Abstract
We used CDK4/hTERT-immortalized normal human bronchial epithelial cells (HBECs) from several individuals to study lung cancer pathogenesis by introducing combinations of common lung cancer oncogenic changes (p53, KRAS, MYC) and followed the stepwise transformation of HBECs to full malignancy. This model demonstrated that: 1) the combination of five genetic alterations (CDK4, hTERT, sh-p53, KRASV12, and c-MYC) is sufficient for full tumorigenic conversion of HBECs; 2) genetically-identical clones of transformed HBECs exhibit pronounced differences in tumor growth, histology, and differentiation; 3) HBECs from different individuals vary in their sensitivity to transformation by these oncogenic manipulations; 4) high levels of KRASV12 are required for full malignant transformation of HBECs, however prior loss of p53 function is required to prevent oncogene-induced senescence; 5) over-expression of c-MYC greatly enhances malignancy but only in the context of sh-p53+KRASV12; 6) growth of parental HBECs in serum-containing medium induces differentiation while growth of oncogenically manipulated HBECs in serum increases in vivo tumorigenicity, decreases tumor latency, produces more undifferentiated tumors, and induces epithelial-to-mesenchymal transition (EMT); 7) oncogenic transformation of HBECs leads to increased sensitivity to standard chemotherapy doublets; 8) an mRNA signature derived by comparing tumorigenic vs. non-tumorigenic clones was predictive of outcome in lung cancer patients. Collectively, our findings demonstrate this HBEC model system can be used to study the effect of oncogenic mutations, their expression levels, and serum-derived environmental effects in malignant transformation, while also providing clinically translatable applications such as development of prognostic signatures and drug response phenotypes.
Keywords: p53, KRAS, c-MYC, immortalized human bronchial epithelial cell, in vitro transformation model of lung cancer, epithelial mesenchymal transition
Introduction
Human lung cancer develops as a multi-step process, usually after prolonged smoke-related tobacco exposure resulting in specific proto-oncogene and tumor suppressor gene alterations in lung epithelial cells (1). Genome-wide analyses have identified multiple genetic and epigenetic alterations in lung tumors (2–5). To translate these findings to the clinic however, it is essential to identify the best targets for early detection and therapeutic intervention by determining which alterations represent “driver” and which represent “passenger” changes.
Functional tests are a critical step in determining “driver” status and are most commonly performed by genetically or pharmacologically “correcting” the defect in a lung cancer line and monitoring its effect. Another approach is to introduce putative oncogenic changes into normal lung epithelial cells and ascertain their contribution to malignancy. To undertake the latter we previously developed an in vitro model of immortalized human bronchial epithelial cells (HBECs) establishing cell lines from over 30 different individuals (6). HBECs were immortalized by over-expressing Cdk4 and human telomerase reverse transcriptase (hTERT) to emulate two of the earliest events and almost universal events in lung cancer pathogenesis: abrogation of the p16/Rb cell cycle checkpoint pathway and bypass of replicative senescence. Importantly, this was the first report of immortalizing lung epithelial cells in the absence of viral oncoproteins, as used previously by other groups (7, 8). Viral oncoproteins such as SV40 early region (containing large T and small t antigens) are known to cause malignant transformation through inactivation of Rb and/or p53, as well inhibit the tumor suppressor gene PP2A phosphatase (9, 10). They also have other less characterized interactions with cellular processes thereby contributing to cancer pathogenesis in unknown ways. Thus, the critical genetic manipulations that lead to malignant transformation of epithelial cells immortalized with viral oncoproteins are not completely “defined”. Moreover, they are likely more oncogenically progressed than non-viral models as shown in mammary epithelial cell systems where full transformation with SV40 early region (ER) required three manipulations (hTERT, SV40 ER, and RAS) (8) compared with six manipulations (hTERT, dominant negative p53, CDK4, c-MYC, and cyclin D1) in the absence of SV40 ER (11).
While HBECs used in our study are immortalized, a phenotype they share with cancer cells, they do not display other cancer cell phenotypes such as disruption of the p53 pathway, extensive copy number changes, lack of contact-inhibition, anchorage-independent growth and the ability to form tumors in immunodeficient mice (6, 12). Furthermore, we have shown immortalized HBECs retain the ability to differentiate into structures found in the normal bronchial epithelium (13, 14). These features make HBECs a physiologically appropriate in vitro model for studying the transformation process of bronchial epithelial cells to lung cancer.
Transformation of lung epithelial cells to full malignancy using defined genetic manipulations has only been described in two studies (8, 15). One study using viral oncoproteins transformed tracheobronchial and small airway epithelial cells with hTERT, SV40 ER and oncogenic HRAS or KRAS (8). The second study showed fully transformed small airway epithelial cells (HSAEC) occurred with CDK4, hTERT, mutant p53, mutant KRAS and either c-MYC, PIK3CA, cyclin D1, or LKB1 manipulations (15). In our bronchial epithelial cell model we have previously shown the combination of CDK4, hTERT, p53 knockdown with mutant EGFR or moderate levels of mutant KRASV12 progressed cells partially but not completely to malignancy, as evidenced by the failure to form tumors in immunodeficient mice (12). As lung cancer develops in both the central bronchus and peripheral small airways, development of an in vitro model of malignant transformation in bronchial epithelial cells is essential.
In the present study we aimed to confirm the genetic tractability of HBECs and fully transform cells to malignancy using oncogenic manipulations commonly found in lung tumors. Loss of p53 function and oncogenic KRAS are two well-known genetic alterations in lung cancer occurring in approximately 50% and 30% of non-small cell lung cancer (NSCLC), respectively (16, 17). Aberrant expression of c-MYC, through amplification or over-expression, is found in approximately 20% of NSCLCs (1). While it is known the protein levels of oncogenic RAS can influence its oncogenic ability (18), expression of oncogenic KRASV12 can also lead to premature senescence of normal human epithelial cells (19). The prevalence of KRAS alterations in NSCLC indicates however, that malignant transformation requires the cell to adapt to this oncogenic stress, perhaps assisted through preceding oncogenic transformations (20). Here, we present one of the first reports of full malignant transformation of lung epithelial cells with defined genetic manipulations. Furthermore, we characterize the effect of oncogenic stress and environmental effects such as growth factors upon tumorigenic transformation in HBECs, illustrate divergent clonal heterogeneity, and determine the capability of this in vitro model for developing and testing lung cancer therapeutics.
Materials and Methods
Cells and culture conditions
HBEC3 (HBEC3KT), HBEC4 (HBEC4KT) and HBEC17 (HBEC17KT) immortalized normal human bronchial epithelial cell lines were established by introducing mouse Cdk4 and human TERT into normal human bronchial epithelial cells (6). HBECs were cultured with KSFM (Life Technologies Inc., Carlsbad, CA) media containing 50 µg/mL of Bovine Pituitary Extract (BPE) (Life Technologies Inc.) and 5 ng/mL of EGF (epidermal growth factor) (Life Technologies Inc.). Partially transformed HBECs (soft agar clones) were also cultured with RPMI-1640 (Life Technologies Inc.) media supplemented with 10% fetal bovine serum (R10). Lung cancer cell lines were established by our laboratory, and maintained in RPMI-1640 (Life Technologies Inc.) with 5% fetal bovine serum (21, 22). All cell lines were DNA fingerprinted (PowerPlex 1.2 Kit, Promega, Madison, WI) and mycoplasma-free (e-Myco Kit, Boca Scientific, Boca Raton, FL).
Viral vector construction and viral transduction
Stable p53 knockdown and moderate expression of KRASV12 was achieved as described previously (12). Expression of high KRASV12 levels used a lentiviral vector, pLenti6-KRASV12, as described previously (23). Lentiviral vectors expressing KRASWT, KRASC12, and KRASD12 were constructed from pLenti6-KRASV12 as described previously (24). A c-MYC expressing retroviral vector (designated pMSCV-MYC) was constructed by ligating a BamHI/SalI-digested c-MYC insert from pCTV3K (25) (a gift from Dr J. Michael Ruppert, University of Alabama) into BglII/XhoI-digested pMSCV-hyg (Clontech, Mountain View, CA). Lentivirus and retrovirus containing medium were produced as described previously (12). Transduced cells were selected with zeocin (12.5 µg/mL), blasticidin (2 µg/mL) or hygromycin (20 µg/mL) for 7–14 days. The presence of mutant KRASV12 in stable cell lines was confirmed using a reverse transcription-PCR (RT-PCR)/RFLP assay as described previously (12).
Immunoblotting and Southern blotting
Preparation of total cell lysates and Western blotting were performed as described previously (26). Primary antibodies are listed in Supplementary Table S1 and were detected with HRP-conjugated anti-rabbit or anti-mouse secondary antibodies (1:2,000 dilution, Thermo Fisher Scientific, Waltham, MA). Actin or HSP90 protein levels were used as loading controls. Southern blotting for HBEC clones derived from large soft agar colonies was performed as described previously (27) using Phototope-Star Detection Kit (New England Biolabs, Ipswich, MA). DNA was probed with the Blasticidin resistance gene amplified from pLenti6-KRASV12 vector using 5’-ATGGCCAAGCCTTTGTCTCAAG-3’ and 5’-TTAGCCCTCCCACACATAACC-3’ primers.
Biochemical and in vitro transformation assays
Senescent cells were stained with Senescence β-Galactosidase Staining Kit (Cell Signaling, Danvers, MA) and blue-stained senescent cells were counted under a microscope (20X total magnification). Percent of positively stained cells was averaged across six fields. Cell cycle analysis was performed on sub-confluent populations of cells harvested 48 hours after seeding, unless otherwise stated, and cell cycle analysis was performed as described previously (28). Cell proliferation assays were performed by seeding 5000 cells in 6-well plates and counting cells every four days. Cells were expanded when sub-confluent, as necessary. Anchorage-dependent colony formation assays were performed as previously described (12); 200–600 viable cells were seeded in triplicate in 100mm plates and cultured for two weeks before staining colonies with methylene blue. Acute KRASV12 toxicity assays were performed by transducing cells with KRASV12 or LacZ lentivirus and selecting for three days with blasticidin before seeding anchorage-dependent colony formation assays. Anchorage-independent (soft agar) growth assays were performed as previously described (12) seeding 1,000 viable cells in 12-well plates. MTS assays to measure drug response to standard platinum-based doublets (paclitaxel-carboplatin (2/3 wt/wt), gemcitabine-cisplatin (25/2 wt/wt), and pemetrexed-cisplatin (20/3 wt/wt)) were performed as previously described (29). Cells were treated for 96 hours with 4-fold dilutions from a maximum dose of 1000/3501 nM (paclitaxel/carboplatin), 1000/298 µM (pemetrexed-cisplatin) or 2000/140 nM (gemcitabine/cisplatin). Each experiment was performed in quadruplicate with eight replicates per experiment. Fitting of data to dose-response inhibition curves, calculation of ED50 values and comparisons based on one-way ANOVA with Dunnett’s post test were performed using GraphPad Prism version 5.00 for Windows (GraphPad Software, San Diego, CA).
In vivo tumorigenicity assays and histologic analysis
In vivo tumorigenicity was evaluated by injection of cells in female 5- to 6-week-old NOD/SCID mice. Each mouse was given a subcutaneous injection on its flank of 3−5×106 viable cells in 0.2 mL of PBS. Mice were monitored every 2–3 days for tumor formation for up to six months. All animal care was in accord with institutional guidelines and approved IACUC protocols. Formalin-fixed, paraffin-embedded (FFPE) xenograft tumor tissue was sectioned and stained with hematoxylin and eosin (H&E), Alcian Blue-PAS, PAS and mucicarmine for histologic analysis. Immunohistochemical (IHC) staining for p63 (BioCare, Concord, CA; clone BC4A4), Napsin A, CK5 and CK7 was performed commercially by ProPath (Dallas, TX). TUNEL (TdT-mediated dUTP Nick-End Labeling) staining was performed using DeadEnd™ Fluorometric TUNEL System (Promega).
Statistical testing and microarray analysis
For comparison of colony formation and senescent cells between different genetically manipulated cell strains we used a two-tailed student’s t-test and P < 0.05 was considered significant. mRNA microarray profiling was performed with Illumina HumanHT-12 v4 Expression BeadChips (Illumina Inc., San Diego, CA) following the manufacturer’s guidelines and analyzed with in-house Visual Basic software MATRIX V1.483. Functional analysis of differentially expressed genes was performed using Ingenuity Pathway Analysis (IPA) (Ingenuity Systems, Inc. Redwood City, CA). The data discussed in this publication have been made available in the National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) public repository (http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession number GSE40828. The predictive ability of the soft agar gene signature was tested using two independent mRNA microarray lung tumor datasets; 209 primary lung adenocarcinomas and squamous cell carcinomas (SPORE dataset) GSE41271 and 442 primary lung adenocarcinomas (NCI Director’s Challenge Consortium dataset) (30). A detailed description of these two datasets and the prediction analysis is given in the supplementary methods.
Results
Oncogenic transformation of HBECs correlates with level of exogenous KRASV12 expression
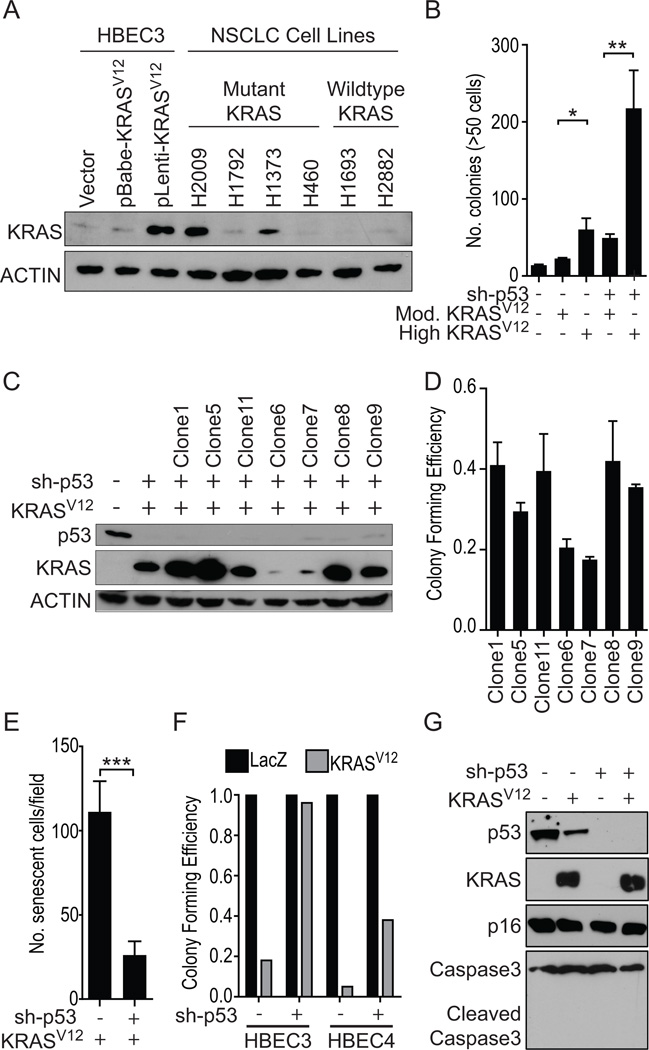
NSCLC cell lines harboring a mutation in KRAS express wide-ranging endogenous levels of the protein (Figure 1A). Thus, to create HBECs that express comparable levels of oncogenic KRAS protein we used two different expression vectors; a retroviral vector, which resulted in modest levels of expression and a lentiviral vector that resulted in high levels of expression (Figure 1A). Compared with HBEC3 expressing moderate levels (retroviral-mediated) of KRASV12 protein, HBEC3 expressing high levels of KRASV12 (lentiviral-mediated) exhibited a significant increase in soft agar colonies in the background of both wild-type p53 (HBEC3) and p53 knockdown (HBEC3p53) (Figure 1B). To confirm the contribution of the level of KRASV12 expression towards HBEC transformation we examined seven clonal populations of HBEC3p53,KRAS (sh-p53 and lentiviral-delivered KRASV12) (Figure 1C). Clones with high levels of KRAS expression had increased anchorage-dependent (liquid) colony forming ability (Figure 1D, Supplementary Figure S1A).
Figure 1. High exogenous levels of KRASV12, comparable to endogenous levels found in mutant KRAS NSCLC cell lines, increases transformation of HBECs and induces senescence, which is largely bypassed with p53 knockdown.
A) Immunoblot for KRAS protein expression in HBEC3 cells infected with KRASV12 using either a moderately-expressing retroviral (pBabe-hyg-KRASV12) or high-expressing lentiviral (pL6-KRASV12) vector. Actin was used as loading control. B) Anchorage-independent (soft agar) colony formation in HBEC3 with high (lentiviral) or moderate (retroviral) levels of KRASV12 in the background of both wildtype p53 and p53 knockdown (sh-p53) (t-test). C) Immunoblot of HBEC3p53,KRAS soft agar clones confirming p53 and KRASV12 manipulations. The presence (+) or absence (−) of each manipulation is shown. D) Anchorage-dependent (liquid) colony formation ability of HBEC3p53,KRAS soft agar clones. E) Quantification of SA-β-gal staining found KRASV12-induced senescence in HBEC3 cells was significantly lower in cells with p53 knockdown compared with p53 wildtype (t-test). F) Anchorage-dependent colony formation assay to compare acute KRASV12-induced toxicity in HBEC3 and HBEC4 with wildtype p53 or p53 knockdown. G) Immunoblot of HBEC3 cell lysates harvested seven days after infection with KRASV12 or LacZ lentivirus. *P < 0.05, **P < 0.01, ***P < 0.001. Full-length blots are presented in Supplementary Figure S8).
High levels of mutant KRASV12 induce senescence in HBECs, in a p53-dependent manner
Lentiviral-mediated introduction of high levels of KRASV12 caused a large subset of HBEC3 cells to display significant morphologic changes, including flattened, enlarged shape and a vacuole-rich cytoplasm, suggestive of oncogene-induced senescence. This effect was not observed following exogenous expression of moderate levels of KRASV12 (Supplementary Figure S1B). Senescence-associated beta-galactosidase (SA-β-gal) staining confirmed the morphologic changes corresponded with senescence (Supplementary Figure S1C). Significantly less RAS-induced senescence was observed in HBEC3 with stable p53 knockdown (HBEC3p53) compared with HBEC3 with wildtype p53 (Figure 1E and Supplementary Figure S1C). An acute, senescence-associated p53-mediated KRASV12 toxicity was shown in anchorage-dependent colony formation assays seeded four days after transduction with KRASV12 lentivirus. Significant toxicity was observed in HBECs with wildtype p53 (HBEC3 and another HBEC line, HBEC4) following transduction with KRASV12 lentivirus compared with control LacZ lentivirus but not in HBECs with p53 knockdown (Figure 1F, and Supplementary Figures S1D and S1E), suggesting induced senescence was p53 dependent. p53-mediated mitigation of oncogenic KRAS stress was also observed with other codon 12 KRAS mutants (KRASC12 and D12) (Supplementary Figure S1F–G) while over-expression of wildtype KRAS in HBEC3 did not induce a senescence phenotype (Supplementary Figure S1G). Immunoblotting for cleaved Caspase-3 confirmed KRASV12-induced toxicity was not apoptosis-associated (Figure 1G), while cell cycle analysis found no increased accumulation of cells in G1 phase in HBEC3p53 cells infected with KRASV12 compared to the control vector (Supplementary Figure S1H).
These results demonstrate oncogenic KRAS mediates a potent cellular stress response in CDK4/hTERT-immortalized HBECs whereby the cells resist oncogenic transformation by engaging cellular senescence. Loss of p53 function however impedes this cellular senescence response, indicating that in these cells, p53 signaling is a primary mediator of RAS-induced senescence. Furthermore, as high levels of oncogenic KRAS expression are required for malignant transformation it demonstrates loss of p53 function is a critical co-oncogenic step in the malignant transformation of the large majority of human bronchial epithelial cells.
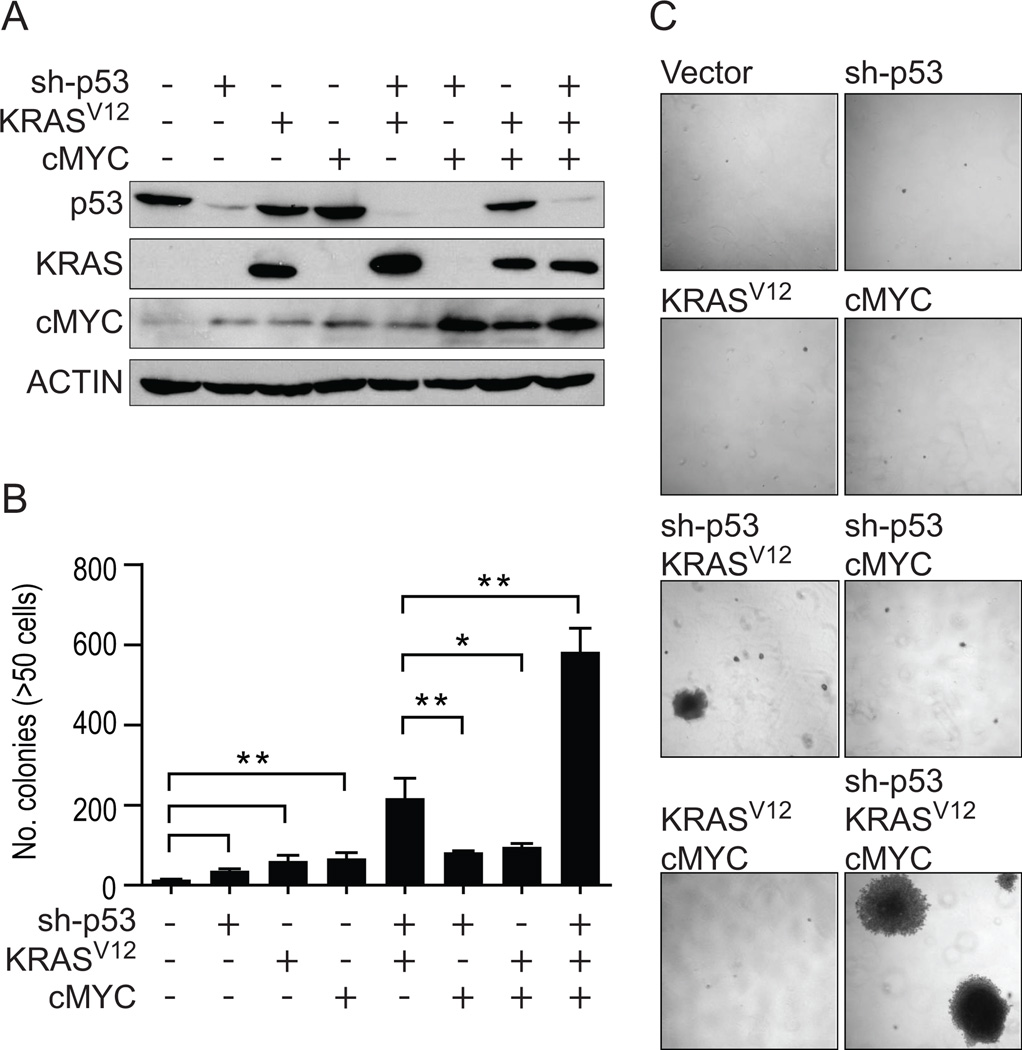
The combination of p53 knockdown and KRASV12 in HBECs significantly increases in vitro transformation, which is further augmented with c-MYC overexpression
The single introduction of p53 knockdown, mutant KRASV12 or c-MYC over-expression resulted in quantitatively modest but significant increases in soft agar colony number (Figure 2A, 2B, 2C). The combination of p53 knockdown and KRASV12 (HBEC3p53,KRAS) resulted in a significant increase in transformation not observed in other dual combinations (p53 knockdown+c-MYC [HBEC3p53,MYC] or KRASV12+c-MYC [HBEC3KRAS,MYC]), while introduction of all three manipulations (HBEC3p53,KRAS,MYC) resulted in the most transformed phenotype (Figure 2B). Higher expression of c-MYC was achieved if over-expressed in HBEC3p53 (p53 knockdown), HBEC3KRAS (KRASV12) or HBEC3p53,KRAS (p53 knockdown and KRASV12) cells (Figure 2A), suggesting partial transformation of immortalized HBEC3 with either p53 knockdown or addition of mutant KRASV12 is required for the cells to tolerate high levels of c-MYC.
Figure 2. Stepwise in vitro transformation of HBEC3 with sh-p53, KRASV12 and c-MYC.
A) Immunoblot of isogenic derivatives of HBEC3 with sh-p53, or over-expression of KRASV12 or c-MYC, alone or in combination. The presence (+) or absence (−) of each manipulation is shown.B) Transformation as defined by anchorage-independent growth in soft agar assays for HBEC3 with each manipulation alone or in combination. C) Representative photographs of soft agar assays showing the formation of large, macroscopic (>1mm) colonies in HBEC3p53,KRAS and HBEC3p53,KRAS,MYC (4X magnification). Full-length blots are presented in Supplementary Figure S8.
Combination of sh-p53+KRASV12 or sh-p53+KRASV12+c-MYC oncogenically transforms HBEC3 in vivo
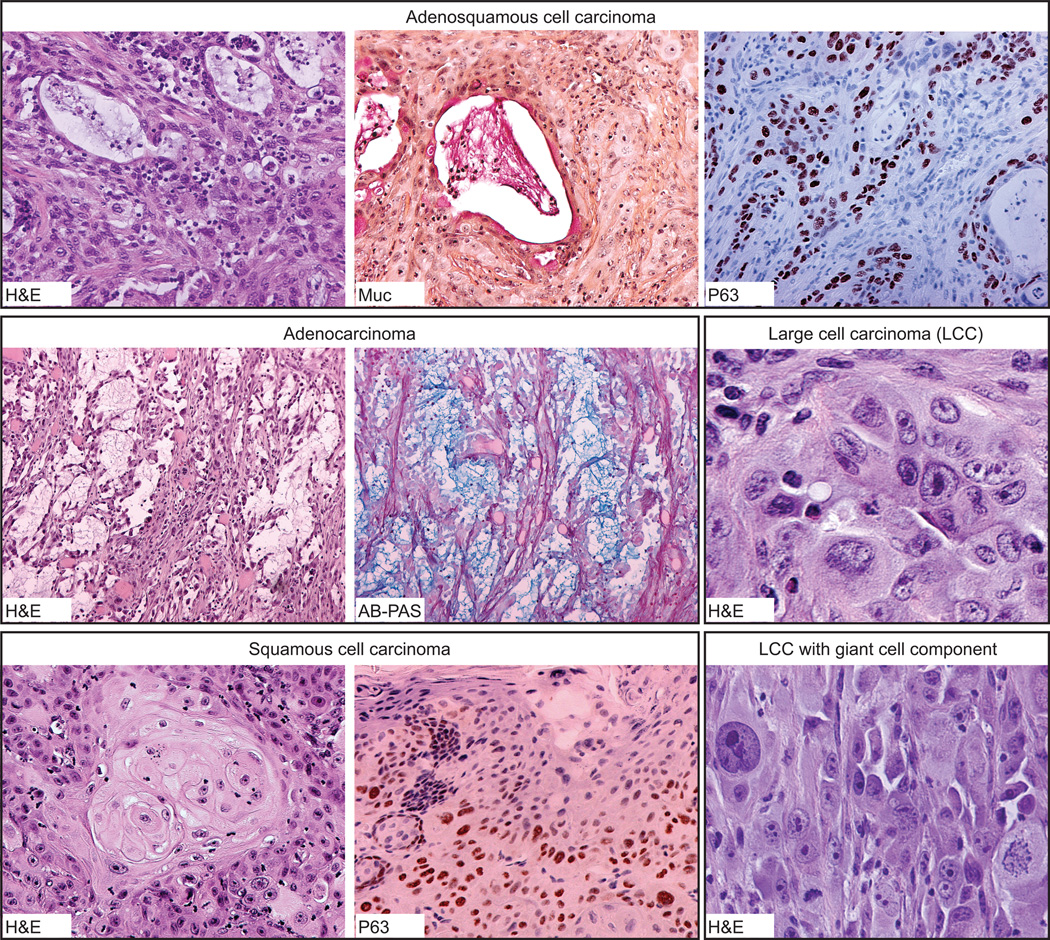
To test the tumor forming ability of HBEC3p53,KRAS and HBEC3p53,KRAS,MYC, cells grown in defined KSFM media were injected subcutaneously into NOD/SCID mice. We had previously found HBEC3p53 with moderate levels of oncogenic KRASV12 (using a retroviral expression vector) failed to form tumors when injected into immunodeficient mice (12). By contrast, transforming with higher levels of KRASV12 resulted in tumor growth in 5/24 (21%) injections (Table 1). Despite the significant increase in in vitro anchorage-independent growth observed in HBEC3p53,KRAS,MYC compared with HBEC3p53,KRAS (Figure 2B), HBEC3p53,KRAS,MYC was only slightly more tumorigenic in vivo with tumors in 3/10 (30%) injections (Table 1). Histopathological analysis showed the oncogenically manipulated HBEC populations formed squamous cell carcinomas, adenocarcinomas, adenosquamous carcinomas, and poorly differentiated carcinomas with typical morphologic features of each histology (Table 1 and Figure 3). Squamous and adenocarcinoma differentiation was confirmed by p63 and mucicarmine/Alcian Blue-PAS positivity, respectively and pathology was verified by an independent lung cancer pathology expert. Adenocarcinomas were found to be strongly cytokeratin 5 positive while negative for Napsin A and the squamous cell marker, cytokeratin 7 (Supplementary Figure S2A). In all cases, adenocarcinoma differentiation was TITF1(NKX2-1)-negative (data not shown), most likely a reflection of the HBECs being derived from central airway cells. The fact that glandular cells are negative for both TITF-1 and Napsin A is not unexpected as the vast majority of adenocarcinomas (and glandular component in adenosquamous carcinomas) in whole tissue sections are either positive for both or negative for both. The development of different tumor histologies from the same HBEC manipulated population suggests either clonal selection or the cells respond to differentiation signals in vivo.
Table 1.
In vivo tumorigenicity of manipulated HBECs
| Cell Line | Mediuma | Tumor Formation Rateb |
Latency (days)c |
Histology |
|---|---|---|---|---|
| HBEC3 p53,KRAS | ||||
| Population | KSFM | 5/24 (21%) | 152 | Squamous cell carcinoma (2) Adenosquamous (2) Poorly differentiated carcinoma (1) |
| Population | R10 | 7/17 (41%) | 128 | Large cell/giant cell carcinoma (3) Adenocarcinoma (2) Squamous cell carcinoma (1) Adenosquamous (1) |
| Clone1 | R10 | 8/8 (100%) | 38 | Large cell/giant cell carcinoma (7) Squamous cell carcinoma (1) |
| Clone5 | R10 | 7/8 (88%) | 57 | Large cell/giant cell carcinoma (5) Papillary adenocarcinoma (1) Poorly differentiated carcinoma (1) |
| Clone11 | R10 | 6/8 (75%) | 117 | Large cell/giant cell carcinoma (3) Poorly differentiated carcinoma (2) Adenocarcinoma (1) |
| Clone6 | R10 | 0/6 (0%) | - | - |
| Clone7 | R10 | 0/7 (0%) | - | - |
| Clone8 | R10 | 0/8 (0%) | - | - |
| Clone9 | R10 | 0/9 (0%) | - | - |
| HBEC3 p53,KRAS,MYC | ||||
| Population | KSFM | 3/10 (30%) | 115 | Adenosquamous (2) Squamous cell carcinoma (1) |
| Population | R10 | 12/12 (100%) | 27 | Large cell/giant cell carcinoma (5) Poorly differentiated carcinoma (4) Squamous cell carcinoma (2) |
| Clone1 | R10 | 9/9 (100%) | 119 | Large cell/giant cell carcinoma (7) Adenocarcinoma (2) |
| Clone2 | R10 | 3/8 (38%) | 90 | Squamous cell carcinoma (2) Large cell/giant cell carcinoma (1) |
| Clone7 | R10 | 10/10 (100%) | 37 | Large cell/giant cell carcinoma (7) Squamous cell carcinoma (3) |
| Clone8 | R10 | 1/6 (17%) | 78 | Large cell/giant cell carcinoma (1) |
| HBEC17 p53,KRAS | ||||
| Population | KSFM | 0/9 (0%) | - | - |
| Population | R10 | 0/8 (0%) | - | - |
| HBEC17 p53,KRAS,MYC | ||||
| Population | KSFM | 0/10 (0%) | - | - |
| Population | R10 | 3/8 (38%) | n.d. | Squamous cell carcinoma (2) Sarcomatoid carcinoma features (1) |
KSFM, Keratinocyte Serum Free Medium; R10, RPMI1640 supplemented with 10% FBS
Number of subcutaneous tumors/number of injections (percentage)
Median time (days) for s.c. xenografts to reach 250mm3; n.d., not determined
Figure 3. Representative formalin-fixed paraffin-embedded sections of subcutaneous xenografts derived from HBEC3p53,KRAS and HBEC3p53,KRAS,MYC.
HBEC3p53,KRAS and HBEC3p53,KRAS,MYC formed subcutaneous tumor reflective of naturally arising lung carcinomas with adenosquamous differentiation (top panel), adenocarcinoma (middle panel on left), and squamous differentiation (lower panel on left), as well as undifferentiated large cell carcinomas, some of which also exhibited a giant cell component (middle and lower panels on right). Squamous and adenocarcinoma differentiation was confirmed with p63 and mucicarmine and/or alcian-blue PAS staining, respectively. The example of adenosquamous cell carcinoma (top panel) clearly shows dual differentiation of peripheral squamous/basal-like cells (p63+/mucin−) and central glandular cells (p63−/mucin+). H&E, hematoxylin and eosin; Muc, mucicarmine; AB-PAS, alcian-blue PAS. Original magnification of images at 10X except adenosquamous H&E and P63 (20X); Large cell carcinoma with giant cell component H&E (20X); and Large cell carcinoma H&E (40X).
Exploration of inter-individual differences in HBEC transformation
To compare inter-individual differences in malignant transformation using the same combination of oncogenic changes, we tested HBEC17, derived from a different individual than HBEC3. We observed a similar transformed phenotype in vitro (Supplementary Figure S2B) however, in contrast to HBEC3, HBEC17 was significantly more resistant to full in vivo transformation (Table 1). The difference in tumor formation rate between HBECs derived from different donors suggests the existence of inter-individual differences in susceptibility to specific oncogene induced malignant transformation.
Growth in serum-containing medium enhances full tumorigenic transformation of HBECs
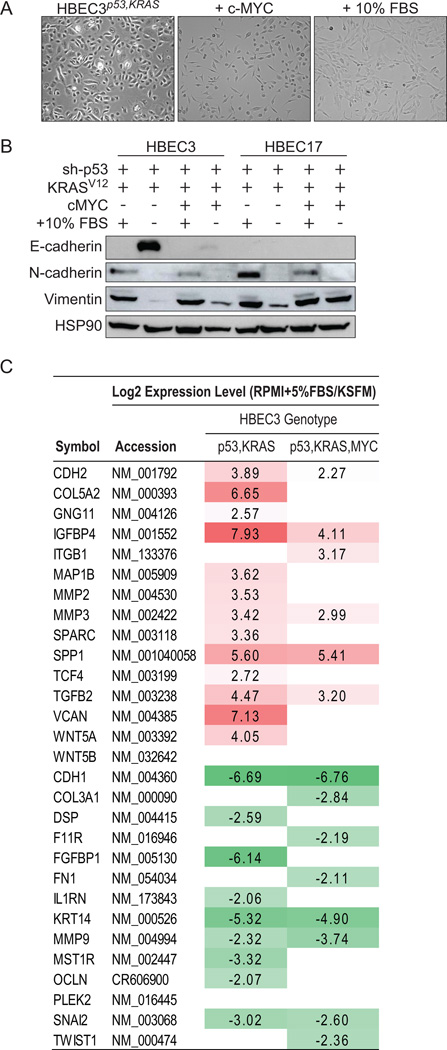
In addition to viral oncoproteins, many previous studies that succeeded in transforming normal cells to malignancy also used serum containing medium instead of defined serum-free medium (31–33). We have previously shown that CDK4/hTERT-immortalized HBECs with no additional oncogenic manipulations require EGF, a supplement present in KSFM medium, for cell growth but oncogenic transformation with p53 knockdown and KRASV12 allows the cells become EGF independent (12). In the present study we show while HBEC3 cannot tolerate growth in RPMI1640 supplemented with serum (FBS), media commonly used for growth of cancer cell lines, HBEC3p53,KRAS is adaptable and will proliferate (Supplementary Figure S3A). Serum-supplemented media has been shown to induce differentiation of epithelial cells in culture (34). Therefore, to further delineate the differences between HBEC3 compared with HBEC3p53,KRAS and HBEC3p53,KRAS,MYC following growth in serum supplemented media, cells grown in either serum-free KSFM or serum-supplemented (5% FBS) KSFM for 96 hr then analyzed for expression of a panel of lung differentiation and cancer stem cell (CSC) markers by qRT-PCR. All three cell lines expressed high levels of basal markers (KRT5 and KRT14), low levels of central airway epithelial cell markers (MUC1 and TUBB4), with undetectable expression of peripheral airway markers (CC10 and SPC) in line with cells derived from bronchial epithelial cells (Supplementary Figure S3B). ALDH activity has been shown to be a marker of CSCs in the lung (28). We therefore measured the expression of two ALDH isozymes, ALDH1A1 and ALDH1A3, to find all three cell lines expressed ALDH1A3 with much lower expression of ALDH1A1. Growth in serum-containing medium resulted in all three cell lines exhibiting significant decreases in the expression of basal markers (KRT5 and KRT14), most notably in HBEC3 (Supplementary Figure S3C). Expression of central airway markers, particularly MUC1, increased more than 10-fold in HBEC3 when cells were grown in serum-supplemented medium whereas HBEC3p53,KRAS and HBEC3p53,KRAS,MYC showed little if any increase in these markers. Expression of CSC markers ALDH1A1 and ALDH1A3 increased in HBEC3 and HBEC3p53,KRAS but not in HBEC3p53,KRAS,MYC. Morphologically, growth in serum-supplemented medium resulted in HBEC3 cells developing a flattened morphology representative of a differentiated state (Supplementary Figure S3D) whereas this was largely absent in HBEC3p53,KRAS cells and completely absent in HBEC3p53,KRAS,MYC. Furthermore, HBEC3p53,KRAS,MYC and to a lesser extent HBEC3p53,KRAS developed an elongated mesenchymal morphology after short-term growth in serum (Supplementary Figure S3D). Together, these data show serum-supplemented medium induces differentiation in parental HBEC3 cells however oncogenic transformation enables the cells to resist serum-induced differentiation and instead undergo EMT.
We reasoned that partially transformed HBECs that have adapted to growth in serum-supplemented RPMI1640 may differ in tumorigenicity compared to cells grown in KSFM. Therefore, to compare the effect of genetic and environmental manipulations, HBEC3p53,KRAS and HBEC3p53,KRAS,MYC were grown in either defined KSFM (serum-free) medium or RPMI medium supplemented with 10% FBS (R10) for at least three weeks, then injected subcutaneously in NOD/SCID mice (Table 1). Growth in serum-containing medium markedly increased in vivo tumorigenicity, decreased tumor latency, and tumors in general were more undifferentiated (e.g. large cell and giant cell carcinomas), particularly in HBEC3p53,KRAS,MYC (Table 1 and Figure 3). This effect is similar to what is observed in patients where poorly differentiated lung tumors are generally associated with aggressive tumor growth. To determine if the rate of apoptosis differed between poor and well differentiated xenograft tumors, FFPE sections were analyzed with a TUNEL (TdT-mediated dUTP Nick-End Labeling) assay. There was no significant difference in the rate of apoptosis in relation to differentiation although a moderate-well differentiated SCC and giant cell carcinoma showed the greatest amount of TUNEL staining (Supplemental Figure S3E). Overall, the increase in tumorigenicity of oncogenically progressed HBECs after growth in serum demonstrates the influential role of exogenous serum-derived factors in the malignant progression of lung cancer.
Malignant transformation of HBECs is enhanced by epithelial to mesenchymal transition (EMT), induced by either c-MYC or growth in serum
In HBEC3p53,KRAS cells, over-expression of c-MYC (in defined KSFM medium) or growth in serum-containing RPMI1640 medium led to increased oncogenic transformation, as shown by soft agar colony formation and in vivo tumor growth. Both of these manipulations also led to HBEC3p53,KRAS cells exhibiting a more mesenchymal-like morphology (Figure 4A). Loss of E-cadherin (epithelial marker) and gain of vimentin and N-cadherin (mesenchymal markers) confirmed an epithelial-mesenchymal transition (EMT) (Figure 4B). c-MYC- or serum-induced EMT was also seen in HBEC17 cells, derived from another individual, with p53 and KRASV12 manipulations (HBEC17p53,KRAS) (Figure 4B). Whole-genome mRNA profiling of HBEC3p53,KRAS and HBEC3p53,KRAS,MYC cells grown in KSFM or R10 confirmed a significant over-representation of EMT-related genes (P = 1.04 × 10−16 and P = 1.38 × 10−09 for HBEC3p53,KRAS and HBEC3p53,KRAS,MYC, respectively) (Figure 4C), with up-regulation of EMT-promoting genes following growth in serum-containing media.
Figure 4. c-MYC over-expression or growth in serum-containing media induces EMT in HBEC3p53,KRAS.
A) Phase contrast photomicrographs showing the morphological effect observed in HBEC3p53,KRAS (left) following over-expression of c-MYC (middle) or switching from defined serum-free media to serum-containing media (right) (20X magnification). B) Immunoblot for EMT markers in oncogenically manipulated HBEC3 and HBEC17 grown in either KSFM (serum free) or serum-containing (R10) media. The presence (+) or absence (−) of serum is shown. C) EMT-related genes altered 4-fold or greater in pairwise analysis of HBEC3p53,KRAS and HBEC3p53,KRAS,cMYC comparing cells grown in serum or defined medium (KSFM). Values log2 transformed. *P < 0.05, **P < 0.01 (t-test). Full-length blots are presented in Supplementary Figure S8).
Isogenic soft agar clones of oncogenically manipulated HBEC3 represent independent genetic events with distinct in vivo growth, tumor histology and differentiation
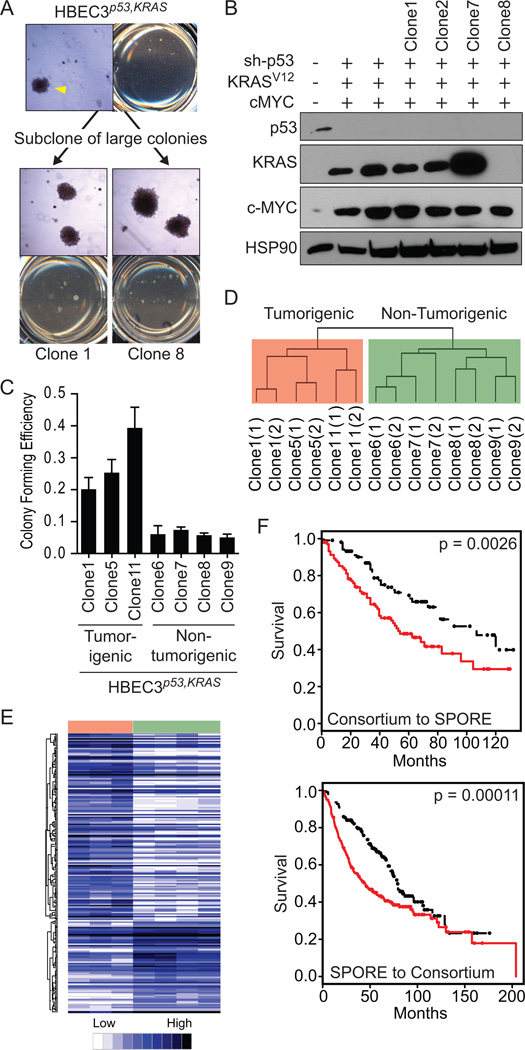
The genetic combinations of shp53+KRASV12 or shp53+KRASV12+c-MYC in HBEC3 led to the formation of a small subset (0.5–1.5% of all soft agar colonies) of very large, macroscopically visible (>1mm) colonies when grown in soft agar, which was not observed with any single manipulation (shp53, KRASV12 or c-MYC) nor dual combination of sh-p53 or KRASV12 with c-MYC (Figure 2C). Seven HBEC3p53,KRAS clones and four HBEC3p53,KRAS,MYC clones were isolated from these large colonies and repeat soft agar assays confirmed the large colony phenotype was maintained (Figure 5A). Southern blotting demonstrated the clones represent independent transformation events as indicated by discrete digestion patterns (Supplementary Figure S4A) and immunoblotting confirmed HBEC3p53,KRAS and HBEC3p53,KRAS,MYC clones maintained their exogenously introduced oncogenic manipulations (Figure 1C and 5B, respectively) (summarized in Supplementary Table S2). While immortalized but non-transformed HBECs preferentially grow in serum-free conditions, they require serum for anchorage-independent growth (Supplementary Figure S4B). Remarkably, following isolation from soft agar (in KSFM+20% FBS) nine of the eleven clones were serum-growth factor dependent as they could no longer tolerate growth in serum-free KSFM medium (Supplementary Table S2). Henceforth, all clones were grown in RPMI-1640 medium containing 10% FBS (R10).
Figure 5. Isolation of large soft agar clones from HBEC3p53,KRAS and HBEC3p53,KRAS,MYC identifies tumorigenic and non-tumorigenic clones and genome-wide mRNA expression profiling of HBEC3p53,KRAS soft agar clones identifies a clinically-applicable signature of prognosis.
A) Uncloned, parental populations of HBEC3p53,KRAS and HBEC3p53,KRAS,MYC form very large (>1mm diameter) soft agar colonies (arrowhead). These large colonies were isolated, expanded, and retested for soft agar colony formation where they maintained the ability to form large soft agar colonies (representative soft agar pictures of two clones) (4X magnification). B) Immunoblot of HBEC3p53,KRAS,MYC soft agar clones confirming p53, KRASV12 and c-MYC manipulations. The presence (+) or absence (−) of each manipulation is shown. C) Anchorage-independent soft agar colony formation ability of HBEC3p53,KRAS soft agar clones. D) Unsupervised hierarchical clustering of whole-genome mRNA expression profiles of HBEC3p53,KRAS soft agar clones harvested at two time points (denoted “(1)” and “(2)”) spanning a three week interval. E) A supervised analysis comparing HBEC3p53,KRAS tumorigenic (Clone1, Clone5, and Clone11) with HBEC3p53,KRAS non-tumorigenic (Clone6, Clone7, Clone8, and Clone9) clones identified 203 probes, representing 171 unique genes, significantly differentially expressed (SAM, FDR = 5%). Samples (represented horizontally: red, tumorigenic clones; green, non-tumorigenic clones) and probes (represented vertically) were clustered using centered Pearson clustering. F) Kaplan-Meier log-rank analysis of overall survival in lung cancer patients predicted to have good (black) or poor (red) outcome using the 171 probe signature HBEC3p53,KRAS soft agar signature. A supervised principal component analysis was used to train the model in one dataset (Consortium) and test in a second dataset (SPORE) (above) then the datasets were reversed to test for model robustness (below). Full-length blots are presented in Supplementary Figure S8).
Injection of the large, soft agar clones into NOD/SCID mice found all four HBEC3p53,KRAS,MYC clones were tumorigenic whereas only three of seven HBEC3p53,KRAS clones formed tumors (Table 1). Thus, even with four oncogenic changes and biologic selection there is dramatic clonal heterogeneity. This also showed the tumorigenicity of the clones reflected the tumorigenicity of the parental population (when grown in R10) where approximately 40% of mice injected with HBEC3p53,KRAS grew tumors compared with 100% of mice injected with HBEC3p53,KRAS,MYC (Table 1). Growth rate and tumor histology differed between tumorigenic clones. While subcutaneous tumors were largely undifferentiated (e.g. large cell and giant cell carcinomas), some xenograft tumors exhibited squamous and adenocarcinoma differentiation (Table 1). This differentiation was clone specific, as no clone differentiated into multiple histologies.
Oncogenic transformation of HBECs increases sensitivity to standard lung cancer chemotherapies
To determine if oncogenic transformation of HBECs altered their sensitivity to standard lung cancer chemotherapies we tested three platinum-based doublets (paclitaxel-carboplatin, gemcitabine-cisplatin, and pemetrexed-cisplatin) currently in use for NSCLC treatment. Oncogenic manipulation of HBEC3 with sh-p53 and KRASV12 resulted in a significant increase in sensitivity to gemcitabine-cisplatin and pemetrexed-cisplatin doublet therapy in vitro (approximately 6-fold and 10-fold, respectively), but not paclitaxel-carboplatin (Supplementary Table S3) (Supplementary Figure S5) (one-way ANOVA, P < 0.001). Overall, the soft agar clones of HBEC3p53,KRAS demonstrated sensitivities comparable to the parental HBEC3p53,KRAS cell line with Clone5 and Clone7 showing intermediate sensitivity. Thus, these tumorigenically progressed HBECs could provide a cell context appropriate, isogenic model system for identifying genetic differences regulating sensitivity and resistance to platinum doublet chemotherapy used in the treatment of NSCLCs.
mRNA profiles of tumorigenic versus non-tumorigenic HBEC3p53,KRAS soft agar clones predict prognosis and histology in clinical lung tumor specimens
The identification of tumorigenic and non-tumorigenic clones of HBEC3p53,KRAS that share the same genetic manipulations (sh-p53 and KRASV12) and biologic selection (large, soft agar colonies) presents a unique cell model to characterize spontaneous transformation events that progress HBECs to full malignancy. Biochemical assays suggest the expression level of KRASV12 is a major contributor towards full transformation (Figure 1C, 1D, 5C) whereas other tumorigenic events such as dysregulation of cell cycle did not differ between clones (Supplementary Figure S6) (summarized in Supplementary Table S2). To analyze molecular differences between tumorigenic versus non-tumorigenic clones the clones were profiled with whole-genome mRNA expression microarrays. The mRNA expression profile of each clone remained stable in culture as shown by comparing mRNAs collected at a three week interval in unsupervised clustering (Figure 5D). Tumorigenic and non-tumorigenic clones separated into two distinct clusters suggesting the clones exhibit a strong expression profile associated with their tumorigenic phenotype, and supervised analysis comparing tumorigenic with non-tumorigenic clones found 171 unique genes (203 probes) that were differentially expressed (SAM, FDR = 5%) (Supplementary Table S4) (Figure 5E).
We tested the ability of the expression patterns of the 171 genes to predict overall survival and disease-free survival in two independent lung tumor cohorts; the SPORE dataset of resected early stage NSCLCs (adeno- and squamous carcinomas) (n=209) and a second dataset of primary lung adenocarcinoma samples (n=442) (NCI Director’s Challenge Consortium dataset). Prediction models were built using supervised principal component analysis and datasets were interchanged as training and testing datasets to test for robustness. Irrespective of dataset, the soft agar clone tumorigenic versus non-tumorigenic signature was able to identify patients with significantly worse overall (Figure 5F) and recurrence-free (Supplementary Figure S7) survival. The successful application of a gene signature derived from an isogenic in vitro model of human bronchial epithelial cell transformation in predicting outcome in clinical lung tumor samples indicates the power of such a system and provides a preclinical model for testing the functional importance of the genes in the signature.
Discussion
In this study we sought to characterize the stepwise progression of lung cancer pathogenesis by introducing defined genetic manipulations commonly found in lung cancer into an in vitro human bronchial epithelial cell model system (Figure 6). We found the expression level of mutant KRAS is a critical transformative factor in HBECs, however inactivation of p53 signaling is required to evade the tumor suppressive barrier of RAS-induced senescence. We also show EMT is an important oncogenic step, where c-MYC or growth in serum-containing medium both spontaneously induced an EMT and led to increased tumorigenicity. In HBECs derived from multiple individuals we demonstrate five genetic changes (hTERT and Cdk4 to immortalize the cells, followed by p53 knockdown, mutant KRASV12, and c-MYC overexpression) together with serum-induced EMT are able to transform cells to a fully malignant state. Tumor xenografts of transformed HBECs were typical of lung cancer but varied in histology, suggesting histologically distinct lung cancers from the central bronchus may originate from a multipotent stem-like cell. Interestingly, clonal analysis of sh-p53+KRASV12-transformed HBECs found the isogenic cells exhibited distinct phenotypes in terms of in vivo tumorigenicity, xenograft histology and drug response. The mRNA profile that distinguished tumorigenic from non-tumorigenic clones was also able to identify a subset of primary lung tumor patients with significantly poorer survival. Together, this demonstrates the utility of the HBEC system as a preclinical model of lung cancer to understand transformation events.
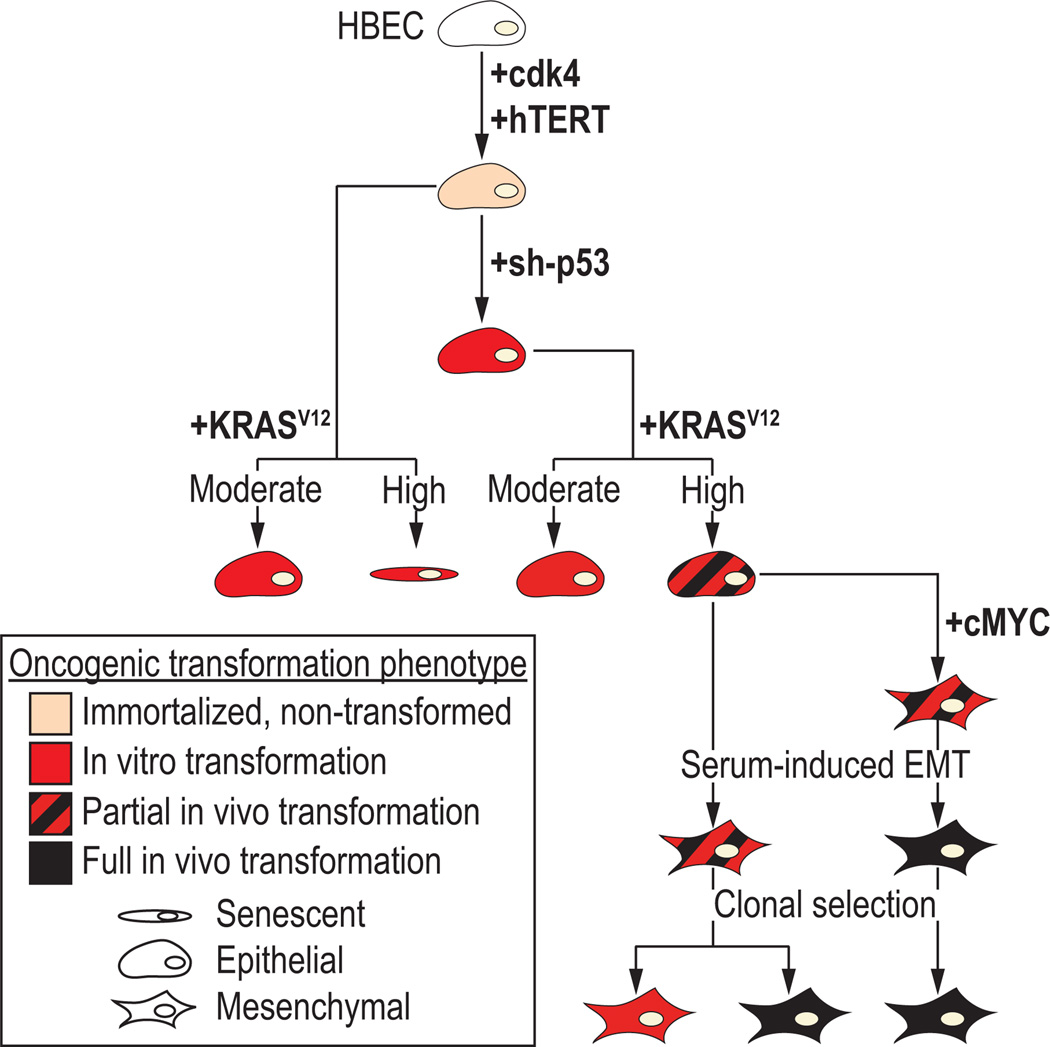
Figure 6. Model of malignant transformation of in vitro human bronchial epithelial cells (HBECs) following stepwise introduction of common lung cancer mutations.
The experimental data presented in this paper identify the following steps: Step 1, CDK4 and hTERT immortalized, human bronchial epithelial cells (HBECs) are non-transformed and lack of anchorage-independent growth in soft agar; Step 2, in vitro transformation as defined by anchorage-independent growth in soft agar is achieved with the single manipulation of loss of p53, moderate KRASV12 expression or both, while expression of high levels of KRASV12 expression leads to in vitro transformation with significant cellular senescence; Step 3, partial in vivo transformation with subcutaneous tumor growth in immunocompromised mice in 30–80% of injections is observed with the combination of p53 loss and high KRASV12; Step 4, an epithelial to mesenchymal transition (EMT) occurs following over-expression of cMYC or growth in serum-containing media; Step 5, combination of cMYC over-expression and growth in serum-containing media results in complete oncogenic transformation of HBECs with tumor growth in vivo observed in >80% of injections in immunocompromised mice. Clonal selection of partially transformed HBECs identifies tumorigenic and non-tumorigenic clones.
Mutant KRAS lung cancers exhibit marked differences in the expression level of KRAS. We found the expression level of oncogenic KRASV12 was an important factor in tumorigenic transformation of HBECs suggesting it exerts its oncogenic effect in a dose-dependent manner, similar to HRASV12 in a human mammary epithelial cell (HMEC) transformation model (18). While higher levels of oncogenic KRAS enhanced HBEC transformation, it also triggered oncogene-induced senescence. Oncogene-induced senescence serves as an important tumor-suppressive barrier in response to persistent oncogenic insult by engaging proliferative arrest through p53 or p16/Rb (35). We found that immortalized HBECs, where the p16/Rb pathway is bypassed as a result of over-expression of CDK4, could largely evade KRASV12-induced senescence with knockdown of p53. Loss of p53 function, therefore, is a key step in the malignant transformation of HBECs by allowing the cells to tolerate high levels of oncogenic KRAS, a driver of malignant transformation. Transformation studies in HMECs, which undergo spontaneous methylation-mediated p16 silencing, also report high levels but not low levels of oncogenic RAS engage senescence machinery (36, 37). However in HMECs, HRAS-induced senescence is mediated through TGF-β signaling in a p53-independent manner. Thus, the mechanism of RAS-induced senescence differs between in vitro epithelial cell models, most likely a consequence of immortalization methods or the cell of origin.
Our data shows micro-environmental signals such as those provided by growth media can strongly influence the transformation of HBECs. Switching immortalized, but non-transformed HBECs from defined, serum-free culture medium to serum-containing medium induced inhibition of cell growth with induction of central airway differentiation markers. When immortalized HBEC with additional oncogenic manipulations are switched to serum-containing medium however, the cells are able to bypass serum-induced differentiation and instead become mesenchymal and more tumorigenic, with a greater frequency of undifferentiated tumors. Over-expression of c-MYC in HBEC3p53,KRAS also induced an EMT and increased tumorigenicity. c-MYC has been shown to induce EMT in TERT-immortalized HMECs (38). In our study, c-MYC induced EMT only in the presence of p53 and KRASV12 alterations and not with c-MYC over-expression alone (data not shown). While c-MYC over-expression or growth in serum-containing media both caused HBEC3p53,KRAS to undergo EMT and increase tumorigenicity, the presence of both manipulations had a synergistic effect resulting in full malignant transformation in HBECs from two individuals. This suggests their tumor-promoting effects signal through mutually exclusive pathways.
In the present study we found inter-individual differences in HBEC transformation suggesting HBECs derived from different donors can vary in their susceptibility to specific oncogene induced malignant transformation. In terms of in vitro anchorage-independent transformation, neither cell line demonstrated anchorage-independent growth following immortalization (CDK4 and hTERT) yet the combination of five genes (CDK4, hTERT, sh-p53, KRASV12, and cMYC) resulted in a colony forming efficiency of ~60% in HBEC3 compared with <15% in HBEC17. These differences could potentially stem from multiple factors including germline polymorphisms (such as SNPs), somatically-acquired mutations derived from either the patient (such as age or environmental exposures) or laboratory practices (such as time in culture), or epigenetic mechanisms all of which may predispose the cells to oncogenic transformation. The patients from which HBEC3 and HBEC17 were established differ in respect to known risk factors for lung cancer such as age and smoking history and it is likely they also differ in respect to unknown germline and/or somatic alterations. Thus, a comprehensive survey of genomic alterations present in HBECs prior to genetic manipulation (such as by whole-genome sequencing) would provide better indication of the level of existing pre-malignant susceptibilities, but these experiments are beyond the scope of the current study.
The lung can be divided into central and peripheral compartments (39). Squamous cell carcinomas usually arise from the central compartment, while adenocarcinomas may arise from either compartment, illustrating the importance of studying oncogenic transformation in both central and peripheral lung epithelial cell models. Many HBEC-derived tumors in our study were p63 positive with squamous differentiation, which likely reflects a stem/basal cell origin. A fewer number of tumors demonstrated distinct dual squamous and adenocarcinoma differentiation or adenocarcinoma differentiation alone. A study using SV40-immortalized tracheobronchial and small airway epithelial cells found both cell types could be fully transformed with oncogenic RAS (8). Another study using non-viral oncoproteins immortalized human small airway epithelial cells (HSAECs) with CDK4, hTERT and a dominant-negative form of p53 (p53CT) and transformed the cells using low levels of KRASV12 plus c-MYC, PIK3CAH1047R, cyclin D1, or LKB1D194A (15). In HBECs, we have shown p53 mutation is not required for immortalization (6) and moreover, it increases oncogenic transformation (6, 12). This disparity may signify intrinsic differences between centrally- and peripherally-derived immortalized epithelial cell models. Taken together, however, our study of defined oncogenic transformation of HBECs (both in the present study and previously (12)) and the study in HSAECs by Sasai et al (15) largely correspond with respect oncogenic transformation of bronchial epithelial cells. We previously showed transformation of HBECs with low levels of KRASV12 (with CDK4, hTERT and p53 knockdown) resulted in a modest increase in anchorage-independent growth but no tumor formation in vivo (12). Sasai et al reported that transformation of HSAECs with low levels of KRASV12 (with CDK4, hTERT and p53CT) failed to yield anchorage-independent or in vivo growth. The authors were able to fully transform HSAECs using low levels of KRASV12 only with additional genetic alterations. In the present study we used a different approach by increasing the level of KRASV12 expression to simulate the high amplification-associated expression often observed in lung cancer (40). We found higher levels of the oncogene resulted in increased in vitro and in vivo transformation compared with lower levels of KRASV12, which could be further increased with a fifth genetic alteration such as cMYC over-expression. Thus, studies in HBECs and HSAECs show increasing the number of oncogenic alterations will increase cellular transformation whereas in the present study, we also show that increasing the level of certain oncogenic alterations can also increase transformation.
In conclusion, using the HBEC system as a progression model of lung cancer we were able to study early transformative events in bronchial epithelial cells and the mechanisms employed to overcome tumor-suppressive barriers. We show HBECs can be transformed to full malignancy by introducing defined genetic manipulations to produce histologically similar lung tumors in xenografts, indicating our in vitro HBEC model retains characteristics of the tissue of origin. Furthermore, we demonstrate HBECs can model preneoplastic changes and spontaneous transformation events such as oncogene-induced senescence and EMT and have clinically translatable applications as shown in isogenic clones exhibiting distinct drug response and tumorigenic phenotypes. Thus, genetically defined in vitro models such as HBECs will be an invaluable tool to assess the contribution of specific genes towards lung cancer pathogenesis, pertinent to recent whole-genome sequencing efforts, and to screen for novel therapeutic compounds directed at oncogenically acquired, tumor-specific vulnerabilities.
Supplementary Material
Acknowledgments
This work was supported by the National Cancer Institute Lung Cancer Specialized Program of Research Excellence (SPORE) (P50CA70907), NASA NSCOR (NNJ05HD36G). JEL supported by NHMRC Biomedical Fellowship (494511) and TSANZ/Allen & Hanburys Respiratory Research Fellowship. We thank Natasha Rekhtman, MD, PhD (Memorial Sloan-Kettering Cancer Center) for her kind assistance in analyzing immunohistochemical sections. We thank the many current and past members of the Minna lab for their technical assistance and manuscript comments, particularly Luc Girard, PhD; Suzie Hight, MS; and Michael Peyton, PhD.
Footnotes
The authors disclose no potential conflicts of interest.
References
- 1.Larsen JE, Minna JD. Molecular biology of lung cancer: clinical implications. Clin Chest Med. 2011;32:703–740. doi: 10.1016/j.ccm.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rudin CM, Durinck S, Stawiski EW, Poirier JT, Modrusan Z, Shames DS, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet. 2012 doi: 10.1038/ng.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Govindan R, Ding L, Griffith M, Subramanian J, Dees ND, Kanchi KL, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell. 2012;150:1121–1134. doi: 10.1016/j.cell.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramirez RD, Sheridan S, Girard L, Sato M, Kim Y, Pollack J, et al. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res. 2004;64:9027–9034. doi: 10.1158/0008-5472.CAN-04-3703. [DOI] [PubMed] [Google Scholar]
- 7.Reddel RR, Ke Y, Gerwin BI, McMenamin MG, Lechner JF, Su RT, et al. Transformation of human bronchial epithelial cells by infection with SV40 or adenovirus-12 SV40 hybrid virus, or transfection via strontium phosphate coprecipitation with a plasmid containing SV40 early region genes. Cancer Res. 1988;48:1904–1909. [PubMed] [Google Scholar]
- 8.Lundberg AS, Randell SH, Stewart SA, Elenbaas B, Hartwell KA, Brooks MW, et al. Immortalization and transformation of primary human airway epithelial cells by gene transfer. Oncogene. 2002;21:4577–4586. doi: 10.1038/sj.onc.1205550. [DOI] [PubMed] [Google Scholar]
- 9.Cheng J, DeCaprio JA, Fluck MM, Schaffhausen BS. Cellular transformation by Simian Virus 40 and Murine Polyoma Virus T antigens. Semin Cancer Biol. 2009;19:218–228. doi: 10.1016/j.semcancer.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arroyo JD, Hahn WC. Involvement of PP2A in viral and cellular transformation. Oncogene. 2005;24:7746–7755. doi: 10.1038/sj.onc.1209038. [DOI] [PubMed] [Google Scholar]
- 11.Kendall SD, Linardic CM, Adam SJ, Counter CM. A network of genetic events sufficient to convert normal human cells to a tumorigenic state. Cancer Res. 2005;65:9824–9828. doi: 10.1158/0008-5472.CAN-05-1543. [DOI] [PubMed] [Google Scholar]
- 12.Sato M, Vaughan MB, Girard L, Peyton M, Lee W, Shames DS, et al. Multiple oncogenic changes (K-RAS(V12), p53 knockdown, mutant EGFRs, p16 bypass, telomerase) are not sufficient to confer a full malignant phenotype on human bronchial epithelial cells. Cancer Res. 2006;66:2116–2128. doi: 10.1158/0008-5472.CAN-05-2521. [DOI] [PubMed] [Google Scholar]
- 13.Vaughan MB, Ramirez RD, Wright WE, Minna JD, Shay JW. A three-dimensional model of differentiation of immortalized human bronchial epithelial cells. Differentiation. 2006;74:141–148. doi: 10.1111/j.1432-0436.2006.00069.x. [DOI] [PubMed] [Google Scholar]
- 14.Delgado O, Kaisani AA, Spinola M, Xie XJ, Batten KG, Minna JD, et al. Multipotent capacity of immortalized human bronchial epithelial cells. PLoS One. 2011;6:e22023. doi: 10.1371/journal.pone.0022023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sasai K, Sukezane T, Yanagita E, Nakagawa H, Hotta A, Itoh T, et al. Oncogene-mediated human lung epithelial cell transformation produces adenocarcinoma phenotypes in vivo. Cancer Res. 2011;71:2541–2549. doi: 10.1158/0008-5472.CAN-10-2221. [DOI] [PubMed] [Google Scholar]
- 16.Toyooka S, Tsuda T, Gazdar AF. The TP53 gene, tobacco exposure, and lung cancer. Hum Mutat. 2003;21:229–239. doi: 10.1002/humu.10177. [DOI] [PubMed] [Google Scholar]
- 17.Sato M, Shames DS, Gazdar AF, Minna JD. A translational view of the molecular pathogenesis of lung cancer. J Thorac Oncol. 2007;2:327–343. doi: 10.1097/01.JTO.0000263718.69320.4c. [DOI] [PubMed] [Google Scholar]
- 18.Elenbaas B, Spirio L, Koerner F, Fleming MD, Zimonjic DB, Donaher JL, et al. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 2001;15:50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 20.Heighway J, Hasleton PS. c-Ki-ras amplification in human lung cancer. Br J Cancer. 1986;53:285–287. doi: 10.1038/bjc.1986.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gazdar AF, Girard L, Lockwood WW, Lam WL, Minna JD. Lung cancer cell lines as tools for biomedical discovery and research. J Natl Cancer Inst. 2010;102:1310–1321. doi: 10.1093/jnci/djq279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phelps RM, Johnson BE, Ihde DC, Gazdar AF, Carbone DP, McClintock PR, et al. NCI-Navy Medical Oncology Branch cell line data base. J Cell Biochem Suppl. 1996;24:32–91. doi: 10.1002/jcb.240630505. [DOI] [PubMed] [Google Scholar]
- 23.Vikis H, Sato M, James M, Wang D, Wang Y, Wang M, et al. EGFR-T790M is a rare lung cancer susceptibility allele with enhanced kinase activity. Cancer Res. 2007;67:4665–4670. doi: 10.1158/0008-5472.CAN-07-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ihle NT, Byers LA, Kim ES, Saintigny P, Lee JJ, Blumenschein GR, et al. Effect of KRAS oncogene substitutions on protein behavior: implications for signaling and clinical outcome. J Natl Cancer Inst. 2012;104:228–239. doi: 10.1093/jnci/djr523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Louro ID, Bailey EC, Li X, South LS, McKie-Bell PR, Yoder BK, et al. Comparative gene expression profile analysis of GLI and c-MYC in an epithelial model of malignant transformation. Cancer Res. 2002;62:5867–5873. [PubMed] [Google Scholar]
- 26.Sato M, Girard L, Sekine I, Sunaga N, Ramirez RD, Kamibayashi C, et al. Increased expression and no mutation of the Flap endonuclease (FEN1) gene in human lung cancer. Oncogene. 2003;22:7243–7246. doi: 10.1038/sj.onc.1206977. [DOI] [PubMed] [Google Scholar]
- 27.Sato M, Sekido Y, Horio Y, Takahashi M, Saito H, Minna JD, et al. Infrequent mutation of the hBUB1 and hBUBR1 genes in human lung cancer. Jpn J Cancer Res. 2000;91:504–509. doi: 10.1111/j.1349-7006.2000.tb00974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sullivan JP, Spinola M, Dodge M, Raso MG, Behrens C, Gao B, et al. Aldehyde dehydrogenase activity selects for lung adenocarcinoma stem cells dependent on notch signaling. Cancer Res. 2010;70:9937–9948. doi: 10.1158/0008-5472.CAN-10-0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greer RM, Peyton M, Larsen JE, Girard L, Xie Y, Gazdar AF, et al. SMAC mimetic (JP1201) sensitizes non-small cell lung cancers to multiple chemotherapy agents in an IAP-dependent but TNF-alpha-independent manner. Cancer Res. 2011;71:7640–7648. doi: 10.1158/0008-5472.CAN-10-3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shedden K, Taylor JM, Enkemann SA, Tsao MS, Yeatman TJ, Gerald WL, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. 2008;14:822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW, Weinberg RA. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 32.Boehm JS, Hession MT, Bulmer SE, Hahn WC. Transformation of human and murine fibroblasts without viral oncoproteins. Mol Cell Biol. 2005;25:6464–6474. doi: 10.1128/MCB.25.15.6464-6474.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Campbell PM, Groehler AL, Lee KM, Ouellette MM, Khazak V, Der CJ. K-Ras promotes growth transformation and invasion of immortalized human pancreatic cells by Raf and phosphatidylinositol 3-kinase signaling. Cancer Res. 2007;67:2098–2106. doi: 10.1158/0008-5472.CAN-06-3752. [DOI] [PubMed] [Google Scholar]
- 34.Masui T, Wakefield LM, Lechner JF, LaVeck MA, Sporn MB, Harris CC. Type beta transforming growth factor is the primary differentiation-inducing serum factor for normal human bronchial epithelial cells. Proc Natl Acad Sci U S A. 1986;83:2438–2442. doi: 10.1073/pnas.83.8.2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Courtois-Cox S, Jones SL, Cichowski K. Many roads lead to oncogene-induced senescence. Oncogene. 2008;27:2801–2809. doi: 10.1038/sj.onc.1210950. [DOI] [PubMed] [Google Scholar]
- 36.Cipriano R, Kan CE, Graham J, Danielpour D, Stampfer M, Jackson MW. TGF-beta signaling engages an ATM-CHK2-p53-independent RAS-induced senescence and prevents malignant transformation in human mammary epithelial cells. Proc Natl Acad Sci U S A. 2011;108:8668–8673. doi: 10.1073/pnas.1015022108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin S, Yang J, Elkahloun AG, Bandyopadhyay A, Wang L, Cornell JE, et al. Attenuation of TGF-beta signaling suppresses premature senescence in a p21-dependent manner and promotes oncogenic Ras-mediated metastatic transformation in human mammary epithelial cells. Mol Biol Cell. 2012;23:1569–1581. doi: 10.1091/mbc.E11-10-0849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cowling VH, Cole MD. E-cadherin repression contributes to c-Myc-induced epithelial cell transformation. Oncogene. 2007;26:3582–3586. doi: 10.1038/sj.onc.1210132. [DOI] [PubMed] [Google Scholar]
- 39.Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers--a different disease. Nat Rev Cancer. 2007;7:778–790. doi: 10.1038/nrc2190. [DOI] [PubMed] [Google Scholar]
- 40.Sunaga N, Kaira K, Imai H, Shimizu K, Nakano T, Shames DS, et al. Oncogenic KRAS-induced epiregulin overexpression contributes to aggressive phenotype and is a promising therapeutic target in non-small-cell lung cancer. Oncogene. 2012 doi: 10.1038/onc.2012.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.