Abstract
Colon cancer is one of the deadliest cancers worldwide because of its metastasis to other essential organs. Metastasis of colon cancer involves a complex set of events, including epithelial to mesenchymal transition (EMT) that increases invasiveness of the tumor cells. Here we show that the xeroderma pigmentosum group E (XPE) gene product DDB2 is down-regulated in high-grade colon cancers, and it plays a dominant role in the suppression of EMT of the colon cancer cells. Depletion of DDB2 promotes mesenchymal phenotype, whereas expression of DDB2 promotes epithelial phenotype. DDB2 constitutively represses genes that are the key activators of EMT, indicating that DDB2 is a master regulator of EMT of the colon cancer cells. Moreover, we observed evidence that DDB2 functions as a barrier for EMT induced by hypoxia and TGF-β. Also, we provide evidence that DDB2 inhibits metastasis of colon cancer. The results presented here identify a transcriptional regulatory pathway of DDB2 that is directly linked to the mechanisms that suppress metastasis of colon cancer.
Introduction
Epithelial to mesenchymal transition (EMT) is a conserved developmental process that is usurped often by epithelial tumors during metastatic progression. Several oncogenic pathways activated by Src, Ras, Ets, Integrin, Wnt, Notch, hypoxia and TGF-β signaling have been shown to induce EMT (1, 2). Moreover, a number of EMT-inducing transcriptional regulators such as Snail1, Slug, Twist, Zeb1 and Zeb2, which are downstream of the aforementioned signaling pathways, have been characterized (3–9). These transcriptional regulators inhibit expression of E-cadherin, which is considered to be a key event in EMT (3). For example, the TGF-β effectors Smads associate with the Zeb proteins to repress expression of E-cadherin (9–13). TGF-β and Wnt/β-catenin-mediated EMT also involves activation of Snail1, which represses expression of E-cadherin (3, 10, 14, 15). Likewise, the hypoxia activated transcription factor HIF-1 induces EMT by activating expression of Twist, Snail and VEGF-A (16–19). Moreover, extracellular matrix (ECM) degrading matrix metalloproteinases (MMPs) also have been found to alter intracellular signaling pathway to initiate the EMT process (20). Thus, the pathways that lead to an activation of the EMT-inducing transcription factors (Snail1, Zeb1, Zeb2 and Twist) have been characterized, and they have been implicated in metastasis of the epithelial tumors. However, except for some microRNA studies, the regulators that inhibit expression of the EMT-inducing transcription factors in the epithelial tumor cells and block mesenchymal transition remain poorly understood.
DDB2 (Damaged DNA binding protein 2), a protein encoded by the nucleotide excision repair (NER) gene XPE, is a multi-functional protein (21, 22). Several studies indicated a role of DDB2 in remodeling damaged-chromatin in the early steps of NER (21). However, in normal mouse fibroblasts and keratinocytes, the NER function of DDB2 is related to the regulation of the cell cycle inhibitor p21 because deletion of p21 reversed the repair deficiency in the Ddb2−/− cells (23). Proteolysis of p21 is inefficient in the Ddb2 −/− cell, and that causes accumulation of high levels of p21 following DNA damage (23). It is possible that the chromatin remodeling activity of DDB2 is linked to the mechanism by which it induces proteolysis of p21 after DNA damage. DDB2 associates with Cul4-DDB1 E3 ligase and functions as a substrate adapter for that ligase (24, 25). In addition to NER, DDB2 is involved in DNA damage-induced apoptosis. High-level accumulation of p21 in the DDB2 deficient cells was found to be the cause of the deficiency in apoptosis. Deletion of p21 in the Ddb2−/− background reversed the apoptosis deficiencies of the Ddb2−/− cells (26, 27).
Interestingly, the Ddb2−/− cells are deficient also in premature senescence (28). The deficiency in senescence results from a lack of reactive oxygen species (ROS) accumulation (28). Studies on the mechanism indicated a distinct role of DDB2 in transcriptional repression. DDB2 represses expression of the anti-oxidant genes Sod2 (29) and Catalase to allow persistent accumulation of ROS and thereby induces premature senescence (28). In this study, we explain the functional significance of the loss of DDB2 expression in high-grade colon cancers and reveal a novel role of DDB2 as a constitutive repressor of the EMT-inducing transcription factors in colon cancer.
Materials and methods
Immunohistochemistry/Immunofluorescence
Tumor sections were processed for immunohistochemistry, as described before (27). The sections were incubated with the DDB2, PCNA or SMA antibody 1:200 dilution overnight, and then washed three times with 1xPBS and incubated with anti-mouse/rabbit AP (VectorLabs AP-2000) and further developed with Alkaline Phosphatase Substrate (VectorLabs SK-5300) following manufacturer protocol. Nuclei were counterstained with hematoxylin. For immunofluorescence of the mouse tumor sections, blocking was performed using 5% horse serum in PBS for 1 hour at room temperature. Sections were incubated overnight at 4°C using following primary antibody. After washing with PBS, sections were incubated with anti-rabbit/anti-mouse immunoglobulin conjugated with FITC/TRITC.
For immunofluorescence of cells, permeabilized cells were incubated with E-cadherin (1:250), vimentin (1:200) or smooth muscle actin (1:200) antibody overnight. Cells were washed for five times with PBS followed by incubation with FITC/TRITC tagged goat anti-mouse antibody (1:500) for 1h at room temperature. Cell nuclei were labeled with DAPI in PBS for five minutes at room temperature. After a final wash with PBS, cells were mounted on slides and photographed under microscope.
Tissue Microarray
Human tissue microarray of normal and colon carcinoma samples were obtained from US Biomax (CO726, CO802, BC050112, CO811, CO801, CO482, CO701 and CO805). Immunohistochemical assay was performed as described above with antibodies against DDB2 (Abcam). Tissues were counterstained with hematoxylin. Intensity of staining was blind scored from 0 (no staining) to 4 (highest intensity of staining). Graphs represent the average intensity of staining and paired T-test of colon carcinoma versus normal colon scores of intensity of the staining.
siRNA transfection and western blots
The siRNA transfection and western blots were performed following previously described procedures (28).
Cell culture
Human colon carcinoma cell lines were cultured in DMEM (HCT 116) or RPMI 1640 (SW 480 and SW 620) medium supplemented with 10% FBS and Penicillin/Streptomycin. The cell lines were obtained from ATCC, and were used within six months. We did not perform any additional authentification after receiving the cell lines. Stable clones of HCT 116 cells expressing control shRNA or DDB2shRNA were selected using Puromycin. Stable clones of SW620 cells expressing empty vector or vector expressing DDB2 were selected using G418.
RT-PCR assays
Semi-quantitative RT-PCR assays were performed following a previously described procedure (28). The primers have been described in the supplemental information.
Tumorigenicity and metastasis experiment
Tumorigenicity studies were carried out using mouse xenografts (30). For lung metastasis, 1×106 cells were injected into the tail vein of 8 weeks old male nude mice. Four weeks after the injection, mice were euthanized by CO2 followed by cervical dislocation. Lung was removed and were fixed in 10% buffer formalin and paraffin embedded. Serial sections of lung tissue was made and examined by hematoxylin and eosin staining. To study metastasis using an orthotopic model, 1×106 cells were injected into the peritoneal cavity of immunocompromised SCID Beige mice. The cells were allowed to grow into a tumor over the course of two weeks. This allowed a tumor of roughly a centimeter to grow and be dissected. Tumor sections were then taken and implanted on the cecum of a host SCID beige mouse using surgical glue. As the liver is the common first site of metastasis of colorectal cancer, the livers were examined after four weeks of implantation. Liver sections were stained with hematoxylin and eosin. Micro-metastases were then counted and statistically evaluated.
Chromatin Immunoprecipitation Assay
Chromatin immunoprecipitation experiments were carried out following a previously described procedure (28).
Results
DDB2-deficiency increases invasiveness and tumorigenicity of colon cancer cells
Analyses of the publicly available database revealed a down regulation of the Ddb2-mRNA in majority of the colon carcinoma datasets. Out of 30 available datasets, 24 datasets showed significant down regulation (Supplemental Fig. 1A). We confirmed the observation at the protein level by analyzing tissue microarrays from a commercial source (US BIOMAX) that contained samples corresponding to the various grades of colon cancer. Immunohistochemical staining of the samples revealed a strong correlation between the loss of DDB2 expression and the high-grade, metastatic colon cancers (Figs.1A, B and Supplemental Fig. 2). DDB2 is a nuclear protein (25). Therefore, we compared the nuclear signal-intensities of DDB2 in our analyses of the tumor microarray. Matched normal colon and colon carcinoma tissues confirmed the observations (Supplemental Fig. 1B). The high-grade specimens also exhibited a loss of E-cadherin expression (Figs. 1A and C).
Figure 1. Loss of DDB2 expression in high grade colon cancer.
(A) DDB2 immunohistochemistry of human tissue microarray of normal colon (n=41) and colon carcinoma grade I (n=59), II (n=96), III (n=51) and metastatic colon carcinoma (n=63) (Left). Representative images of normal human colon section and colon carcinoma grade I, II, III and metastatic colon carcinoma are shown. E-cadherin immunohistochemistry of human tissue microarray of normal colon (n=13) and colon carcinoma grade I (n=33), II (n=58), III (n=31) and metastatic colon carcinoma (n=61) (Right). Scale bar for all the images = 10 um. (B) Intensity of DDB2 staining was scored from 0 to 4. Graph representing the average intensity of DDB2 staining for normal colon versus colon carcinoma grade I, II and III. p value is calculated by one way ANOVA: ***P<0.001. With a significance level of 0.05, F value is calculated: F =18.22 that is significantly greater than the critical F ratio. (C) Intensity of E-cadherin staining was scored from 0 to 4. Graph representing the average intensity of E-cadherin staining for normal colon versus colon carcinoma grade I, II and III. p value is calculated by one way ANOVA: ***P<0.001. With a significance level of 0.05, F value is calculated: F=14.62 that is significantly greater than the critical F ratio.
To further investigate the significance of DDB2 down regulation in colon cancer, we compared the colon cancer cell line HCT116 expressing control-shRNA with that expressing DDB2-shRNA (Fig. 2A). First, the DDB2-shRNA expressing HCT116 cells were found to be significantly more invasive compared to the control cell line, as judged by matrigel migration assay (Fig. 2B). The invasiveness was confirmed also by scratch-healing assay (Supplemental Fig. 3A). Moreover, in soft agar colony formation assay, the DDB2-shRNA expressing cells produced significantly more colonies than the control HCT116 cells (Fig. 2C). The DDB2-deficient cells were resistant to anoikis (Supplemental Fig. 3B). Consistent with that, the DDB2-deficient cells showed higher expression of p-ERK and p-AKT compared to the normal cells (Supplemental Fig. 3B). To confirm the increased tumorigenicity in the absence of DDB2, we compared tumor growth using mouse xenograft experiments. Two million HCT116 cells expressing control shRNA or DDB2-shRNA were injected into nude mice subcutaneously and tumor growth was measured. Clearly, the DDB2-deficient cells generated a larger tumor mass in a much shorter time (Fig. 2D and E). That was evident also in a tumor growth curve analysis (Fig. 2F). The experiment in Fig. 2E was performed with a different pair of HCT116-clones expressing shRNA against DDB2 or control. Also, polyclonal cells stably expressing shRNA for DDB2 formed greater number of soft agar colonies and aggressive xenograft tumors (Supplemental Fig. 4 A and B).
Figure 2. DDB2 deficiency increases invasiveness and tumorigenecity of colon cancer cells.
(A) Protein extracts from HCT 116 expressing control shRNA or DDB2 shRNA were subjected to Western Blot analysis with DDB2 (50ug protein) antibody. (B) Transwell invasion assay of HCT116 cells expressing control shRNA or DDB2 shRNA (mean +/− standard deviation; n=3). (C) HCT116 cells expressing control shRNA or DDB2 shRNA were grown on soft agar for 10 days. Top panel shows representative field of soft agar colonies. Bottom panel shows the bar graph representing quantification of soft agar colonies (mean +/− standard deviation; n=3). (D) HCT116 cells expressing control shRNA or DDB2 shRNA were injected subcutaneously into nude mice. Representative picture of xenograft tumors induced in nude mice by HCT 116 cells expressing control shRNA or DDB2 shRNA 7 weeks post-inoculation (n=5). (E) Bar graph representing tumor mass in nude mice from HCT 116 cells expressing control shRNA or DDB2 shRNA at the point of sacrifice of the mice (mean +/− standard error of mean; n=10). (F) Tumor volume induced in nude mice by HCT 116 cells expressing control shRNA or DDB2 shRNA (n=5). The asterisk in Panel B, C and E indicates statistically significant differences, with the following P value calculated by Student’s t-test: *P<0.05; **P<0.01; ***P<0.001; ns - not significant.
DDB2 inhibits epithelial to mesenchymal transition of the colon cancer cells
Closer analyses of the DDB2-knockdown cells indicated a change in morphology of the cells. Unlike the epithelial cuboid appearance of the parental cells, the DDB2-deficient HCT116 cells exhibited elongated mesenchymal-like morphology, a change that is observed during epithelial to mesenchymal transition (EMT) (Fig. 3A). Also, there was a clear loss of surface expression of E-cadherin and increase in expression of the mesenchymal markers vimentin in the DDB2-deficient cells (Fig. 3A and B). In western blot assays (Fig. 3B), the E-cadherin level was decreased upon depletion of DDB2. Polyclonal cell line stably expressing shRNA for DDB2 also exhibited similar phenotype (Supplemental Fig. 5 A and B). We further investigated whether DDB2 inhibits EMT of colon cancer cells, in vivo. Analyses of the xenograft tumor sections revealed aggressive nature of the tumor from DDB2 knockdown cells, as evidenced by increased expression of smooth muscle actin, vimentin and PCNA, as well as reduced apoptosis (Fig. 3C, Supplemental Fig. 6 and 7). The increased proliferation, as judged by PCNA staining, most likely reflects increased angiogenesis that is induced by the mesenchymal-like cells (31). The tumor samples also exhibited a loss of E-cadherin expression (Fig. 3C).
Figure 3. DDB2 inhibits epithelial to mesenchymal transition of colon cancer cells.
(A) Representative phase contrast images of HCT 116 cells expressing control shRNA or DDB2 shRNA. HCT 116 cells expressing control shRNA or DDB2 shRNA were subjected to immunocytochemical analysis using E-cadherin, Vimentin or Smooth Muscle Actin antibody. Scale bar for all the images= 5 um. (B) Protein extracts from HCT 116 cells expressing control shRNA or DDB2 shRNA were subjected to Western blot analysis with E-cadherin (20ug protein), Vimentin (50ug protein) and DDB2 (50ug protein) antibody. Tubulin was used as a loading control. (C) HCT116 cells expressing control shRNA or DDB2 shRNA were injected subcutaneously into nude mice. Four weeks post inoculation mice were sacrificed and tumor sections were fixed in 10% Formalin, processed and embedded with paraffin for sectioning. Prepared tumor section slides were then subjected to immunohistochemical analysis using H & E staining, Smooth muscle actin, PCNA, E-cadherin and Vimentin antibody. Representative images are shown. Scale bar for all the images= 10 um.
Next, we compared the human colon cancer lines SW480 and SW620, which were derived from the same patient. SW480 corresponds to an early stage, whereas SW620 corresponds to a later metastatic stage (32). The SW620 cells are mainly mesenchymal, whereas SW480 cells are epithelial (33). Knockdown of DDB2 in the SW480 cells resulted in EMT-like morphological changes of these cells (Fig. 4A). Moreover, expression of DDB2 in the SW620 cells increased population of cells with epithelial morphology (Fig. 4B). Interestingly, there was about a 3.5-fold decrease in the DDB2 expression during progression from SW480 to SW620 (Fig. 4C). Knockdown of DDB2 in the SW480 cells caused a significant reduction (about 5-fold) in the level of E-cadherin (Fig. 4D), whereas expression of DDB2 in the SW620 cells caused a significant increase in the epithelial phenotype, as measured by increased E-cadherin and reduced vimentin expression (Fig. 4E and F). Together, these observations suggest that DDB2 is a regulator of EMT in colon cancer cells.
Figure 4. Loss of DDB2 expression in mesenchymal colon cancer cells.
(A) Representative phase contrast images of SW480 cells expressing control siRNA or DDB2 siRNA. Scale bar for all the images= 5 um. (B) Representative phase contrast images of SW620 cells expressing empty vector or vector expressing DDB2. Scale bar for all the images= 5 um. (C) Protein extracts from SW480 and SW620 cells were subjected to western blot analysis using DDB2 (50ug protein), E-cadherin (20ug protein), Vimentin (50ug protein) antibody. Tubulin was used as a loading control. (D) Protein extracts from SW480 cells expressing control siRNA or DDB2 siRNA were subjected to Western blot analysis using E-cadherin (20ug protein), DDB2 (50ug protein) or Vimentin (50ug protein) antibody. Tubulin was used as a loading control. (E) Protein extracts from SW620 cells expressing empty vector or vector expressing DDB2 were subjected to Western blot analysis using E-cadherin (20ug protein), DDB2 (50ug protein) or Vimentin (50ug protein) antibody. Tubulin was used as a loading control. (F) SW620 cells expressing empty vector or vector expressing DDB2 were subjected to immunocytochemical analysis using E-cadherin, Vimentin or Smooth Muscle Actin antibody. Scale bar for all the images= 10 um.
To determine whether DDB2 inhibits EMT induced by TGF-β and hypoxia, we over-expressed DDB2 in SW480 cells. The cells were then subjected to treatments with TGF-β or hypoxia. Extracts of the treated cells were analyzed for expression of E-cadherin. Treatments with TGF-β or hypoxia caused an inhibition in the levels of E-cadherin in the SW480 cells, whereas the cells over-expressing DDB2 exhibited an attenuated inhibition of E-cadherin expression by TGF-β or hypoxia (Supplemental Fig. 8A and B). Unlike the TGF-β treated cells, DDB2 expression did not inhibit the level of vimentin in the hypoxia treated cell (Supplemental Fig. 8A), which is likely due to the shorter timeframe (24h instead of 48h) of the hypoxia experiment. The DDB2-expressing cells undergo apoptosis at the longer time-points in hypoxia.
DDB2 transcriptionally represses expression of the genes that induce EMT
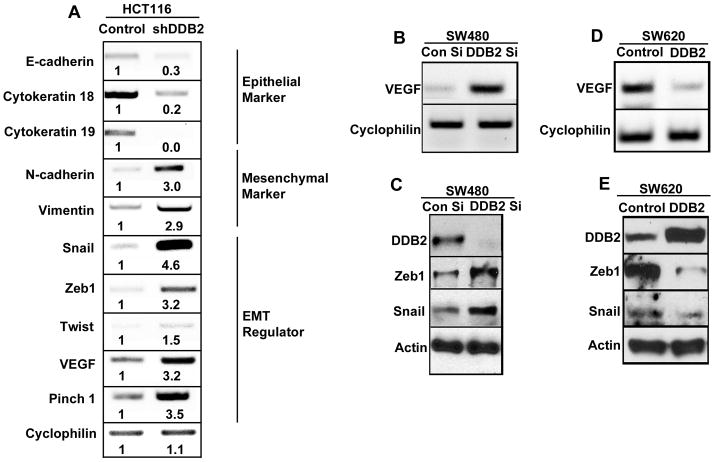
RNA expression analyses further confirmed that the DDB2-deficient cells express lower levels of the epithelial markers E-cadherin, Cytokeratin 18 and Cytokeratin 19 (Fig. 5A), and increased expression of the mesenchymal markers vimentin and N-cadherin (Fig. 5A). Several transcription factors, which have been implicated in the process of EMT such as Snail and Zeb 1 were up regulated at the mRNA-level in the absence of DDB2 (Fig. 5A). VEGF is known to be an important player in the EMT process (18, 34). We observed significant increase in the level of VEGF in the DDB2-knockdown cells (Fig. 5A). Moreover, there was increased expression of Pinch 1, a protein that has been implicated in EMT (35). Also, siRNA mediated knockdown of DDB2 in the SW480 cells increased expression of VEGF, Zeb1 and Snail (Fig. 5B and C). Furthermore, expression of DDB2 in the SW620 cells inhibited expression of VEGF, Snail and Zeb1 (Fig. 5D and E).
Figure 5. DDB2 acts as a transcriptional repressor of VEGF, Zeb1 and Snail.
(A) Total RNA from HCT 116 cells expressing control shRNA or DDB2 shRNA was analyzed for the expression of E-cadherin, Cytokeratin 18, Cytokeratin 19, Vimentin, N-cadherin, Snail, Zeb1, Twist1, VEGF, Pinch 1. Cyclophilin was used as a loading control. Quantification was performed using Image J software. (B) Total RNA from SW480 cells expressing control siRNA or DDB2 siRNA was analyzed for VEGF expression. Cyclophilin was used as a loading control. (C) Protein extracts were made from SW480 cells expressing control siRNA or DDB2 siRNA and Western blot analysis was performed with Zeb1 (50ug protein), Snail (100ug protein) and DDB2 (50ug protein) antibody. (D) Total RNA from SW620 cells expressing empty vector or DDB2 expressing vector was analyzed for VEGF expression. Cyclophilin was used as a loading control. (E) Protein extracts were made from SW620 cells expressing empty vector or DDB2 expressing vector and Western blot analysis was performed with Zeb1 (20ug protein), Snail (100ug protein) and DDB2 (50ug protein) antibody. Actin was used as a loading control.
Our previous studies on the regulation of the antioxidant genes by DDB2 identified a transcriptional repression function of DDB2. To examine whether DDB2 regulates expression of these genes directly, we performed Chromatin IP experiments to look at DDB2 occupancy at 3000 bp upstream from the transcription start sites of these genes. The PCR-amplicons on the three promoters are indicated schematically in supplemental Figure. 9B. DDB2 interacted with the VEGF promoter in chromatin-IP (ChIP) assays at two distinct sites (Fig. 6A). Moreover, DDB2 interacted with the Zeb1 and the Snail promoters at specific sites (Fig. 6B and C). The binding of DDB2 to those sites was confirmed further using a monoclonal antibody against the epitope tag T7 in chromatin-IP experiments with cells expressing T7-tagged DDB2 (Supplemental Fig. 9). The DDB2-associated protein Cul4a binds to Suv39h, a histone H3K9 methylase that participates in repression (28). Therefore, we examined whether DDB2 functions as a transcriptional repressor by recruiting Suv39h onto the promoters of VEGF, Zeb1 and Snail. Chromatin IP was performed with antibodies against DDB1, Cul4a, Suv39h and H3K9Me3 using the HCT116 cells or the HCT116 cells expressing the DDB2-shRNA. There was no significant difference in the levels of these proteins between the two cell-types (Supplemental Fig. 11). The ChIP-DNAs were probed for VEGF (probe set V), Zeb1 (probe set VIII) and Snail (probe set IX), and the bindings in the HCT116 control cells were compared with those in the HCT116-DDB2shRNA cells. Clearly, there was reduced occupancy of DDB1, Cul4a and Suv39h on the promoter of all three genes in the cells expressing shRNA for DDB2 (Fig. 6D). Consistent with that, those cells also exhibited a reduced H3K9Me3 interaction with the promoters (Fig. 6D). Although we employed a semi-quantitative method to assess the binding, the results presented are averages from three different batches of chromatin-preparations. There was also an increase in the histones H3 and H4 acetylation in the HCT116-DDB2-shRNA cells (Fig. 6E). Previously, it was shown that the NER factors assemble with RNA polymerase II on the promoter involving the XPC protein (36). However, we did not detect association of XPC with the DDB2-interaction sites in the promoters of VEGF, Snail and Zeb1 (Fig. 6F). Together, the observations suggest that DDB2 recruits Suv39h through its interaction with Cul4a to bring about histone H3K9 tri-methylation in the promoters of VEGF, Zeb1 and Snail, leading to repression.
Figure 6. DDB2 epigenetically regulates expression of VEGF, Zeb1 and Snail.
(A), (B) and (C) HCT 116 cells were subjected to ChIP assays on VEGF, Zeb1 or Snail promoter. 2 μg Antibody against DDB2 or IgG was used for immunoprecipitation. (D) HCT 116 cells expressing control shRNA or DDB2 shRNA were subjected to ChIP assays using 2 μg antibody against Cul4a, DDB1, Suv39h, H3K9Me3 or IgG. The VEGF, Zeb1 and Snail promoter fragments in the immunoprecipitated chromatin were quantified with the primer pairs V (for VEGF), VIII (for Zeb1) and IX (for Snail). Bar graph represents immunoprecipitation as percentages to the input material (mean +/− standard deviation; n=3) normalized to IgG control. (E) HCT 116 cells expressing control shRNA or DDB2 shRNA were subjected to ChIP assays using 2 μg antibody against H3Ac, H4Ac, H3K14Ac or IgG. The VEGF and Zeb1 promoter fragments in the immunoprecipitated chromatin were quantified with the primer pairs V (for VEGF) and VIII (for Zeb1). Bar graph represents immunoprecipitation as percentages of the input material (mean +/− standard deviation; n=3) normalized to IgG control. (F) HCT 116 cells were subjected to ChIP assays with 2 μg antibody against XPC or IgG. The VEGF, Zeb1 and Snail promoter fragments in the immunoprecipitated chromatin were quantified with the primer pairs V (for VEGF), VIII (for Zeb1) and IX (for Snail). The asterisk in panel D, E and F indicates statistically significant differences, with the following P value calculated by Student’s t-test: *P<0.05; **P<0.01; ***P<0.001; ns - not significant.
DDB2 regulates metastasis of colon cancer cells
Next, we performed functional metastasis assays to examine whether the DDB2-knockdown cells are more metastatic. First, we carried out metastasis experiment using an orthotopic mouse model. The control HCT116 or the DDB2-knockdown HCT116 cells were subcutaneously injected in SCID beige mice. Once the tumors reached 0.5 mm in diameter, the tumors were excised and pieces of tumors were surgically implanted onto the cecum of host mice. Four weeks following implantation, the mice were analyzed for metastatic growth in the liver. We observed extensive metastasis with the tumor generated from the DDB2-knockdown cells in the liver (Fig. 7A). The tumor implant generated with the control HCT116 cells did not undergo any visible metastasis in any organ under these experimental conditions.
Figure 7. DDB2 regulates metastasis of colon cancer cells.
(A) HCT116 cells expressing control shRNA or DDB2 shRNA were injected subcutaneously into SCID beige mice. Once xenografts were established, they were excised and orthotopically implanted into other SCID beige mice using microsurgical techniques (n=3). Animals were sacrificed 4 weeks post-implantation and examined for liver metastasis. Representative H & E staining of liver sections from mice injected with HCT 116 cells expressing control shRNA or DDB2 shRNA are shown (left panel). Quantification of percentage of metastasis occurrence in mice injected with HCT116 cells expressing control shRNA or DDB2 shRNA (right panel). (B) HCT116 cells expressing control shRNA or DDB2 shRNA were injected into 8 weeks old male nude mice intravenously via the tail vein (n=3). Mice were sacrificed after four weeks and lung tissues were harvested. Representative H & E staining of lung from mice injected with HCT 116 cells expressing control shRNA or DDB2 shRNA (left panel). Quantification of percentage of micro-metastasis occurrence in mice injected with HCT116 cells expressing control shRNA or DDB2 shRNA (right panel). (C) SW620 cells expressing empty vector or DDB2 expressing vector were injected into 8 weeks old male nude mice intravenously via tail vein (n=3). Mice were sacrificed after six weeks and lung tissues were harvested. Representative H & E staining of lung from mice injected with SW620 cells expressing empty vector or DDB2 expressing vector (left panel). Quantification of percentage of metastasis occurrence in mice injected with SW620 cells expressing empty vector or DDB2 expressing vector (right panel). Scale bar for all the images= 10 um. (D) Schematic diagram indicating the mechanism by which DDB2 inhibits EMT. EMT inducing signals (hypoxia or TGF-b) increases expression of VEGF, Snail1 and Zeb1 to reduce expression of E-cadherin and bring about EMT-like changes in colon cancer cells. The XPE gene product DDB2, on the other hand, binds to the promoters of VEGF, Snail1 and Zeb1 to inhibit their expression, and thus, supports MET (mesenchymal to epithelial transition).
To further confirm the observation, we performed tail vein injection of the DDB2-depleted cells in athymic nude mice. The parental or the DDB2-knockdown cells were injected into the tail vein of mice. To detect acceleration of metastasis by the loss of DDB2, we looked at the lung tumor metastasis after 4 weeks. The mice were sacrificed and examined for lung nodules. The mice injected with the DDB2 knockdown cell line clearly exhibited evidence of lung tumor micro-metastasis within that time, whereas the control HCT116 injected mice exhibited fewer micro-metastatic nodules (Fig. 7B). Polyclonal cells stably expressing shRNA for DDB2 also exhibited increased metastatic frequency (Supplemental Fig. 12). Because expression of DDB2 inhibited expression of the EMT genes and blocked EMT, we sought to determine whether expression of DDB2 inhibits metastasis. To investigate that, we compared the metastatic line SW620 with SW620 stably expressing DDB2. Expression of DDB2 had only marginal effects on the proliferation of the SW620 cells, in vitro (Supplemental Fig. 13). Cells were injected in nude mice via tail vein. Six-weeks following injection, the mice were sacrificed and the metastatic colonies in the lung were compared. There was a significant reduction in the metastatic colonies in the lung tissue from mice injected with DDB2-expressing SW620 cells (Fig. 7C). These observations provide evidence that DDB2 is a potent inhibitor of colon cancer metastasis.
Discussion
The work presented here is significant in several ways. First, the observations demonstrate a role of DDB2, an NER protein, in the regulation of colon cancer metastasis. We show that it is a potent regulator of EMT, as it transcriptionally inhibits expression of the key genes required for EMT and tumor invasion. Moreover, we show that DDB2 stands as a barrier downstream of the signaling pathways that induce EMT. Together, the results identify a new tumor suppression function of DDB2 that inhibits metastasis of colon cancers.
Mutations in the nucleotide repair genes are rare in CRC. However, in a study with small groups of CRC patients, it was shown that 25% (2/8) exhibited LOH at the XPE loci, 11q12-13 (37). Our analyses of the publicly available database indicated a reduced expression of DDB2 in a much greater population of the CRC patients. Moreover, the reduction of DDB2 expression coincides with the appearance of high-grade colon cancers. Therefore, progression of colon cancer associates with the activation of mechanisms that reduce DDB2 expression. It is noteworthy that DDB2 is a p53-induced gene (38), and p53 mutations are common in colon cancer, and that might explain the loss of expression. The loss of DDB2 expression is expected also to reduce the repair (NER) activity, which may contribute to the evolution of the high-grade colon cancer. In this study, however, we show that the transcriptional repressor function of DDB2 regulates EMT. The observation that DDB2 inhibits EMT, and over-expression of DDB2 could reverse the mesenchymal phenotype to epithelial phenotype could not be explained by its NER activity. Moreover, expression of DDB2 inhibited the metastatic activity of the SW620 cells, which could not be explained by its NER function. Transcriptional repression of VEGF, Snail and Zeb1 expression, on the other hand, will readily explain the inhibitions of EMT and metastasis.
DDB2 was shown to bind the Sod2 gene promoter through a specific sequence-element and constitutively repress that gene (29). However, we observed that DDB2 could repress expression of the Catalase gene in the absence of that sequence-element (28). Therefore, we suspect that, in addition to sequence-specific binding, DDB2 functions also as a co-repressor through interactions with other DNA binding factors. Previously, we showed that the association of DDB2 induced an increase in the histone H3K9 tri-methylation of the Sod2 and the Catalase promoters, leading to transcriptional repression (28). In this study, we observed a similar phenomenon. For example, DDB2 enhanced the recruitment of Cul4, DDB1 and Suv39h onto the promoters of VEGF, Snail, and Zeb1. Recruitment of Suv39h, a histone H3K9-methylase, correlated with an increase in H3K9-trimethylation of the target genes. Expression of DDB2-shRNA inhibited the interaction of Suv39h, coinciding with a reduction in the levels of H3K9-trimethylation (Fig. 6). The ChIP regions in the VEGF, Snail and Zeb1 promoters contain anywhere between 30 and 50 DNA-elements corresponding to sequence-specific DNA-binding proteins, and a large number of them are present in all three promoters (See supplemental Figure 10A). Therefore, it remains possible that DDB2 is recruited to these promoters through interactions with one or more DNA-binding proteins that commonly regulate these promoters. Nonetheless, our observations indicate a clear role of DDB2 in the constitutive repression of the EMT genes, involving H3K9 tri-methylation of the promoters of VEGF, Snail1 and Zeb1.
The decrease in expression of DDB2 was observed also in the colon cancer cell line SW620 compared to SW480. These two lines are derived from one patient at an early point in the tumor progression, SW480, and at a later metastatic stage of progression, SW620 (32). Both lines harbor the same mutation in the DNA-binding domain of p53 (39). It is noteworthy that DDB2 expression is not completely dependent upon p53 because other mechanisms could increase its expression in p53−/− cells (28, 40). Therefore, it is not surprising that the SW480 cells express DDB2 at a higher level compared to the SW620 cells. Interestingly, re-expression of DDB2 in the SW620 cells induced expression of the epithelial markers associated with inhibition of the mesenchymal morphology. These observations also indicate that DDB2 is one of the major regulators in epithelial tumor cells that resist conversion to a mesenchymal-like morphology, and that induction of EMT occurs mainly after a loss of DDB2 expression. Consistent with that, we show that a reduced expression of DDB2 enhances metastasis of colon cancer cells in both experimental metastasis assays and in an orthotopic xenograft model. Moreover, re-expression of DDB2 inhibits metastasis. Taken together, the results presented here explain why loss of DDB2 expression coincides with the high-grade progression of colon cancers. Moreover, the results, for the first time, demonstrate an important tumor suppression function of the xeroderma pigmentosum gene DDB2 in colon cancer metastasis.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Cancer Institute to PR and SB (CA77637 and CA 156164).
References
- 1.Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–29. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 2.Huber MA, Kraut N, Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr Opin Cell Biol. 2005;17:548–58. doi: 10.1016/j.ceb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 3.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–28. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 4.Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, et al. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 5.Bolos V, Peinado H, Perez-Moreno MA, Fraga MF, Esteller M, Cano A. The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressors. J Cell Sci. 2003;116:499–511. doi: 10.1242/jcs.00224. [DOI] [PubMed] [Google Scholar]
- 6.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–39. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 7.Eger A, Aigner K, Sonderegger S, Dampier B, Oehler S, Schreiber M, et al. DeltaEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene. 2005;24:2375–85. doi: 10.1038/sj.onc.1208429. [DOI] [PubMed] [Google Scholar]
- 8.Vandewalle C, Comijn J, De Craene B, Vermassen P, Bruyneel E, Andersen H, et al. SIP1/ZEB2 induces EMT by repressing genes of different epithelial cell-cell junctions. Nucleic Acids Res. 2005;33:6566–78. doi: 10.1093/nar/gki965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Comijn J, Berx G, Vermassen P, Verschueren K, van Grunsven L, Bruyneel E, et al. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol Cell. 2001;7:1267–78. doi: 10.1016/s1097-2765(01)00260-x. [DOI] [PubMed] [Google Scholar]
- 10.Xu J, Lamouille S, Derynck R. TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 2009;19:156–72. doi: 10.1038/cr.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Postigo AA, Depp JL, Taylor JJ, Kroll KL. Regulation of Smad signaling through a differential recruitment of coactivators and corepressors by ZEB proteins. EMBO J. 2003;22:2453–62. doi: 10.1093/emboj/cdg226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Postigo AA. Opposing functions of ZEB proteins in the regulation of the TGFbeta/BMP signaling pathway. EMBO J. 2003;22:2443–52. doi: 10.1093/emboj/cdg225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y, El-Naggar S, Darling DS, Higashi Y, Dean DC. Zeb1 links epithelial-mesenchymal transition and cellular senescence. Development. 2008;135:579–88. doi: 10.1242/dev.007047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peinado H, Quintanilla M, Cano A. Transforming growth factor beta-1 induces snail transcription factor in epithelial cell lines: mechanisms for epithelial mesenchymal transitions. J Biol Chem. 2003;278:21113–23. doi: 10.1074/jbc.M211304200. [DOI] [PubMed] [Google Scholar]
- 15.Yook JI, Li XY, Ota I, Hu C, Kim HS, Kim NH, et al. A Wnt-Axin2-GSK3beta cascade regulates Snail1 activity in breast cancer cells. Nat Cell Biol. 2006;8:1398–406. doi: 10.1038/ncb1508. [DOI] [PubMed] [Google Scholar]
- 16.Forsythe JA, Jiang BH, Iyer NV, Agani F, Leung SW, Koos RD, et al. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol Cell Biol. 1996;16:4604–13. doi: 10.1128/mcb.16.9.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang MH, Wu MZ, Chiou SH, Chen PM, Chang SY, Liu CJ, et al. Direct regulation of TWIST by HIF-1alpha promotes metastasis. Nat Cell Biol. 2008;10:295–305. doi: 10.1038/ncb1691. [DOI] [PubMed] [Google Scholar]
- 18.Mak P, Leav I, Pursell B, Bae D, Yang X, Taglienti CA, et al. ERbeta impedes prostate cancer EMT by destabilizing HIF-1alpha and inhibiting VEGF-mediated snail nuclear localization: implications for Gleason grading. Cancer Cell. 2010;17:319–32. doi: 10.1016/j.ccr.2010.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imai T, Horiuchi A, Wang C, Oka K, Ohira S, Nikaido T, et al. Hypoxia attenuates the expression of E-cadherin via up-regulation of SNAIL in ovarian carcinoma cells. Am J Pathol. 2003;163:1437–47. doi: 10.1016/S0002-9440(10)63501-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Radisky DC, Levy DD, Littlepage LE, Liu H, Nelson CM, Fata JE, et al. Rac1b and reactive oxygen species mediate MMP-3-induced EMT and genomic instability. Nature. 2005;436:123–7. doi: 10.1038/nature03688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stoyanova T, Roy N, Kopanja D, Raychaudhuri P, Bagchi S. DDB2 (damaged-DNA binding protein 2) in nucleotide excision repair and DNA damage response. Cell Cycle. 2009;8:4067–71. doi: 10.4161/cc.8.24.10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wittschieben BO, Iwai S, Wood RD. DDB1-DDB2 (xeroderma pigmentosum group E) protein complex recognizes a cyclobutane pyrimidine dimer, mismatches, apurinic/apyrimidinic sites, and compound lesions in DNA. J Biol Chem. 2005;280:39982–9. doi: 10.1074/jbc.M507854200. [DOI] [PubMed] [Google Scholar]
- 23.Stoyanova T, Yoon T, Kopanja D, Mokyr MB, Raychaudhuri P. The xeroderma pigmentosum group E gene product DDB2 activates nucleotide excision repair by regulating the level of p21Waf1/Cip1. Mol Cell Biol. 2008;28:177–87. doi: 10.1128/MCB.00880-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nag A, Bondar T, Shiv S, Raychaudhuri P. The xeroderma pigmentosum group E gene product DDB2 is a specific target of cullin 4A in mammalian cells. Mol Cell Biol. 2001;21:6738–47. doi: 10.1128/MCB.21.20.6738-6747.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shiyanov P, Nag A, Raychaudhuri P. Cullin 4A associates with the UV-damaged DNA-binding protein DDB. J Biol Chem. 1999;274:35309–12. doi: 10.1074/jbc.274.50.35309. [DOI] [PubMed] [Google Scholar]
- 26.Stoyanova T, Roy N, Kopanja D, Bagchi S, Raychaudhuri P. DDB2 decides cell fate following DNA damage. Proc Natl Acad Sci U S A. 2009;106:10690–5. doi: 10.1073/pnas.0812254106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stoyanova T, Roy N, Bhattacharjee S, Kopanja D, Valli T, Bagchi S, et al. p21 cooperates with DDB2 protein in suppression of ultraviolet ray-induced skin malignancies. J Biol Chem. 2012;287:3019–28. doi: 10.1074/jbc.M111.295816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roy N, Stoyanova T, Dominguez-Brauer C, Park HJ, Bagchi S, Raychaudhuri P. DDB2, an essential mediator of premature senescence. Mol Cell Biol. 2010;30:2681–92. doi: 10.1128/MCB.01480-09. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Minig V, Kattan Z, van Beeumen J, Brunner E, Becuwe P. Identification of DDB2 protein as a transcriptional regulator of constitutive SOD2 gene expression in human breast cancer cells. J Biol Chem. 2009;284:14165–76. doi: 10.1074/jbc.M808208200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roy N, Elangovan I, Kopanja D, Bagchi S, Raychaudhuri P. Tumor regression by phenethyl isothiocyanate involves DDB2. Cancer Biol Ther. 2012;14 doi: 10.4161/cbt.22631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collino F, Revelli A, Massobrio M, Katsaros D, Schmitt-Ney M, Camussi G, et al. Epithelial-mesenchymal transition of ovarian tumor cells induces an angiogenic monocyte cell population. Exp Cell Res. 2009;315:2982–94. doi: 10.1016/j.yexcr.2009.06.010. [DOI] [PubMed] [Google Scholar]
- 32.Leibovitz A, Stinson JC, McCombs WB, 3rd, McCoy CE, Mazur KC, Mabry ND. Classification of human colorectal adenocarcinoma cell lines. Cancer Res. 1976;36:4562–9. [PubMed] [Google Scholar]
- 33.Kubens BS, Zanker KS. Differences in the migration capacity of primary human colon carcinoma cells (SW480) and their lymph node metastatic derivatives (SW620) Cancer Lett. 1998;131:55–64. doi: 10.1016/s0304-3835(98)00201-8. [DOI] [PubMed] [Google Scholar]
- 34.Gonzalez-Moreno O, Lecanda J, Green JE, Segura V, Catena R, Serrano D, et al. VEGF elicits epithelial-mesenchymal transition (EMT) in prostate intraepithelial neoplasia (PIN)-like cells via an autocrine loop. Exp Cell Res. 2010;316:554–67. doi: 10.1016/j.yexcr.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 35.Li Y, Dai C, Wu C, Liu Y. PINCH-1 promotes tubular epithelial-to-mesenchymal transition by interacting with integrin-linked kinase. J Am Soc Nephrol. 2007;18:2534–43. doi: 10.1681/ASN.2007030315. [DOI] [PubMed] [Google Scholar]
- 36.Le May N, Mota-Fernandes D, Velez-Cruz R, Iltis I, Biard D, Egly JM. NER factors are recruited to active promoters and facilitate chromatin modification for transcription in the absence of exogenous genotoxic attack. Mol Cell. 2010;38:54–66. doi: 10.1016/j.molcel.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 37.Takebayashi Y, Nakayama K, Kanzaki A, Miyashita H, Ogura O, Mori S, et al. Loss of heterozygosity of nucleotide excision repair factors in sporadic ovarian, colon and lung carcinomas: implication for their roles of carcinogenesis in human solid tumors. Cancer Lett. 2001;174:115–25. doi: 10.1016/s0304-3835(01)00690-5. [DOI] [PubMed] [Google Scholar]
- 38.Tan T, Chu G. p53 Binds and activates the xeroderma pigmentosum DDB2 gene in humans but not mice. Mol Cell Biol. 2002;22:3247–54. doi: 10.1128/MCB.22.10.3247-3254.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodrigues NR, Rowan A, Smith ME, Kerr IB, Bodmer WF, Gannon JV, et al. p53 mutations in colorectal cancer. Proc Natl Acad Sci U S A. 1990;87:7555–9. doi: 10.1073/pnas.87.19.7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nichols AF, Itoh T, Zolezzi F, Hutsell S, Linn S. Basal transcriptional regulation of human damage-specific DNA-binding protein genes DDB1 and DDB2 by Sp1, E2F, N-myc and NF1 elements. Nucleic Acids Res. 2003;31:562–9. doi: 10.1093/nar/gkg152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.