Abstract
Purpose
Promoter hypermethylation has been recently proposed as a mean for HNSCC detection in salivary rinses. In a prospective study of a high-risk population, we showed that EDNRB promoter methylation in salivary rinses is a useful biomarker for oral cancer and premalignancy.
Experimental Design
Using that cohort, we evaluated EDNRB methylation status and 8 additional genes. Clinical risk assessment by expert clinicians was performed and compared with biomarker performance in the prediction of premalignant and malignant disease. Methylation status of 9 genes was analyzed in salivary rinses of 191 patients by Quantitative methylation-specific PCR.
Results
HOXA9, EDNRB and DCC methylation were associated (p= 0.012; p<0.0001; p=0.0005) with premalignant or malignant disease. On multivariable modeling, histological diagnosis was only independently associated with EDNRB (p=0.0003) or DCC (p=0.004) methylation. A subset of patients received clinical risk classification (CRC) by expert clinicians based on lesion examination. CRC, DCC and EDNRB were associated with diagnosis of dysplasia/cancer on univariate (p=0.008; p=0.026; p=0.046) and multivariate analysis (p=0.012; p=0.037; p=0.047). CRC identified dysplasia/cancer with56% of sensitivity and 66% of specificity with a similar AUC (0.61, 95%CI=0.60-0.81) when compared to EDNRB and DCC combined AUC (0.60, 95%CI=0.51-0.69), sensitivity of 46% and specificity of 72%. A combination of EDNRB, DCC and CRC was optimal AUC (0.67, 95%CI=0.58-0.76).
Conclusion
EDNRB and/or DCC methylation in salivary rinses compares well to examination by an expert clinician in CRC of oral lesions. These salivary biomarkers may be particularly useful in oral premalignancy and malignancy screening in clinical care settings in which expert clinicians are not available
Keywords: HNSCC, hypermethylation, saliva, DCC, EDNRB
INTRODUCTION
Currently, in the United States, there will be over 50,000 estimated cases of head and neck cancer in 2013(1). However, there has been modest improvement in survival of head and neck cancer patients in the past three decades (2), only 50% of patients are cured with initial therapy (2). Early detection of oral cancer has been shown to lead to significantly reduced morbidity and mortality (3, 4). Studies show that visual inspection may be cost-effective for oral cancer screening (5), but also has limited sensitivity and specificity (6) and fails to detect occult disease (7, 8) as well as recurrence and second primary tumors. For these reasons new methods for risk evaluation have been proposed. The use of body fluids, such as serum, plasma, urine and saliva, in screening for different types of tumors, has been shown as an effective method for detecting cancer related genetic and epigenetic alterations, including aberrant promoter methylation (9-11). Many authors have shown correlations with diagnosis and prognosis, as well treatment response using protein, DNA, and RNA obtained from body fluids (10-12). Our group previously showed the ability of an expanded panel of CpG-rich promoters to detect HNSCC-specific promoter hypermethylation using serum and saliva rinses and real time quantitative methylation-specific PCR (QMSP) assays (11). We also demonstrated that promoter hypermethylation of KIF1A and EDNRB is a frequent event in primary HNSCC and that these genes are potential biomarkers for salivary rinse detection of oral cancer (13). In this study, we evaluate the performance of a modified and expanded methylation biomarker panel for salivary rinses from a previously reported cohort of patients presenting with oral lesions, including premalignant and malignant disease. A clinical risk assessment was performed and correlated with histological diagnosis and biomarker status.
METHODS
Samples
The cohort salivary rinse specimens were collected via dental clinics associated with the NYU College of Dentistry in collaboration with the Department of Otolaryngology- Head and Neck Surgery at Johns Hopkins Medical Institutions. Patients were enrolled prospectively into a trial to assess clinical and molecular determinants of risk for malignancy and premalignancy. Institutional review board approval was obtained prior to collection and a written informed consent was obtained from each subject. Patients were evaluated using a clinical risk assessment tool based upon history, visual and physical, considering lesion site, size, and appearance. Each lesion was assigned a primary Enrollment Risk Classification of Low Risk, High Risk, or Known Cancer, as defined by the WHO Classification (14) in Table 1. If a subject had multiple oral lesions, each lesion was classified separately. Subject enrollment criteria were: 1) English and/or Spanish speaking, 2) over 18 years of age, 3) the presence of a candidate oral epithelial lesion (WHO Classification criteria), 4) the absence of a medical condition which would prevent scalpel biopsy; and exclusion criteria were: significant bleeding disorder; lactating or pregnant. Salivary rinses were obtained by rinsing and gargling with 20 cc of normal saline solution and also using a cotton swab to perform a brushing of oral cavity surfaces to include exfoliated cells. The collected material was then centrifuged, supernatant discarded and the cell pellet saved for analysis. Appropriate demographic information was collected, including gender, age, race and risk factors for malignancy, such as tobacco and alcohol consumption. Formal incisional biopsies were performed on any presenting lesions with clinical suspicion for oral cancer or premalignancy and after histopathological examination the lesions were diagnosed as: benign (with or without atypia), premalignant with epithelial dysplasia classification (mild, moderate, severe or carcinoma in-situ), or malignant with squamous cell carcinoma.
Table 1.
Clinical risk assessment criteria
| Study Group | Case Definitions |
|---|---|
|
Low Risk
for malignancy |
Leukoplakia. Adherent white patch on any oral mucosal surface which has no apparent explanation or etiology (i.e., frictional trauma, infection). Such lesions will NOT have associated erythroplakia, ulceration, erosion, or submucosal extension / induration. |
|
High Risk
for malignancy |
Any of the following features, alone or in combination. May be associated with other high-risk signs and symptoms such as history of oral cancer or other cancer with metastatic potential, regional adenopathy, submucosal extension / induration, ulceration. Leukoplakia Any adherent white patch on any oral mucosal surface which has no apparent explanation or etiology but DOES HAVE associated, ulceration, erosion, or submucosal extension / induration.. Erythroplakia: Red patch on any mucosal surface which has no apparent explanation or etiology. Erythroleukoplakia: Mixed red and white patch on any oral mucosal surface which has no apparent explanation or etiology Ulceration: Any break in the oral epithelial surface which has no apparent explanation or etiology. This may or may not be symptomatic and may be of undetermined duration. There is increased risk for malignancy with increased duration. |
| Known Cancer | Biopsy-proven oral squamous cell carcinoma prior to study enrollment |
DNA extraction
DNA obtained from salivary rinses was extracted by digestion with 50 μg/mL proteinase K (Boehringer) in the presence of 1% SDS at 48°C overnight followed by phenol/chloroform extraction and ethanol precipitation.
Bisulfite Treatment
Sodium Bisulfite conversion of DNA extracted from saliva was conducted using the EpiTect Bisulfite kit (Qiagen, Inc.), according to the manufacturer’s instructions. Bissulfite-converted DNA was stored at −80°C.
Gene selection
The genes used in this study are a result of a prior published biomarker discovery (11). A total of 21 informative genes were considered for this first study and were selected from three different sources: (a) genes with promoters that are reported as hypermethylated in HNSCC; (b) genes with promoters that are reported as hypermethylated in other solid tumors; and (c) gene discovery using expression microarray-based approach via unmasking of expression. The first step involved a screening evaluation, designed to eliminate targets that had an inappropriately high frequency of promoter hypermethylation in normal, control samples. It was performed by comparing tumor samples (cases) with salivary rinses or serum (from controls) in a limited, random subset of both patient and controls. A salivary rinse compartments screening evaluation to compare salivary rinses (case) and salivary rinses (control) in additional limited sets of HNSCC patients and controls was also performed. Proportions of gene methylation were compared between tumor samples (from cases) and salivary rinses or serum samples (from controls) using Fisher’s exact test. Sensitivity and specificity of each individual gene in detecting HNSCC were calculated along with 95% confidence intervals. We could note distinct methylation patterns as follows: (a) methylation was detected only in HNSCC but not in control; (b) a higher frequency and higher level of methylation was noted in HNSCC compared with controls with absent methylation in a subset of control samples; (c) a higher frequency of methylation was noted in HNSCC compared with controls but levels of methylation in specimens were similar in both groups; (d) a similar frequency of methylation was noted in both groups (tumor and salivary rinses); however, a quantitative difference between groups was noted, and (e) methylation was noted in both HNSCC and controls at a similar frequency with no difference in methylation levels. Finally, promoter hypermethylation can be associated with age, race, or tobacco and alcohol exposure. The results included the frequency distributions AUC, sensitivity, and specificity for each gene. Based on the above results, genes that could distinguish tumor samples (case) from salivary rinse samples (control) for binary results (either presence or absence of methylation) and an AUC >0.60 and at least 90% specificity or sensitivity were selected for further testing on salivary rinses in a limited cohort of HNSCC patients. Receiver operating characteristic curves for some selected panels based on the method of multivariable logistic regression modeling were constructed for salivary rinse samples, where the single point represented the performance of the panel with a positive panel being defined as at least one gene of the panel presented methylation. From the initial screening of 21 genes for salivary rinses, ultimately 8 genes were selected as part of a panel to distinguish salivary rinses from HNSCC patients and healthy controls DAPK, DCC, MINT-31, TIMP-3, p16, MGMT, CCNA1, and PGP 9.5. Of note PGP9.5 and TIMP-3 have an identical methylation pattern, so PGP9.5 will be omitted from the panel to simplify analysis.
Quantitative Methylation Specific PCR
Bisulfite-modified DNA was used as a template for fluorescence-based real-time PCR, as previously described (15). In brief, primers and probes were designed to specifically amplify the bisulfite-converted DNA for the ACTB gene and all genes of interest (Table 2). Amplification reactions were carried out in triplicate in a final volume of 10 μL containing 1,5 μL of bisulfite-modified DNA; 600 nmol/L of each primer; 200 μmol/L of probe; 0.75 unit of platinum Taq polymerase (Invitrogen); 200 μmol/L of each dATP, dCTP, dGTP, and dTTP; 200 nmol/L of ROX Reference Dye (Invitrogen); 16.6 mmol/L ammonium sulfate; 67 mmol/L Trizma (Sigma); 6.7 mmol/L magnesium chloride; 10 mmol/L mercaptoethanol; and 0.1% DMSO. Thermal cycling started with denaturation step at 95°C for 3 min followed by 50 cycles at 95°C for 15 s and 60°C for 1 min. Amplification reactions were carried out in 384-well plates in a 7900H sequence detector (Perkin- Elmer Applied Biosystems) and analyzed by a sequence detector system (SDS 2.3; Applied Biosystems). Leukocyte DNA from a healthy individual was methylated in vitro with excess SssI methyltransferase (New England Biolabs, Inc.) to generate completely methylated DNA, and serial dilutions of this DNA were used to construct a calibration curve for each plate. Normal leukocyte DNA or DNA from a known unmethylated cell line, bisulfite treated, was used as negative control. The relative quantity of methylation in a particular sample was determined by the ratios between the values of the gene of interest and the internal reference gene (ACTB), (average value of triplicates of the gene of interest divided by the average value of triplicates of ACTB × 100).
Table 2.
Primers sequences for genes used in the study
| Gene | Forward 5′-3′ | Probe 6FAM 5′-3′ TAMRA | Reverse 5′-3′ |
|---|---|---|---|
| DAPK | GGATAGTCGGATCGAGTTAACGTC | TTCGGTAATTCGTAGCGGTAGGGTTTGG | CCCTCCCAAACGCCGA |
| MGMT | CGAATATACTAAAACAACCCGCG | AATCCTCGCGATACGCACCGTTTACG | GTATTTTTTCGGGAGCGAGGC |
| P16 | TTATTAGAGGGTGGGGCGGATCGC | TTATTAGAGGGTGGGGCGGATCGC | GACCCCGAACCGCGACCGTAA |
| TIMP3 | GCGTCGGAGGTTAAGGTTGTT | AACTCGCTCGCCCGCCGAA | CTCTCCAAAATTACCGTACGCG |
| DCC | CGCGATTTTTGGTTTCGAAGG | GGTTTTTGTATTTTTCGGAGTTTTTTTG | TACCGATTACTTAAAAATACGCG |
| MINT31 | GAGTGATTTATTAGGTTTCGTC | ACGCCGAAAAACACTTCCCCAAC | CGAAAACGAAACGCCGCGA |
| CCNA1 | TCGCGGCGAGTTTATTCG | CGTTATGGCGATGCGGTTTCGG | CCGACCGCGACAAACG |
| EDNRB | GGGAGTTGTAGTTTAGTTAGTTAGGGAGTAG | TTTTTATTCGTCGGGAGGAG | CCCGCGATTAAACTCGAAAA |
| HOXA9 | AATAAATTTTATCGTAGAGCGGTAC | ACGCGTACGGTTAATGGGGGCGC | TTCGGTGTTATTAAGTTGTTATATG |
| ACTB | TGGTGATGGAGGAGGTTTAGTAAGT | ACCACCACCCAACACACAATAACAAACACA | AACCAATAAAACCTACTCCTCCCTTAA |
Statistical analysis
Gene methylation was dichotomized at zero (i.e., no methylation vs. any methylation), since we did not find any improvement of change in the performance of this test based on modeling with a continuous or binary variable. The cohort was divided into subcategories of histologic outcome including benign, premalignant and malignant. Predictors associated with head and neck cancers were evaluated as well, including age, gender, race, smoking status, and alcohol consumption. Age was analyzed as a continuous variable, whereas all other variables were considered as categorical variables. Univariate and multivariate proportional odds modeling were constructed sequentially to explore the association of the variables with histologic outcome. Variables of significance based on the univariate models (p<0.20) along with those deemed to be biologically and clinically important were retained for further analysis. Simultaneous effects expressed by these variables were studied using the multivariate proportional odds model. Odds ratio were reported with 95% confidence intervals, which indicated the strength of the association and its uncertainty.
In a second analysis, we explored the independent association of methylation with histology, by excluding patients with known cancer at initial presentation (n=30). The remaining 161 patients were categorized as having benign (with or without atypia) or dysplastic/cancerous lesions. Univariate and multivariate logistic regression analyses were performed using the same biologically and clinically important covariates as described above. Receiver operating characteristics (ROC) analysis was performed to estimate classification accuracy, sensitivity and specificity of the predictor along with 95% confidence intervals, and also Area Under Curve (AUC) as an index of predictive power. A similar analysis was made categorizing lesions as either benign/mild grade dysplasia or moderate or severe grade dysplasia/carcinoma in situ/cancer (Supplementary data).
Statistical analyses were performed using SAS (v 9.2, SAS Institute, Cary, NC) and STATA software (v 8.2, College Station, Texas), and all statistical tests were two-sided with p<0.05 considered statistically significant. Except for the univariate analysis where we accepted any p< 0.20, and then subjected those variables to multivariate analysis
RESULTS
Cohort
A total of 191 patients were included in this study. Most of them were males (69.9%) and Caucasian (69.6%) with a mean age of 54.1 years (ranging from 18 to 90 years). Alcohol or tobacco consumption (current or past) was found in 49.4% and 72.2%, respectively. When comparing baseline characteristics, benign, dysplasia, and cancer groups were similar.) 67.3% of patients presenting with a benign lesion were tobacco users (former and current), 69.8% of patients with epithelial dysplasia used tobacco, and 74.3% of patients with invasive cancer were tobacco users. For alcohol consumption these values were 70.8%, 74.4% and 77.1% for the histologic categories, respectively.
Risk Classification
Dentists, based on WHO classification, performed clinical risk assessment and lesions were categorized as low-risk and high-risk for dysplasia/cancer. After biopsy, risk classification was compared to histopathological diagnosis (Table 3).
Table 3.
Clinical Risk Assessement vs. Histological Diagnosis
| Low Risk | High risk | Cancer | Total | |
|---|---|---|---|---|
| Benign | 75 | 38 | 0 | 113 |
| Mild Dysplasia | 16 | 11 | 0 | 27 |
| Mod. Dysplasia | 2 | 7 | 1 | 10 |
| Severe Dysplasia | 3 | 3 | 0 | 6 |
| Cancer | 0 | 6 | 29 | 35 |
| Total | 96 | 65 | 30 | 191 |
9-gene methylation status
A univariate analysis was performed for association between individual genes and histopathology. At least one methylated gene was detected in 28.3% and 32.6% of benign and premalignant lesions respectively, while 57.1% of the malignant lesions had at least one methylated gene. When analyzing genes separately, EDNRB demonstrated the highest relative risk of association with diagnosis of malignancy as a single biomarker (OR=3.6, 95%CI=2.0-6.4; p<0.0001), followed by DCC (OR=3.3, 95%CI=1.7-6.6; p=0.0005) and HOXA9 (OR=2.1, 95%CI=1.2-3.7; p=0.12). Also CCNA1, P16 and MINT31 demonstrated associations with histopathology, when analyzed separately (p=0.0003; p=0.031; p=0.019), however with large CIs (OR=6.4; 95%CI=2.4-17.4; OR=6.9, 95%CI=1.2-39.9; OR=16.5, 95%CI=1.6-171.9). DAPK, TIMP3 and MGMT were not associated with histopathology in this cohort.
Predictive factors
Univariate analysis of predictors showed age as the only variable associated with histopathologic diagnosis in this cohort (OR=1.3, 95%CI= 1.1-1.6; p=0.014). Neither tobacco nor alcohol consumption (p=0.372 and p=0.435, respectively) were significant predictors. In a multivariate analysis, after adjusting for covariates (age, race, gene methylation in any gene from the 7 gene panel, and tobacco and alcohol consumption), age remained associated with histopathologic diagnosis (OR=1.3, 95%CI= 1.0-1.6; p=0.034). DCC and HOXA9 were analyzed as individual biomarkers in the multivariate analysis adjusted for age, sex, race, tobacco, alcohol, along with DCC/EDNRB/HOXA9, and these markers decreased the significance of age (p=0.051 and p=0.055, respectively). While HOXA9 was no longer significantly associated with histopathologic diagnosis on multivariate analysis, DCC and EDNRB show strong independent associations (OR=2.8, 95%CI=1.4-5.7; p=0.004; OR=3.1, 95%CI=1.7-5.8; p=0.0003), with histopathologic diagnosis.
DCC and EDNRB exhibit similar performance to risk classification
To analyze the predictive power of expert risk classification, the 161 samples were analyzed according to histopathological diagnosis (benign n= 113 vs. epithelial dysplasia/cancer n= 48) having excluded patients that had known cancer at first presentation (n=30). None of the predictors (e.g. age, sex, tobacco, and alcohol consumption) reached statistical significance on univariate analysis. Risk classification, described as low risk or high risk, was associated with histopathological diagnosis (OR=2.5, 95%CI= 1.3-5.1; p=0.008). Again, DCC and EDNRB, as single biomarkers, were associated with histopathological diagnosis (OR= 2.6, 95%CI= 1.1-6.1; p= 0.026; OR=2.1, 95%CI=1.0-4.4; p= 0.046; respectively).
A multivariate model analyzed risk classification and EDNRB, HOXA9 and DCC methylation status. Risk classification was again independently associated with histopathological diagnosis in 3 genes analysis (OR=2.6, 95%CI=1.3-5.2;p=0.008, OR=2.5, 95%CI=1.2-5.0; p=0.010 and OR=2.5, 95%CI=1.2-5.0;p=0.012, respectively) after adjusting either of these three genes. DCC and EDNRB methylation status had again a significant association with a dysplasia/cancer diagnosis (OR=2.5, 95%CI=1.1-6.0; p=0.037 and OR=2.1, 95%CI=1.0-4.6; p=0.047, respectively) after adjusting for risk classification.
To predict the accuracy of risk classification and biomarkers, sensitivity and specificity were calculated using ROC analysis (Table 4). The AUC was also calculated with a 95% CI. DCC as a sole biomarker, had 27% (95% CI= 15-42) sensitivity and 88% (95% CI= 80-93) specificity, with AUC of 0.57 (95% CI= 0.50-0.64) and EDNRB had 38% (95% CI=24-53) sensitivity and 78% (95% CI=69-85) specificity, with AUC of 0.58 (95% CI=0.50-0.66) when treated as a binary variable (methylation versus no methylation). The combination of both genes, EDNRB and DCC, improved performance somewhat (sensitivity 46%; 95% CI= 31-61 and specificity 72%; 95% CI= 62-80), with AUC of 0.60 (95% CI= 0.51-0.69). Risk classification, when analyzed as a single predictor for histopathologic diagnosis, had 56% (95% CI= 41-71) sensitivity and 66% (95% CI= 57-75) specificity, with AUC of 0.61 (95% CI= 0.53-0.70). Using logistic regression analysis, we combined risk classification and EDNRB methylation status, and separately risk classification and DCC, (sensitivity 73%; 95%CI= 58-85 and 69%; 95%CI= 54-81, respectively; specificity 51%; 95%CI= 42-61 and 59%; 95%CI= 50-68, respectively).
Table 4.
Predictive accuracy of risk classification and markers (n=161)
| Predictor | Sensitivity (%, 95%CI) | Specificity (%, 95%CI) | AUC (95%CI) |
|---|---|---|---|
| Risk Classification | 56 (41-71) | 66 (57-75) | 0.61 (0.53-0.70) |
| EDNRB | 38 (24-53) | 78 (69-85) | 0.58 (0.50-0.66) |
| DCC | 27 (15-42) | 88 (80-93) | 0.57 (0.50- 0.64) |
|
EDNRB and risk classification |
73 (58-85) | 51 (42-61) | 0.65 (0.56-0.74) |
|
DCC and risk classification |
69 (54-81) | 59 (50-68) | 0.65 (0.57-0.74) |
| DCC and EDNRB | 46 (31-61) | 72 (62-80) | 0.60 (0.51-0.69) |
|
EDNRB, DCC and risk classification |
75 (60-86) | 48 (38-57) | 0.67 (0.58-0.76) |
Finally the combination of risk classification, DCC and EDNRB showed 75% of sensitivity (95%CI=60-86) and 48% of specificity (95%CI= 38-57) with AUC of 0.67 (95%CI= 0.58- 0.76) (Figure 1).
Figure 1.
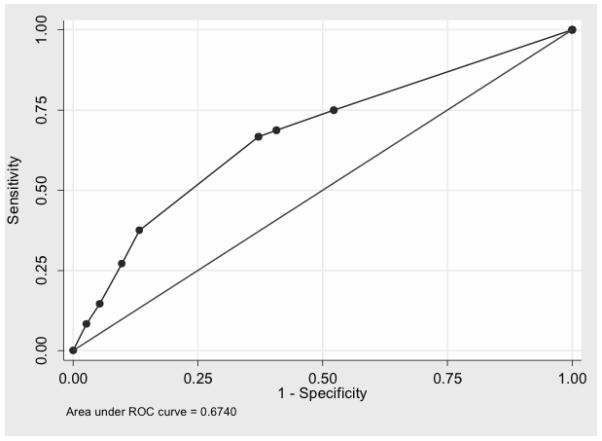
ROC curve showing performance of EDNRB, DCC and risk classification.
ROC curves corresponding to the use of gene signatures, clinical exam and a combination of these are included (Supplementary Figure S1).
Results categorizing lesions as either benign/mild grade dysplasia or moderate or severe grade dysplasia/carcinoma in situ/cancer are presented in supplementary tables 1 and 2.
Discussion
Late diagnosis with advanced-stage disease is the main cause of head and neck cancer morbidity and mortality (4). Although the oral cavity is an easy site for physical exam, often a delay in seeking medical care leads to the advanced-stage of disease at time of diagnosis (16).
Detection of DNA methylation in salivary rinse samples is a potential non-invasive method for early diagnosis of head and neck cancer. Our group, has previously shown it is possible to correlate methylation status with overall survival and prognosis using salivary rinses (11). Also, promoter CpG islands of KIF1A and EDNRB were shown to be methylated in primary HNSCC. These highly specific salivary biomarkers, and were shown to be potential biomarkers for HNSCC detection (13) as well as predictors of risk in oral cavity cancer and premalignancy (17). Furthermore, salivary rinse includes microorganisms, residual food, enucleated orthokeratinized cells and for that reason, extraction of nucleic acids, and purification must be carried out carefully for stable results. QMSP sensitivity allowed us to define methylated genes that were highly specific for tumor, and rarely or never present in any of the oral cavity sites that shed cells in salivary rinses. Also, the presence of cells from all epithelial surfaces may be helpful as potential predictor of malignant risk, as many studies show methylation as an early event on HNSCC carcinogenesis.
For this study, genes were selected using three criteria: (i) genes already reported in literature to be hypermethylated in head and neck cancers, (ii) genes reported as hypermethylated in other solid tumors, and (iii) genes identified via a methylation microarray based approach. The genes were tested in a pilot study with a limited cohort. Based upon specificity and sensitivity of tested genes, a panel was selected for analysis in an expanded cohort.
Our study used a cohort with mean age of 54.1 years with a history of tobacco and alcohol consumption, presenting with lesions epithelial oral lesions deemed suspicious for epithelial dysplasia or malignancy to study correlations between methylation status of select genes and cancer progression. Using uni- and multivariate analyses, only age, among predictors, was associated with histology (p=0.014 and p=0.034, respectively), although tobacco and alcohol exposure are well known risk factors for oral cancer.
Although scalpel biopsy is the gold standard method for diagnosis, it may only represent a portion of the lesion and may not be representative of all pathologic changes. Many of the molecular alterations that may indicate early stages of malignant transformation cannot be seen in the morphological analysis (18). Such diagnostic testing requires training and proper equipment. For this study, salivary rinse samples were collected by untrained personnel without previous experience in this collection protocol. The amount of DNA collected was sufficient to perform the analysis with several genes. The results obtained in this study support an easy and efficient method for oral cancer screening and potentially for prevention.
Clinical risk assessment was performed by dentists, based on WHO classification (14), classifying lesions as high-risk or low-risk. The risk classification assessment associated with methylation status was statistically significant, although not all histolopathological diagnoses matched the clinical risk classification (Table 3). Also in multivariate analysis risk classification was associated with histology, showing that the features of a detected oral lesion is important for early diagnosis.
Our study was able to find a significant correlation between histopathologic diagnosis and methylation status for DCC, EDNRB and HOXA9, as single biomarkers. Deleted in colorectal cancer (DCC) is a putative tumor-suppressor gene at 18q21 that encodes a transmembrane protein with structural similarity to neural cell adhesion molecule (NCAM) (19), and is involved in both epithelial and neuronal cell differentiation (20). DCC hypermethylation has been detected in oral squamous cell carcinoma, other head and neck cancers, breast, gastric and colon cancer (21-23). Our group has previously shown that DCC is epigenetically inactivated by promoter hypermethylation in a majority of HNSCC cases (21). In this study, DCC and EDNRB were hypermethylated in 40% of malignant salivary rinses samples and associated with malignant histopathologic diagnosis, independent of other predictors factors such as age and tobacco/ethanol exposure. These results suggest that they can be used as single biomarkers of malignancy.
EDNRB (Endothelin receptor type B) is a G protein-coupled receptor, which activates a phosphatidylinositol-calcium second messenger system. Its ligand, endothelin, consists of a family of three potent vasoactive peptides: ET1, ET2, and ET3. Studies suggest that the multigenic disorder, Hirschsprung disease type 2, can be due to mutations in EDNRB. Pattani et al. (2010) showed that EDNRB promoter hypermethylation in salivary rinses is associated with increased risk of oral cancer and premalignancy.
The HOXA9 gene is part of the A cluster on chromosome 7 and encodes a DNA-binding transcription factor which may regulate gene expression, morphogenesis, and differentiation. A specific translocation event, which causes a fusion between this gene and the NUP98 gene, has been associated with myeloid leukemogenesis. HOXA9 was found to be methylated in high-grade gliomas (24) and reported as a potential biomarker for prevention and early detection in oral squamous cell carcinoma using saliva samples (25).
Association of DCC and EDNRB hypermethylation with histopathologic diagnosis was discovered from a different approach in the attempt to identify a novel panel of promoter hypermethylation markers to improve the ability to detect epigenetic changes associated with HNSCC in salivary rinses (11, 13, 17). In combining the results from both DCC and EDNRB methylation assays, we observed improvement in performance, indicating their potential as biomarkers for HNSCC.
Clinical risk assessment also appeared as an important variable in this study. When we analyzed according to histopathology (benign vs. dysplasia/cancer), risk classification presented a strong association on univariate analysis and also when associated with the three genes, DCC, EDNRB and HOXA9, in the multivariate analysis. Also using the logistic regression model, clinical risk classification had better performance than EDNRB and DCC combined (AUC= 0.61 and 0.60, respectively), showing that a highly trained professional can be as efficient as the two biomarkers combined. The combination of the three predictive variables (DCC, EDNRB and risk classification) improved performance (AUC= 0.67), suggesting that visual inspection when associated with a reliable biomarker, gives optimal results on disease detection and evaluation.
Clinical assessment can be good for initial risk analysis and treatment indication, helping to prevent and detect malignant or premalignant lesions in a very cost effective manner. The use of biomarkers can have a complementary role when clinical evaluation is prejudiced and also can help to detect occult disease, analyze surgical margins, predict patient outcome and sensitivity to oncologic therapies, and reveal disease status.
Salivary rinses have shown to be useful in diagnosing infectious and autoimmune diseases, and have been demonstrated to be very effective in the detection of aberrant methylation, gene expression, HPV detection and also miRNA expression (13, 26-28). Our study showed that salivary rinses can be obtained by untrained professionals and also that QMSP provides a cost-effective method that allows high-throughput and rapid analysis. The use of this technique as a means of early detection of premalignant and malignant lesions reinforces its usefulness as a screening and surveillance strategy. This low invasive approach allows easier high-risk population screening that may facilitate preventive medicine, therapeutic planning, and prognostic counseling.
The presence of EDNRB and/or DCC promoter methylation in salivary rinses compares well to examination by an expert clinician in risk classification of oral premalignant and malignant lesions. Given the current costs and availability of an expert health care provider trained and experienced in oral cancer diagnosis, it is simply not feasible to screen at risk populations via expert physical examination. Therefore, risk assessment with a salivary biomarker is attractive as a cost effective means to identify higher risk patients that should be referred for expert exam and biopsy. Also, these salivary biomarkers may be particularly useful in identifying patients with lesions that appear low risk by physical examination, but are identified as high risk by epigenetic salivary biomarkers.
Supplementary Material
TRANSLATIONAL RELEVANCE.
Late Head and Neck Squamous Cell Carcinoma (HNSCC) diagnosis is responsible for disease morbidity and mortality. Oral screenings have been proposed as means of prevention. Our study compares oral lesion clinical risk assessment and molecular biomarkers. The presence of gene promoter methylation in salivary rinses compares well to examination by an expert clinician in risk classification of oral premalignant and malignant lesions. Given the current costs and availability of an expert health care provider trained and experienced in oral cancer diagnosis, risk assessment with a salivary biomarker is attractive as a cost effective means to identify higher risk patients that should be referred for expert exam and biopsy. Also, these salivary biomarkers may be particularly useful in identifying patients with lesions that appear low risk by physical examination, but are identified as high risk by epigenetic salivary biomarkers.
Acknowledgments
Grant Support National Institute of Dental and Craniofacial Research (NIDCR) and NIH Specialized Program of Research Excellence grant 5P50DE019032, NIDCR/NIH grant U54 DE14257, and Early Detection Research Network grant U01-CA084986. The funding agencies had no role in the design of the study, data collection or analysis, the interpretation of the results, the preparation of the manuscript, or the decision to submit the manuscript for publication. J.L. Schussel has a Coordenação de Aperfeiçoamento de Pessoal de Nível Superior scholarship (BEX 2363/09-7).
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Goodwin WJ., Jr. Salvage surgery for patients with recurrent squamous cell carcinoma of the upper aerodigestive tract: when do the ends justify the means? Laryngoscope. 2000;110:1–18. doi: 10.1097/00005537-200003001-00001. [DOI] [PubMed] [Google Scholar]
- 3.Patton LL. The effectiveness of community-based visual screening and utility of adjunctive diagnostic aids in the early detection of oral cancer. Oral Oncol. 2003;39:708–23. doi: 10.1016/s1368-8375(03)00083-6. [DOI] [PubMed] [Google Scholar]
- 4.Mignogna MD, Fedele S, Lo Russo L. The World Cancer Report and the burden of oral cancer. Eur J Cancer Prev. 2004;13:139–42. doi: 10.1097/00008469-200404000-00008. [DOI] [PubMed] [Google Scholar]
- 5.Subramanian S, Sankaranarayanan R, Bapat B, Somanathan T, Thomas G, Mathew B, et al. Cost-effectiveness of oral cancer screening: results from a cluster randomized controlled trial in India. Bull World Health Organ. 2009;87:200–6. doi: 10.2471/BLT.08.053231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagao T, Warnakulasuriya S, Ikeda N, Fukano H, Fujiwara K, Miyazaki H. Oral cancer screening as an integral part of general health screening in Tokoname City, Japan. J Med Screen. 2000;7:203–8. doi: 10.1136/jms.7.4.203. [DOI] [PubMed] [Google Scholar]
- 7.Califano J, Westra WH, Koch W, Meininger G, Reed A, Yip L, et al. Unknown primary head and neck squamous cell carcinoma: molecular identification of the site of origin. J Natl Cancer Inst. 1999;91:599–604. doi: 10.1093/jnci/91.7.599. [DOI] [PubMed] [Google Scholar]
- 8.Cianchetti M, Mancuso AA, Amdur RJ, Werning JW, Kirwan J, Morris CG, et al. Diagnostic evaluation of squamous cell carcinoma metastatic to cervical lymph nodes from an unknown head and neck primary site. Laryngoscope. 2009;119:2348–54. doi: 10.1002/lary.20638. [DOI] [PubMed] [Google Scholar]
- 9.Hoque MO, Begum S, Topaloglu O, Jeronimo C, Mambo E, Westra WH, et al. Quantitative detection of promoter hypermethylation of multiple genes in the tumor, urine, and serum DNA of patients with renal cancer. Cancer Res. 2004;64:5511–7. doi: 10.1158/0008-5472.CAN-04-0799. [DOI] [PubMed] [Google Scholar]
- 10.Hoque MO, Feng Q, Toure P, Dem A, Critchlow CW, Hawes SE, et al. Detection of aberrant methylation of four genes in plasma DNA for the detection of breast cancer. J Clin Oncol. 2006;24:4262–9. doi: 10.1200/JCO.2005.01.3516. [DOI] [PubMed] [Google Scholar]
- 11.Carvalho AL, Jeronimo C, Kim MM, Henrique R, Zhang Z, Hoque MO, et al. Evaluation of promoter hypermethylation detection in body fluids as a screening/diagnosis tool for head and neck squamous cell carcinoma. Clin Cancer Res. 2008;1:97–107. doi: 10.1158/1078-0432.CCR-07-0722. [DOI] [PubMed] [Google Scholar]
- 12.Nunes DN, Kowalski LP, Simpson AJ. Detection of oral and oropharyngeal cancer by microsatellite analysis in mouth washes and lesion brushings. Oral Oncol. 2000;36:525–8. doi: 10.1016/s1368-8375(00)00045-2. [DOI] [PubMed] [Google Scholar]
- 13.Demokan S, Chang X, Chuang A, Mydlarz WK, Kaur J, Huang P, et al. KIF1A and EDNRB are differentially methylated in primary HNSCC and salivary rinses. Int J Cancer. 2010 doi: 10.1002/ijc.25248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kramer IR, Pindborg JJ, Bezroukov V, Infirri JS. Guide to epidemiology and diagnosis of oral mucosal diseases and conditions. World Health Organization. Community Dent Oral Epidemiol. 1980;8(1):1–26. doi: 10.1111/j.1600-0528.1980.tb01249.x. [DOI] [PubMed] [Google Scholar]
- 15.Harden SV, Guo Z, Epstein JI, Sidransky D. Quantitative GSTP1 methylation clearly distinguishes benign prostatic tissue and limited prostate adenocarcinoma. J Urol. 2003;169:1138–42. doi: 10.1097/01.ju.0000049627.90307.4d. [DOI] [PubMed] [Google Scholar]
- 16.Ragin CC, Modugno F, Gollin SM. The epidemiology and risk factors of head and neck cancer: a focus on human papillomavirus. J Dent Res. 2007;86:104–14. doi: 10.1177/154405910708600202. [DOI] [PubMed] [Google Scholar]
- 17.Pattani KM, Zhang Z, Demokan S, Glazer C, Loyo M, Goodman S, et al. Endothelin receptor type B gene promoter hypermethylation in salivary rinses is independently associated with risk of oral cavity cancer and premalignancy. Cancer Prev Res (Phila) 2010;3:1093–103. doi: 10.1158/1940-6207.CAPR-10-0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warnakulasuriya S, Reibel J, Bouquot J, Dabelsteen E. Oral epithelial dysplasia classification systems: predictive value, utility, weaknesses and scope for improvement. J Oral Pathol Med. 2008;37:127–33. doi: 10.1111/j.1600-0714.2007.00584.x. [DOI] [PubMed] [Google Scholar]
- 19.Fearon ER, Cho KR, Nigro JM, Kern SE, Simons JW, Ruppert JM, et al. Identification of a chromosome 18q gene that is altered in colorectal cancers. Science. 1990;247:49–56. doi: 10.1126/science.2294591. [DOI] [PubMed] [Google Scholar]
- 20.Hedrick L, Cho KR, Fearon ER, Wu TC, Kinzler KW, Vogelstein B. The DCC gene product in cellular differentiation and colorectal tumorigenesis. Genes Dev. 1994;8:1174–83. doi: 10.1101/gad.8.10.1174. [DOI] [PubMed] [Google Scholar]
- 21.Carvalho AL, Chuang A, Jiang WW, Lee J, Begum S, Poeta L, et al. Deleted in colorectal cancer is a putative conditional tumor-suppressor gene inactivated by promoter hypermethylation in head and neck squamous cell carcinoma. Cancer Res. 2006;66:9401–7. doi: 10.1158/0008-5472.CAN-06-1073. [DOI] [PubMed] [Google Scholar]
- 22.Tamura G. Promoter methylation status of tumor suppressor and tumor-related genes in neoplastic and non-neoplastic gastric epithelia. Histol Histopathol. 2004;19:221–8. doi: 10.14670/HH-19.221. [DOI] [PubMed] [Google Scholar]
- 23.Miyamoto K, Fukutomi T, Akashi-Tanaka S, Hasegawa T, Asahara T, Sugimura T, et al. Identification of 20 genes aberrantly methylated in human breast cancers. Int J Cancer. 2005;116:407–14. doi: 10.1002/ijc.21054. [DOI] [PubMed] [Google Scholar]
- 24.Di Vinci A, Casciano I, Marasco E, Banelli B, Ravetti GL, Borzi L, et al. Quantitative methylation analysis of HOXA3, 7, 9, and 10 genes in glioma: association with tumor WHO grade and clinical outcome. J Cancer Res Clin Oncol. 2012;138:35–47. doi: 10.1007/s00432-011-1070-5. [DOI] [PubMed] [Google Scholar]
- 25.Guerrero-Preston R, Soudry E, Acero J, Orera M, Moreno-Lopez L, Macia-Colon G, et al. NID2 and HOXA9 promoter hypermethylation as biomarkers for prevention and early detection in oral cavity squamous cell carcinoma tissues and saliva. Cancer Prev Res (Phila) 2011;4:1061–72. doi: 10.1158/1940-6207.CAPR-11-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosas SL, Koch W, da Costa Carvalho MG, Wu L, Califano J, Westra W, et al. Promoter hypermethylation patterns of p16, O6-methylguanine-DNA-methyltransferase, and death-associated protein kinase in tumors and saliva of head and neck cancer patients. Cancer Res. 2001;61:939–42. [PubMed] [Google Scholar]
- 27.Zhao M, Rosenbaum E, Carvalho AL, Koch W, Jiang W, Sidransky D, et al. Feasibility of quantitative PCR-based saliva rinse screening of HPV for head and neck cancer. Int J Cancer. 2005;117:605–10. doi: 10.1002/ijc.21216. [DOI] [PubMed] [Google Scholar]
- 28.Lallemant B, Evrard A, Combescure C, Chapuis H, Chambon G, Raynal C, et al. Clinical relevance of nine transcriptional molecular markers for the diagnosis of head and neck squamous cell carcinoma in tissue and saliva rinse. BMC Cancer. 2009;9:370. doi: 10.1186/1471-2407-9-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


