Abstract
Vertebrate ALDH2 genes encode mitochondrial enzymes capable of metabolizing acetaldehyde and other biological aldehydes in the body. Mammalian ALDH1B1, another mitochondrial enzyme sharing 72% identity with ALDH2, is also capable of metabolizing acetaldehyde but has a tissue distribution and pattern of activity distinct from that of ALDH2. Bioinformatic analyses of several vertebrate genomes were undertaken using known ALDH2 and ALDH1B1 amino acid sequences. Phylogenetic analysis of many representative vertebrate species (including fish, amphibians, birds and mammals) indicated the presence of ALDH1B1 in many mammalian species and in frogs (Xenopus tropicalis); no evidence was found for ALDH1B1 in the genomes of birds, reptiles or fish. Predicted vertebrate ALDH2 and ALDH1B1 subunit sequences and structures were highly conserved, including residues previously shown to be involved in catalysis and coenzyme binding for human ALDH2. Studies of ALDH1B1 sequences supported the hypothesis that the ALDH1B1 gene originated in early vertebrates from a retrotransposition of the vertebrate ALDH2 gene. Given the high degree of similarity between ALDH2 and ALDH1B1, it is surprising that individuals with an inactivating mutation in ALDH2 (ALDH2*2) do not exhibit a compensatory increase in ALDH1B1 activity. We hypothesized that the similarity between the two ALDHs would allow for dominant negative heterotetramerization between the inactive ALDH2 mutants and ALDH1B1. Computational-based molecular modeling studies examining predicted protein-protein interactions indicated that heterotetramerization between ALDH2 and ALDH1B1 subunits was highly probable and may partially explain a lack of compensation by ALDH1B1 in ALDH2*2 individuals.
Keywords: Aldehyde dehydrogenases, ALDH1B1, ALDH2, ALDH2*2, Heterotetramerization, Retrotransposition
1. Introduction
The aldehyde dehydrogenase (ALDH; EC 1.2.1.3) gene superfamily (http://aldh.org/superfamily.php) encodes ALDHs which participate in metabolic pathways involving aldehydes in the body, examples of which include alcohol, retinoids, neurotransmitters, lipids, amino acids, drugs and xenobiotics [1]. The human ALDH2 gene is localized on chromosome 12 [2] and encodes liver mitochondrial ALDH2 which functions in acetaldehyde and peroxidic aldehyde metabolism [1,3,4]. The physiological importance of ALDH2 is illustrated best in East Asian human subjects possessing a dominant inactive genetic variant, ALDH2*2 (E487 K). This variant results in alcohol ‘flushing’ and a lowered acetaldehyde clearance capacity, arising from lowered coenzyme (NAD+) binding affinity and is generally considered inactive (greater than 90% loss of enzyme activity) [5–9]. Typically, ALDH2 forms homotetramers in vitro, but the dominant negative effect of ALDH2*2 on ALDH2 activity has been shown to be due to the formation of ALDH2/ALDH2*2 heterotetramers which have poor activity in vitro[10]. ALDH1B1 is another mitochondrial ALDH and was previously designated as ALDHx or ALDH5. It is encoded by an intronless coding region gene (ALDH1B1) on chromosome 9 [11]. ALDH1B1 shares 72 percent amino acid identity with ALDH2, and like ALDH2, it forms homotetramers and is capable of metabolizing acetaldehyde in the body, in addition to other short chain aldehydes [12]. ALDH1B1 is present in the liver, as well as in the stomach and small intestines, where it likely plays a role in first-pass metabolism of alcohol-derived acetaldehyde. Immunohistochemical staining of ALDH1B1 in human liver shows a strong prevalence of ALDH1B1 [13], although these are likely lower than ALDH2 levels based on reported relative transcript levels [12]. With a km of 3.2 μM for acetaldehyde, ALDH2 plays a major role in acetaldehyde metabolism in “normal” ALDH2*1/*1 individuals who have ingested moderate amounts of ethanol [14]. In subjects in whom ALDH2 has been pharmacologically inhibited, the aversive effects of acetaldehyde typically begin around 40–60 μM [15]. In an alcoholic population, consumption of a 0.5 g/kg dose of ethanol (equivalent to 3–4 drinks in an average weight man), individuals with a normal ALDH2 (ALDH2*1/*1) resulted in a blood acetaldehyde concentration of 1.8 μM [9]. However, under the same conditions, alcoholic ALDH2 heterozygous (ALDH2*1/*2) and mutant (ALDH2*2/*2) individuals achieved a blood acetaldehyde concentration of 57.5 and 108.7 μM, respectively. Values were similar for non-alcoholic populations of ALDH2*1/*1 and ALDH2*1/*2 individuals, but the ethanol dose was poorly tolerated by ALDH2*2/*2 individuals so no data are available for that group. At these elevated concentrations, ALDH1B1 (Km – 55 μM) would be expected to participate in acetaldehyde metabolism. There is also some evidence that ALDH1B1 participates in the ethanol detoxification pathway. Epidemiological data has revealed that mutations in ALDH1B1 result in physiological effects which are consistent with increased acetaldehyde toxicity (i.e., ethanol aversion and ethanol hypersensitivity reactions) [16,17]. Finally, in a newly developed ALDH1B1 mouse knockout model, preliminary data suggests that acetaldehyde levels are higher in ALDH1B1 deficient mice than in wild-type mice upon ethanol feeding [Manuscript in preparation]. Given the acetaldehyde metabolizing capacity of ALDH1B1, it is surprising that ALDH2*2 individuals are so susceptible to acetaldehyde toxicity in that their reduced metabolic activity is not compensated for by ALDH1B1 activity to a greater extent. We hypothesize that part of the reason that this does not occur may be due to ALDH2*2 forming heterotetramers with ALDH1B1 and thereby exerting a similar negative effect on ALDH1B1 activity as has been seen for ALDH2. This possibility was first suggested by the authors (Vasiliou) in 2009 [18].
Given the similarities between ALDH1B1 and ALDH2, it is important to understand their similarities and differences at a sequence and phylogenetic level to better identify what unique roles ALDH1B1 may play in physiology and pathophysiology. Furthermore, given their structural similarity and the mechanism by which ALDH2*2 monomers lower the activity of ALDH2, the possibility of a similar heterotetramerization-based suppression of ALDH1B1 warrants examination. This study identifies and describes the predicted sequences, structures and phylogeny of vertebrate ALDH2 and ALDH1B1 genes and enzymes and compares these results with those previously reported for human (Homo sapiens) and mouse (Mus musculus) ALDH2 and ALDH1B1. Phylogenetic analyses describe the relationships and potential origins of ALDH2 and ALDH1B1 genes during mammalian and vertebrate evolution. This paper also describes molecular modeling studies of protein–protein interactions between ALDH1B1 and ALDH2 subunits to examine whether dominant negative heteromerization of ALDH2*2 with ALDH1B1 may contribute to a lack of compensation by ALDH1B1 in ALDH2*2 individuals.
2. Materials and methods
2.1. Vertebrate ALDH2 and ALDH1B1 gene and enzyme identification and phylogenetic analysis
ALDH1B1 and ALDH2 sequences for multiple representative vertebrate species were retrieved from major databases (UniProt, NCBI) using single and iterative HMMER profile searches (phmmer and jackhmmer) using human ALDH1B1 and ALDH2 to seed searches (http://hmmer.janelia.org/search/phmmer). Confirmation of the presence or absence of ALDH2 and ALDH1B1 genes in vertebrate genomes was based on several methods, including HMMER profile searches of completed proteomes (as listed by UniProt), and protein BLAT searches (UCSC Genome Browser – http://genome.ucsc.edu) [19]. Human ALDH1A1, ALDH1A2 and ALDH1A3 were used as the outgroup as these sequences are the most closely related to ALDH1B1 and ALDH2. In addition, all sequences that were considered ALDH2 or ALDH1B1 by the automated analysis of NCBI homologene were retrieved (http://www.ncbi.nlm.nih.gov/homologene). All sequences were aligned using the most accurate version of T-Coffee (http://tcoffee.crg.cat/) and phylogenetic trees were constructed using neighbor joining methods with 1000 replicate bootstrap in PHYLIP 3.69 (http://evolution.genetics.washington.edu/phylip.html). Sequences were identified as members of either the ALDH1B1 or ALDH2 group. Gene locations, predicted gene structures, and protein sequences were observed for each ALDH examined. Comparative structures for human and mouse ALDH2 and ALDH1B1 genes were derived from the AceView website (http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/) [20]; the major isoform was used in each case, with capped 5′-and 3′-ends for the predicted mRNA sequences including introns and coding exons. For analysis of secondary structure and oligomerization residues, alignments of human ALDH2 [21], ALDH1B1 [12,22], mouse ALDH2 [23] and ALDH1B1 [12] and of other predicted vertebrate ALDH2 and ALDH1B1 sequences were calculated using a ClustalW-technique (http://www.ebi.ac.uk/clustalw/) [24].
2.2. Subunit docking studies and comparative binding energies and stabilities for vertebrate ALDH2 and ALDH1B1 subunits
Web tools were used to predict secondary structures and mitochondrial leader sequences for vertebrate ALDH2 and ALDH1B1 subunits (http://swissmodel.expasy.org/workspace). The crystal structure for human ALDH2 (PDB ID: 1O01) [25], ALDH2*2 (PDB ID: 2ONM) [26], and sheep ALDH1A1 (PDB ID: 1BXS) [27] were downloaded from the RCSB Protein Data Bank (http://www.rcsb.org/). A homology model was created for human ALDH1B1 from ALDH2 and for human ALDH1A1 from sheep ALDH1A1. As controls to evaluate the homology modeling and docking studies, a human ALDH2 homology model was created from the human ALDH1A1 structure and a human ALDH1A1 homology model was created from the ALDH2 structure. Computational studies were conducted using Discovery Studio 3.1 (Accelrys, San Diego; http://accelrys.com). All proteins were prepared using the prepare protein function with a CHARMM force field [28]. For each docking study, a single subunit of ALDH1B1, ALDH2, or ALDH2*2 was minimized and then docked against a homotrimer consisting of three subunits of ALDH1B1 or ALDH2. Minimization was performed using a steepest descent method followed by the conjugate gradient method (using 10,000 steps and an RMS gradient 0.01) utilizing the Generalized Born implicit solvent model [29] to approximate the shape of a formed tetramer and remove steric overlaps. Docking was completed using the ZDOCK [30] and ZRANK [31] algorithms to obtain the most likely pose and to calculate the interaction energy, i.e., the energy of interaction between the monomer and trimer. The best pose was minimized (as noted above) to calculate the protein stability parameter, i.e., the total stability of the final protein tetramer in solution. Individual weak interactions between amino acids were measured by calculating hydrogen bonds (cutoff of 3.5 Å).
3. Results and discussion
3.1. Phylogeny and evolution of vertebrate ALDH2 and ALDH1B1
The predicted locations, sizes and number of coding exons for vertebrate ALDH1B1 and ALDH2 genes examined are presented in Tables 1 and 2, respectively. The ALDH1B1 sequences were found in frogs (Xenopus tropicalis) and mammals, including marsupials – opossum (Monodelphis domestica) and Tasmanian devil (Sarcophilus harrisii) – and a monotreme species – platypus (Ornithorhynchus anatinus), but notably absent from birds (Gallus gallus, Taeniopygia guttata) and lower species, such as zebrafish (Danio rerio) (Table 1). In contrast, ALDH2 sequences were identified in all mammalian, bird, lizard, frog and fish genomes examined, although there were three such ALDH2 genes identified in the zebrafish (D. rerio) genome (Table 2). The phylogram demonstrates the separation of the sequences into three distinct groups during vertebrate evolution, namely ALDH1B1, ALDH2 and the ‘outgroup’ ALDH1A-like sequences (Fig. 1), and suggests that ALDH1B1 genes have been derived from an ancestral vertebrate ALDH2 gene. The phylogenetic distribution of ALDH1B1 genes have not been described previously in this detail, and the gap distribution (namely the lack of ALDH1B1 in birds) suggests that either multiple duplication events occurred or ALDH1B1 was lost early in the avian lineage.
Table 1.
ALDH1B1 and ALDH1A genes and enzymes in selected vertebrate species.
| Animal | Species | ALDH gene | RefSeq ID1 Ensembl/NCBI |
GenBank ID |
UNIPROT ID |
Amino acids | Chromosome location (coding exons) |
Coding exons (strand) |
Gene size coding exon(s) |
Subunit MW | Gene expression levels |
Mitochondrial leader sequence |
% amino acid sequence identity with human isozyme |
||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALDH2 | ALDH1B1 | ALDH1A1 | |||||||||||||
| Human | Homo sapiens | 1B1 | NM_000692 | BC001619 | P30837 | 517 | 9:38,385,746–38,387,296 | 1 (+ve) | 1551 | 57,206 | 2.2 | 1…17 | 72 | 100 | 64 |
| Chimpanzee | Pan troglodytes | 1B1 | XP_001170562a | AK306189 | na | 517 | 9:38,899,005–38,900,555 | 1 (+ve) | 1551 | 57,276 | na | 1…17 | 72 | 99 | 64 |
| Gorilla | Gorilla gorilla | 1B1 | ENSGGOT13385b | Na | na | 517 | 9:39,239,653–39,241,203 | 1 (+ve) | 1551 | 57,265 | na | 1…17 | 71 | 98 | 64 |
| Orangutan | Pongo abelii | 1B1 | NM_001134104 | CR860576 | Q5R6B5 | 517 | 9:23,179,163–23,180,713 | 1 (−ve) | 1551 | 57,261 | na | 1…17 | 71 | 98 | 64 |
| Gibbon | Nomascus leucogenys | 1B1 | XP_003263524a | na | na | inc | GL397291:19,434,443–19,436,340^ | 1 (−ve) | inc | inc | na | 1…17 | 71 | 98 | 63 |
| Rhesus | Macaca mulatta | 1B1 | XP_001114412a | EHH23951 | F7H5N9 | 517 | 15:39,015,754–39,017,304 | 1 (−ve) | 1551 | 57,175 | na | 1…17 | 71 | 98 | 64 |
| Marmoset | Callithrix jacchus | 1B1 | XP_002743154a | Na | na | 514 | 1:140,345,262–140,346,964 | 2 (+ve) | 1703 | 56,987 | na | 1…12 | 71 | 96 | 64 |
| Mouse | Mus musculus | 1b1 | NM_028270 | BC020001 | Q9CZS1 | 519 | 4:45,815,336–45,816,892 | 1 (+ve) | 1557 | 57,553 | 2.8 | 1…19 | 72 | 94 | 65 |
| Rat | Rattus norvegicus | 1b1 | NM_001011975 | BC081884 | Q66HF8 | 519 | 5:62,314,156–62,315,712 | 1 (+ve) | 1557 | 57,625 | 0.1 | 1…19 | 71 | 93 | 65 |
| Pig | Sus scrofa | 1B1 | XP_003353634a | AK347304 | na | 517 | 1: 250,617,521–250,619,071 | 1 (+ve) | 1551 | 57,535 | na | 1…17 | 71 | 94 | 64 |
| Dog | Canis lupus familiaris | 1B1 | XP_538742a | na | na | 520 | 11:57,607,718–57,609,277 | 1 (+ve) | 1560 | 57,693 | na | 1…19 | 71 | 94 | 64 |
| Panda | Ailuropoda melanoleuca | 1B1 | XP_002914982a | na | na | 517 | GL192407.1:2,469,008–2,470,558^ | 1 (−ve) | 1551 | 57,604 | na | 1…16 | 69 | 93 | 64 |
| Horse | Equus caballus | 1B1 | XP_001915212a | na | na | inc | 25:3,341,847–3,343,364 | 1 (+ve) | 1518 | inc | na | inc | 63 | 84 | 58 |
| Cow | Bos taurus | 1B1 | XP_599364a | na | na | inc | 8:62,891,068–62,892,600 | 1 (+ve) | 1533 | inc | na | inc | 65 | 85 | 59 |
| Opossum | Monodelphis domestica | 1B1 | XP_003341640a | na | na | 517 | 6:31,302,337–31,303,887 | 1 (+ve) | 1551 | 57,208 | na | 1…12 | 70 | 88 | 66 |
| Tasmanian devil | Sarcophilis harrisii | 1B1 | XP_003761415a | na | na | 517 | GL841485:1,411,647–1,413,197 | 1 (+ve) | 1551 | 57,180 | na | 1…16 | 71 | 89 | 66 |
| Platypus | Ornithorynchus anatinus | 1B1 | XP_001519422a | na | na | 517 | Contig15042:5,629–7,179* | 1 (−ve) | 1551 | 57,667 | na | 1…18 | 70 | 90 | 65 |
| Frog | Xenopus tropicalis | 1B1 | XP_002940309a | na | na | 520 | GL173,226 ++ 17640–19,124^ | 1 (+ve) | 1560 | 57,008 | na | 1…17 | 67 | 76 | 63 |
| Human | Homo sapiens | 1A1 | NM_000689 | AK293176 | P00352 | 501 | 9:74,705,947–74,757,736 | 13 (−ve) | 52,656 | 54,862 | 4.4 | na | 68 | 65 | 100 |
| Human | Homo sapiens | 1A2 | NM_003888 | BC030589 | O94788 | 518 | 15:56,034,690–56,145,140 | 13 (−ve) | 112,500 | 56,724 | 2.6 | na | 67 | 67 | 73 |
| Human | Homo sapiens | 1A3 | NM_000693 | BC069274 | P47895 | 512 | 15:101,420,009–101,456,830 | 13 (+ve) | 36,822 | 56,108 | 3.5 | na | 66 | 64 | 71 |
na: Not available; +ve indicates positive strand; −ve indicates negative strand.
RefSeq refers to the NCBI reference sequence.
Predicted NCBI sequence.
Gorilla sequence was derived from BLAT using the UCSC web browser [19].
Gene scaffold ID.
Refers to Contig ID; bps refers to base pairs of nucleotide sequence; Inc. an incomplete genome sequence was available for analysis; gene expression levels for ALDH1B1 and ALDH1A genes are taken from [20].
Table 2.
Aldehyde dehydrogenase (ALDH2) genes and enzymes in selected vertebrate species.
| Animal | Species | ALDH2 gene | RefSeq ID1 Ensembl/NCBI |
GenBank ID |
UNIPROT ID |
Amino acids | Chromosome location (coding exons) |
Coding exons (strand) |
Gene size coding exon(s) |
Subunit MW | Gene expression levels |
Mitochondrial leader sequence |
% amino acid sequence identity with human isozyme |
||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALDH2 | ALDH1B1 | ALDH1A1 | |||||||||||||
| Human | Homo sapiens | 2 | NM_000690 | BC002967 | P05091 | 517 | 12:110,689,170–110,726,169 | 13 (+ve) | 37,000 | 56,381 | 2.8 | 1…24 | 100 | 72 | 68 |
| Chimpanzee | Pan troglodytes | 2 | XP_5059379a | na | na | inc | 12:112,226,948–112,248,403 | inc (+ve) | inc | inc | na | inc | 100 | 72 | 68 |
| Gorilla | Gorilla gorilla | 2 | BLAT derived | na | na | 517 | 12:112,226,948–112,248,403 | 13 (+ve) | *21,456 | 56,283 | na | 1…24 | 99 | 72 | 68 |
| Orangutan | Pongo abelii | 2 | NP_001124747 | na | Q5RF00 | 517 | 12:113,595,100–113,634,788 | 13 (+ve) | 39,689 | 56,381 | na | 1…25 | 99 | 72 | 68 |
| Gibbon | Nomascus leucogenys | 2 | XP_003274503a | na | na | 517 | GL397363:46,368–133,282 | 13 (+ve) | 86,915 | 56,333 | na | 1…25 | 99 | 71 | 67 |
| Rhesus | Macaca mulatta | 2 | EHH21190 | JU474666 | na | 517 | 11:112,707,165–112,740,223 | 13 (+ve) | 33,059 | 56,375 | na | 1…25 | 98 | 71 | 68 |
| Marmoset | Callithrix jacchus | 2 | XP_002753080a | na | na | inc | 9:101,943,540–101,982,296 | inc (+ve) | inc | inc | na | 1…25 | 90 | 66 | 61 |
| Mouse | Mus musculus | 2 | NM_009656 | BC005476 | P47738 | 519 | 5:122,018,932–122,043,490 | 13 (−ve) | 24,559 | 56,538 | 5.8 | 1…26 | 95 | 74 | 68 |
| Rat | Rattus norvegicus | 2 | NP_115792 | BC062081 | P11884 | 519 | 12:36,081,803–36,114,854 | 13 (+ve) | 33,052 | 56,516 | 1.3 | 1…27 | 94 | 71 | 68 |
| Pig | Sus scrofa | 2 | NP_001038076 | DQ266356 | Q2XQV4 | 521 | 14:39,812,513–39,840,428 | 13 (+ve) | 27,916 | 56,921 | na | 1…29 | 92 | 71 | 67 |
| Dog | Canis lupus familiaris | 2 | XP_853628a | na | na | 521 | 26:12,337,006–12,370,934 | 13 (+ve) | 33,929 | 56,763 | na | 1…29 | 93 | 72 | 67 |
| Panda | Ailuropoda melanoleuca | 2 | XP_002913100a | na | na | 521 | GL192354:77,348–103,761^ | 13 (−ve) | 26,414 | 56,604 | na | 1…29 | 93 | 72 | 67 |
| Horse | Equus caballus | 2 | XP_001490960a | na | P12762 | 521 | 8:20,270,525–20,285,138 | 13 (−ve) | *14,614 | 56,660 | na | 1…29 | 93 | 71 | 66 |
| Cow | Bos taurus | 2 | NP_001068835 | BC116084 | P20000 | 520 | 17:64,552,475–64,577,817 | 13 (−ve) | 25,343 | 56,653 | na | 1…21 | 93 | 74 | 67 |
| Opossum | Monodelphis domestica | 2 | XP_003340827a | na | na | 518 | 3:482,516,060–482,539,080 | 13 (−ve) | 23,021 | 56,689 | na | 1…17 | 90 | 75 | 70 |
| Tasmanian devil | Sarcophilis harrisii | 2 | XP_003761072a | na | na | inc | GL841441:112,474–132,708^ | inc (−ve) | inc | inc | na | 1…25 | 81 | 66 | 58 |
| Platypus | Ornithorynchus anatinus | 2 | XP_001506461a | na | na | 518 | 2:1,211,623–1,240,836 | 13 (+ve) | 29,214 | 56,675 | na | 1…29 | 85 | 71 | 66 |
| Chicken | Gallus gallus | 2 | XP_415171a | na | E1BT93 | 519 | 15:6,260,328–6,267,619 | 13 (+ve) | 7292 | 56,791 | na | 1…27 | 87 | 74 | 68 |
| Zebra finch | Taeniopygia guttata | 2 | XP_002196279a | na | na | 520 | 15:3,870,030–3,877,271 | 13 (+ve) | 7242 | 56,782 | na | 1…12 | 83 | 71 | 66 |
| Lizard | Anolis carolensis | 2 | XP_003225175a | na | na | 527 | GL343282:19,405–31,883 | 13 (+ve) | 12,479 | 57,706 | na | 1…32 | 81 | 69 | 65 |
| Frog | Xenopus tropicalis | 2 | NM_001004907 | CR762336 | Q28EU7 | 521 | GL172947:177,785–195,692^ | 13 (+ve) | 17,908 | 57,146 | na | 1…25 | 83 | 73 | 69 |
| Zebrafish | Danio rerio | 2a | XP_002662252a | AF260121 | Q8QGQ2 | 516 | 5:72,536,565–72,561,417 | 13 (−ve) | 24,853 | 56,563 | na | 1…17 | 75 | 68 | 64 |
| Zebrafish | Danio rerio | 2b | NM_213301 | AY398308 | Q6TH48 | 516 | 5:72,465,282–72,486,769 | 13 (−ve) | 21,488 | 56,550 | na | 1…17 | 75 | 68 | 64 |
| Zebrafish | Danio rerio | 2c | CAM13323 | BX510999 | Q7SXU3 | 516 | 5:72,441,576–72,461,572 | 13 (−ve) | 19,997 | 56,483 | na | 1…18 | 76 | 69 | 65 |
na: not available. Gene expression levels for ALDH2 genes are taken from [20]; Note the presence of three ALDH2 genes on zebrafish (D. rerio) in chromosome 5.
RefSeq refers to the NCBI reference sequence.
Predicted NCBI sequence.
Gene scaffold ID; bps refers to base pairs of nucleotide sequence; Inc.
Refer to an incomplete genome sequence being available.
Fig. 1.
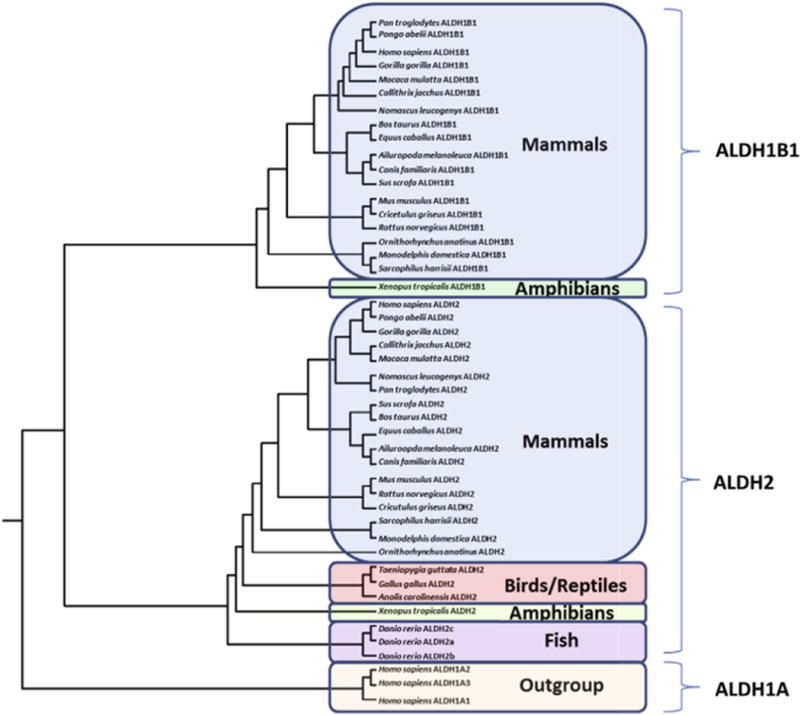
Phylogenetic tree for vertebrate ALDH2 and ALDH1B1 protein sequences. The tree is labeled with the protein name and the species name of the vertebrate. Note the major clusters for the vertebrate ALDH1B1, ALDH2 and human ALDH1A sequences. The tree is ‘rooted’ with human ALDH1A1, ALDH1A2 and ALDH1A3 sequences. See Tables 1 and 2 for details of sequences and gene locations. Note the absence of ALDH1B1 sequences from birds, reptiles and fish, whereas ALDH2 sequences were observed for all vertebrate genomes examined, including three sequences for the zebrafish genome.
3.2. Predicted gene locations and exonic structures for vertebrate ALDH2 and ALDH1B1 genes
Each vertebrate ALDH2 gene examined had 13 coding exons (Fig. 2 and Table 2), whereas the vertebrate ALDH1B1 genes consistently contained only a single coding exon with the single exception that marmoset ALDH1B1 has two coding exons (Fig. 2 and Table 1). In contrast to all of the other vertebrate genomes, the zebrafish (D. rerio) genome contained three predicted ALDH2 genes, designated as ALDH2a, ALDH2b and ALDH2c, which are closely localized on chromosome 5 [32]. This is indicative of an origin of these multiple genes from successive unequal crossover events in the ancestral fish ALDH2 gene. These gene duplication events may have arisen from rapid, lineage-specific expansion of the zebrafish genome mediated by recent tandem duplications, as reported for other genes of this teleost species [33].
Fig. 2.

Comparative structures for human and mouse ALDH2 and ALDH1B1 genes. Data are derived from the AceView website (http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/). The major isoform for each gene is shown, with capped 5′- and 3′-ends for the predicted mRNA sequences. Introns and coding exons are shown. Note the differences in scale for the ALDH2 (44 or 28 kbs) and ALDH1B1 (both 6 kbs) genes. Coding exons are depicted as pink (or shaded) bars and untranslated 5′ and 3′ regions are shown as white bars. CpG islands located in the 5′-promoter regions are labeled. The location of predicted binding sites for the transcription factors Myb, Myc and PPARG in the promoter region for the human genes are also presented. Comparative expression levels are shown as (×) times the average expression level; NM refers to the NCBI RefSeq sequence. Flags depict the 3′UTR polyadenylation sites. Black flags indicate the typical AATAAA signal, and blue flags indicate a SNP variant of this signal. Filled flags correspond to events with many supporting cDNAs, and empty flags indicate fewer supporting cDNAs.
3.3. Alignments of human, mouse and frog ALDH2 and ALDH1B1 amino acid sequences
Amino acid alignments for human (H. sapiens)[21], mouse (M. musculus)[23] and frog (X. tropicalis) ALDH2 and ALDH1B1 sequences [12] are shown in Fig. 3. Comparisons of the ALDH2 and ALDH1B1 sequences with the human ALDH2 sequence, for which the tertiary structure has been described (PDB ID: 1CW3A) [25,26], enabled identification of key residues contributing to catalysis, structure and function. Mitochondrial ALDH2 N-terminal leader sequences (human ALDH2 residues 1–24), which enable ALDH2 uptake into the mitochondrion [34], were predicted for each of the other vertebrate ALDH2 and ALDH1B1 sequences examined. These sequences were highly divergent and varied in length from 12 residues (opossum and marmoset ALDH1B1) to 32 residues (lizard ALDH2) (Table 1). Active site residues (human ALDH2 numbers used) binding the substrate (Glu285; Cys319) or stabilizing the transition state for the catalyzed reaction (Asn186) were conserved in all ALDH2 and ALDH1B1 sequences examined (Fig. 3). Within the NAD+ binding domain, a dinucleotide-binding motif near the N-terminal end of the αG helix (262Gly-Ser-Thr-Glu-Val-267Gly) [25] was predominantly conserved in the vertebrate ALDH2 and ALDH1B1 sequences examined with the exception of a 266Ile/Val substitution in the human ALDH2 sequence (Fig. 3). Glu487 was also conserved among all vertebrate ALDH2 and ALDH1B1 sequences examined. This residue is subject to genetic variation (ALDH2*2) in East Asian human populations. Specifically, the substitution with Lys487 lowers coenzyme (NAD+) binding affinity and results in greater than a 90% loss of enzyme activity. The consequent reduced catalytic activity of ALDH2*2 lowers acetaldehyde clearance capacity, rendering the subject vulnerable to alcohol ‘flushing’ and other unpleasant physiological symptoms attending acetaldehyde accumulation following alcohol consumption [5–9].
Fig. 3.
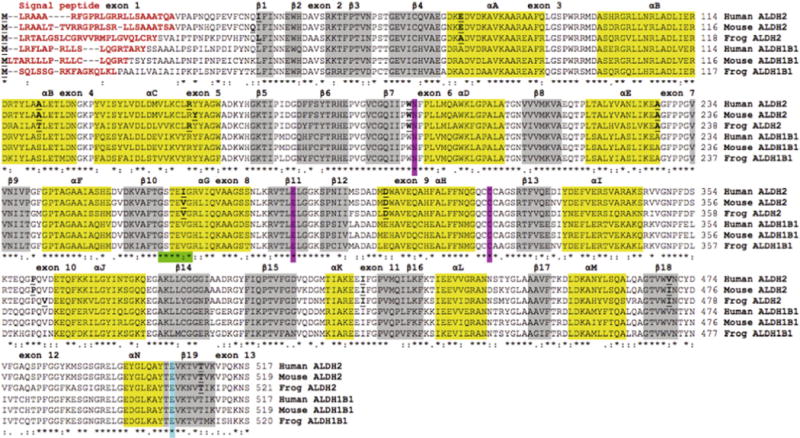
Amino acid sequence alignments for human, mouse and frog ALDH2 and ALDH1B1 sequences. See Tables 1 and 2 for sources of ALDH1B1 and ALDH2 sequences, respectively. Symbols below all of the sequences describe the similarities in the amino acids across the sequences at each position and denotes them as being identical (*), highly similar (:) or less similar (.). Residues with no symbols have no similarity. Key functional residues include N-signal peptide residues (red text or bold) and the active site triad residues Asn; Glu; and Cys (pink shading or AS). Predicted secondary structural regions, shown as α-helix (yellow shading or black shading) and β-sheet (grey shading), are based on [25,26] for human ALDH2. Known or predicted exon junctions are identified by underlined bold font, with exon numbers referring to the human ALDH2 gene. Other important enzyme regions, including the NAD+ binding domain (green shading or shaded ****:*) and the ALDH2*2 variant position (blue shading or #) are shown.(For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Predicted secondary structures for mouse and frog ALDH2 sub-units and human, mouse and frog ALDH1B1 subunits were compared with those previously reported structure for human ALDH2 [25] (Fig. 3). Alpha-helix and β-sheet locations (consistent with experimental three-dimensional structures for human ALDH2) were observed for each of the ALDH2 and ALDH1B1 sequences examined, suggesting strong conservation of amino acid sequences and secondary structure [25]. Three distinct domains for each subunit of tetrameric ALDH2 have been previously identified: (i) the coenzyme (NAD+) binding domain (β strands 1–4 and 7–11; helices A to G and helix N), (ii) the catalytic domain (β strands 12–18; helices H to M), and (iii) the oligomerization domain (β strands 5, 6 and 19). Major changes in structure have been reported for the ALDH2*2 tetramer, specifically disruptions in a disordered region surrounding the Lys487 substitution (which is located at the dimer interface involving αG, β10 and β11) and an alteration in the loop consisting of residues 463–478, all of which are involved in NAD+ binding [26]. These changes in structure cause a 3 Å shift of the αG helix towards the active site cleft which may significantly contribute to the dramatically reduced affinity of ALDH2*2 towards NAD+, and thereby reduce ALDH2 activity.
3.4. Proposed retroviral origin for frog and mammalian ALDH1B1 genes
The evolutionary origin of the vertebrate ALDH1B1 gene may have occurred in a two stage process, namely the retroviral integration of an ALDH2 cDNA segment into an ancestral amphibian chromosome, and a second retroviral integration of an ALDH2 cDNA segment into an ancestral mammalian chromosome, which was retained throughout subsequent monotreme, marsupial and eutherian mammalian evolution. This proposed origin for the vertebrate ALDH1B1 genes is supported by the following evidence: (i) the high levels of sequence identities observed for vertebrate ALDH2 and ALDH1B1 amino acid sequences (Figs. 3 and 4; Tables 1 and 2), (ii) the phylogenetic clustering observed for vertebrate ALDH2 and ALDH1B1 sequences (Fig. 1), and (iii) the presence of identical or similar CpG islands within the genomic structures for human and mouse ALDH2 and ALDH1B1 genes (Fig. 2). Moreover, the generation of single exon coding genes from a retroviral integration process derived from a cDNA has been described for many vertebrate genes, which serves as a precedent for such a mechanism. More than 400 retrogenes have been reported to contribute to the appearance of novel coding sequences during mammalian evolution where the mRNA transcript is reverse-transcribed and integrated into genomic DNA [35,36]. These include several retrotransposon-encoding enzymes differentially expressed in the body, examples of which are phosphoglycerate kinase 2 (PGK2) which encodes a sperm-specific enzyme functioning in glycolysis [37], ribosomal protein genes that have been retrotransposed from an X-chromosome gene [38], and a testis-specific form of the human pyruvate dehydrogenase E1 alpha subunit [39]. It appears likely that a retroviral origin for the frog and mammalian ALDH1B1 genes is consistent with these and other examples for such events during vertebrate evolution (Fig. 5).
Fig. 4.
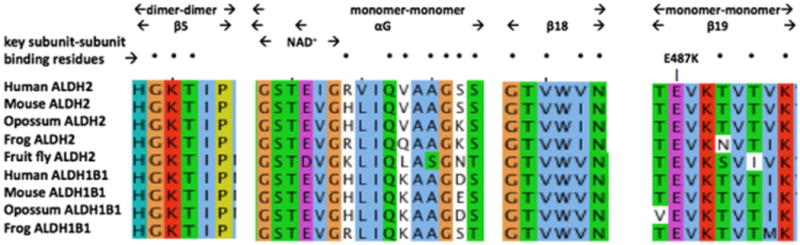
Comparative amino acid sequences alignments for ALDH2 and ALDH1B1 subunit binding domains. See Tables 1 and 2 for sources of ALDH1B1 and ALDH2 sequences, respectively. Three regions for vertebrate ALDH2 and ALDH1B1 sequences are shown, including dimer–dimer (β5); NAD+ binding site, monomer–monomer αG and β18, and monomer–monomer (β19). Amino acid residues are color coded: yellow for P (proline); green for hydrophilic amino acids, S (serine), Q (glutamine), N (asparagine), and T (threonine); brown for glycine (G); light blue for hydrophobic amino acids, L (leucine), I (isoleucine), V (valine), M (methionine), W (tryptophan); dark blue for amino acids, T (tyrosine) and H (histidine); purple for acidic amino acids, E (glutamate) and D (aspartate); and red for basic amino acids, K (lysine) and R (arginine). The site of the ALDH2*2 variant (E487K) is identified by the arrow.
Fig. 5.
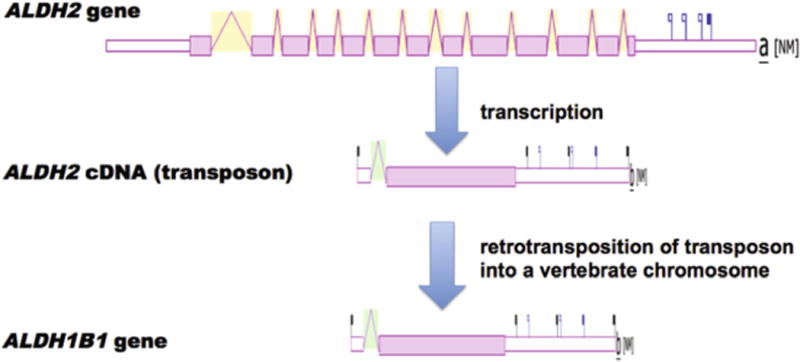
A proposal for the evolutionary appearance of the mammalian and frog ALDH1B1 genes by retroviral integration of ancestral ALDH2 cDNA sequences. Proposed evolutionary appearance for the single coding exon ALDH1B1 gene derived from a retrovirally induced integration of ALDH2 cDNAs and the subsequent integration into another chromosome within an ancestral vertebrate genome; ALDH2 and ALDH1B1 gene structures were derived from AceView.
3.5. Human ALDH2 and ALDH1B1 subunit docking studies
Fig. 4 compares aligned amino acid sequences for the oligomer-forming residues (previously identified for human ALDH2 [23,25]) in human, mouse, opossum, frog and fruit fly ALDH2 sequences and in human, mouse, opossum, and frog ALDH1B1 sequences. Monomer–monomer ALDH2 and ALDH1B1 binding amino acid residues located in the αG, β18 and β19 secondary structures for these enzymes were predominantly identical in sequence, whereas the dimer–dimer ALDH2 and ALDH1B1 binding residues located in the β5 secondary structure were identical for all of the vertebrate ALDH2 and ALDH1B1 sequences examined. This demonstrates a high degree of conservation for the oligomer-forming residues for vertebrate ALDH2 and ALDH1B1 subunits and suggests that heterotetramers between these subunits may be formed, particularly given their colocalization within mitochondria.
To evaluate variations due to the homology modeling process, a homology model of ALDH2 was created based on the structure of ALDH1A1, we created a homology model of ALDH1A1 based on the structure of ALDH2 (66% amino acid identity). The ALDH2 homology model was docked against a trimer of ALDH2, and the ALDH1A1 homology model was docked against a trimer of ALDH2. Interaction energy and protein stability were calculated for each (Table 3), and an RMSD (root mean squared distance) was calculated between the original and the homology model e.g., ALDH1A1 vs. ALDH1A1 homology model. The difference in interaction energy between the original homotetramer and homology model for ALDH1A1 and ALDH2 was 9.3% and 22.5%, respectively. The difference between protein stability for ALDH1A1 and ALDH2 was 0.26% and 0.25%, respectively. The RMSD between the control and homology model for ALDH1A1 and ALDH2 was 0.98 and 1.49 Å, respectively. This indicates that the models are broadly comparable, especially in respect to protein stability and RMSD (in most cases < 2.0 Å is considered highly successful), but care must be taken comparing binding energy differences under approximately 20%.
Table 3.
Comparative docking interaction energies and protein stabilities for ALDH2 and ALDH1B1 subunits.a
| Trimer | Monomer | Interaction energy (kcal/mol) | % change from homotetramer | Protein stability (kcal/mol) | % change from homotetramer |
|---|---|---|---|---|---|
| ALDH1A1 | ALDH1A1 | −352.75 | – | −102570.56 | – |
| ALDH1A1 | ALDH1A1 homology from ALDH2 | −320.00 | 9.28 | −102306.17 | 0.26 |
| ALDH2 | ALDH2 | −363.90 | – | −109639.68 | – |
| ALDH2 | ALDH2 homology from ALDH1A1 | −281.96 | 22.52 | −109915.10 | −0.25 |
| ALDH2 | ALDH2*2 | −323.77 | 11.03 | −109298.92 | 0.31 |
| ALDH1B1 | ALDH1B1 | −308.87 | – | −115686.15 | – |
| ALDH1B1 | ALDH2 | −445.60 | −44.27 | −114127.71 | 1.35 |
| ALDH1B1 | ALDH2*2 | −367.23 | −18.89 | −114295.66 | 1.20 |
Energies for the binding of an ALDH monomer to an ALDH trimer were calculated in silico. The interaction energy parameter for each indicates the energy of interaction between the monomer and the trimer, whereas the protein stability parameter represents the calculated total stability of the final protein tetramer in solution.
Computational protein–protein docking studies were undertaken for human ALDH2 and ALDH1B1 subunits to investigate whether these subunits were capable of forming heterotetramers in silico. In these studies, a single monomer of protein (ALDH1B1, ALDH2*2 or ALDH2) was bound to a homotrimer of either ALDH1B1 or ALDH2. Comparison of the most energetically favorable conformation of each tetramer suggested that all tetramers use a similar binding modality to known ALDH2 crystal structures (Fig. 6). Analysis of the interaction energy and protein stability (Table 3) suggests that all of the complexes (hetero- and homo-tetramers) are energetically favorable and may be assembled in nature. The binding of either ALDH2 or ALDH2*2 to an ALDH2 trimer was used as a positive control. Previous in vitro studies have indicated that ALDH2*2 has very little catalytic activity and that heteromerization between ALDH2 and ALDH2*2 accounts for the lack of sufficient active ALDH2 in heterozygous individuals [10]. The in silico docking studies indicated that the binding of ALDH2*2 to ALDH2 is less favorable than binding of the wild-type ALDH2 to an ALDH2 trimer by 11%, although it should be noted that this occurs with a high enough frequency in vivo to strongly reduce total ALDH2 activity in heterozygotes. In the present study, all calculated protein stability values were broadly similar between homo- and heterotetramers of the same trimer, with variations from 0.25% to 1.35%. This indicates that no gross deformations are caused by heterotetramer formation. Interestingly, the binding of either ALDH2 or ALDH2*2 to the ALDH1B1 trimer is more energetically favorable (44% and 19%, respectively) than is ALDH1B1 binding to an ALDH1B1 trimer.
Fig. 6.
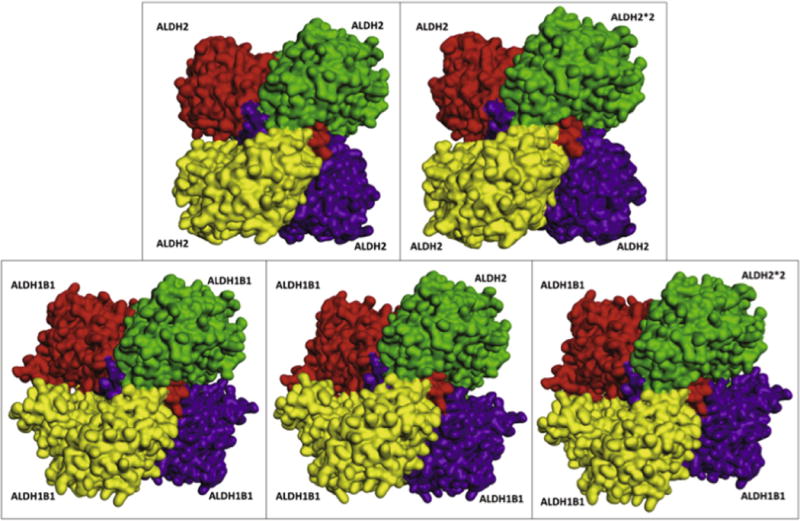
ALDH2 and ALDH1B1 subunit-subunit docking results. Predicted three-dimensional ALDH2 and ALDH1B1 subunit-containing tetrameric structures. Trimer (ALDH2 and ALDH1B1) subunits are shown in yellow, red and purple. The monomer (ALDH2, ALDH2*2 or ALDH1B1), which completes the tetramer in the docking studies, is shown in green. The top-right and bottom-left subunits form a dimer (green and yellow), as do the top-left and bottom-right subunits.
Given that amino acids in the oligomerization domains were not completely conserved, hydrogen bonds made between the docked and minimized ALDH monomers and trimers were calculated and recorded (Table 4). Overall, there were 102–115 hydrogen bond interactions made between the monomer and trimers upon binding. More than half of these occurred in the monomer–monomer interface, with the remainder concentrated in the dimer–dimer interface (specifically between the two subunits which make β5:β5 interactions), and a few interactions were made between the monomer and the subunit diagonal from it (which does not participate in any canonical monomer–monomer or dimer–dimer binding). To determine which amino acid residues participate in subunit interactions, we tallied hydrogen bonds made by each residue in the monomer–monomer interface (αG:αG, β18:β19) and the dimer–dimer interface (β5:β5). These were counted if they were made by either the monomer or its homologous residue on the trimer. Typically interactions were symmetrical (each interaction was made with the same donor / acceptor residues from the monomer to the trimer and from the trimer to the monomer), but this was not always the case. These interactions were summed between the monomer and trimer to show trends, but individual variation in binding patterns (data not shown) likely contributes to differences seen in binding energies between different tetramer assemblies. Additional analyses were conducted to determine if the residues participating in interactions were identical between ALDH1B1 and ALDH2. Of the residues making interactions between subunits, those on beta sheets were fully conserved (three residues in β5, four residues in β18, and three residues in β19). The interactions made were also highly conserved, with the exception that ALDH2 or ALDH2*2 monomers did not make a Lys142 interaction when binding to an ALDH1B1 trimer. Further, an Asn454 interaction was only seen when ALDH2 was bound to an ALDH1B1 trimer. Although the interactions between the two αG helices were less conserved than the interactions between the β sheets, about half (4/7) of the residues were conserved (same amino acid) and about half (4/7) of the positions were conserved (interaction at the homologous amino acid position). However, these small differences in helix–helix binding do not appear to disrupt overall binding – these bonds represent a small fraction of the total weak bonds measured across the monomer–monomer and dimer–dimer interfaces, which is measured in aggregate by the interaction energy parameter. None of the individual substitutions observed appears to cause large disruptions in electrostatic forces.
Table 4.
Specific interactions made by ALDH homo- and heterotetramersa.
| Trimer | Monomer | β5 sheet | αG helix | β18 sheet | β19 sheet | H-bond interactions by subunit | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
||||||||||||||||||
| 141 | 142 | 143 | 251 | 254 | 255 | 257 | 258 | 259 | 260 | 449 | 451 | 453 | 454 | 490 | 492 | 494 | B | C | D | total | ||
| ALDH2 | ALDH2 | P | P | P | P | P | P | P | P | P | P | P | P | P | P | 68 | 19 | 28 | 115 | |||
| ALDH2 | ALDH2*2 | P | P | P | P | P | P | P | P | P | P | P | P | P | P | 65 | 15 | 33 | 113 | |||
| ALDH1B1 | ALDH1B1 | P | P | P | P | P | P | P | P | P | P | P | P | P | P | 65 | 5 | 32 | 102 | |||
| ALDH1B1 | ALDH2 | P | P | P | P | P | P | P | P | P | P | P | P | P | P | 61 | 8 | 37 | 106 | |||
| ALDH1B1 | ALDH2*2 | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | 66 | 16 | 30 | 112 | ||
| Residue in ALDH2 | GLY | LYS | THR | ARG | GLN | LYS | ALA | GLY | SER | SER | GLY | VAL | VAL | ASN | THR | THR | LYS | |||||
| Conserved residue | Y | Y | Y | N | Y | N | Y | Y | N | Y | Y | Y | Y | Y | Y | Y | Y | |||||
Hydrogen bonds made between the ALDH monomers and trimers were calculated and recorded. Specific interactions between the monomer–monomer interface (αG:αG, β18:β19) and the dimer–dimer interface (β5:β5) were compared between the different assemblies. If a specific residue made a H-bond interaction between the correct secondary structure interface (either on the monomer or the trimer), then it is counted as present (P). The identity of the amino acid residue in ALDH2 is listed and whether it is conserved between ALDH2 and ALDH1B1 (Y) or not (N). Total H-bond interactions by subunit are also recorded (interaction cutoff of 3.5 Å) by subunit. The monomer is designated the A subunit, its dimer is designated the B subunit, the subunit opposite A in the dimer–dimer interface that participates in β5:β5 interactions is the D subunit, and the diagonal subunit which participates in no canonical interactions is the C subunit.
Given the favorable binding energies, similar overall protein stabilities and the similar weak interactions between the monomer and trimers, the present results suggest that either ALDH2 or ALDH2*2 may be included in an ALDH1B1 heterotetramer. Because of the effect of ALDH2*2 subunits on the catalytic activity of ALDH2 tetramers, it is reasonable to expect that the binding of ALDH2*2 to an ALDH1B1 trimer would similarly repress ALDH1B1 catalytic activity. This may explain why the presence of ALDH1B1 does not compensate for a lack of ALDH2 activity in ALDH2*2 individuals, despite favorable enzyme kinetics for substrates such as ethanol-derived acetaldehyde. It will be important that these in silico observations be verified under in vitro and in vivo conditions.
4. Conclusions
BLAST and BLAT analyses of several vertebrate genome databases were undertaken using amino acid sequences reported for human ALDH2 and ALDH1B1 to interrogate vertebrate genomes. Predicted amino acid sequences and structures for the vertebrate ALDH2 and ALDH1B1 subunits showed a high degree of similarity with human ALDH2, which served as a reference structure for these ALDHs. Three ALDH2 genes were observed for the zebrafish (D. rerio) genome and were closely located on chromosome 5. Amino acid sequences for vertebrate ALDH2 and ALD1B1 subunit-subunit binding regions showed a high degree of sequence identity. The present results support the molecular evolution of vertebrate ALDH2 and ALDH1B1 genes with the mammalian and frog ALDH1B1 genes as being generated by retrotranspositional duplication events of ALDH2 genes at two distinct stages of vertebrate evolution: in an ancestral gene leading to the evolutionary appearance of frogs and in an ancestral genome leading to the evolutionary appearance of monotreme, marsupial and eutherian mammals.
This is the first study to describe evidence that suggests that human ALDH2*2 and ALDH1B1 subunits may be capable of forming heterotetramers. Accordingly, through a mechanism similar to that documented for human mitochondrial ALDH2, the ALDH2*2 genetic variant may reduce the catalytic activity of ALDH1B1. This novel hypothesis may explain why the presence of ALDH1B1 does not compensate for a lack of ALDH2 activity in ALDH2*2 individuals. Further, it opens the possibility that reduced ALDH1B1 activity could be involved in the pathogenesis of ALDH2*2-related disorders.
Acknowledgments
The authors wish to thank the Computational Chemistry and Biology Core Facility at the University of Colorado Anschutz Medical Campus for their contributions to these studies. This work was supported, in part, by the following NIH Grants; EY17963 and EY11490. Fellowship support to B.C.J. (F31 AA020728) is also acknowledged.
Footnotes
Conflict of interest statement
The authors declare that there are no conflicts of interest.
References
- 1.Marchitti SA, Brocker C, Stagos D, Vasiliou V. Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Expert Opin Drug Metab Toxicol. 2008;4(6):697–720. doi: 10.1517/17425250802102627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hsu LC, Bendel RE, Yoshida A. Genomic structure of the human mitochondrial aldehyde dehydrogenase gene. Genomics. 1988;2(1):57–65. doi: 10.1016/0888-7543(88)90109-7. [DOI] [PubMed] [Google Scholar]
- 3.Wang RS, Nakajima T, Kawamoto T, Honma T. Effects of aldehyde dehydrogenase-2 genetic polymorphisms on metabolism of structurally different aldehydes in human liver. Drug Metab Dispos. 2002;30(1):69–73. doi: 10.1124/dmd.30.1.69. [DOI] [PubMed] [Google Scholar]
- 4.Ohta S, Ohsawa I, Kamino K, Ando F, Shimokata H. Mitochondrial ALDH2 deficiency as an oxidative stress. Ann N Y Acad Sci. 2004;1011:36–44. doi: 10.1007/978-3-662-41088-2_4. [DOI] [PubMed] [Google Scholar]
- 5.Yoshida A, Ikawa M, Hsu LC, Tani K. Molecular abnormality and cDNA cloning of human aldehyde dehydrogenases. Alcohol. 1985;2(1):103–106. doi: 10.1016/0741-8329(85)90024-2. [DOI] [PubMed] [Google Scholar]
- 6.Goedde HW, Agarwal DP. Polymorphism of aldehyde dehydrogenase and alcohol sensitivity. Enzyme. 1987;37(1–2):29–44. doi: 10.1159/000469239. [DOI] [PubMed] [Google Scholar]
- 7.Higuchi S, Muramatsu T, Shigemori K, Saito M, Kono H, Dufour MC, Harford TC. The relationship between low Kmaldehyde dehydrogenase phenotype and drinking behavior in Japanese. J Stud Alcohol. 1992;53(2):170–175. doi: 10.15288/jsa.1992.53.170. [DOI] [PubMed] [Google Scholar]
- 8.Cook TA, Luczak SE, Shea SH, Ehlers CL, Carr LG, Wall TL. Associations of ALDH2 and ADH1B genotypes with response to alcohol in Asian Americans. J Stud Alcohol. 2005;66(2):196–204. doi: 10.15288/jsa.2005.66.196. [DOI] [PubMed] [Google Scholar]
- 9.Chen YC, Peng GS, Tsao TP, Wang MF, Lu RB, Yin SJ. Pharmacokinetic and pharmacodynamic basis for overcoming acetaldehyde-induced adverse reaction in Asian alcoholics, heterozygous for the variant ALDH2*2 gene allele. Pharmacogenet Genomics. 2009;19(8):588–599. doi: 10.1097/FPC.0b013e32832ecf2e. [DOI] [PubMed] [Google Scholar]
- 10.Wang X, Sheikh S, Saigal D, Robinson L, Weiner H. Heterotetramers of human liver mitochondrial (class 2) aldehyde dehydrogenase expressed in Escherichia coli. A model to study the heterotetramers expected to be found in Oriental people. J Biol Chem. 1996;271(49):31172–31178. doi: 10.1074/jbc.271.49.31172. [DOI] [PubMed] [Google Scholar]
- 11.Hsu LC, Chang WC. Cloning and characterization of a new functional human aldehyde dehydrogenase gene. J Biol Chem. 1991;266(19):12257–12265. [PubMed] [Google Scholar]
- 12.Stagos D, Chen Y, Brocker C, Donald E, Jackson BC, Orlicky DJ, Thompson DC, Vasiliou V. Aldehyde dehydrogenase 1B1: molecular cloning and characterization of a novel mitochondrial acetaldehyde-metabolizing enzyme. Drug Metab Dispos. 2010;38(10):1679–1687. doi: 10.1124/dmd.110.034678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Y, Orlicky DJ, Matsumoto A, Singh S, Thompson DC, Vasiliou V. Aldehyde dehydrogenase 1B1 (ALDH1B1) is a potential biomarker for human colon cancer. Biochem Biophys Res Commun. 2011;405(2):173–179. doi: 10.1016/j.bbrc.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klyosov AA, Rashkovetsky LG, Tahir MK, Keung WM. Possible role of liver cytosolic and mitochondrial aldehyde dehydrogenases in acetaldehyde metabolism. Biochemistry. 1996;35(14):4445–4456. doi: 10.1021/bi9521093. [DOI] [PubMed] [Google Scholar]
- 15.Johnsen J, Stowell A, Morland J. Clinical responses in relation to blood acetaldehyde levels. Pharmacol Toxicol. 1992;70(1):41–45. doi: 10.1111/j.1600-0773.1992.tb00423.x. [DOI] [PubMed] [Google Scholar]
- 16.Husemoen LL, Fenger M, Friedrich N, Tolstrup JS, BeenfeldtFredriksen S, Linneberg A. The association of ADH and ALDH gene variants with alcohol drinking habits and cardiovascular disease risk factors. Alcohol Clin Exp Res. 2008;32(11):1984–1991. doi: 10.1111/j.1530-0277.2008.00780.x. [DOI] [PubMed] [Google Scholar]
- 17.Linneberg A, Gonzalez-Quintela A, Vidal C, Jorgensen T, Fenger M, Hansen T, Pedersen O, Husemoen LL. Genetic determinants of both ethanol and acetaldehyde metabolism influence alcohol hypersensitivity and drinking behaviour among Scandinavians. Clin Exp Allergy. 2010;40(1):123–130. doi: 10.1111/j.1365-2222.2009.03398.x. [DOI] [PubMed] [Google Scholar]
- 18.Endo J, Sano M, Katayama T, Hishiki T, Shinmura K, Morizane S, Matsuhashi T, Katsumata Y, Zhang Y, Ito H, Nagahata Y, Marchitti S, Nishimaki K, Wolf AM, Nakanishi H, Hattori F, Vasiliou V, Adachi T, Ohsawa I, Taguchi R, Hirabayashi Y, Ohta S, Suematsu M, Ogawa S, Fukuda K. Metabolic remodeling induced by mitochondrial aldehyde stress stimulates tolerance to oxidative stress in the heart. Circ Res. 2009;105(11):1118–1127. doi: 10.1161/CIRCRESAHA.109.206607. [DOI] [PubMed] [Google Scholar]
- 19.Karolchik D, Bejerano G, Hinrichs AS, Kuhn RM, Miller W, Rosenbloom KR, Zweig AS, Haussler D, Kent WJ. Comparative genomic analysis using the UCSC genome browser. Methods Mol Biol. 2007;395:17–34. doi: 10.1007/978-1-59745-514-5_2. [DOI] [PubMed] [Google Scholar]
- 20.Thierry-Mieg D, Thierry-Mieg J. AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006;7(Suppl. 1):S12, 11–14. doi: 10.1186/gb-2006-7-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hempel J, Kaiser R, Jornvall H. Mitochondrial aldehyde dehydrogenase from human liver. Primary structure, differences in relation to the cytosolic enzyme, and functional correlations. Eur J Biochem. 1985;153(1):13–28. doi: 10.1111/j.1432-1033.1985.tb09260.x. [DOI] [PubMed] [Google Scholar]
- 22.Stewart MJ, Malek K, Crabb DW. Distribution of messenger RNAs for aldehyde dehydrogenase 1, aldehyde dehydrogenase 2, and aldehyde dehydrogenase 5 in human tissues. J Investig Med. 1996;44(2):42–46. [PubMed] [Google Scholar]
- 23.Chang C, Yoshida A. Cloning and characterization of the gene encoding mouse mitochondrial aldehyde dehydrogenase. Gene. 1994;148(2):331–336. doi: 10.1016/0378-1119(94)90708-0. [DOI] [PubMed] [Google Scholar]
- 24.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steinmetz CG, Xie P, Weiner H, Hurley TD. Structure of mitochondrial aldehyde dehydrogenase: the genetic component of ethanol aversion. Structure. 1997;5(5):701–711. doi: 10.1016/s0969-2126(97)00224-4. [DOI] [PubMed] [Google Scholar]
- 26.Larson HN, Zhou J, Chen Z, Stamler JS, Weiner H, Hurley TD. Structural and functional consequences of coenzyme binding to the inactive asian variant of mitochondrial aldehyde dehydrogenase: roles of residues 475 and 487. J Biol Chem. 2007;282(17):12940–12950. doi: 10.1074/jbc.M607959200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore SA, Baker HM, Blythe TJ, Kitson KE, Kitson TM, Baker EN. Sheep liver cytosolic aldehyde dehydrogenase: the structure reveals the basis for the retinal specificity of class 1 aldehyde dehydrogenases. Structure. 1998;6(12):1541–1551. doi: 10.1016/s0969-2126(98)00152-x. [DOI] [PubMed] [Google Scholar]
- 28.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. Charmm – a program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem. 1983;4(2):187–217. [Google Scholar]
- 29.Feig M, Brooks CL., 3rd Recent advances in the development and application of implicit solvent models in biomolecule simulations. Curr Opin Struct Biol. 2004;14(2):217–224. doi: 10.1016/j.sbi.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 30.Chen R, Weng Z. Docking unbound proteins using shape complementarity, desolvation, and electrostatics. Proteins. 2002;47(3):281–294. doi: 10.1002/prot.10092. [DOI] [PubMed] [Google Scholar]
- 31.Pierce B, Weng Z. ZRANK: reranking protein docking predictions with an optimized energy function. Proteins. 2007;67(4):1078–1086. doi: 10.1002/prot.21373. [DOI] [PubMed] [Google Scholar]
- 32.Jackson B, Brocker C, Thompson DC, Black W, Vasiliou K, Nebert DW, Vasiliou V. Update on the aldehyde dehydrogenase gene (ALDH) superfamily. Hum Genomics. 2011;5(4):283–303. doi: 10.1186/1479-7364-5-4-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu J, Peatman E, Tang H, Lewis J, Liu Z. Profiling of gene duplication patterns of sequenced teleost genomes: evidence for rapid lineage-specific genome expansion mediated by recent tandem duplications. BMC Genomics. 2012;13:246. doi: 10.1186/1471-2164-13-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou J, Bai Y, Weiner H. Proteolysis prevents in vivo chimeric fusion protein import into yeast mitochondria. Cytosolic cleavage and subcellular distribution. J Biol Chem. 1995;270(28):16689–16693. doi: 10.1074/jbc.270.28.16689. [DOI] [PubMed] [Google Scholar]
- 35.Yu Z, Morais D, Ivanga M, Harrison PM. Analysis of the role of retrotransposition in gene evolution in vertebrates. BMC Bioinformatics. 2007;8:308. doi: 10.1186/1471-2105-8-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10(10):691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCarrey JR, Kumari M, Aivaliotis MJ, Wang Z, Zhang P, Marshall F, Vandeberg JL. Analysis of the cDNA and encoded protein of the human testis-specific PGK-2 gene. Dev Genet. 1996;19(4):321–332. doi: 10.1002/(SICI)1520-6408(1996)19:4<321::AID-DVG5>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 38.Uechi T, Maeda N, Tanaka T, Kenmochi N. Functional second genes generated by retrotransposition of the X-linked ribosomal protein genes. Nucleic Acids Res. 2002;30(24):5369–5375. doi: 10.1093/nar/gkf696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dahl HH, Brown RM, Hutchison WM, Maragos C, Brown GK. A testis-specific form of the human pyruvate dehydrogenase E1 alpha subunit is coded for by an intronless gene on chromosome 4. Genomics. 1990;8(2):225–232. doi: 10.1016/0888-7543(90)90275-y. [DOI] [PubMed] [Google Scholar]


