Abstract
Metastatic prostate cancers generally rely on androgen receptor (AR) signaling for growth and survival, even following systemic androgen deprivation therapy (ADT). However, recent evidence suggests that some advanced prostate cancers escape ADT by utilizing signaling programs and growth factors that bypass canonical AR ligand-mediated mechanisms. We utilized an in vitro high-throughput RNAi screen to identify pathways in androgen-dependent prostate cancer cell lines whose loss of function promotes androgen ligand-independent growth. We identified 40 genes where knockdown promoted proliferation of both LNCaP and VCaP prostate cancer cells in the absence of androgen. Of these, 14 were down-regulated in primary and metastatic prostate cancer, including two subunits of the protein phosphatase 2 (PP2A) holoenzyme complex: PPP2R1A, a structural subunit with known tumor-suppressor properties in several tumor types; and PPP2R2C, a PP2A substrate-binding regulatory subunit that has not been previously identified as a tumor suppressor. We demonstrate that loss of PPP2R2C promotes androgen ligand depletion-resistant prostate cancer growth without altering AR expression or canonical AR-regulated gene expression. Furthermore, cell proliferation induced by PPP2R2C loss was not inhibited by the AR antagonist MDV3100, indicating that PPP2R2C loss may promote growth independently of known AR-mediated transcriptional programs. Immunohistochemical analysis of PPP2R2C protein levels in primary prostate tumors determined that low PPP2R2C expression significantly associated with an increased likelihood of cancer recurrence and cancer-specific mortality. These findings provide insights into mechanisms by which prostate cancers resist AR-pathway suppression, and support inhibiting PPP2R2C complexes or the growth pathway(s) activated by PPP2R2C as a therapeutic strategy.
Keywords: PP2A, PPP2R2C, castration-resistant prostate cancer, androgen-pathway independence
INTRODUCTION
Prostate cancer is the second leading cause of cancer-related mortality in American men (1). Most deaths are attributable to metastatic disease, a stage that exhibits high initial response rates to systemic androgen deprivation therapy (ADT) (2). Unfortunately, nearly all metastatic tumors recur as castration-resistant prostate cancers (CRPC) (2). Currently available treatments for CRPC provide limited improvements in survival, though recent developments of novel immunotherapies and more potent antiandrogens have resulted in additional modest increases in patient longevity (3-5).
Importantly, the vast majority of castration-resistant prostate cancers reactivate androgen receptor (AR)-mediated transcriptional networks that appear to be necessary for prostate cancer survival and growth. The maintenance of AR-pathway activity has been shown to occur through a variety of mechanisms, including but not limited to transcriptional and genomic amplification of AR, mutations of AR that allow promiscuous ligand binding, generation of constitutively-active AR splice variants, de novo synthesis of intratumoral androgens, and transactivation of the AR by intracellular signal transduction programs (6, 7). However, there is mounting evidence that alternative mechanisms can promote CRPC progression independent of AR activation. Several recent in vitro and in vivo studies provide evidence that PI3K pathway signaling is sufficient for CRPC survival in the setting of low or absent AR activity (8, 9). With clinical efforts focused on extinguishing AR activity through enhanced AR blockade and the elimination of AR ligands, additional androgen pathway-independent resistance mechanisms are likely to emerge.
To identify genes and pathways that modify prostate cancer growth in the context of suppressed AR signaling we performed in vitro high-throughput RNAi screening (HTRS) using a siRNA library designed to target 6650 individual genes representing several major ontology classes including cellular kinases, phosphatases, transcription factors, and growth factor receptors. We hypothesized that a subset of genes and gene networks could confer a castration-resistant phenotype in prostate cancer cells previously dependent upon androgen-mediated signaling for growth and survival. To prioritize candidate hits for clinical relevance, the HTRS results were cross-referenced with gene expression datasets collected from primary prostate cancers and cases of metastatic CRPC. This evaluation revealed that a subset of transcripts encoding proteins comprising the protein phosphatase 2A (PP2A) complex were enriched for a phenotype of AR ligand-independent growth promotion in the HTRS experiments and were also downregulated in a subset of primary and metastatic prostate carcinomas.
PP2A is a highly conserved serine/threonine phosphatase that has a broad spectrum of biologic roles including the negative regulation of signal transduction, cell cycle progression, and gene expression (10, 11). The PP2A holoenzyme is comprised of a core dimer – consisting of a catalytic subunit (PPP2CA/PPP2CB) and a structural subunit (PPP2R1A/PPP2R1B) – which form a heterotrimeric complex with a “B” subunit. The B subunit originates from one of 18 genes grouped into four structurally unrelated families and is thought to dictate substrate specificity, cellular localization, and enzymatic activity of the PP2A complex (12). Loss of PP2A activity is often a transforming event, which is perhaps most obvious in cells expressing the SV40 small t antigen (SV40ST) viral oncogene. SV40ST competes with B subunits to bind the PPP2C-PPP2R1 heterodimer (13); SV40ST binding is required for viral transformation (14). Additionally, non-viral mechanisms of PP2A inhibition are well documented in a wide variety of human cancers, and de novo inactivating mutations have been identified in each subunit class of the PP2A holoenzyme (10).
In prostate cancer, decreased expression of the catalytic subunit PPP2CA and the B-regulatory subunit PPP2R2A have been observed in a subset of primary tumors (15, 16). Studies of PP2A in ADT-resistant prostate cancers determined that loss of PPP2CA function is sufficient to confer AR ligand-depleted growth in vitro through a mechanism that sustained AR transcriptional activity (17). In the present report, we identified four PP2A components that, when downregulated, induce ADT-resistant prostate cancer cell growth. Of these, transcripts encoding the PP2A A and B regulatory family subunits, PPP2R1A and PPP2R2C, respectively, were significantly decreased in a subset of ADT-resistant primary and metastatic prostate cancers. Loss of PPP2R2C associated with increased prostate cancer metastasis and cancer-specific mortality. Importantly, we determined that loss of PPP2R2C promoted prostate cancer cell growth in the absence of AR ligands without upregulating AR expression or activating AR target genes. The ADT-independent growth conferred by PPP2R2C suppression did not occur via activation of cSRC, PI3K, or ERK1/2 signaling. Collectively, these studies provide insights into mechanisms capable of conferring androgen-independent prostate cancer progression that does not rely on AR activation and suggests that strategies capable of promoting PP2A activity, or modulating signaling programs regulated via PP2A, should be evaluated for therapeutic effects in patients with castration resistant prostate cancers.
MATERIALS AND METHODS
Cell Culture and siRNA Transfection
LNCaP and VCaP cell lines were obtained from ATCC (Manassas, VA, USA) and maintained in androgen-replete phenol red-free RPMI1640 + 10% Fetal Bovine Serum (FBS) (Life Technologies, Grand Island, NY, USA) + 1%PenStrep (Life Technologies). Cells were used within 6 months of resuscitation. One day prior to transfection cells were passaged into androgen-deplete phenol red-free Optimem (Life Technologies) supplemented with 3% CSS onto Matrix Screen Mate tissue culture-treated 384 well plates (Thermo Scientific, Hudson, NH, USA) using the densities determined during the feasibility study on a on a FluidX XRD384 peristaltic liquid handler (Boston, MA, USA) in a total well volume of 50μL.
Experimental working plates were prepared by first stamping 10μM siRNA library stocks into dry 384 well Abgene 1055-384 well microarray plates (Thermo Scientific) using a Vario workstation (Cybio, Jena, Germany) equipped with a 25μL 384-well tip head. A sidecar plate containing UNI1 non-targeting control, Kif11, and assay specific controls (Sigma-Aldrich, St. Louis, MO, USA) was then replicated on each experimental working plate on the Cybio Vario workstation. Optimem and Lipofectamine RNAiMax (Life Technologies) were added to the experimental working plates on a FluidX XRD384 peristaltic liquid handler using predefined metrics determined via the Quellos HTS Feasibility assay. Transfections were carried out by adding 5μL for VCAP and 0.625μL for LNCAP respectively from each experimental working plate onto 3 replicate cell plates using the CyBio Vario workstation.
Cell viability was assessed after the 96-hour incubation with the addition of Cell Titer-Glo (Promega, Madison, WI, USA) on the FluidX 384 liquid handler. Luminescence was read using an Envision HTS multilabel reader (Perkin Elmer, Waltham, MA, USA).
Deconvoluted siRNA experiments were performed in 96-well plates using transfection conditions scaled-up from 384-well experiments in a total volume of 125μl. Kif11, UNI1 non-targeting control, and PPP2R2C-targeting siRNA were purchased from Sigma-Aldrich. 96 hours after transfection cell viability was assessed by adding Cell Titer-Glo and measuring luminescence on a Synergy2 multiwell plate reader (BioTek, Winooski, VT, USA). siRNA sequences can be found in Supplementary Table 2.
Dasatinib was purchased from Chemitek (Indianapolis, IN, USA), R1881 was purchased from PerkinElmer, U0126 was purchased from Cell Signaling Technology (Beverly, MA, USA), and LY294002 was purchased from Promega. MDV3100 was obtained as a gift from Medivation Inc (San Francisco, CA, USA).
Statistical analysis of siRNA experiments
The data from the HTRS experiments were defined as the raw signal luminescence measured by the Envision HTS multilabel reader. For each sample, the raw intensities were transformed to Z-scores within each plate. The median standardized value for each gene was then used as an input for the downstream analysis. To downweight the influence of siRNAs undergoing large changes on the parameters used to define the Z-score transformation, the estimated means and variances were defined after removing the top and bottom 2.5% of the data within each plate.
Analysis of follow-up siRNA screens and qRT-PCR data was performed in Prism 4.0 software (GraphPad, La Jolla, CA, USA). Raw luminescence was normalized to the mean luminescence value of siUNI for each cell line. Treatment effect was measured by normalizing raw luminescence values in treatment groups to the median luminescence value in the vehicle-treated group. Statistical analysis was performed using an unpaired two-sided Student’s T-test using Welch’s correction to determine significance.
The statistical significance of PPP2R2C expression in gene expression microarray data and TMA nuclear intensity staining was performed using a Student’s T-Test. Whereas, multivariate analysis assessing the association between PPP2R2C staining intensity and patient survival data was performed using the Proc-Mixed procedure in SAS (SAS Institute, Cary, NC, USA) adjusting for the random effects of pre-operative PSA and tumor stage. The mean nuclear staining intensity (MNI) value was calculated by averaging the values for all TMAs cores derived from each individual patient. Kaplan-Meier survival estimates were generated with the Proc-Lifetest procedure in SAS utilizing a logrank significance threshold of p <0.05. Patients were grouped based on high PPP2R2C expression in primary tumors (mean nuclear staining intensity ≥1) and low PPP2R2C expression in primary tumors (mean nuclear staining intensity <1).
Quantitative Real-Time PCR (qRT-PCR)
RNA was isolated from cells 24 hours post-transfection using the RNeasy kit (Qiagen Inc, Valencia, CA) and cDNA was generated with Superscript II (Life Sciences) as per protocol. qRT-PCR reactions were performed in triplicate using an Applied Biosystems 7900 sequence detector with SYBR Green PCR master mix (Life Technologies). Primers were designed using PrimerQuest (IDT, San Diego, CA, USA), and all reactions were normalized to the expression of the housekeeping gene Rpl13a. A water negative control did not produce significant amplification product. PCR primer sequences can be found in Supplementary Table 3. Statistical analysis was performed using an unpaired two-sided Student’s T-test using Welch’s correction to determine significance.
Gene Expression Microarrays
Agilent 44K whole human genome expression oligonucleotide microarrays (Agilent Technologies, Inc., Santa Clara, CA) were used to profile 14 human primary prostate tumors and 54 castration-resistant prostate metastases from eleven patients (the patient cohort is described in detail by Holcomb et al(18)). The tumor samples were laser-capture microdissected, and total RNA isolated and amplified as described previously(19). Probe labeling and hybridization was performed following the Agilent suggested protocols and fluorescent array images were collected using the Agilent DNA microarray scanner G2565BA. Agilent Feature Extraction software was used to grid, extract, and normalize data. The resulting data were then combined with a publicly available dataset consisting of 15 normal prostate samples from untreated cancer-free patients(20). Expression ratios were log2 scaled and mean-centered across each gene.
Protein Collection and Immunoblotting
Protein was collected from tissue culture by lysing adherent cells with a cell lysis buffer (1.5M Urea, 1%SDS, 1%NP-40, 2%Tween20, 250nM NaCl, PBS) supplemented with 1x phosphatase inhibitors (PhosStop, Roche Diagnostics) and a 1x protease inhibitor cocktail (Complete Mini, Roche Diagnostics, Basel, Switzerland). Protein was quantified per protocol using a bicinchoninic acid assay (Thermo Scientific). Normalized cell lysates were loaded onto a 12% NuPAGE Bis-Tris gel (Life Technologies) in MES buffer. Protein was transferred to nitrocellulose membranes using a semi-dry transfer apparatus and Tris/CAPS buffer. Immunoblots were probed with primary antibodies targeting AKT (Cell Signaling Technology,#9272), phospho-AKT (Cell Signaling Technology, #4058), Erk1/2 (Sigma, M5670), diphosphorylated-Erk1/2 (Sigma, M9692), PP2R2C (Abnova, Taipei City, Taiwan; clone 6D1), Src (Cell Signaling Technology, #2110), and phospho-Src (Cell Signaling Technology, #2101). Horseradish-peroxidase conjugated secondary antibodies (Thermo Scientific) were used in conjugation with Supersignal West Pico Chemiluminescent substrate (Thermo Scientific) to visualize protein targets. Membranes were then stripped for 15 minutes in Stripping Buffer (Thermo Scientific) and re-probed with anti-Actin antibody (Santa Cruz Biotechnology, Santa Cruz, CA; sc-1616) as a loading control.
Tissue Microarray Construction
Prostate specimens were collected from a cohort of one hundred men who underwent radical prostatectomy at Virginia Mason Medical Center in Seattle, WA between 1954 and 1997 (Supplementary Table 1). Tissue specimens were formalin-fixed and embedded in paraffin following surgical resection. Patients were monitored for both biochemical recurrence (PSA>0.2 ng/ml following radical prostatectomy) and pathologic recurrence (pelvic disease, regional lymph node metastasis, distant metastasis, or prostate cancer-specific mortality (PCSM)). Tissue diagnosis or overt radiographic signs confirmed the presence of disease recurrence. To classify patients as developing prostate cancer-specific mortality, death certificates were obtained and prostate cancer had to be the first listed cause of death. Thirty-four patients were identified with known adverse outcomes of either distant metastasis or PCSM. These patients were matched by age, PSA and time since surgery at a 1:2 ratio with 66 patients who had undergone radical prostatectomy and had no evidence of recurrence. Fred Hutchinson Cancer Research Center’s Internal Review Office granted approval for the use of patient tissue samples and de-identified clinical data for research purposes according to the requirements of the Human Subjects Institutional Review Board guidelines (IRB #7624, Approval dates: 11/18/11-11/16/12).
Representative formalin-fixed, paraffin-embedded blocks of radical prostatectomy specimens were selected. A genitourinary pathologist reviewed hematoxylin and eosin-stained sections from representative FFPE blocks collected from each patient. Prostate cancer was confirmed and assigned a Gleason score. The H&E-stained sections were marked to identify the regions of cancer and benign glandular epithelia and three tissue microarrays (TMA) were constructed with 38 × 25 × 12 mm3 paraffin recipient blocks utilizing a manual tissue arrayer (Beecher Instrument, Silver Spring, MD, USA). Patients were represented in the TMAs by four 0.6 mm diameter cores of tissue, one benign and three cancer, for a total of 400 biopsy cores.
Immunohistochemistry
FFPE TMAs were deparaffinized and rehydrated. 3% Hydrogen peroxide was used to quench endogenous tissue peroxidases and an avidin/biotin blocking system (Dako, Glostrop, Denmark) was used to quench endogenous avidin/biotin. Antigen retrieval was performed with 10mM citrate buffer, pH=6.0 in a pressure cooker. Tissue was blocked with normal serum specific to the secondary antibody (horse) and the primary antibody for PPP2R2C (clone 6D1, Abnova) was applied for 30 min at room temp, followed by biotinylated anti-mouse secondary antibodies (Dako) and ABC reagent (Vector Laboratories, Burlingame, CA, USA). DAB (Dako) was used as the chromogen and the TMA was counterstained with Meyer’s hematoxylin (Dako). PPP2R2C nuclear staining was assessed by a genitourinary pathologist and graded on a 0-3 scale, with a score of 3 representing intense nuclear staining.
RESULTS
siRNA screen identifies modifiers of castration-resistant prostate cancer proliferation
To identify genes capable of promoting the growth of prostate cancer cells in the absence of exogenous AR ligands, we performed a high-throughput RNAi screen (HTRS) using two androgen-sensitive prostate cancer cell lines; LNCaP and VCaP. Briefly, cells were cultured in charcoal-dextran stripped fetal bovine serum (CSS) in order to simulate androgen deprivation therapy. Optimal transfection conditions were determined as previously described (21, 22). A library of siRNAs designed to inhibit the translation of 6650 individual genes was used in an arrayed screening strategy whereby a pool of three siRNAs targeting each specific gene was separately introduced into replicate cell cultures (one gene per well) in 384-well culture plates. After 96 hours of growth, cell numbers were estimated using the Cell Titer-Glo luminescence reagent. Raw luminescence signal intensity from the HTRS screen was transformed into Z-scores within each plate, and median standardized Z-scores for each gene were used in downstream analysis. To prioritize genes for further study, we arbitrarily set a significance threshold of Z ≥ 1.96. At this cut-point, suppression of 380 unique genes in LNCaP and 240 unique genes in VCaP induced cell growth in the absence of exogenous androgens (Figure 1), of which 40 induced growth in both cell lines (Table 1). Of the 40 genes whose suppression induced growth in both LNCaP and VCaP cells, two encoded components of the PP2A serine/threonine phosphatase complex: PPP2R2C and PPP2R1A.
Figure 1. High throughput RNAi screening identifies suppressors of CRPC growth.
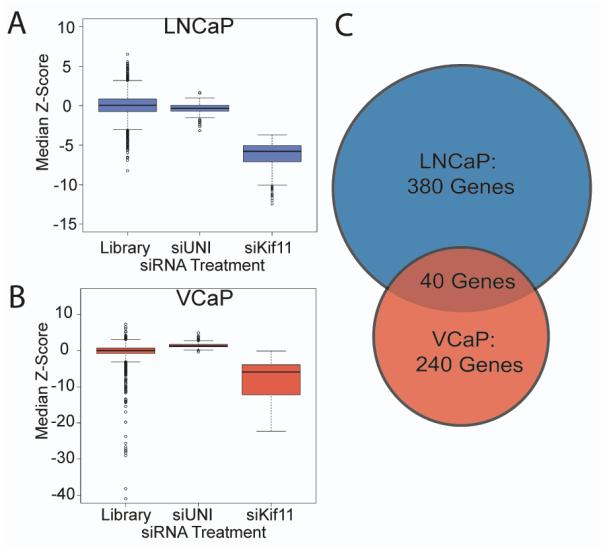
Pools of siRNAs targeting 6650 genes (Library) were used to knockdown individual gene targets in LNCaP and VCaP cells grown in androgen-depleted medium. HTRS Z-score results are plotted for the LNCaP (A) and VCaP (B) cell lines. siUNI is a non-targeting scrambled control siRNA. siKif11 is a positive control cell death-inducing siRNA targeting Kif11. (C) In the library screen, 380 siRNAs and 240 siRNAs induced castration-resistant growth in LNCaP or VCaP, respectively. Of these, 40 genes induced growth in androgen depleted conditions in both cell lines (Significance threshold: Z≥1.96).
Table 1.
Genes with growth-promoting effects following siRNA suppression.
| Official Gene Symbol |
Gene ID |
Gene Name | Median Z-Score |
|
|---|---|---|---|---|
|
| ||||
| LNCaP | VCaP | |||
| APOB | 338 | Apolipoprotein B (including Ag(x) antigen) | 2.43 | 1.97 |
| AQP8 | 343 | Aquaporin 8 | 2.40 | 2.26 |
| ART4 | 420 | ADP-ribosyltransferase 4 | 2.03 | 2.72 |
| CAPN13 | 92291 | Calpain 13 | 2.11 | 2.29 |
| CYP2A6 | 1548 | Cytochrome P450, family 2, subfamily A, polypeptide 6 | 2.19 | 2.15 |
| DNAJB2 | 3300 | DnaJ (Hsp40) homolog, subfamily B, member 2 | 3.79 | 2.09 |
| DPEP1 | 1800 | Dipeptidase 1 | 3.22 | 2.14 |
| ESRRB | 2103 | Estrogen-related receptor beta | 2.51 | 2.81 |
| GABRA2 | 2555 | Gamma-aminobutyric acid A receptor, alpha 2 | 2.44 | 2.53 |
| GBP7 | 388646 | Guanylate binding protein 7 | 3.07 | 2.86 |
| IDS | 3423 | Iduronate 2-sulfatase | 2.00 | 2.25 |
| KCND2 | 3751 | Potassium voltage-gated channel, Shal-related subfamily, 2 | 2.20 | 3.07 |
| KCTD5 | 54442 | Potassium channel tetramerisation domain containing 5 | 3.13 | 2.47 |
| KCTD12 | 115207 | potassium channel tetramerisation domain containing 12 | 3.54 | 3.81 |
| KIF2C | 11004 | Kinesin family member 2C | 2.01 | 2.01 |
| KLHL3 | 26249 | Kelch-like 3 | 2.39 | 2.54 |
| KLHL13 | 90293 | Kelch-like 13 | 3.16 | 2.42 |
| LAMB2 | 3913 | Laminin, beta 2 | 3.48 | 3.46 |
| MGC26694 | 284439 | Hypothetical protein MGC26694 | 3.49 | 7.26 |
| MIPEP | 4285 | Mitochondrial intermediate peptidase | 2.27 | 2.67 |
| NPM2 | 10361 | Nucleophosmin/nucleoplasmin, 2 | 2.26 | 2.43 |
| NR2F1 | 7025 | Nuclear receptor subfamily 2, group F, member 1 | 2.34 | 3.45 |
| PAFAH1B1 | 5048 | Platelet-activating factor acetylhydrolase, isoform Ib, alpha | 2.70 | 2.20 |
| PAQR4 | 124222 | Progestin and adipoQ receptor family member IV | 2.38 | 2.25 |
| PLOD2 | 5352 | Procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 | 2.32 | 3.09 |
| PPP2R1A | 5518 | Protein phosphatase 2 regulatory subunit A (PR65 alpha) | 2.20 | 3.29 |
| PPP2R2C | 5522 | Protein phosphatase 2 regulatory subunit B (PR55 gamma) | 2.81 | 2.10 |
| PYGM | 5837 | Phosphorylase, glycogen; muscle | 3.29 | 2.25 |
| RAD23A | 5886 | RAD23 homolog A | 3.21 | 2.04 |
| RARG | 5916 | Retinoic acid receptor, gamma | 2.04 | 2.05 |
| RHD | 6007 | Rhesus blood group, D antigen | 2.41 | 2.03 |
| RNF20 | 56254 | Ring finger protein 20 | 3.01 | 2.03 |
| RNF144 | 9781 | Ring finger protein 144 | 2.39 | 2.77 |
| SRM | 6723 | Spermidine synthase | 2.34 | 2.07 |
| TNXB | 7148 | Tenascin XB | 2.32 | 2.57 |
| TOPBP1 | 11073 | Topoisomerase (DNA) II binding protein | 2.48 | 2.22 |
| UBA6 | 55236 | Ubiquitin-like modifier activating enzyme 6 | 2.01 | 2.09 |
| UNC5C | 8633 | Unc-5 homolog C | 3.68 | 3.18 |
| USP21 | 27005 | Ubiquitin specific protease 21 | 2.13 | 2.17 |
| ZFAND3 | 60685 | Zinc finger, AN1-type domain 3 | 3.83 | 3.65 |
PP2A subunit expression is reduced in primary and metastatic prostate cancers
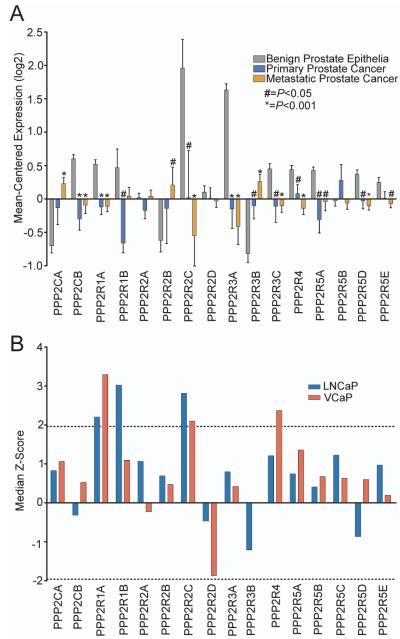
To prioritize HTRS hits exhibiting potential tumor-suppressor expression patterns, we cross-referenced the screen results with transcript abundance levels determined by microarray measurements of laser-capture microdissected benign prostate epithelia (n=15), ADT-resistant primary prostate tumors (n=14), and CRPC metastases (n=54). Out of the 40 proliferation-suppressor genes identified in the in vitro HTRS, 14 genes were significantly downregulated in primary and metastatic CRPC samples compared to benign prostate epithelia, including PPP2R2C and PPP2RA1 (Figure 2). Specifically, the mean expression of PPP2R2C in these primary and metastatic cancers was 3.85-fold lower (P=0.034) and 5.69-fold lower (P<0.001) than benign prostate epithelia, respectively. PPP2R1A expression was also decreased in metastatic prostate tumors (1.55-fold, P<0.001).
Figure 2. PPP2R2C is downregulated in primary and metastatic prostate cancer.
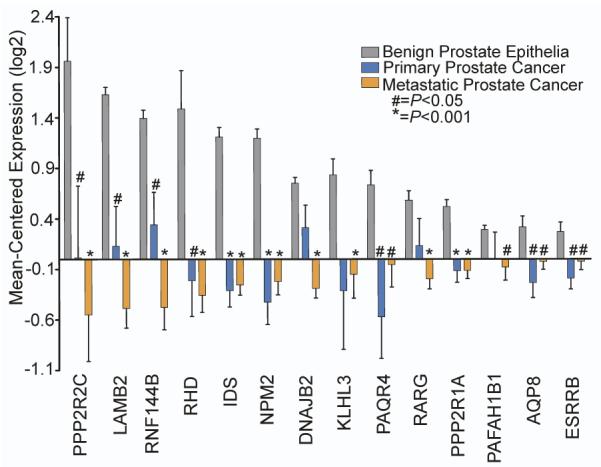
Gene expression microarray data from 16 benign prostate samples, 15 ADT-resistant primary tumors, and 55 CRPC metastases was cross-referenced with results from the HTRS experiments. Of the 40 overlapping hits identified in the VCaP and LNCaP HTRS experiments, 14 were significantly downregulated in primary tumors and CRPC metastases compared to benign prostate epithelia. Statistical significance was determined by the Student’s T-test.
We next evaluated HTRS experimental results and human prostate gene expression data for evidence of other PP2A components behaving as tumor suppressors. Previous studies have reported that suppression of the PP2A constituents PPP2CA and PPP2R2A can promote castration-resistant prostate cancer cell growth (15, 16). However, transcripts encoding these PP2A subunits were not down-regulated in the primary prostate cancers we evaluated (Figure 3a). Further, transcripts encoding PPP2CA were significantly higher in metastatic CRPC (1.9-fold, p<0.001; Figure 3a). In the HTRS experiments, siRNAs targeting PPP2CA or PPP2R2A did not induce significant castration-resistant growth in either LuCAP or VCaP cells (Figure 3b).
Figure 3. High-throughput siRNA screening results and corresponding patient gene expression data for PP2A subunits.
(A) PP2A subunit expression in benign prostate epithelia, ADT-resistant primary prostate tumors, and CRPC metastases. (B) Knockdown of PPP2R1A and PPP2R2C induce sufficient growth to reach the significance threshold in both cell lines. Knockdown of other PP2A B-regulatory subunits induced variable effects on proliferation. Dotted lines represent Z=+/−1.96.
As four of the 16 PP2A subunits tested in the HTRS experiments induced AR ligand-independent proliferation in at least one cell line (Figure 3b), we sought to further assess the mechanism(s) by which PP2A activity contributes to this phenotype. We selected PPP2R2C for further investigation based on the findings that PPP2R2C induced significant androgen depletion-resistant proliferation in both cell lines evaluated, and PPP2R2C was the HTRS hit found to be most down-regulated in castration-resistant metastatic prostate cancer compared to benign epithelium (Figure 2).
Knockdown of PPP2R2C promotes prostate cancer growth in the absence of androgens without inducing AR activity in vitro
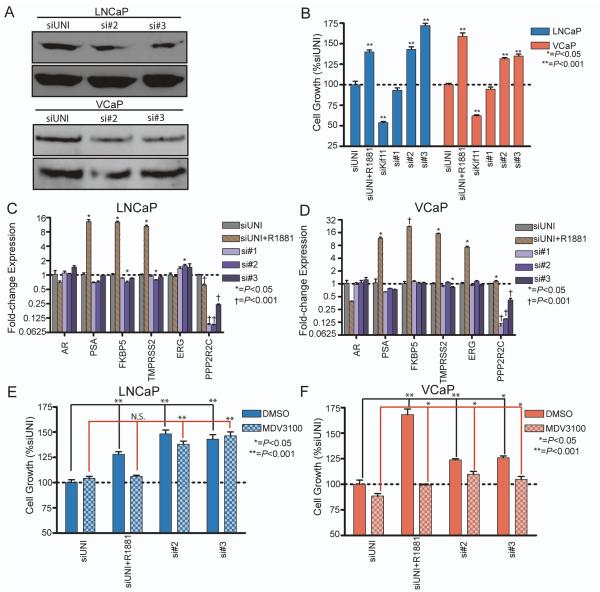
Because the siRNA screens were performed using pools of three siRNAs targeting each gene, we evaluated individual siRNA efficacy by transfecting LNCaP and VCaP with the deconvoluted pool of PPP2R2C siRNAs, designated siRNA #1-3. Suppression of PPP2R2C by gene-specific siRNA was confirmed by qRT-PCR and correlated with assays of cell proliferation. PPP2R2C knockdown by si#2 and si#3 were confirmed at the protein level in both cell lines (Figure 4a). Scrambled control siRNAs (siUNI) did not alter PPP2R2C expression or influence cell proliferation (Figure 4b). The positive control for cell death, siKiff11, substantially reduced the number of tumor cells in both LNCaP and VCaP experiments (Figure 4b). As expected, exposing LNCaP or VCaP cells to androgen (R1881) stimulated proliferation and induced the AR target genes PSA, FKBP5, TMRPSS2, and ERG (in VCaP; Figure 4b-d). siRNA#1 suppressed PPP2R2C transcripts by 10-fold in both cell lines, but did not influence cell proliferation in either line. However, both siRNA #2 and siRNA #3 reduced PPP2R2C mRNAs in LNCaP by 11.1-fold and 4.2-fold (P<0.001), respectively (Figure 4b-c), and induced cell proliferation in androgen-depleted medium by 42% (siRNA #2, P<0.001) and 72% (siRNA #3, P<0.001). This increase in cell number approximated the influence of adding the androgen R1881 (Figure 4b). In VCaP cells PPP2R2C transcripts were reduced 6.7-fold by siRNA #2 and 4.2-fold by siRNA #3 (P<0.001; Figure 4d), and these siRNAs increased cell proliferation by approximately 33% in androgen depleted growth conditions (P<0.001; Figure 4b). Knockdown of PPP2R2C in the presence of androgens did not significantly increase growth in either cell line (Supplemental Figure 1). PPP2R2C suppression did not alter AR or PSA expression in either cell line (Figure 4c-d). Suppression of PPP2R2C with siRNA #2 in LNCaP cells did slightly decrease the expression of FKBP5 (1.4-fold, P=0.02) and TMPRSS2 (1.3-fold, P=0.02) and resulted in a 1.6-fold increase in ERG expression (P=0.01; Figure 4c). Suppression of PPP2R2C with siRNA #3 in VCaP resulted in decreased TMPRSS2 expression (1.2-fold, P=0.01; Figure 4d). PPP2R2C downregulated with si#3 in LNCaP and si#2 in VCaP did not affect androgen-regulated gene expression. These data indicate that the growth advantages attained from PPP2R2C knockdown were not mediated by an increase in canonical AR transcriptional activity.
Figure 4. siRNA knockdown of PPP2R2C induces growth in LNCaP and VCaP through pathways independent of the Androgen Receptor.
LNCaP and VCaP were transfected with the deconvoluted pool of PPP2R2C siRNAs (si#1-3), in addition to a scrambled control (siUNI) and a positive control for transfection (siKif11). siUNI + 1nM R1881 is a positive control for androgen-induced cell growth and gene expression. (A) Western immunoblot analysis of PPP2R2C protein with siRNAs targeting PPP2R2C. (B) Growth of LNCaP and VCaP cells in androgen-depleted medium is induced by PPP2R2C-targeted siRNA #2 and siRNA #3, but not siRNA #1 (C – D) qRT-PCR analysis of cDNA collected from LNCaP (C) and VCaP (D) demonstrates successful suppression of PPP2R2C expression. PPP2R2C knockdown does not increase the expression of AR or AR-regulated genes (PSA, FKBP5, TMPRSS2, ERG). Co-treatment with non-targeting siRNA + 1nM R1881 induced AR-regulated gene expression. Statistical comparisons of gene expression were performed between siUNI and gene-specific siRNA within each cell line. (E – F) LNCaP (E) and VCaP (F) were transfected with non-targeting siRNA or siRNA targeting PPP2R2C and co-treated with 5μM MDV3100. Raw luminescence values in each cell line were normalized to siUNI. Statistical comparisons between siRNA targeting PPP2R2C and siUNI were performed within each cell line.
To further assess the potential role of the AR in PPP2R2C mediated prostate cancer cell growth in the context of androgen depletion, LNCaP and VCaP cells were treated with the AR antagonist MDV3100. In the absence of exogenous androgens, MDV3100 had a slight effect in reducing VCaP cell numbers after 96 hours (12%, p=0.05) and no discernable effect on LNCaP growth (Figure 4e-f). As expected, MDV3100 effectively suppressed the proliferative effects induced by the synthetic androgen R1881 in both lines (Figure 4e-f). Suppression of PPP2R2C induced cell proliferation in VCaP cells (si#2: 24%, p<0.001; si#3: 18%, p=0.002) and LNCaP cells (si#2: 32%, p<0.001; si#3: 40%, p<0.001) in androgen-depleted medium despite treatment with MDV3100 (Figure 4e-f). Collectively, these experiments indicate that PPP2R2C loss is sufficient to induce AR pathway-independent proliferation in prostate cancer cells.
PPP2R2C knockdown does not mediate proliferation through activation of SRC, PI3K, or ERK1/2 signal transduction in prostate carcinoma
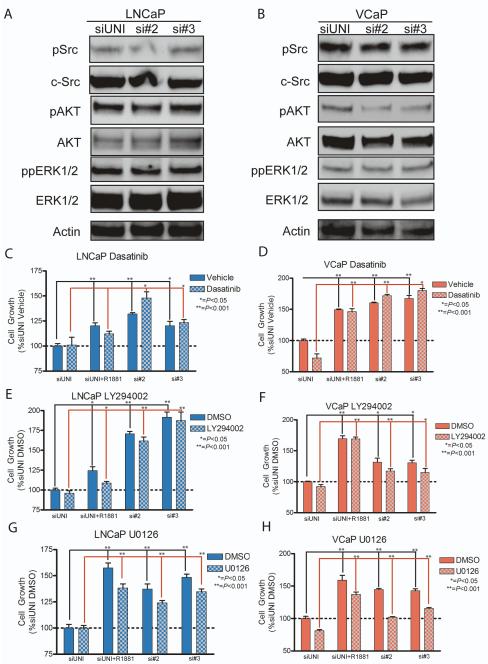
Previous experimentation in non-epithelial cancer models indicate that PPP2R2C may negatively regulate the activity of the v-src sarcoma viral oncogene homolog SRC (23). As SRC has been postulated to promote CRPC growth (24, 25), we hypothesized that PPP2R2C loss may promote castration-resistant growth in LNCaP and VCaP cells by modulating SRC phosphorylation and consequent activity. However, knockdown of PPP2RC did not result in SRC phosphorylation (Figure 5a-b) and treatment with the SRC inhibitor Dasatinib failed to inhibit castration-resistant growth induced by PPP2R2C knockdown (Figure 5c-d). Therefore, these experiments indicate that androgen depletion-resistant growth of LNCaP and VCaP cell lines following PPP2R2C knockdown is not mediated through enhanced SRC activity.
Figure 5. PPP2R2C knockdown does not activate SRC, PI3K, or ERK1/2 signal transduction pathways.
(A – B) Western immunoblot analysis following PPP2R2C knockdown. No changes in the quantity or phosphorylation status of SRC, AKT, or ERK1/2 in LNCaP (a) or VCaP (b) cells cultured in androgen-depleted conditions was observed. (C – D) Cell growth assays of LNCaP and VCaP cells. Treatment with 1μM Dasatinib, a SRC kinase inhibitor, does not significantly inhibit growth induced by PPP2R2C knockdown when compared to the scrambled control siRNA. (E – F) Treatment of LNCaP and VCaP cells with 2μM LY294002, a PI3K inhibitor, does not significantly inhibit growth induction by PPP2R2C knockdown in androgen depleted medium. (G – H) Treatment of LNCaP and VCaP cells with 10μM U0126, an inhibitor of MEK, does not inhibit growth induced by PPP2R2C knockdown in androgen depleted medium. Raw luminescence values were normalized to untreated control cells transfected with a non-targeting siRNA (siUNI).
We next evaluated the influence of PPP2R2C in other signal transduction pathways postulated to influence CRPC growth. Specifically, we investigated the phosphorylation of extracellular signal regulated kinase 1/2 (ERK1/2) and Protein Kinase B/AKT (AKT) (8, 26). Depletion of PPP2R2C did not result in observable changes in the phosphorylation of ERK1/2 when compared to the scrambled control siRNA. AKT phosphorylation was slightly decreased in VCaP, but was unchanged in LNCaP (Figure 5a-b). To confirm that PPP2R2C knockdown did not induce growth through PI3K or ERK1/2 activation, cells were co-treated with pharmacological inhibitors of these pathways and siRNAs targeting PPP2R2C. Inhibition of PI3K with LY294002, or MEK with U0126, failed to abrogate growth-induction mediated by PPP2R2C knockdown (Figure 5e-h). Therefore, it appears that PPP2R2C knockdown influences signaling pathways capable of promoting prostate cancer cell growth that are distinct from PI3K, ERK1/2, or SRC in the setting of androgen depletion.
Decreased PPP2R2C expression is primary prostate cancers is associated with disease relapse and prostate cancer-specific mortality
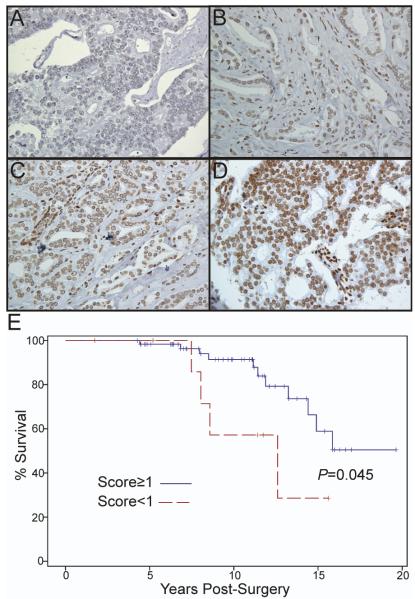
Because PPP2R2C downregulation promotes castration-resistant growth, we hypothesized that PPP2R2C expression might influence long-term disease outcome in men with prostate carcinoma. To explore this hypothesis, we performed immunohistochemical (IHC) staining for PPP2R2C on radical prostatectomy tissues from a cohort of 100 patients with documented long-term clinical outcomes that included biochemical relapse, local recurrence, distant metastases, and PCSM (Supplementary Table 1). A range of PPP2R2C protein expression was noted between individual cores, yet staining within each tumor core was largely homogenous. PPP2R2C staining of LNCaP and VCaP cell lines also demonstrated nuclear expression (Supplementary Figure 2). Each TMA core was categorized based on IHC nuclear intensity according to a scale from 0-3, with a score of 0 representing no PPP2R2C staining and a score of 3 representing high-intensity nuclear staining (Figure 6a-d). We analyzed the impact of nuclear PPP2R2C expression on prostate cancer morbidity and mortality using the Proc Mixed model in SAS (SAS 9.3, Cary, NC) adjusted for patient pre-surgical serum PSA levels and tumor stage at the time of prostatectomy. Lower PPP2R2C staining was significantly correlated with the risk of biochemical relapse (P≤0.01) and the development of distant metastases (P<0.001). We also observed that PCSM was correlated with low PPP2R2C protein expression (P=0.048; Table 2). Kaplan-Meier survival curves for patients with tumors expressing high PPP2R2C (mean staining intensity ≥ 1) and those with low PPP2R2C (mean staining intensity <1) revealed a survival advantage in those patients whose tumors expressed high levels of PPP2R2C (logrank test P=0.045) (Figure 7e).
Figure 6. PPP2R2C protein expression is downregulated in primary prostate tumors and associates with prostate cancer-specific mortality.
(A – D) IHC for PPP2R2C was performed on a tissue microarray comprised of primary prostate adenocarcinomas acquired by radical prostatectomy. Representative primary tumor cores with mean nuclear intensity (MNI) PPP2R2C staining of 0 (a), 1 (b), 2 (c), and 3 (d) were photographed at 400× original magnification. (E) Patients were subdivided into groups based on the mean nuclear staining intensity (MNI) of PPP2R2C in representative tumor cores. Kaplan-Meier analysis demonstrated a significant difference in survival between patients with high PPP2R2C mean nuclear staining intensity (MNI ≥ 1; blue) and those patients with low PPP2R2C expression (MNI<1; red) (log-rank test; P=0.045).
Table 2.
Multivariate analysis of PPP2R2C staining and patient outcomes
| Clinical Outcome | PPP2R2C Mean Nuclear Intensity |
P-Value | |
|---|---|---|---|
| Biochemical Relapse | No | 2.25 | |
| Yes | 1.74 | P=0.006 | |
| Distant Metastases | No | 2.32 | |
| Yes | 1.55 | P<0.001 | |
| PCSM | No | 2.21 | |
| Yes | 1.79 | P=0.048 | |
DISCUSSION
The development and clinical use of increasingly potent therapeutics targeting the AR and androgenic steroid production in prostate cancer (4, 5) is likely to select against those tumor cells relying on AR-mediated pathways for survival. Therefore, identifying AR-pathway independent prostate cancer growth and survival mechanisms will become increasingly relevant in clinical disease. Here, we provide evidence that decreased expression of the regulatory PP2A subunit, PPP2R2C, promotes CRPC growth. Importantly, growth induction was not mediated by the upregulation of the AR or androgen-regulated genes, and growth was not inhibited by treatment with the potent AR antagonist MDV3100. These data strongly suggest that PPP2R2C loss activates growth and survival pathways that bypass traditional approaches to suppress prostate cancer growth via the AR. Therefore, we expected to observe PPP2R2C loss in a subset of clinical CRPC. Multivariate regression analysis demonstrated a statistically significant association between decreased PPP2R2C expression, disease relapse, and increased PCSM. Kaplan-Meier survival analysis indicated that low PPP2R2C expression in primary tumors is associated with decreased long-term survival. To our knowledge, this is the first evidence suggesting that PPP2R2C loss may influence cancer-specific mortality.
PP2A is a known tumor suppressor and loss of PP2A phosphatase activity or altered substrate specificity has been shown to occur frequently in several human malignancies (10). Decreased PP2A function is sufficient to promote castration-resistant growth of the LNCaP cell line, and studies of the castration-resistant LNCaP derivative C4-2b, have reported lowered levels of the catalytic subunit PPP2CA (17). However, complete loss of PPP2CA expression can be detrimental to cancer cell growth and result in cell death (13), suggesting that PPP2CA expression plays a dual role in promoting survival and negatively regulating proliferation. Considering that the SV40ST oncogene promotes cell transformation by inhibiting B-subunit binding without initiating apoptosis (14), it is likely that loss of select PP2A B-subunits could alter PP2A substrate specificity and promote tumorigenesis while avoiding the apoptotic effect of total PPP2CA loss. Knock-down of individual PP2A regulatory B-subunits has demonstrated differential effects on cell transformation (10, 13), an effect replicated in the HTRS experiments conducted in this study. Investigation into transforming B-subunit loss has demonstrated patterns of growth pathway deregulation similar to those observed with inhibition of PP2A catalytic activity and those induced by SV40ST oncogene expression (13).
The effect of PPP2R2C expression on PP2A substrate specificity is largely unknown. However, PPP2R2C has been shown to negatively regulate the activity of the SRC oncogene in osteosarcoma cells (23). Surprisingly, PPP2R2C downregulation did not alter SRC pathway activity in prostate cancer cells. This discrepancy may be due to tissue-specific differences in subcellular localization of the PP2A complex. In osteosarcoma cells, investigators detected perinuclear expression of PPP2R2C (23). In contrast, PPP2R2C demonstrated distinct nuclear localization in prostatic tissue. Tissue-specific variations in the subcellular localization of other PP2A B-regulatory subunits are widely reported; for example, PPP2R2A is detected in the cytoplasm of neural extracts (27), but expression is nuclear in the Jurkat cell line and rat epidermis (28, 29).
Further mechanistic studies are necessary to identify specific PP2A targets mediated by PPP2R2C expression in prostate carcinoma. Because PP2A directly regulates the activity of more than 50 kinases and other known proto-oncogenes (10, 11), this will likely require extensive proteomics-based experiments with an emphasis on deregulated survival and growth pathways found in clinical CRPC. The promotion of CRPC growth via decreased PPP2R2C expression provides an important example, supported by in vivo patient outcomes, of prostate cancer survival pathways that do not function through increased AR activity, but instead through the deregulation of existing signal transduction pathways. The identification of pharmacological inhibitors of PPP2R2C complexes, or the growth pathway(s) activated by PPP2R2C loss may prove useful as a therapeutic in men with CRPC, or as neoadjuvant therapy in men with downregulated PPP2R2C in primary disease.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Phil Febbo and Robin Tharakan for helpful discussions. We thank Jared Lucas for assistance with immunohistochemistry. This research was supported by 5RC1CA146849, the Pacific Northwest SPORE in Prostate Cancer P50CA097186 and CDMRP Award PC09337P1. EGB was supported by Department of Defense Prostate Cancer Research Program Predoctoral Training Award (W81XWH-10-1-0133). MDV3100 was received as a gift from Medivation Inc., San Francisco, CA.
Footnotes
CONFLICT OF INTEREST The authors report no conflict of interest.
REFERENCES
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Pagliarulo V, Bracarda S, Eisenberger MA, Mottet N, Schroder FH, Sternberg CN, et al. Contemporary role of androgen deprivation therapy for prostate cancer. Eur Urol. 2012;61:11–25. doi: 10.1016/j.eururo.2011.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Small EJ, Schellhammer PF, Higano CS, Redfern CH, Nemunaitis JJ, Valone FH, et al. Placebo-controlled phase III trial of immunologic therapy with sipuleucel-T (APC8015) in patients with metastatic, asymptomatic hormone refractory prostate cancer. J Clin Oncol. 2006;24:3089–94. doi: 10.1200/JCO.2005.04.5252. [DOI] [PubMed] [Google Scholar]
- 4.Scher HI, Beer TM, Higano CS, Anand A, Taplin ME, Efstathiou E, et al. Antitumour activity of MDV3100 in castration-resistant prostate cancer: a phase 1-2 study. Lancet. 2010;375:1437–46. doi: 10.1016/S0140-6736(10)60172-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Bono JS, Logothetis CJ, Molina A, Fizazi K, North S, Chu L, et al. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med. 2011;364:1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bluemn EG, Nelson PS. The androgen/androgen receptor axis in prostate cancer. Curr Opin Oncol. 2012;24:251–7. doi: 10.1097/CCO.0b013e32835105b3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nelson PS. Molecular states underlying androgen receptor activation: a framework for therapeutics targeting androgen signaling in prostate cancer. J Clin Oncol. 2012;30:644–6. doi: 10.1200/JCO.2011.39.1300. [DOI] [PubMed] [Google Scholar]
- 8.Mulholland DJ, Tran LM, Li Y, Cai H, Morim A, Wang S, et al. Cell autonomous role of PTEN in regulating castration-resistant prostate cancer growth. Cancer Cell. 2011;19:792–804. doi: 10.1016/j.ccr.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S, et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell. 2011;19:575–86. doi: 10.1016/j.ccr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eichhorn PJ, Creyghton MP, Bernards R. Protein phosphatase 2A regulatory subunits and cancer. Biochim Biophys Acta. 2009;1795:1–15. doi: 10.1016/j.bbcan.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 11.Millward TA, Zolnierowicz S, Hemmings BA. Regulation of protein kinase cascades by protein phosphatase 2A. Trends Biochem Sci. 1999;24:186–91. doi: 10.1016/s0968-0004(99)01375-4. [DOI] [PubMed] [Google Scholar]
- 12.Sablina AA, Hahn WC. The role of PP2A A subunits in tumor suppression. Cell Adh Migr. 2007;1:140–1. doi: 10.4161/cam.1.3.4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sablina AA, Hector M, Colpaert N, Hahn WC. Identification of PP2A complexes and pathways involved in cell transformation. Cancer Res. 2010;70:10474–84. doi: 10.1158/0008-5472.CAN-10-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arroyo JD, Hahn WC. Involvement of PP2A in viral and cellular transformation. Oncogene. 2005;24:7746–55. doi: 10.1038/sj.onc.1209038. [DOI] [PubMed] [Google Scholar]
- 15.Singh AP, Bafna S, Chaudhary K, Venkatraman G, Smith L, Eudy JD, et al. Genome-wide expression profiling reveals transcriptomic variation and perturbed gene networks in androgen-dependent and androgen-independent prostate cancer cells. Cancer Lett. 2008;259:28–38. doi: 10.1016/j.canlet.2007.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mao X, Boyd LK, Yanez-Munoz RJ, Chaplin T, Xue L, Lin D, et al. Chromosome rearrangement associated inactivation of tumour suppressor genes in prostate cancer. Am J Cancer Res. 2011;1:604–17. [PMC free article] [PubMed] [Google Scholar]
- 17.Bhardwaj A, Singh S, Srivastava SK, Honkanen RE, Reed E, Singh AP. Modulation of protein phosphatase 2A activity alters androgen-independent growth of prostate cancer cells: therapeutic implications. Mol Cancer Ther. 2011;10:720–31. doi: 10.1158/1535-7163.MCT-10-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holcomb IN, Young JM, Coleman IM, Salari K, Grove DI, Hsu L, et al. Comparative analyses of chromosome alterations in soft-tissue metastases within and across patients with castration-resistant prostate cancer. Cancer research. 2009;69:7793–802. doi: 10.1158/0008-5472.CAN-08-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.True L, Coleman I, Hawley S, Huang CY, Gifford D, Coleman R, et al. A molecular correlate to the Gleason grading system for prostate adenocarcinoma. Proc Natl Acad Sci U S A. 2006;103:10991–6. doi: 10.1073/pnas.0603678103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Page ST, Lin DW, Mostaghel EA, Marck BT, Wright JL, Wu J, et al. Dihydrotestosterone administration does not increase intraprostatic androgen concentrations or alter prostate androgen action in healthy men: a randomized-controlled trial. J Clin Endocrinol Metab. 2011;96:430–7. doi: 10.1210/jc.2010-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartz SR, Zhang Z, Burchard J, Imakura M, Martin M, Palmieri A, et al. Small interfering RNA screens reveal enhanced cisplatin cytotoxicity in tumor cells having both BRCA network and TP53 disruptions. Mol Cell Biol. 2006;26:9377–86. doi: 10.1128/MCB.01229-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toyoshima M, Howie HL, Imakura M, Walsh RM, Annis JE, Chang AN, et al. Functional genomics identifies therapeutic targets for MYC-driven cancer. Proc Natl Acad Sci U S A. 2012;109:9545–50. doi: 10.1073/pnas.1121119109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eichhorn PJ, Creyghton MP, Wilhelmsen K, van Dam H, Bernards R. A RNA interference screen identifies the protein phosphatase 2A subunit PR55gamma as a stress-sensitive inhibitor of c-SRC. PLoS Genet. 2007;3:e218. doi: 10.1371/journal.pgen.0030218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kobayashi T, Inoue T, Shimizu Y, Terada N, Maeno A, Kajita Y, et al. Activation of Rac1 is closely related to androgen-independent cell proliferation of prostate cancer cells both in vitro and in vivo. Mol Endocrinol. 2010;24:722–34. doi: 10.1210/me.2009-0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mendiratta P, Mostaghel E, Guinney J, Tewari AK, Porrello A, Barry WT, et al. Genomic strategy for targeting therapy in castration-resistant prostate cancer. J Clin Oncol. 2009;27:2022–9. doi: 10.1200/JCO.2008.17.2882. [DOI] [PubMed] [Google Scholar]
- 26.Drake JM, Graham NA, Stoyanova T, Sedghi A, Goldstein AS, Cai H, et al. Oncogene-specific activation of tyrosine kinase networks during prostate cancer progression. Proc Natl Acad Sci U S A. 2012;109:1643–8. doi: 10.1073/pnas.1120985109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strack S, Zaucha JA, Ebner FF, Colbran RJ, Wadzinski BE. Brain protein phosphatase 2A: developmental regulation and distinct cellular and subcellular localization by B subunits. J Comp Neurol. 1998;392:515–27. [PubMed] [Google Scholar]
- 28.Guo CY, Brautigan DL, Larner JM. ATM-dependent dissociation of B55 regulatory subunit from nuclear PP2A in response to ionizing radiation. J Biol Chem. 2002;277:4839–44. doi: 10.1074/jbc.M110092200. [DOI] [PubMed] [Google Scholar]
- 29.O’Shaughnessy RF, Welti JC, Sully K, Byrne C. Akt-dependent Pp2a activity is required for epidermal barrier formation during late embryonic development. Development. 2009;136:3423–31. doi: 10.1242/dev.037010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.