Abstract
Replication stress and DNA damage activate the ATR-CHK1 checkpoint signaling pathway that licenses repair and cell survival processes. In this study, we examined the respective roles of the ATR and CHK1 kinases in ovarian cancer cells using genetic and pharmacological inhibitors of in combination with cisplatin, topotecan, gemcitabine and the poly(ADP-ribose)-polymerase (PARP) inhibitor veliparib (ABT-888), four agents with clinical activity in ovarian cancer. RNAi-mediated depletion or inhibition of ATR sensitized ovarian cancer cells to all four agents. In contrast, while cisplatin, topotecan and gemcitabine each activated CHK1, RNAi-mediated depletion or inhibition of this kinase in cells sensitized them only to gemcitabine. Unexpectedly, we found that neither the ATR kinase inhibitor VE-821 or the CHK1 inhibitor MK-8776 blocked ATR-mediated CHK1 phosphorylation or autophosphorylation, two commonly used readouts for inhibition of the ATR-CHK1 pathway. Instead, their ability to sensitize cells correlated with enhanced CDC25A levels. Additionally, we also found that VE-821 could further sensitize BRCA1-depleted cells to cisplatin, topotecan and veliparib beyond the potent sensitization already caused by their deficiency in homologous recombination. Taken together, our results established that ATR and CHK1 inhibitors differentially sensitize ovarian cancer cells to commonly used chemotherapy agents, and that CHK1 phosphorylation status may not offer a reliable marker for inhibition of the ATR-CHK1 pathway. A key implication of our work is the clinical rationale it provides to evaluate ATR inhibitors in combination with PARP inhibitors in BRCA1/2-deficient cells.
Keywords: Poly(ADP-ribose) polymerase, veliparib, cisplatin, topotecan, gemcitabine, ovarian cancer, homologous recombination, BRCA1, BRCA2
INTRODUCTION
Epithelial ovarian cancers are initially treated with platinum-based therapies, which induce very high response rates. Despite this initial chemoresponsiveness, more than 70% of patients will die of this disease. Accordingly, there is intense interest in identifying approaches to enhance the initial responses and/or to counter the emergence of resistance (1).
One possible approach to increase sensitivity to chemotherapy is the pharmacological inhibition of the replication checkpoint signaling pathway (Reviewed in ref. 2). This pathway, which promotes cell survival, is activated by inhibition of DNA replication, as occurs when dNTP levels are disrupted or the replication fork encounters DNA damage. When such genotoxic stress blocks DNA replication, the continued action of helicases that unwind the DNA in front of the advancing DNA polymerases causes the accumulation of extensive regions of single-stranded DNA, which is coated with replication protein A. The replication protein A-coated single-stranded DNA attracts the kinase ATR and promotes the loading of the Rad9-Hus1-Rad1 (9-1-1) complex onto DNA. The 9-1-1 complex and its associated protein, TopBP1, then activate ATR, which phosphorylates hundreds of substrates (3–6). Although the effects of most of these phosphorylations have not been characterized, one ATR substrate that has been intensely studied is Chk1, a kinase that phosphorylates CDC25A to block the firing of replication origins, stabilizes stalled replication forks, and regulates DNA repair.
Since the identification of the ATR pathway and the demonstration that it helps cells survive genotoxic stresses, there has been much interest in developing small molecules to target components of this pathway, especially the kinases ATR and Chk1 (7). Chk1 inhibitors have received the most attention, likely because this enzyme has a ‘conventional’ kinase domain that resembles the domains of many other kinases for which effective inhibitors have been identified (8, 9). In contrast, because ATR possesses a phosphatidylinositol 3-kinase-like kinase domain, development of potent and selective inhibitors for this family of kinases has proceeded at a slower pace (10). It is also possible that development of ATR inhibitors has been discouraged by the notion that Chk1, which is the target of a number of inhibitors already in development (8, 9), relays the majority of the ATR signal that promotes cell survival. However, recent studies suggest that the effects of disabling ATR versus Chk1 may differ in that Chk1 inhibition might not uniformly sensitize to genotoxic drugs (11–13). These emerging results raise questions about the relative roles of ATR and Chk1 in tumor cells treated with chemotherapy agents.
Accordingly, the present studies were designed to comprehensively compare the roles of the ATR and Chk1 in ovarian cancer cell lines treated with classes of agents that, despite diverse mechanisms of action, have activity in this disease. Specifically, these studies were designed to address three issues. First, using small inhibitory (si)RNAs and highly selective small molecule inhibitors, we compared the effects of disabling ATR versus Chk1 in ovarian cancer cells exposed to cisplatin, gemcitabine, topotecan, and veliparib. Second, we examined the ATR/Chk1 signaling pathway looking for reliable markers of sensitization that could potentially be used in future clinical trials. Finally, given the hypersensitivity of homologous recombination (HR) deficient ovarian cancers to cisplatin, topotecan and PARP inhibitors (14), we investigated whether inhibition of the ATR/Chk1 pathway could further sensitize BRCA1- or BRCA2-disabled cells. Our results indicate that Chk1 inhibitors robustly sensitize to gemcitabine but not the other agents, whereas ATR inhibition sensitizes to a much broader range of chemotherapy. Importantly, interruption of ATR signaling (but not Chk1 signaling) strikingly further sensitized BRCA1- and BRCA2-deficient ovarian cancer cells to PARP inhibition, providing a potential approach for making PARP inhibitors even more effective in HR-deficient tumors.
MATERIALS AND METHODS
Materials
Veliparib (ABT-888) was purchased from Enzo Life Sciences, Selleck Chemicals, or Chemietek; VE-821 and MK-8776 were from ChemieTek; LY 2603618 was from Selleck Chemicals; and gemcitabine and cisplatin were from Sigma-Aldrich. Topotecan was provided by the Drug Synthesis Branch of the National Cancer Institute (Bethesda, MD).
Antibodies to various antigens were as follows: phospho-Ser345-Chk1, phospho-Ser296-Chk1, BRCA1, and horseradish peroxidase-linked rabbit and mouse immunoglobulin Gs from Cell Signaling Technology; Chk1 and Rad51 from Santa Cruz Biotechnology; phospho-Ser139-H2AX from Millipore; CDC25A from Abcam; ATR from Genetex; and heat shock protein 90 (HSP90) from D. Toft (Mayo Clinic).
Tissue culture
SKOV3 cells (V. Shridhar, Mayo Clinic) and OVCAR-8 cells (D. Scudiero, National Cancer Institute, Frederick, MD) were cultured in RPMI 1640 containing 8% fetal bovine serum and 1 mM glutamine. PEO1 and PEO4 cells (F. Couch, Mayo Clinic) were cultured in DMEM containing 10% heat-inactivated fetal bovine serum, 100 µM nonessential amino acids, 10 µg/ml insulin, 40 units/ml penicillin G, 40 µg/ml streptomycin, and 1 mM glutamine. Lines were genotyped shortly before acquisition and were reinitiated every 2 to 3 months from stocks that were cryopreserved immediately after receipt from the indicated sources.
To assess colony formation in non-transfected OVCAR-8 and SKOV3 cells, 200 cells per well (in 6-well dishes) were plated, allowed to adhere 4–6 h, treated with the indicated agents, and allowed to form colonies for 7–9 days. For OVCAR-8 cells transfected with siRNAs, the indicated numbers of cells were plated. PEO1 and PEO4 cells were plated at 1000 cells per dish in 60-mm dishes, allowed to adhere overnight, treated with the indicated agents continuously, and cultured for 14 days. Following incubation, plates were stained with Coomassie Brilliant Blue and scored for colony formation (≥ 50 cells) manually. For clonogenic assays using non-transfected cells, percent survivals of all individual and combination treatments were normalized to cells treated with vehicle only. For clonogenic assays using cells transfected with siRNA, percent survivals at each drug concentration were normalized to the vehicle-treated control for the given siRNA.
Transfection
siRNAs (400 nmol/transfection) were mixed with 5 × 106 cells in 0.2 ml RPMI-1640 containing 8% fetal bovine serum in a 0.4-cm electroporation cuvette and electroporated with two 10-mS, 280-V pulses in a BTX ECM830 square wave electroporator (Harvard Apparatus, Holliston, MA) on two consecutive days. The transfected cells were cultured for 48 h before use. Rad51 SMARTpool siRNA was from Thermo Scientific. Sequences of other siRNAs (from Thermo Scientific) were: ATR-2, 5’-CCUCCGUGAUGUUGCUUGA-3’ (15); Chk1, 5’-AAGCGUGCCGUAGACUGUCCA-3’ (16); BRCA1, 5’-GUGGGUGUUGGACAGUGUA-3’ (17); and luciferase, 5’-CUUACGUGAGUACUUCGA-3’ (18).
Immunoblotting and Cell Cycle Analysis
Logarithmically proliferating cells were exposed to the indicated drugs for 4 h, washed with PBS, and lysed in 2X SDS-PAGE sample buffer (1 × 107 cells/ml). Lysates (2 × 105 cells/lane) were separated by SDS-PAGE, transferred to Immobilon P, and blotted for the indicated antigens. For cell cycle analyses, Logarithmically proliferating OVCAR-8 cells were incubated with one or both drugs for 24 h, released by trypsinization, and analyzed as described (19).
Homologous Recombination Assay
OVCAR-8 cells with stable integration of pDR-GFP, an HR substrate that generates a functional green fluorescent protein (GFP) upon successful HR by I-SceI cleavage, were generated as described (20). For studies with siRNAs, OVCAR-8-DR-GFP cells were electroporated on day one with siRNA (as described above), on day two with siRNA plus 40 µg pCβASceI plasmid (encoding I-SceI), and analyzed for GFP fluorescence on day five.
RESULTS
ATR depletion sensitizes to genotoxic chemotherapy more broadly than Chk1 depletion
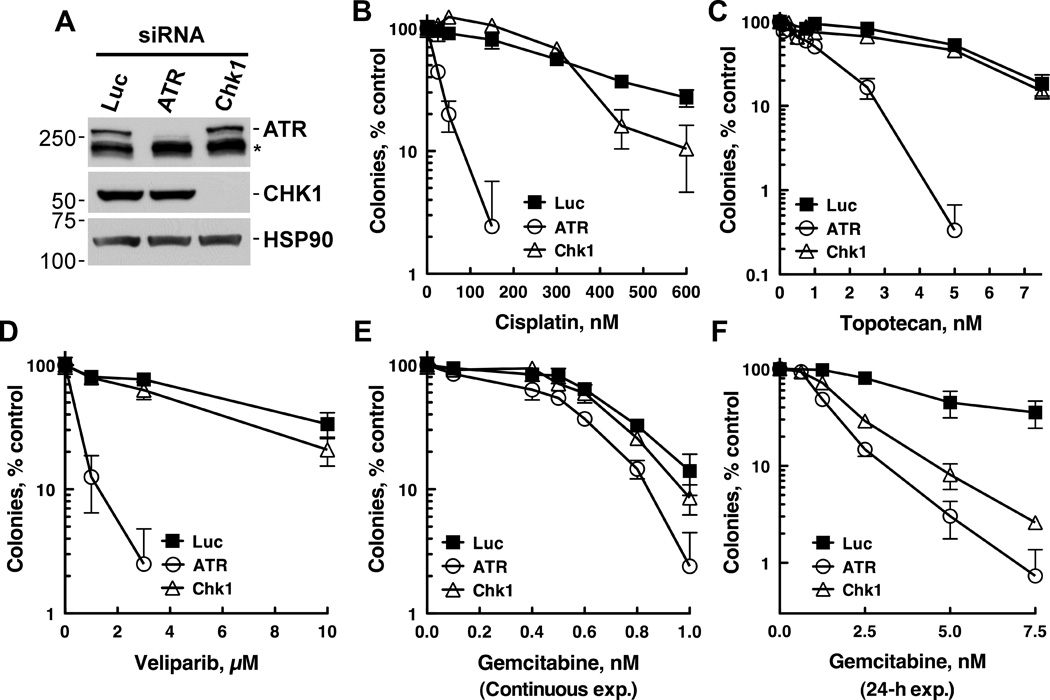
Ovarian cancers are responsive to multiple genotoxic agents, including cisplatin, topotecan, gemcitabine, and veliparib, all of which act by disparate mechanisms. These mechanisms include DNA crosslinking (cisplatin), topoisomerase I poisoning (topotecan), DNA synthesis inhibition by dNTP disruption and DNA polymerase stalling (gemcitabine), and PARP inhibition (veliparib). To address how disabling Chk1 versus ATR affects the sensitivity of ovarian cancer cells to these agents, we initially used siRNAs to deplete ATR and Chk1. As shown in Fig. 1, depletion of ATR (Fig. 1A) sensitized OVCAR-8 cells to continuous cisplatin (Fig. 1B), topotecan (Fig. 1C), and veliparib (Fig. 1D) exposure. In contrast, Chk1 depletion did not affect the cytotoxicity of these agents (Fig. 1B, C, D). Interestingly, neither ATR nor Chk1 depletion sensitized OVCAR-8 cells to gemcitabine under these continuous exposure conditions (Fig. 1E), possibly because gemcitabine metabolites remain trapped in the cells longer than ATR remains suppressed (about 72 h after siRNA transfection, data not shown). In accord with this possibility, ATR and Chk1 depletion effectively sensitized the cells to a 24-h gemcitabine exposure (Fig. 1F).
Figure 1. ATR depletion broadly sensitizes to multiple chemotherapy agents, whereas Chk1 depletion selectively sensitizes to gemcitabine.
OVCAR-8 cells were transfected with control (Luc), ATR, or Chk1 siRNAs. 48 h after transfection, cells were trypsinized and used to analyze ATR and Chk1 expression (A) or in clonogenic assays (B–F). For clonogenic assays, cells (250 per well) were plated, allowed to adhere for 4–6 h, and treated with cisplatin (B), topotecan (C), veliparib (D), or gemcitabine (E) for 8 d. In panel F, after cells were allowed to adhere for 4 h, they were treated with gemcitabine for 24 h, washed, and cultured for 8 days. A representative experiment from 3 independent experiments is shown.
The ATR inhibitor VE-821 also sensitizes more broadly to chemotherapy
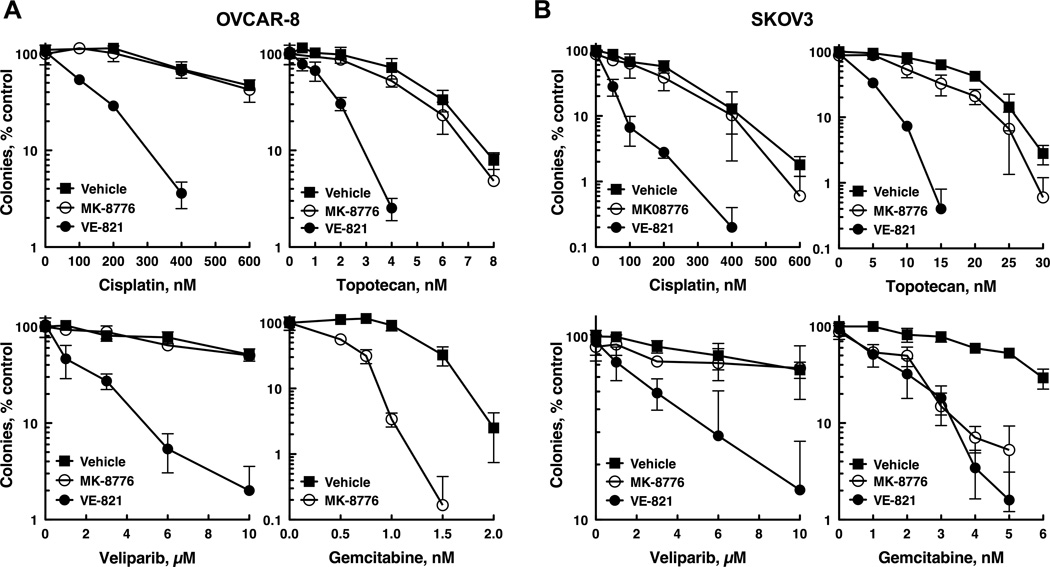
In further experiments, we explored whether ATR and Chk1 inhibitors caused effects similar to those seen with ATR and Chk1 siRNAs. For these studies we used VE-821, a potent ATR inhibitor (Ki ~ 13 nM) with high selectively for ATR versus other phosphoinositol 3-kinase-like kinases, including ATM (21). To inhibit Chk1, we used MK-8776 (SCH 900776), which effectively inhibits Chk1 (Ki ~ 3 nM) and sensitizes cells to antimetabolites but does not affect the closely related kinase Chk2 (13, 22, 23). As was observed in cells depleted of ATR, VE-821 sensitized OVCAR-8 (Fig. 2A), SKOV3 (Fig. 2B), and PEO1 (Supp. Fig. 1) ovarian cancer cells to cisplatin, topotecan and veliparib. MK-8776, on the other hand, selectively sensitized these cell lines to gemcitabine but not the other agents (Figs. 2A, B and Supp. Fig. 1), just as was observed with Chk1 siRNA. Consistent with these findings, parallel studies with another Chk1 inhibitor, LY 2603618, showed that this agent also robustly sensitized SKOV3, OVCAR-8, and PEO1 cells to gemcitabine (Supp. Fig. 2). Taken together, the findings in Figs. 1 and 2 indicate that 1) disruption of ATR signaling broadly sensitizes ovarian cancer cells to genotoxic chemotherapies that act by disparate mechanisms; 2) disabling Chk1 selectively sensitizes to gemcitabine; and 3) VE-821 and MK-8776 phenocopy the effects of depleting ATR and Chk1, respectively, suggesting that these agents are sensitizing cells by inhibiting the intended checkpoint kinases.
Figure 2. The ATR inhibitor VE-821 and the Chk1 inhibitor MK-8776 phenocopy the effects of ATR and Chk1 depletion.
OVCAR-8 cells were trypsinized, plated as single cells, allowed to adhere 4 h, treated with 0.3 µM MK-8776 or 1 µM VE-821 plus cisplatin, topotecan, veliparib, or gemcitabine for 8 d. The experiment shown is representative of 4 (SKOV3) and 5 (OVCAR-8) independent experiments.
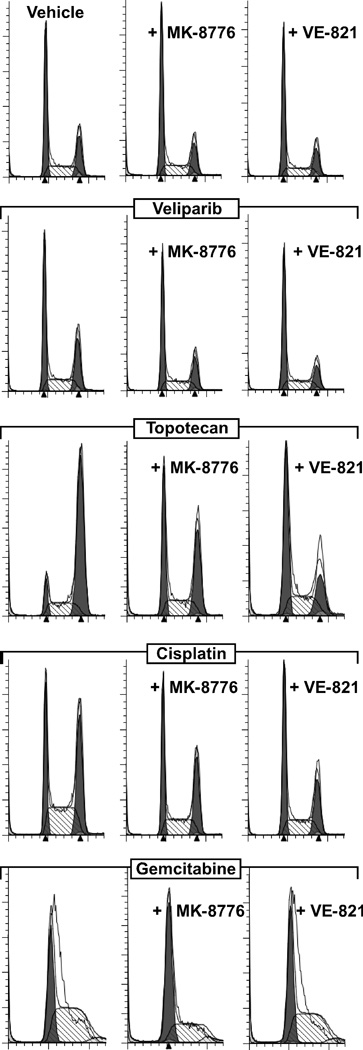
VE-821 and MK-8776 abrogate chemotherapy-induced cell cycle arrest
We next tested whether these checkpoint inhibitors could override the cell cycle arrests induced by these chemotherapy agents. Consistent with the lack of effect of PARP inhibition in cells with functional homologous recombination (HR), veliparib minimally affected the cell cycle of OVCAR-8 cells, and co-treatment with MK-8776 or VE-821 had little additional impact (Fig. 3). In contrast, in cells exposed to cisplatin or topotecan, the addition of MK-8776 or VE-821 reduced the S phase (cisplatin) and G2/M (cisplatin and topotecan) accumulations induced by these agents, whereas these checkpoint inhibitors modesty increased the G1 arrest induced by gemcitabine. Collectively, these results indicate that both checkpoint inhibitors effectively override the arrest induced by topotecan and cisplatin but do not allow gemcitabine-treated cells to bypass the disruption of replication caused by this antimetabolite.
Figure 3. MK-8776 and VE-821 disrupt chemotherapy-induced cell cycle checkpoints.
OVCAR-8 cells were co-treated with vehicle, 0.1 µM MK-8776, or 1 µM VE-821 plus 10 µM veliparib, 20 nM topotecan, 0.6 µM cisplatin, or 5 nM gemcitabine for 24 h and analyzed by flow cytometry.
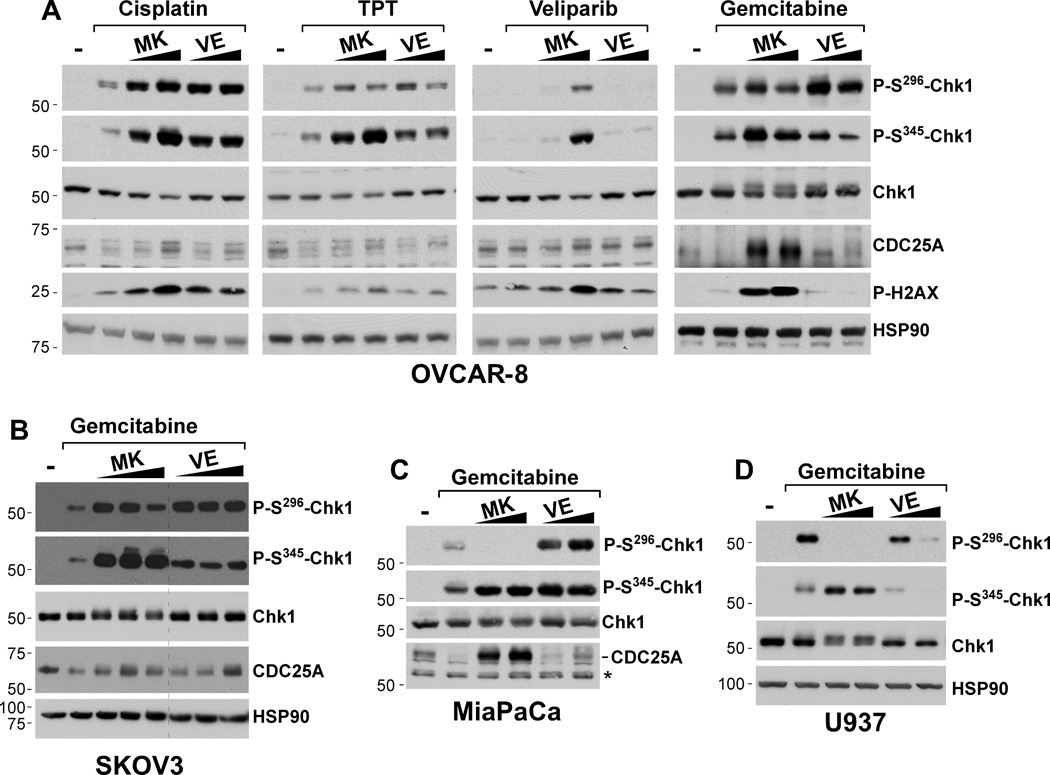
VE-821 and MK-8776 do not effectively block ATR-mediated Chk1 phosphorylation and Chk1 autophosphorylation in ovarian cancer cells
The observation that VE-821 and MK-8776 abrogate the cell cycle arrest induced by cisplatin and topotecan suggests that they are inhibiting the ATR-Chk1 signaling pathway. To further evaluate the impacts of these agents on this pathway, we next assessed their effects on ATR-mediated Chk1 phosphorylation (Ser345) and Chk1 autophosphorylation (Ser296). Consistent with previous studies of Chk1 inhibitors (9), MK-8776 (0.3 and 1 µM) caused increased Chk1 Ser345 phosphorylation and H2AX Ser139 phosphorylation, a marker of DNA damage, in OVCAR-8 cells co-treated with the Chk1 inhibitor plus cisplatin, topotecan, veliparib, or gemcitabine (Fig. 4A) and in SKOV3 ovarian cells treated with gemcitabine (Fig. 4B). This increased Ser345 phosphorylation has been attributed to disruption of PP2A-mediated dephosphorylation on this site and increased DNA damage that accumulates when Chk1 cannot regulate replication (9). In contrast, the effects of MK-8776 on Chk1 autophosphorylation (Ser296) revealed unexpected results. Previous work showed that Chk1 Ser296 autophosphorylation is blocked by MK-8776 and other Chk1 inhibitors (13, 22, 23). In agreement with these earlier results, we observed that MK-8776 (0.3 and 1 µM) effectively blocked gemcitabine-induced Chk1 Ser296 phosphorylation in MiaPaCa pancreatic cancer cells (Fig. 4C) and U937 leukemia cells (Fig. 4D). Surprisingly, however, MK-8776 did not prevent Chk1 Ser296 autophosphorylation in OVCAR-8 cells treated with cisplatin, and this effect was seen over a wide range of cisplatin concentrations that spanned from twice (1 µM) to ten times the IC50 (50 µM)(Supp. Fig. 3). Similarly, MK-8776 did not blunt Ser296 autophosphorylation in cells exposed to gemcitabine, and topotecan (Fig. 4A). Indeed, with all of the agents tested, MK-8776 actually increased genotoxin-induced Chk1 phosphorylation. MK-8776 likewise caused increased gemcitabine-induced Chk1 Ser296 phosphorylation in SKOV3 ovarian cancer cells (Fig. 4B). Taken together, the results in Fig. 4 demonstrate that MK-8776 blocks Chk1 autophosphorylation in some cells but not others.
Figure 4. MK-8776 and VE-821 do not block Chk1 phosphorylation.
(A) OVCAR-8 cells were pretreated with vehicle (−), MK-8776 (0.3 and 1.0 µM), or VE-821 (1.0 or 4.0 µM) for 15 min and then exposed to cisplatin (4 µM), topotecan (TPT, 20 nM), veliparib (10 µM), or gemcitabine (Gem, 20 nM) for 4 h in the continued presence of MK-8776 or VE-821. (B) SKOV3 cells were pretreated with vehicle (−), MK-8776 (0.3, 1.0, and 4 µM), or VE-821 (1.0, 4.0, and 6.0 µM) for 15 min and then exposed to 20 nM gemcitabine for 4 h. (C, D) MiaPaCa (C) or U937 (D) cells were pretreated with vehicle (−), MK-8776 (0.3 and 1.0 µM), or VE-821 (1.0 or 4.0 µM) for 15 min and then exposed to gemcitabine (40 nM, MiaPaCa cells; 20 nM U937 cells) for 4 h. Cell lysates were then immunoblotted for the indicated antigens.
In parallel analyses, we also evaluated the effects of the ATR inhibitor VE-821 on the ATR-Chk1 pathway in ovarian cancer cells. As reported previously (and similar to what we observed with MK-8776), VE-821 (1 and 4 µM) enhanced H2AX phosphorylation on Ser139 induced by topotecan, and cisplatin in OVCAR-8 cells (Fig. 4A), suggesting that ATR inhibition caused the accumulation of additional DNA damage. Surprisingly, VE-821 did not block ATR-mediated Ser345 Chk1 or Ser296 autophosphorylation triggered by gemcitabine, topotecan, or cisplatin (Fig. 4A). Comparable results were also seen in gemcitabine-treated SKOV3 cells, even at concentrations up to 6 µM VE-821 (Fig. 4B). Analyses of the effects of VE-821 in other cell lines revealed additional complexity. Whereas, VE-821 (1 and 4 µM) did not diminish Chk1 Ser345 (or Ser296) phosphorylation in MiaPaCa cells (Fig. 4C), the higher VE-821 concentration did disrupt these phosphorylation events in U937 cells (Fig. 3D). These results demonstrate that VE-821 does not effectively disrupt ATR-mediated Chk1 phosphorylation in several cell types, including ovarian cancer cells.
VE-821 and MK-8776 disrupt chemotherapy-induced CDC25A degradation
To further examine the impact of ATR and Chk1 inhibitors on this signaling pathway, we assessed the effects of MK-8776 and VE-821 on levels of CDC25A, a Chk1 substrate that is targeted for proteasomal degradation following Chk1-mediated phosphorylation. As expected for agents that activate Chk1, gemcitabine, topotecan, and cisplatin caused decreases in CDC25A levels (Fig. 4A). These genotoxin-induced reductions of CDC25A were blocked by MK-8776 and VE-821, thus demonstrating that even though these checkpoint inhibitors did not block (and in some cases stimulated) Chk1 phosphorylation, they still disrupted the checkpoint signal.
Disabling ATR disrupts HR repair, a pathway that protects cells from cisplatin, topotecan, and veliparib, and further sensitizes cells with disabled HR to these agents
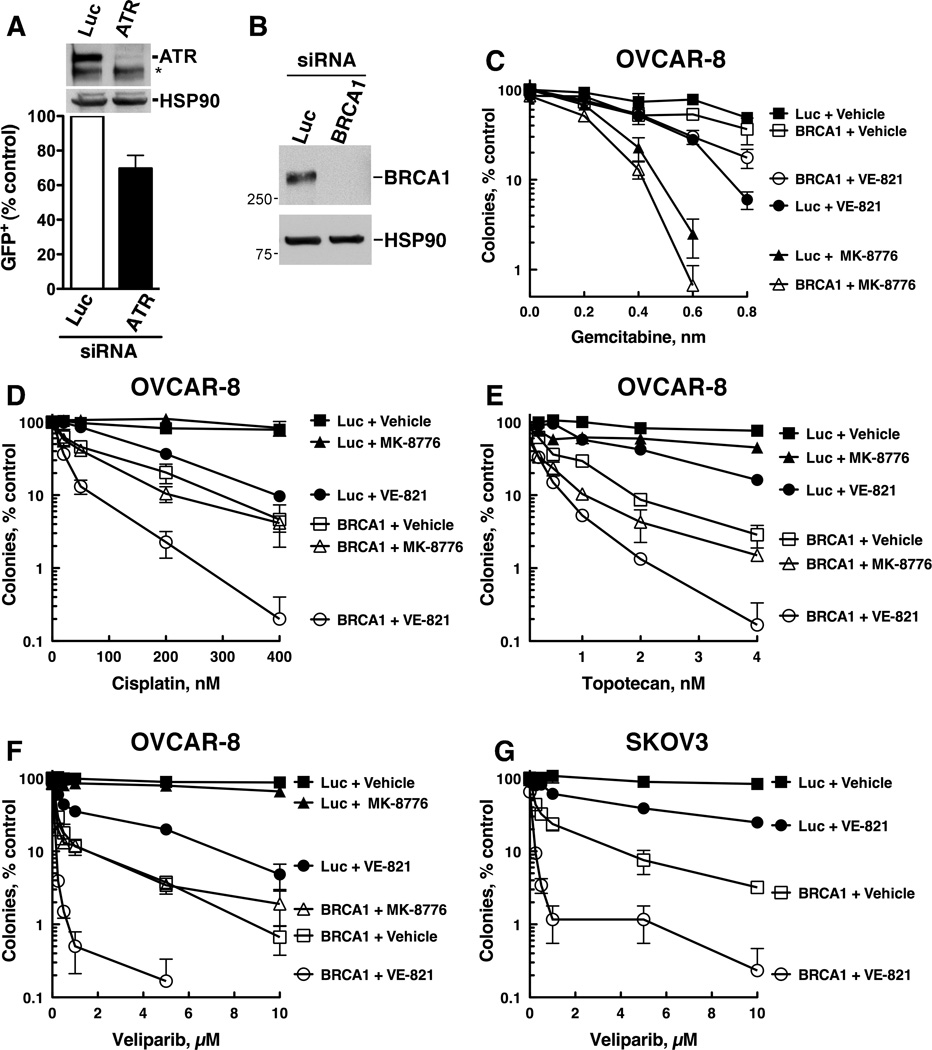
Our finding that disabling Chk1 did not sensitize to cisplatin, topotecan, or veliparib indicates that other ATR substrates help protect cells from the lesions induced by these agents. Because ATR also phosphorylates and regulates proteins that participate in HR repair, such as BRCA1 (Reviewed in ref. 24)], and because cisplatin, topotecan, and veliparib cause damage that is repaired by HR (25–28), we reasoned that ATR might participate in HR. Consistent with this idea, ATR depletion reduced HR-mediated repair of DR-GFP, a stably integrated HR substrate (Fig. 5A), following transfection of the I-SceI nuclease that cleaves between non-functional GFP repeats, thus promoting HR repair.
Figure 5. ATR inhibition further sensitizes cells with defective HR to cisplatin, topotecan, and veliparib.
(A) OVCAR-8 cells that have stably integrated DR-GFP HR substrate were transfected with pCβASceI plasmid plus control (Luc) or ATR siRNA and examined for GFP fluorescence 72 h after plasmid transfection. Mean +/− S.D; n = 3; *P = 0.02 by paired t-test. * indicates nonspecific band. OVCAR-8 (B–F) or SKOV3 (G) cells were transfected with control (Luc) or BRCA1 siRNA. 48 h after transfection, cells were trypsinized and used to analyze BRCA1 expression (B, OVCAR-8 cells) and for clonogenic assays (C–G). For clonogenic assays, cells were plated, allowed to adhere for 6 h, and treated 0.3 µM MK-8776 or 1 µM VE-821 plus gemcitabine (C) cisplatin (D), topotecan (E), or veliparib (F, G) for 8 d. A representative experiment from 3 independent experiments is shown.
To examine potential interactions between ATR and HR, we next, asked how disabling HR by depleting BRCA1 (Fig. 5B), alone and in combination with ATR or Chk1 inhibition, affected responses to these agents. These studies revealed several noteworthy findings. First, BRCA1 depletion did not sensitize to gemcitabine (Fig. 5C), consistent with a previous report (26), but did robustly sensitize to cisplatin, topotecan, and veliparib (Fig. 5D–G). Interestingly, these results show that ATR depletion—but not Chk1 depletion— sensitizes to the same agents that cause damage repaired by HR (i.e., cisplatin, topotecan, and veliparib—see Fig. 1). These results, therefore, suggest that ATR regulation of HR contributes to cell survival more than ATR-mediated activation of Chk1 in cells treated with agents that induce lesions repaired by HR. Second, even when BRCA1 was depleted, MK-8776 did not further sensitize cells to any of the agents (Fig. 5C–F), indicating that even when HR was disabled, Chk1 did not facilitate survival. Third, MK-8776 could still robustly sensitize BRCA1-depleted cells to gemcitabine, although this sensitization was no greater than in control (Luc) cells (Fig. 5C). Fourth, even when HR was disabled by BRCA1 depletion, VE-821 additionally sensitized cells to cisplatin and topotecan (Fig. 5D and E). Fifth, VE-821 was particularly effective at further sensitizing BRCA1-depleted cells to veliparib (Fig. 5F), a result that was also observed in BRCA1-depleted SKOV3 cells (Fig. 5G and Supp. Fig. 4). Taken together, these results indicate that even in cells with defects in HR, ATR still plays a critical role in promoting the survival and proliferation of cells exposed to cisplatin, topotecan, and especially veliparb, suggesting that in addition to regulating HR, ATR has additional roles in protecting tumor cells from damage inflicted by these agents.
DISCUSSION
These studies were designed to compare the impact of disabling ATR versus Chk1 using siRNA or small molecule inhibitors in ovarian cancer cells exposed to chemotherapy agents that are representatives of four classes of agents with activity in this disease. This analysis demonstrated that the ATR inhibitor VE-821, like ATR siRNA, sensitized to a wide range of genotoxic stresses. In contrast, Chk1 depletion, like Chk1 inhibition, showed a much more restricted sensitization pattern. These observations have important implications for current efforts to develop Chk1 and ATR inhibitors as described in greater detail below.
Initial studies of ATR and Chk1 inhibitors used agents such as caffeine or UCN-01, which inhibit ATR or Chk1, respectively (29–32), but have subsequently been shown to inhibit multiple enzymes (33–37). More recent studies have focused on increasingly selective kinase inhibitors. For example, the Chk1 inhibitor AZD7762 sensitizes to a wide range of anticancer therapies, including gemcitabine, topotecan, cisplatin, ionizing radiation, and even the microtubule disruptor paclitaxel (38–42). Notably, however, in addition to potently inhibiting Chk1 (Ki ~ 4 nM), AZD7762 also inhibits Chk2 with similarly potency and shows less than 10-fold selectivity for multiple members of the CAMK, AGC, and Src families of kinases (38). Thus, some of the effects of this agent may be attributable to inhibition of other kinases. Similarly, VE-821, one of the first selective ATR inhibitors to be reported, also sensitizes cells to multiple agents, including cisplatin, camptothecin, etoposide, and ionizing radiation (21). Therefore, even though these Chk1 and ATR inhibitors sensitize to similar types of genotoxic chemotherapy agents, it remains unclear whether these overlapping sensitization profiles are due solely to Chk1 and ATR inhibition or whether they are caused by inhibition of other kinases. The present studies provide insight into this question by first comparing the effects of ATR and Chk1 depletion (using siRNAs), and then performing a head-to-head comparison of VE-821 with MK-8776, an agent identified based on its ability to selectively inhibit Chk1 relative to Chk2 (22).
When the effects of ATR vs. Chk1 siRNAs were compared, ATR knockdown sensitized cells to cisplatin, topotecan, gemcitabine, and veliparib (Fig. 1). Consistent with the ATR siRNA results, VE-821 also sensitized multiple ovarian cancer cell lines to these same agents (Fig. 2). In marked contrast, Chk1 depletion only sensitized to gemcitabine (Fig. 1). Similarly, even though MK-8776 effectively overrode the cell cycle arrests induced by topotecan and cisplatin (thus demonstrating effective Chk1 inhibition – Fig. 3), this Chk1 inhibitor only sensitized to gemcitabine (Fig. 2). Taken together, these results indicate that ATR protects ovarian cancer cells from multiple genotoxic stresses, whereas the role of Chk1 appears limited to gemcitabine, a result consistent with recent reports suggesting that MK-8776 preferentially sensitizes to the antimetabolites hydroxyurea, gemcitabine and cytarabine (13, 22).
One question that emerges from these studies is why ATR and Chk1 have such different pro-survival effects in cells exposed to genotoxins that act by disparate mechanisms. With the exception of veliparib, all of these agents disrupt DNA replication and activate checkpoints that block cell cycle progression, events that require Chk1 signaling. Nonetheless, disabling Chk1 only sensitized to gemcitabine, suggesting that other ATR-regulated events are important for the other agents. Indeed, our studies raise the possibility that one such event may be the mobilization of the HR machinery because the agents that cause damage repaired by HR (cisplatin, topotecan, and veliparib) all require ATR—but not Chk1—to promote survival. Notably, however, because ATR inhibition further sensitizes cells with disabled HR (i.e., BRCA1 depletion—see Fig. 5F, G– or BRCA2 mutation, Fig. S1), ATR must also control other checkpoint and repair processes that promote survival.
Several studies have addressed how disabling Chk1 sensitizes cells to replication stress, but no unifying picture has emerged. On the one hand, inappropriate progression through S phase, premature exit from G2, and mitotic catastrophe have been proposed as the mechanism by which cells die when Chk1 is inhibited during replication stress, especially when p53 signaling is disabled (Reviewed in ref. 9). In contrast, other studies suggest that override of these checkpoints does not correlate with toxicity (43), and consistent with these prior findings, we observed that disabling Chk1 actually augmented gemcitabine-induced arrest in G1/S (Fig. 3) while at the same time sensitizing to gemcitabine. On the other hand, recent studies found that stalled replication forks were cleaved by the endonucleases MRE11 (44) or MUS81 (45) when Chk1 was disabled. This aberrant cleavage then caused replication fork collapse, the accumulation of double-stranded DNA breaks, and cell death. Given these disparate findings, it remains unclear if these and/or other mechanisms participate in the toxicity of the gemcitabine+MK-8776 combination in ovarian cancers, but future studies that address these questions may help identify potential biomarkers for a clinical trial of such a drug combination.
Our studies to further characterize the effects of these checkpoint inhibitors on ovarian cancer cells revealed several unexpected findings. Previous studies showed that MK-8776 and other Chk1 inhibitors block Chk1 autophosphorylation on Ser296 (38, 46–48) and that VE-821 abrogates ATR-mediated Chk1 Ser345 phosphorylation (21), suggesting that these phosphorylation events may provide an effective way to assess disruption of this signaling pathway in clinical trials (9, 48). The present studies, however, show that even when checkpoint inhibitors override the checkpoint signal (as demonstrated by CDC25A preservation and cell cycle arrest – Figs. 3 and 4), these Chk1 phosphorylation events may not be reliable markers of pathway inhibition. In particular, VE-821 concentrations that sensitized to cisplatin, topotecan, or gemcitabine did not block ATR-mediated Chk1 Ser345 phosphorylation in ovarian cancer cells (Fig. 4A and B) even though VE-821 blocked this phosphorylation in U937 leukemia cells (Fig. 4D). In a similar vein, MK-8776 concentrations that enhanced gemcitabine-induced cytotoxicity in ovarian cancer cells failed to inhibit Chk1 autophosphorylation on Ser296 (Fig. 4A and B) even though the expected effects of MK-8776 on Chk1 Ser296 phosphorylation were readily detected in pancreatic cancer and leukemia cell lines (Fig. 4C and D). Collectively, our observations raise the possibility that these Chk1 sites might not be appropriate biomarkers to assess pathway inhibition in all cell types. Equally important, the ability of VE-821 to sensitize cells to cisplatin and topotecan at concentrations that do not inhibit Chk1 Ser345 phosphorylation suggests that ATR inhibition might sensitize cells by altering phosphorylation of other, currently unappreciated substrates. Whether phosphorylation of these substrates is more sensitive than phosphorylation of Chk1, a situation analogous to differential effects of rapamycin on phosphorylation of substrates by the ATR-related kinase mTOR (49, 50), remains to be explored.
Emerging data suggest that high grade serous ovarian cancer, the most common histological subtype, can be categorized into tumors with defects in HR (which includes mutations in BRCA1 and BRCA2) and tumors that are proficient in HR (14). Importantly, our results demonstrate that although MK-8776 does not further sensitize cells with HR defects to any of the genotoxic chemotherapies tested here, this agent still sensitizes cell deficient in BRCA1 (OVCAR-8 treated with siRNA, Fig. 5C) or BRCA2 (PEO1, Fig. S1) to gemcitabine. In stark contrast, even in cells with defective HR, which are hypersensitive to cisplatin, topotecan, and veliparib, VE-821 further sensitized the cells to these chemotherapy agents (Fig. 5).
Because Chk1 was the first ATR substrate identified and was shown to mediate some of the effects of ATR activation, much of the effort in drug development has focused on Chk1 inhibitors. The present demonstration that VE-821, like ATR siRNA, sensitizes to a much broader range of genotoxic stresses, including highly active anticancer agents such as cisplatin, topoisomerase I poisons, and veliparib, suggests that further investigation of ATR inhibitors and their mechanism of sensitization might also be worthwhile, especially in cancers with defects in HR.
Supplementary Material
ACKNOWLEDGMENTS
We thank Pam Becker and Deb Strauss for assistance with manuscript preparation, and Maria Jasin for the pDR-GFP plasmid.
Financial Support: Funding in part by the Mayo Clinic Ovarian Cancer SPORE P50 CA136393, a grant from the Fred C. and Katherine Andersen Foundation, and a Mayo Clinic Eagles Pilot Grant.
S.H. Kaufmann received a commercial research grant from Schering-Plough for correlative studies of a clinical trial of MK-8776 ($10,000 or more).
Abbreviations
- 9-1-1
Rad9-Hus1-Rad1 complex
- HSP90
heat shock protein 90
- HR
homologous recombination
- PARP
poly(ADP-ribose)-polymerase
- RPA
replication protein A
- siRNA
small inhibitory RNA
Footnotes
Potential Conflicts of Interests: The other authors have no conflicts to report.
Authors Contributions:
Conception and design: C. Huntoon, K. Flatten, A. Wahner-Hendrickson, A. Huehls, S. Kaufmann, L. Karnitz
Development of methodology: C. Huntoon, S. Kaufmann, L. Karnitz
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): C. Huntoon, K. Flatten, A. Wahner-Hendrickson, A. Huehls, S. Sutor, S. Kaufmann, L. Karnitz
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): S. Kaufmann, L. Karnitz
Writing, review, and/or revision of the manuscript: S. Kaufmann, L. Karnitz
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): C. Huntoon, K. Flatten, S. Kaufmann, L. Karnitz
Study supervision: S. Kaufmann, L. Karnitz
REFERENCES
- 1.Vaughan S, Coward JI, Bast RC, Jr, Berchuck A, Berek JS, Brenton JD, et al. Rethinking ovarian cancer: recommendations for improving outcomes. Nat Rev Cancer. 2011;11:719–725. doi: 10.1038/nrc3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 4.Stokes MP, Rush J, Macneill J, Ren JM, Sprott K, Nardone J, et al. Profiling of UV-induced ATM/ATR signaling pathways. Proc Natl Acad Sci U S A. 2007;104:19855–19860. doi: 10.1073/pnas.0707579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mu JJ, Wang Y, Luo H, Leng M, Zhang J, Yang T, et al. A proteomic analysis of ataxia telangiectasia-mutated (ATM)/ATM-Rad3-related (ATR) substrates identifies the ubiquitin-proteasome system as a regulator for DNA damage checkpoints. J Biol Chem. 2007;282:17330–17334. doi: 10.1074/jbc.C700079200. [DOI] [PubMed] [Google Scholar]
- 6.Smolka MB, Albuquerque CP, Chen SH, Zhou H. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc Natl Acad Sci U S A. 2007;104:10364–10369. doi: 10.1073/pnas.0701622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Toledo LI, Murga M, Fernandez-Capetillo O. Targeting ATR and Chk1 kinases for cancer treatment: a new model for new (and old) drugs. Mol Oncol. 2011;5:368–373. doi: 10.1016/j.molonc.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai Y, Grant S. New insights into checkpoint kinase 1 in the DNA damage response signaling network. Clin Cancer Res. 2010;16:376–383. doi: 10.1158/1078-0432.CCR-09-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma CX, Janetka JW, Piwnica-Worms H. Death by releasing the breaks: CHK1 inhibitors as cancer therapeutics. Trends Mol Med. 2011;17:88–96. doi: 10.1016/j.molmed.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wagner JM, Kaufmann SH. Targeting ATR and Chk1 kinases for cancer treatment: a new model for new (and old) drugs. Pharmaceuticals. 2010;3 [Google Scholar]
- 11.Wagner JM, Karnitz LM. Cisplatin-induced DNA damage activates replication checkpoint signaling components that differentially affect tumor cell survival. Mol Pharmacol. 2009;76:208–214. doi: 10.1124/mol.109.055178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilsker D, Bunz F. Loss of ataxia telangiectasia mutated- and Rad3-related function potentiates the effects of chemotherapeutic drugs on cancer cell survival. Mol Cancer Ther. 2007;6:1406–1413. doi: 10.1158/1535-7163.MCT-06-0679. [DOI] [PubMed] [Google Scholar]
- 13.Montano R, Chung I, Garner KM, Parry D, Eastman A. Preclinical development of the novel Chk1 inhibitor SCH900776 in combination with DNA-damaging agents and antimetabolites. Mol Cancer Ther. 2012;11:427–438. doi: 10.1158/1535-7163.MCT-11-0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Romero I, Bast RC., Jr Minireview: human ovarian cancer: biology, current management, and paths to personalizing therapy. Endocrinology. 2012;153:1593–1602. doi: 10.1210/en.2011-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Casper AM, Durkin SG, Arlt MF, Glover TW. Chromosomal instability at common fragile sites in Seckel syndrome. Am J Hum Genet. 2004;75:654–660. doi: 10.1086/422701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao H, Watkins JL, Piwnica-Worms H. Disruption of the checkpoint kinase 1/cell division cycle 25A pathway abrogates ionizing radiation-induced S and G2 checkpoints. Proc Natl Acad Sci USA. 2002;99:14795–14800. doi: 10.1073/pnas.182557299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bartz SR, Zhang Z, Burchard J, Imakura M, Martin M, Palmieri A, et al. Small interfering RNA screens reveal enhanced cisplatin cytotoxicity in tumor cells having both BRCA network and TP53 disruptions. Mol Cell Biol. 2006;26:9377–9386. doi: 10.1128/MCB.01229-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 19.Mesa RA, Loegering D, Powell HL, Flatten K, Arlander SJ, Dai NT, et al. Heat shock protein 90 inhibition sensitizes acute myelogenous leukemia cells to cytarabine. Blood. 2005;106:318–327. doi: 10.1182/blood-2004-09-3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reaper PM, Griffiths MR, Long JM, Charrier JD, Maccormick S, Charlton PA, et al. Selective killing of ATM- or p53-deficient cancer cells through inhibition of ATR. Nature Chem Biol. 2011;7:428–430. doi: 10.1038/nchembio.573. [DOI] [PubMed] [Google Scholar]
- 22.Guzi TJ, Paruch K, Dwyer MP, Labroli M, Shanahan F, Davis N, et al. Targeting the replication checkpoint using SCH 900776, a potent and functionally selective CHK1 inhibitor identified via high content screening. Mol Cancer Ther. 2011;10:591–602. doi: 10.1158/1535-7163.MCT-10-0928. [DOI] [PubMed] [Google Scholar]
- 23.Schenk EL, Koh BD, Flatten KS, Peterson KL, Parry D, Hess AD, et al. Effects of selective checkpoint kinase 1 inhibition on cytarabine cytotoxicity in acute myelogenous leukemia cells in vitro. Clin Cancer Res. 2012;18:5364–5373. doi: 10.1158/1078-0432.CCR-12-0961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet. 2007;8:735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- 25.Bhattacharyya A, Ear US, Koller BH, Weichselbaum RR, Bishop DK. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J Biol Chem. 2000;275:23899–23903. doi: 10.1074/jbc.C000276200. [DOI] [PubMed] [Google Scholar]
- 26.Fedier A, Steiner RA, Schwarz VA, Lenherr L, Haller U, Fink D. The effect of loss of Brca1 on the sensitivity to anticancer agents in p53-deficient cells. Int J Oncol. 2003;22:1169–1173. [PubMed] [Google Scholar]
- 27.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 28.Huehls AM, Wagner JM, Huntoon CJ, Karnitz LM. Identification of DNA Repair Pathways That Affect the Survival of Ovarian Cancer Cells Treated with a Poly(ADP-Ribose) Polymerase Inhibitor in a Novel Drug Combination. Mol Pharmacol. 2012;82:767–776. doi: 10.1124/mol.112.080614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sarkaria JN, Busby EC, Tibbetts RS, Roos P, Taya Y, Karnitz LM, et al. Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res. 1999;59:4375–4382. [PubMed] [Google Scholar]
- 30.Hall-Jackson CA, Cross DA, Morrice N, Smythe C. ATR is a caffeine-sensitive, DNA-activated protein kinase with a substrate specificity distinct from DNA-PK. Oncogene. 1999;18:6707–6713. doi: 10.1038/sj.onc.1203077. [DOI] [PubMed] [Google Scholar]
- 31.Busby EC, Leistritz DF, Abraham RT, Karnitz LM, Sarkaria JN. The Radiosensitizing Agent 7-Hydroxystaurosporine (UCN-01) Inhibits the DNA Damage Checkpoint Kinase hChk1. Cancer Res. 2000;60:2108–2112. [PubMed] [Google Scholar]
- 32.Graves PR, Yu L, Schwarz JK, Gales J, Sausville EA, O'Connor PM, et al. The Chk1 Protein Kinase and the Cdc25C Regulator Pathways are Targets of the Anticancer Agent UCN-01. J Biol Chem. 2000;275:5600–5605. doi: 10.1074/jbc.275.8.5600. [DOI] [PubMed] [Google Scholar]
- 33.Blasina A, Price BD, Turenne GA, McGowan CH. Caffeine inhibits the checkpoint kinase ATM. Curr Biol. 1999;9:1135–1138. doi: 10.1016/s0960-9822(99)80486-2. [DOI] [PubMed] [Google Scholar]
- 34.Nishijima H, Nishitani H, Saito N, Nishimoto T. Caffeine mimics adenine and 2'-deoxyadenosine, both of which inhibit the guanine-nucleotide exchange activity of RCC1 and the kinase activity of ATR. Genes Cells. 2003;8:423–435. doi: 10.1046/j.1365-2443.2003.00644.x. [DOI] [PubMed] [Google Scholar]
- 35.Wharton W, Goz B. Induction of alkaline phosphatase activity in HeLa cells. Inhibition by xanthine derivatives and thermostability studies. Biochem Pharmacol. 1979;28:763–768. doi: 10.1016/0006-2952(79)90356-3. [DOI] [PubMed] [Google Scholar]
- 36.Beavo JA, Rogers NL, Crofford OB, Hardman JG, Sutherland EW, Newman EV. Effects of xanthine derivatives on lipolysis and on adenosine 3',5'-monophosphate phosphodiesterase activity. Mol Pharmacol. 1970;6:597–603. [PubMed] [Google Scholar]
- 37.Kieseier BC, Kiefer R, Clements JM, Miller K, Wells GM, Schweitzer T, et al. Matrix metalloproteinase-9 and-7 are regulated in experimental autoimmune encephalomyelitis. Brain. 1998;121(Pt 1):159–166. doi: 10.1093/brain/121.1.159. [DOI] [PubMed] [Google Scholar]
- 38.Zabludoff SD, Deng C, Grondine MR, Sheehy AM, Ashwell S, Caleb BL, et al. AZD7762, a novel checkpoint kinase inhibitor, drives checkpoint abrogation and potentiates DNA-targeted therapies. Mol Cancer Ther. 2008;7:2955–2966. doi: 10.1158/1535-7163.MCT-08-0492. [DOI] [PubMed] [Google Scholar]
- 39.McNeely S, Conti C, Sheikh T, Patel H, Zabludoff S, Pommier Y, et al. Chk1 inhibition after replicative stress activates a double strand break response mediated by ATM and DNA-dependent protein kinase. Cell Cycle. 2010;9:995–1004. doi: 10.4161/cc.9.5.10935. [DOI] [PubMed] [Google Scholar]
- 40.Mitchell JB, Choudhuri R, Fabre K, Sowers AL, Citrin D, Zabludoff SD, et al. In vitro and in vivo radiation sensitization of human tumor cells by a novel checkpoint kinase inhibitor, AZD7762. Clin Cancer Res. 2010;16:2076–2084. doi: 10.1158/1078-0432.CCR-09-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morgan MA, Parsels LA, Zhao L, Parsels JD, Davis MA, Hassan MC, et al. Mechanism of radiosensitization by the Chk1/2 inhibitor AZD7762 involves abrogation of the G2 checkpoint and inhibition of homologous recombinational DNA repair. Cancer Res. 2010;70:4972–4981. doi: 10.1158/0008-5472.CAN-09-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, McLauchlan H, et al. The selectivity of protein kinase inhibitors: a further update. Biochem J. 2007;408:297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parsels LA, Morgan MA, Tanska DM, Parsels JD, Palmer BD, Booth RJ, et al. Gemcitabine sensitization by checkpoint kinase 1 inhibition correlates with inhibition of a Rad51 DNA damage response in pancreatic cancer cells. Mol Cancer Ther. 2009;8:45–54. doi: 10.1158/1535-7163.MCT-08-0662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thompson R, Montano R, Eastman A. The Mre11 nuclease is critical for the sensitivity of cells to Chk1 inhibition. PLoS ONE. 2012;7:e44021. doi: 10.1371/journal.pone.0044021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Forment JV, Blasius M, Guerini I, Jackson SP. Structure-specific DNA endonuclease Mus81/Eme1 generates DNA damage caused by Chk1 inactivation. PLoS ONE. 2011;6:e23517. doi: 10.1371/journal.pone.0023517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walton MI, Eve PD, Hayes A, Valenti M, De Haven Brandon A, Box G, et al. The preclinical pharmacology and therapeutic activity of the novel CHK1 inhibitor SAR-020106. Mol Cancer Ther. 2010;9:89–100. doi: 10.1158/1535-7163.MCT-09-0938. [DOI] [PubMed] [Google Scholar]
- 47.Walton MI, Eve PD, Hayes A, Valenti MR, De Haven Brandon AK, Box G, et al. CCT244747 Is a Novel Potent and Selective CHK1 Inhibitor with Oral Efficacy Alone and in Combination with Genotoxic Anticancer Drugs. Clin Cancer Res. 2012 doi: 10.1158/1078-0432.CCR-12-1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parsels LA, Qian Y, Tanska DM, Gross M, Zhao L, Hassan MC, et al. Assessment of chk1 phosphorylation as a pharmacodynamic biomarker of chk1 inhibition. Clin Cancer Res. 2011;17:3706–3715. doi: 10.1158/1078-0432.CCR-10-3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choo AY, Yoon SO, Kim SG, Roux PP, Blenis J. Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proc Natl Acad Sci U S A. 2008;105:17414–17419. doi: 10.1073/pnas.0809136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu K, Toral-Barza L, Shi C, Zhang WG, Lucas J, Shor B, et al. Biochemical, cellular, and in vivo activity of novel ATP-competitive and selective inhibitors of the mammalian target of rapamycin. Cancer Res. 2009;69:6232–6240. doi: 10.1158/0008-5472.CAN-09-0299. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.