. Hippocampus. 2012;22:1493–1500. doi: 10.1002/hipo.20990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruana DA, Alexander GM, Dudek SM. New insights into the regulation of synaptic plasticity from an unexpected place: hippocampal area CA2. Learning and Memory. 2012;19:391–400. doi: 10.1101/lm.025304.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassilhas RC, Lee KS, Fernandes J, Oliveira MG, Tufik S, Meeusen R, de Mello MT. Spatial memory is improved by aerobic and resistance exercise through divergent molecular mechanisms. Neuroscience. 2012;202:309–317. doi: 10.1016/j.neuroscience.2011.11.029. [DOI] [PubMed] [Google Scholar]
- Cesca F, Baldelli P, Valtorta F, Benfenati F. The synapsins: key actors of synapse function and plasticity. Progress in Neurobiology. 2010;91:313–348. doi: 10.1016/j.pneurobio.2010.04.006. [DOI] [PubMed] [Google Scholar]
- Chaddock L, Pontifex MB, Hillman CH, Kramer AF. A review of the relation of aerobic fitness and physical activity to brain structure and function in children. Journal of the International Neuropsychological Society. 2011;17:975–985. doi: 10.1017/S1355617711000567. [DOI] [PubMed] [Google Scholar]
- Chang YK, Liu S, Yu HH, Lee YH. Effect of acute exercise on executive function in children with attention deficit hyperactivity disorder. Archives of Clinical Neuropsychology. 2012;27:225–237. doi: 10.1093/arclin/acr094. [DOI] [PubMed] [Google Scholar]
- Coles K, Tomporowski PD. Effects of acute exercise on executive processing, short-term and long-term memory. Journal of Sports Sciences. 2008;26:333–344. doi: 10.1080/02640410701591417. [DOI] [PubMed] [Google Scholar]
- Cotman CW, Berchtold NC. Exercise: a behavioral intervention to enhance brain health and plasticity. Trends in Neurosciences. 2002;25:295–301. doi: 10.1016/s0166-2236(02)02143-4. [DOI] [PubMed] [Google Scholar]
- Delcuve GP, Khan DH, Davie JR. Roles of histone deacetylases in epigenetic regulation: emerging paradigms from studies with inhibitors. Clinical Epigenetics. 2012;4:5. doi: 10.1186/1868-7083-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E, Caruncho H, Liu WS, Smalheiser NR, Grayson DR, Costa E, Guidotti A. A reelin-integrin receptor interaction regulates Arc mRNA translation in synaptoneurosomes. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:5479–5484. doi: 10.1073/pnas.1031602100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer J, Zhao X, van Praag H, Wodtke K, Gage FH, Christie BR. Effects of voluntary exercise on synaptic plasticity and gene expression in the dentate gyrus of adult male Sprague-Dawley rats in vivo. Neuroscience. 2004;124:71–79. doi: 10.1016/j.neuroscience.2003.09.029. [DOI] [PubMed] [Google Scholar]
- Fedewa AL, Ahn S. The effects of physical activity and physical fitness on children’s achievement and cognitive outcomes: a meta-analysis. Research Quarterly for Exercise and Sport. 2011;82:521–535. doi: 10.1080/02701367.2011.10599785. [DOI] [PubMed] [Google Scholar]
- Ferreira AF, Real CC, Rodrigues AC, Alves AS, Britto LR. Moderate exercise changes synaptic and cytoskeletal proteins in motor regions of the rat brain. Brain Research. 2010;1361:31–42. doi: 10.1016/j.brainres.2010.09.045. [DOI] [PubMed] [Google Scholar]
- Garcia PC, Real CC, Ferreira AF, Alouche SR, Britto LR, Pires RS. Different protocols of physical exercise produce different effects on synaptic and structural proteins in motor areas of the rat brain. Brain Research. 2012;1456:36–48. doi: 10.1016/j.brainres.2012.03.059. [DOI] [PubMed] [Google Scholar]
- Gomes da Silva S, Unsain N, Masco DH, Toscano-Silva M, de Amorim HA, Silva Araujo BH, Simoes PS, Naffah-Mazzacoratti Mda G, Mortara RA, Scorza FA, Cavalheiro EA, Arida RM. Early exercise promotes positive hippocampal plasticity and improves spatial memory in the adult life of rats. Hippocampus. 2012;22:347–358. doi: 10.1002/hipo.20903. [DOI] [PubMed] [Google Scholar]
- Gomez-Pinilla F, Zhuang Y, Feng J, Ying Z, Fan G. Exercise impacts brain-derived neurotrophic factor plasticity by engaging mechanisms of epigenetic regulation. European Journal of Neuroscience. 2011;33:383–390. doi: 10.1111/j.1460-9568.2010.07508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green JT, Chess AC, Burns M, Schachinger KM, Thanellou A. The effects of two forms of physical activity on eyeblink classical conditioning. Behavioural Brain Research. 2011;219:165–174. doi: 10.1016/j.bbr.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin EW, Bechara RG, Birch AM, Kelly AM. Exercise enhances hippocampal-dependent learning in the rat: evidence for a BDNF-related mechanism. Hippocampus. 2009;19:973–980. doi: 10.1002/hipo.20631. [DOI] [PubMed] [Google Scholar]
- Guy J, Cheval H, Selfridge J, Bird A. The role of MeCP2 in the brain. Annual Review of Cell and Developmental Biology. 2011;27:631–652. doi: 10.1146/annurev-cellbio-092910-154121. [DOI] [PubMed] [Google Scholar]
- Hellwig S, Hack I, Kowalski J, Brunne B, Jarowyj J, Unger A, Bock HH, Junghans D, Frotscher M. Role for reelin in neurotransmitter release. Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2011;31:2352–2360. doi: 10.1523/JNEUROSCI.3984-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herting MM, Nagel BJ. Aerobic fitness relates to learning on a virtual Morris Water Task and hippocampal volume in adolescents. Behavioural Brain Research. 2012;233:517–525. doi: 10.1016/j.bbr.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins ME, Nitecki R, Bucci DJ. Physical exercise during adolescence versus adulthood: differential effects on object recognition memory and brain-derived neurotrophic factor levels. Neuroscience. 2011;194:84–94. doi: 10.1016/j.neuroscience.2011.07.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima T, Emi K, Yuzaki M. Activity-dependent repression of Cbln1 expression: mechanism for developmental and homeostatic regulation of synapses in the cerebellum. Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2009;29:5425–5434. doi: 10.1523/JNEUROSCI.4473-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinno S, Jeromin A, Kosaka T. Expression and possible role of neuronal calcium sensor-1 in the cerebellum. Cerebellum. 2004;3:83–88. doi: 10.1080/14734220310025187. [DOI] [PubMed] [Google Scholar]
- Jorntell H, Hansel C. Synaptic memories upside down: bidirectional plasticity at cerebellar parallel fiber-Purkinje cell synapses. Neuron. 2006;52:227–238. doi: 10.1016/j.neuron.2006.09.032. [DOI] [PubMed] [Google Scholar]
- Kramer AF, Erickson KI, Colcombe SJ. Exercise, cognition, and the aging brain. Journal of Applied Physiology. 2006;101:1237–1242. doi: 10.1152/japplphysiol.00500.2006. [DOI] [PubMed] [Google Scholar]
- Kwon SE, Chapman ER. Synaptophysin regulates the kinetics of synaptic vesicle endocytosis in central neurons. Neuron. 2011;70:847–854. doi: 10.1016/j.neuron.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KH, Chatila TA, Ram RA, Thompson RF. Impaired memory of eyeblink conditioning in CaMKIV KO mice. Behavioral Neuroscience. 2009;123:438–442. doi: 10.1037/a0014724. [DOI] [PubMed] [Google Scholar]
- Levenson JM, Roth TL, Lubin FD, Miller CA, Huang IC, Desai P, Malone LM, Sweatt JD. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. Journal of Biological Chemistry. 2006;281:15763–15773. doi: 10.1074/jbc.M511767200. [DOI] [PubMed] [Google Scholar]
- Liu F, Day M, Muniz LC, Bitran D, Arias R, Revilla-Sanchez R, Grauer S, Zhang G, Kelley C, Pulito V, Sung A, Mervis RF, Navarra R, Hirst WD, Reinhart PH, Marquis KL, Moss SJ, Pangalos MN, Brandon NJ. Activation of estrogen receptor-beta regulates hippocampal synaptic plasticity and improves memory. Nature neuroscience. 2008;11:334–343. doi: 10.1038/nn2057. [DOI] [PubMed] [Google Scholar]
- Lou SJ, Liu JY, Chang H, Chen PJ. Hippocampal neurogenesis and gene expression depend on exercise intensity in juvenile rats. Brain Research. 2008;1210:48–55. doi: 10.1016/j.brainres.2008.02.080. [DOI] [PubMed] [Google Scholar]
- Lu B. BDNF and activity-dependent synaptic modulation. Learning and Memory. 2003;10:86–98. doi: 10.1101/lm.54603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messa M, Congia S, Defranchi E, Valtorta F, Fassio A, Onofri F, Benfenati F. Tyrosine phosphorylation of synapsin I by Src regulates synaptic-vesicle trafficking. Journal of Cell Science. 2010;123:2256–2265. doi: 10.1242/jcs.068445. [DOI] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Molinari S, Battini R, Ferrari S, Pozzi L, Killcross AS, Robbins TW, Jouvenceau A, Billard JM, Dutar P, Lamour Y, Baker WA, Cox H, Emson PC. Deficits in memory and hippocampal long-term potentiation in mice with reduced calbindin D28K expression. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:8028–8033. doi: 10.1073/pnas.93.15.8028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molteni R, Ying Z, Gomez-Pinilla F. Differential effects of acute and chronic exercise on plasticity-related genes in the rat hippocampus revealed by microarray. European Journal of Neuroscience. 2002;16:1107–1116. doi: 10.1046/j.1460-9568.2002.02158.x. [DOI] [PubMed] [Google Scholar]
- Mukai H, Kimoto T, Hojo Y, Kawato S, Murakami G, Higo S, Hatanaka Y, Ogiue-Ikeda M. Modulation of synaptic plasticity by brain estrogen in the hippocampus. Biochimica et Biophysica Acta. 2010;1800:1030–1044. doi: 10.1016/j.bbagen.2009.11.002. [DOI] [PubMed] [Google Scholar]
- Neeper SA, Gomez-Pinilla F, Choi J, Cotman CW. Physical activity increases mRNA for brain-derived neurotrophic factor and nerve growth factor in rat brain. Brain Research. 1996;726:49–56. [PubMed] [Google Scholar]
- Noh JS, Sharma RP, Veldic M, Salvacion AA, Jia X, Chen Y, Costa E, Guidotti A, Grayson DR. DNA methyltransferase 1 regulates reelin mRNA expression in mouse primary cortical cultures. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:1749–1754. doi: 10.1073/pnas.0409648102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontifex MB, Saliba BJ, Raine LB, Picchietti DL, Hillman CH. Exercise improves behavioral, neurocognitive, and scholastic performance in children with attention-deficit/hyperactivity disorder. Journal of Pediatrics. 2012 doi: 10.1016/j.jpeds.2012.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond JL, Lisberger SG, Mauk MD. The cerebellum: a neuronal learning machine? Science. 1996;272:1126–1131. doi: 10.1126/science.272.5265.1126. [DOI] [PubMed] [Google Scholar]
- Schmidt H. Three functional facets of calbindin D-28k. Frontiers in Molecular Neuroscience. 2012;5:25. doi: 10.3389/fnmol.2012.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segre CV, Chiocca S. Regulating the regulators: the post-translational code of class I HDAC1 and HDAC2. Journal of Biomedicine and Biotechnology. 2011;2011:690848. doi: 10.1155/2011/690848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro M. Plasticity, hippocampal place cells, and cognitive maps. Archives of Neurology. 2001;58:874–881. doi: 10.1001/archneur.58.6.874. [DOI] [PubMed] [Google Scholar]
- Sharma RP, Tun N, Grayson DR. Depolarization induces downregulation of DNMT1 and DNMT3a in primary cortical cultures. Epigenetics. 2008;3:74–80. doi: 10.4161/epi.3.2.6103. [DOI] [PubMed] [Google Scholar]
- Sibley BA, Etnier JL. The relationship between physical activity and cognition in children: a meta-analysis. Pediatric Exercise Science. 2003;15:243–246. [Google Scholar]
- Sinagra M, Gonzalez Campo C, Verrier D, Moustie O, Manzoni OJ, Chavis P. Glutamatergic cerebellar granule neurons synthesize and secrete reelin in vitro. Neuron Glia Biology. 2008;4:189–196. doi: 10.1017/S1740925X09990214. [DOI] [PubMed] [Google Scholar]
- Smith PJ, Blumenthal JA, Hoffman BM, Cooper H, Strauman TA, Welsh-Bohmer K, Browndyke JN, Sherwood A. Aerobic exercise and neurocognitive performance: a meta-analytic review of randomized controlled trials. Psychosomatic Medicine. 2010;72:239–252. doi: 10.1097/PSY.0b013e3181d14633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solum DT, Handa RJ. Estrogen regulates the development of brain-derived neurotrophic factor mRNA and protein in the rat hippocampus. Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2002;22:2650–2659. doi: 10.1523/JNEUROSCI.22-07-02650.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoodley CJ, Schmahmann JD. Evidence for topographic organization in the cerebellum of motor control versus cognitive and affective processing. Cortex. 2010;46:831–844. doi: 10.1016/j.cortex.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui L, Wang Y, Ju LH, Chen M. Epigenetic regulation of reelin and brain-derived neurotrophic factor genes in long-term potentiation in rat medial prefrontal cortex. Neurobiology of learning and memory. 2012;97:425–440. doi: 10.1016/j.nlm.2012.03.007. [DOI] [PubMed] [Google Scholar]
- Sultan FA, Day JJ. Epigenetic mechanisms in memory and synaptic function. Epigenomics. 2011;3:157–181. doi: 10.2217/epi.11.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa T, Meshorer E. Chromatin and nuclear architecture in the nervous system. Trends in Neurosciences. 2008;31:343–352. doi: 10.1016/j.tins.2008.03.005. [DOI] [PubMed] [Google Scholar]
- Thomas AG, Dennis A, Bandettini PA, Johansen-Berg H. The effects of aerobic activity on brain structure. Frontiers in Psychology. 2012;3:86. doi: 10.3389/fpsyg.2012.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomporowski PD, Davis CL, Miller PH, Naglieri JA. Exercise and children’s intelligence, cognition, and academic achievement. Educational Psychology Review. 2008;20:111–131. doi: 10.1007/s10648-007-9057-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong L, Shen H, Perreau VM, Balazs R, Cotman CW. Effects of exercise on gene-expression profile in the rat hippocampus. Neurobiology of Disease. 2001;8:1046–1056. doi: 10.1006/nbdi.2001.0427. [DOI] [PubMed] [Google Scholar]
- Toyoda H, Zhao MG, Mercaldo V, Chen T, Descalzi G, Kida S, Zhuo M. Calcium/calmodulin-dependent kinase IV contributes to translation-dependent early synaptic potentiation in the anterior cingulate cortex of adult mice. Molecular Brain. 2010;3:27. doi: 10.1186/1756-6606-3-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai HW, Grant PA, Rissman EF. Sex differences in histone modifications in the neonatal mouse brain. Epigenetics. 2009;4:47–53. doi: 10.4161/epi.4.1.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uysal N, Tugyan K, Kayatekin BM, Acikgoz O, Bagriyanik HA, Gonenc S, Ozdemir D, Aksu I, Topcu A, Semin I. The effects of regular aerobic exercise in adolescent period on hippocampal neuron density, apoptosis and spatial memory. Neuroscience Letters. 2005;383:241–245. doi: 10.1016/j.neulet.2005.04.054. [DOI] [PubMed] [Google Scholar]
- van Praag H. Exercise and the brain: something to chew on. Trends in Neurosciences. 2009;32:283–290. doi: 10.1016/j.tins.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Praag H, Christie BR, Sejnowski TJ, Gage FH. Running enhances neurogenesis, learning, and long-term potentiation in mice. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:13427–13431. doi: 10.1073/pnas.96.23.13427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaynman S, Ying Z, Gomez-Pinilla F. Interplay between brain-derived neurotrophic factor and signal transduction modulators in the regulation of the effects of exercise on synaptic-plasticity. Neuroscience. 2003;122:647–657. doi: 10.1016/j.neuroscience.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Vaynman S, Ying Z, Gomez-Pinilla F. Hippocampal BDNF mediates the efficacy of exercise on synaptic plasticity and cognition. European Journal of Neuroscience. 2004;20:2580–2590. doi: 10.1111/j.1460-9568.2004.03720.x. [DOI] [PubMed] [Google Scholar]
- Vaynman SS, Ying Z, Yin D, Gomez-Pinilla F. Exercise differentially regulates synaptic proteins associated to the function of BDNF. Brain Research. 2006;1070:124–130. doi: 10.1016/j.brainres.2005.11.062. [DOI] [PubMed] [Google Scholar]
- Veldic M, Caruncho HJ, Liu WS, Davis J, Satta R, Grayson DR, Guidotti A, Costa E. DNA-methyltransferase 1 mRNA is selectively overexpressed in telencephalic GABAergic interneurons of schizophrenia brains. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:348–353. doi: 10.1073/pnas.2637013100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss MW, Nagamatsu LS, Liu-Ambrose T, Kramer AF. Exercise, brain, and cognition across the life span. Journal of Applied Physiology. 2011;111:1505–1513. doi: 10.1152/japplphysiol.00210.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters RE, Rotevatn S, Li P, Annex BH, Yan Z. Voluntary running induces fiber type-specific angiogenesis in mouse skeletal muscle. American Journal of Physiology: Cell physiology. 2004;287:C1342–C1348. doi: 10.1152/ajpcell.00247.2004. [DOI] [PubMed] [Google Scholar]
- Weeber EJ, Beffert U, Jones C, Christian JM, Forster E, Sweatt JD, Herz J. Reelin and ApoE receptors cooperate to enhance hippocampal synaptic plasticity and learning. Journal of Biological Chemistry. 2002;277:39944–39952. doi: 10.1074/jbc.M205147200. [DOI] [PubMed] [Google Scholar]
- Yuzaki M. The delta2 glutamate receptor: 10 years later. Neuroscience Research. 2003;46:11–22. doi: 10.1016/s0168-0102(03)00036-1. [DOI] [PubMed] [Google Scholar]
- Yuzaki M. The delta2 glutamate receptor: a key molecule controlling synaptic plasticity and structure in Purkinje cells. Cerebellum. 2004;3:89–93. doi: 10.1080/14734220410028921. [DOI] [PubMed] [Google Scholar]
- Yuzaki M. Cbln1 and its family proteins in synapse formation and maintenance. Current Opinion in Neurobiology. 2011;21:215–220. doi: 10.1016/j.conb.2011.01.010. [DOI] [PubMed] [Google Scholar]
- Zoladz JA, Pilc A. The effect of physical activity on the brain derived neurotrophic factor: from animal to human studies. Journal of Physiology and Pharmacology. 2010;61:533–541. [PubMed] [Google Scholar]
- Zukin RS, Richter JD, Bagni C. Signals, synapses, and synthesis: how new proteins control plasticity. Frontiers in Neural Circuits. 2009;3:14. doi: 10.3389/neuro.04.014.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]