Abstract
The proteasomal ATPase ring, comprising Rpt1-Rpt6, associates with the heptameric α ring of the proteasome core particle (CP) in the mature proteasome, with the Rpt C-terminal tails inserting into pockets of the α ring1–4. Rpt ring assembly is mediated by four chaperones, each binding a distinct Rpt subunit5–10. We report that the base subassembly of the proteasome, which includes the Rpt ring, forms a high affinity complex with the CP. This complex is subject to active dissociation by the chaperones Hsm3, Nas6, and Rpn14. Chaperone-mediated dissociation was abrogated by a nonhydrolyzable ATP analog, indicating that chaperone action is coupled to nucleotide hydrolysis by the Rpt ring. Unexpectedly, synthetic Rpt tail peptides bound α pockets with poor specificity, except for Rpt6, which uniquely bound the α2/α3 pocket. Although the Rpt6 tail is not visualized within an α pocket in mature proteasomes2–4, it inserts into the α2/α3 pocket in the base-CP complex and is important for complex formation. Thus, the Rpt-CP interface is reconfigured when the lid complex joins the nascent proteasome to form the mature holoenzyme.
Keywords: proteasome, chaperone, single particle cryoEM, ATPase
The proteasome mediates selective protein degradation in eukaryotes1. It is composed of a 19-subunit regulatory particle (the RP, PA700, or 19S complex) and a 28-subunit proteolytic core particle (CP, or 20S complex) composed of four stacked heptameric rings. Ubiquitinated proteins are recognized by the RP, translocated into the CP via a channel in its outer (α) ring, and degraded. The RP comprises the 10-subunit base and 9-subunit lid1–4. Central to the base is the Rpt ring, a heterohexameric ATPase complex that abuts the α ring, with flexible C-terminal tails of multiple Rpt subunits inserting into “α pockets” of the CP11–14.
The Rpt ring is formed from three modules1: Rpt3-Rpt6, Rpt4-Rpt5, and Rpt1-Rpt2. Within these modules are four assembly chaperones. Although unrelated phylogenetically, these “RP chaperones” each bind a CP-proximal C-domain within a specific Rpt subunit5,9, as follows: Rpt1-Hsm3, Rpt3-Nas6, Rpt5-Nas2, and Rpt6-Rpn14.
A debated aspect of RP assembly is whether the CP facilitates the process15–16, although CP-dependent and CP-independent assembly pathways are not mutually exclusive. We have proposed that insertion of Rpt tails into CP α pockets is important for RP assembly in yeast, and that RP chaperones antagonize Rpt tail insertion into CPα pockets by steric hindrance, thus promoting temporal order in assembly5,6 (Supplementary Fig. 1).
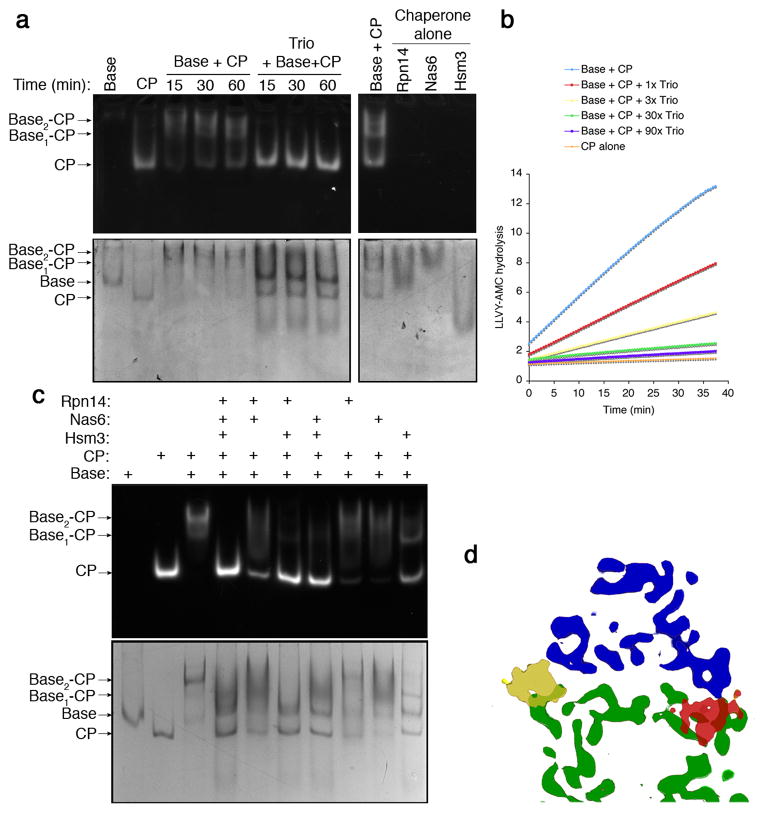
The base is considered a key assembly intermediate of yeast proteasomes5,6,9,10. To investigate the effects of RP chaperones on the RP-CP interface, and to model the behavior of early assembly intermediates, we developed a reconstitution assay for the base-CP complex. When purified base and CP were mixed, they formed complexes (base1-CP and base2-CP; collectively base-CP) with an apparent Kd of ~3 nM (Supplementary Fig. 2). Base-CP was visualized on native PAGE by in-gel assay for hydrolysis of the fluorogenic peptide LLVY-AMC (Fig. 1a). When chaperones Hsm3, Nas6, and Rpn14 were added in excess at time zero, the chaperone trio inhibited complex formation beyond the detection limit. Because tail-pocket contacts mediate base-CP association12, this experiment satisfies a key prediction of the model5,6 that RP chaperones antagonize insertion of Rpt tails into CP α pockets. A fourth RP chaperone, Nas2, dissociates prior to base assembly17 and is accordingly inactive in these assays (data not shown).
Figure 1. Chaperones inhibit base-CP assembly.
a, Purified base (160 nM) and CP (80 nM) were incubated with or without Rpn14, Nas6, and Hsm3 (trio, 1.6 μM each), and resolved via native PAGE. Above, in-gel peptidase assay (0.02% SDS); below, Coomassie stain. For input protein see Supplementary Fig. 2.
b, Base (5 nM) and CP (2nM) were challenged with chaperone trio (amounts in molar excess of base; ATP at 2 mM). In this and all real-time experiments, LLVY-AMC hydrolysis is expressed as relative fluorescence units (r.f.u.) and experiments were performed in triplicate with traces combined for presentation.
c, Native gel analysis of base-CP formation as in a, following addition of chaperones to base (160 nM) singly or in combination at 10-fold molar excess of base.
d, A yeast Rpt hexamer model was built using with the hexameric P97 D1 domain structure as template (see supplementary methods). This model was fit into the EM map2 of yeast Rpt hexamer. Relative positions of Hsm3 (red) and Nas6 (yellow) on the Rpt ring (blue) were assessed by superimposing Hsm3-Rpt1C and Nas6-Rpt3C structures onto the Rpt ring model that had been fit into the EM map. A clipped view of the Rpt ring with bound chaperones and CP (green) is presented. Areas of overlap highlight steric clashes between chaperones and CP.
Antagonism of base-CP association, assessed above with chaperones in excess, remained strong when chaperones were added at 1:1 stoichiometry versus base, indicating potent interference (Supplementary Fig. 3). Base-CP association can be quantified by real-time fluorometric assays that track LLVY-AMC hydrolysis. Free CP hydrolyzes LLVY-AMC slowly, due to closure of its gated channel1; binding of base opens the channel via Rpt-tail-α-pocket interactions12,13. Suppression of base-CP assembly by the chaperone trio is readily observed in this assay (Fig. 1b) at low-nanomolar levels, which are comparable to or below their estimated intracellular abundance (ref. 18 and data not shown). Each of these chaperones was found to individually antagonize base-CP association, though with different potencies (Fig. 1c; Supplementary Fig. 4). In summary, Hsm3, Nas6, and Rpn14 act through a common and coordinate mechanism to antagonize Rpt ring-CP association.
To further assess how RP chaperones regulate proteasomes, we determined the crystal structures of Hsm3 and of Hsm3 complexed with the Rpt1 C-domain (PDB ID 4FP7 and 4JPO; Supplementary Figs. 5–9; see also refs. 19, 20), and modeled this complex into the cryoEM structure of the yeast proteasome holoenzyme2. The Nas6-Rpt3 co-complex21 was similarly modeled into holoenzyme. The results suggest physical clashing between CP and chaperones in holoenzyme (Fig. 1d), consistent with the steric interference hypothesis5,6. Although modeling is not completely predictive, these data agree with previous attempts to model chaperones into holoenzyme5,20. The Rpn14-Rpt6 structure is unsolved and thus the relevance of steric interference to Rpn14 remains conjectural. The RP-CP interface is likely to be dynamic, due to conformational changes in the Rpt ring during cycles of ATP hydrolysis (see below). Consequently, steric interference may apply to a subset of conformational states.
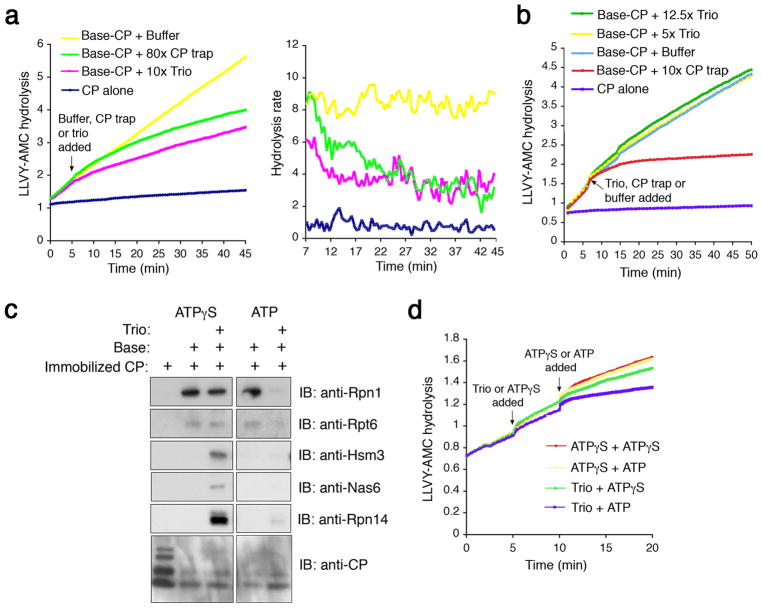
In the base-CP experimental model, RP chaperones may act by binding to free base, thus preventing association of base with CP. Alternatively, or in addition, chaperones might interact transiently with base-CP to actively promote dissociation. To assess these models, we assayed the time course of base-CP dissociation after chaperone addition. As a control, we examined spontaneous dissociation of base-CP using a “CP trap” that is inactivated by the proteasome inhibitor epoxomicin. The trap captures base that had dissociated from base-CP (Supplementary Fig. 10), thus suppressing LLVY-AMC hydrolysis. Trap addition resulted in slow loss of LLVY-AMC hydrolytic activity over more than 15 minutes (Fig. 2a), whereas chaperone addition led to immediate reduction in hydrolytic activity (Fig. 2a, left panel), as expected from active dissociation. Upon chaperone addition, a new, stable steady-state hydrolytic rate was established within approximately 7–8 minutes (Fig. 2a, right panel). Chaperone addition to holoenzyme had only weak dissociative effects (Supplementary Fig. 11a). Thus, RP chaperones may work preferentially on assembly intermediates.
Figure 2. Base-CP association is nucleotide-dependent.
a, Base (5 nM)-stimulated CP (2 nM) activity was monitored over time (2mM ATP, 50 mM KCl). At 5.5 min, chaperone trio or CP trap was added in molar excess of base, or active CP, respectively. CP trap inhibits re-association of base with active CP. Right plot, hydrolysis rate (r.f.u./min) over time.
b, Purified base (5 nM) and CP (2 nM) were assembled in the presence of ATPγS (0.1 mM throughout). At 6 min, chaperone trio or CP trap were added in molar excess.
c, CP (15 nM) was immobilized on IgG resin via ProA tag, and incubated with base (~80 nM) and chaperone trio (160 nM) in the presence of 2 mM ATP or 0.5 mM ATPγS. CP-bound proteins were washed with buffer (50 mM KCl), then eluted with TEV protease while maintaining nucleotide concentration. Immunoblots were probed with indicated antibodies. Images are from the same gel and exposure.
d, CP (2 nM) activity (LLVY-AMC hydrolysis; r.f.u.) was monitored in the presence of base (5 nM) and 0.1 mM ATPγS for 5 min. Chaperone trio (50 nM) or buffer alone containing 0.1 mM ATPγS was then added. At 10 min, buffer containing either ATPγS or ATP + ATPγS was added. Final nucleotide concentrations were either 0.1mM ATPγS or 10mM ATP + 0.1mM ATPγS. See also Supplementary Fig. 12.
To assess the possible relevance of Rpt ring conformational dynamics to base-CP dissociation, we compared the effects of ATP and nonhydrolyzable ATPγS in the base-CP dissociation assay. Base-CP assembled normally with ATPγS (Supplementary Fig. 11b), but subsequent chaperone addition had no detectable effect on LLVY-AMC hydrolysis, indicating failure to dissociate the complex (Fig. 2b). Antagonism of base-CP association by chaperones may therefore be finely tuned to the Rpts’ conformational state. The simplest interpretation is that ATPγS mimics the ATP-bound state of the Rpts, with chaperones inhibiting base-CP association when their cognate Rpts are bound to ADP (or is free of nucleotide), but not to ATP. With ATPγS, RP chaperones could fail to antagonize base-CP simply because they cannot bind Rpts under these conditions. However, chaperone-base complexes form comparably with ATP, ATPγS, or ADP (Supplementary Fig. 11c).
The results described above suggest that a stable base-chaperone-CP co-complex may form in the presence of ATPγS. To test this, we immobilized CP to a resin, added base and chaperones, and assayed resin-bound components after washing. When chaperones were added in the presence of ATP, base dissociated from the complex (Fig. 2c), whereas, with ATPγS, base remained bound to both CP and chaperones. Thus, the chaperones’ lack of effect on LLVY-AMC hydrolysis in the presence of ATPγS reflects failure to dissociate base-CP, and the chaperones’ capacity to compete with CP for occupancy of the base may be dependent on the Rpt nucleotide hydrolytic cycle.
To test whether preformed base-chaperone-CP complex is primed for dissociation, the complex was formed in the presence of ATPγS, and dissociation was then monitored following ATP addition. ATP produced rapid base-CP dissociation; LLVY-AMC hydrolyzing activity decayed with a half-life of approximately one minute (Fig. 2d; Supplementary Fig. 12). In contrast, ATP and ATPγS produced indistinguishable hydrolytic profiles in the absence of chaperones (Fig. 2d). Thus, RP chaperones actively dissociate base-CP.
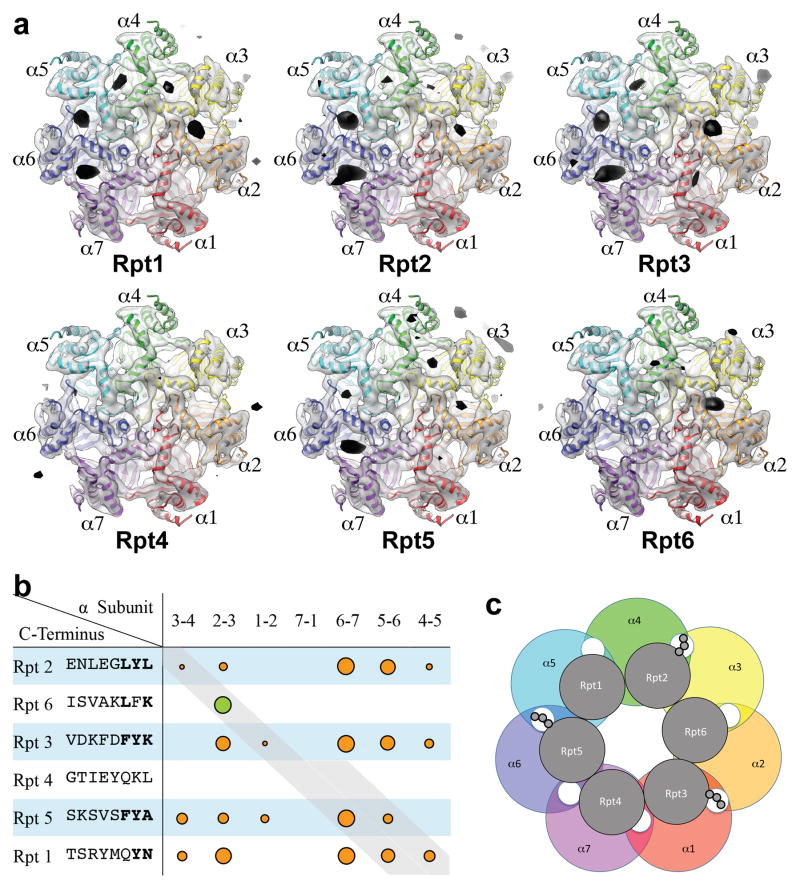
To better understand chaperone action within the base-CP complex, we studied the specificity of insertion of Rpt tails into α pockets, interactions proposed to be under chaperone control5,6. We previously determined by single particle electron cryomicroscopy (cryoEM) that the C termini of the homohexameric archaeal PAN ATPases bind the α pockets of the homoheptameric α ring of archaeal CP13. This approach is used here to assign distinct α pocket binding preferences to each yeast Rpt tail. We determined seven subnanometer resolution three-dimensional (3D) reconstructions of the yeast CP, one from CP alone (Supplementary Figs. 13,14), and the others from CP incubated individually with six different Rpt peptides. Each peptide comprised 8 amino acids from the C-terminus of an Rpt.
The pseudo 7-fold symmetry of the heteroheptameric α and β rings imposes a challenge to single particle cryoEM. To break this pseudo-symmetry, we fused a GST tag to the C terminus of subunit β2 (Supplementary Fig. 13a,e) and verified that the tag does not alter CP function. Differences between maps of peptide-CP and CP alone were calculated as described13 (Supplementary Fig. 15). At proper thresholds, difference densities correspond to peptides bound to α pockets. Because the pocket between subunits α7 and α1 lacks the Lys residue that is required for binding an Rpt’s C terminus11, no specific binding of any peptide to this pocket is expected. Thresholds were therefore set to show no difference density in the α7/α1 pocket, which in fact always had the lowest difference density corresponding to tail peptide (Fig. 3). As controls, difference maps between two independent 3D reconstructions from two separately collected datasets of the same sample showed no significant difference density, indicating that difference densities assigned to each peptide were not generated by image misalignment or random noise (Supplementary Methods).
Figure 3. Difference Maps Reveal Binding Sites of Rpt C terminal peptides to CP α pockets.
a, Top views of 3D density maps of CP superimposed with difference densities corresponding to C-terminal peptides of various Rpts. Peptides were present at 0.5 mM, CP at 1.6 μM. The amount of each peptide bound is reflected by the size of black densities within each pocket. EMD accession numbers are: free CP, 5593; Rpt1-CP, 5611; Rpt2-CP, 5612; Rpt3-CP, 5613; Rpt4-CP, 5614; Rpt5-CP, 5615; Rpt6-CP, 5616.
b, Summary of Rpt tail peptide binding sites and relative intensities. Sizes of circles represent the volume of difference densities generated by peptides. Gray diagonals denote Rpt tail-α pocket mapping of intact proteasomes by crosslinking22. Rpt4, Rpt5, and Rpt1 each crosslink to two α pockets, suggesting ambiguous register.
c, Predominant tail-pocket interactions in yeast holoenzymes as determined by cryoEM2–4.
Fig. 3d compares binding specificities of the six Rpt peptides for the sevenα pockets of the CP. In an individual difference map, the sizes of densities in different pockets correlate with peptide binding affinities for these pockets. Fig. 3e summarizes these data. Figs. 3e and 3f also represent previous mapping of Rpt tails to α pockets in holoenzyme2–4,22. Our data reveal an unexpected lack of specificity in the Rpt tail-α pocket interaction. We therefore suggest that the specificity of tail-pocket interactions within the mature complex is largely guided by constraints on possible tail-pocket interactions that arise from the defined subunit arrangements of the apposed Rpt and α rings. For example, within holoenzyme the Rpt2 and Rpt3 C-termini insert into α3/α4 and α1/α2 pockets, respectively2–4,22. However, free forms of Rpt2 and Rpt3 C-termini show marked preference for noncognate pockets (Fig. 3d).
Our results raise the question of how the register of the RP-CP interface is determined. Only the Rpt6 peptide showed high specificity of binding to an established22 cognate pocket (Fig. 3d,e). Thus, the Rpt6 tail has the specificity to serve as an anchor point for either the mature proteasome or an assembly intermediate.
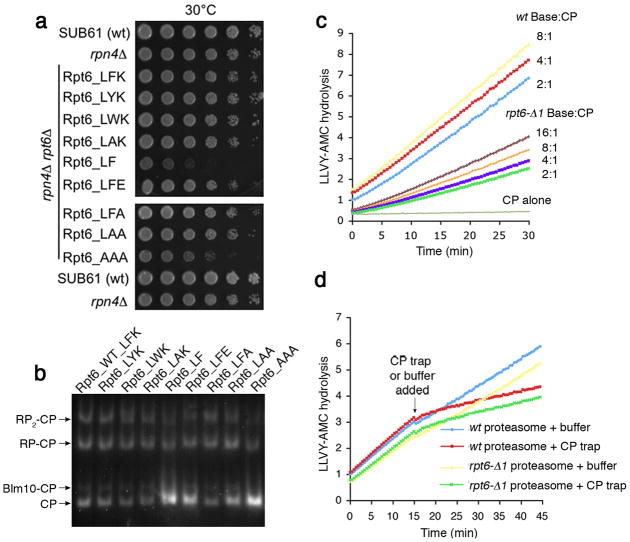
The unique binding specificity of the free Rpt6 tail peptide led us to examine its physiological significance genetically. We generated substitution mutations in Rpt6 tail residues, and assessed effects on proteasome assembly using assays of cell growth (Fig. 4a) and native PAGE (Fig. 4b). Several substitutions resulted in proteasome defects, notably a block substitution of alanines for the terminal LFK sequence (Figs. 4a and 4b; Supplementary Fig. 16). Deletion of one residue from the C-terminus had a similar effect (Figs. 4a and 4b; Supplementary Figs. 16–18).
Figure 4. Rpt6 C-terminal tail promotes formation of base-CP complex.
a, Growth defects of C-terminal rpt6 mutants. Strains were spotted onto plates containing rich media (YPD) in 4-fold serial dilutions. Plates were incubated at 30°C for 2 days.
b, Whole cell extracts (100 μg) from rpt6 mutants as in a were resolved by native PAGE and subject to LLVY-AMC assay in 0.02% SDS.
c, Role of Rpt6 tail in base-CP association. Assembly kinetics of wild-type or rpt6- Δ1 base with CP was measured via LLVY-AMC hydrolysis. Purified CP (2 nM) was mixed with the indicated fold-excess of base (2 mM ATP). LLVY-AMC hydrolysis is indicated in r.f.u.
d, Stability of proteasome holoenzyme (2 nM) from wild-type or rpt6-Δ1 mutants was assessed in 2mM ATP by adding 50-fold molar excess of CP trap or buffer alone at 15 min.
The exceptional specificity of the free Rpt6 tail for the α2/α3 pocket, together with its role in proteasome assembly, appear inconsistent with recent cryoEM studies of the yeast proteasome, which visualize the Rpt2, Rpt3, and Rpt5 tails within α pockets, with Rpt6 apparently not fixed within an α pocket, presumably being too flexible to be visualized2–4. To resolve this paradox, we first employed the base-CP assembly assay to test whether the Rpt6 tail helps to stabilize the base-CP interface. Base complexes were purified from rpt6-Δ1 and wild-type cells, then mixed with CP; activation of LLVY-AMC hydrolysis was assayed. At 16:1, the rpt6-Δ1 base remained deficient in comparison to a two-fold excess of wild-type base over CP (Fig. 4c). The defect in activation can be attributed largely to reduced base-CP association, as shown by native PAGE followed by Coomassie staining (Supplementary Fig. 19).
One scenario to explain the phenotype of rpt6 tail mutants is that their reduced proteasome levels simply reflect a lower affinity between CP and RP. To test this, we purified rpt6-Δ1 proteasome holoenzymes and incubated them in the presence or absence of CP trap to follow their dissociation over time. Mutant and wild-type proteasomes were comparable in stability (Fig. 4d). Thus, the Rpt6 tail influences biosynthetic proteasome assembly rather than holoenzyme stability.
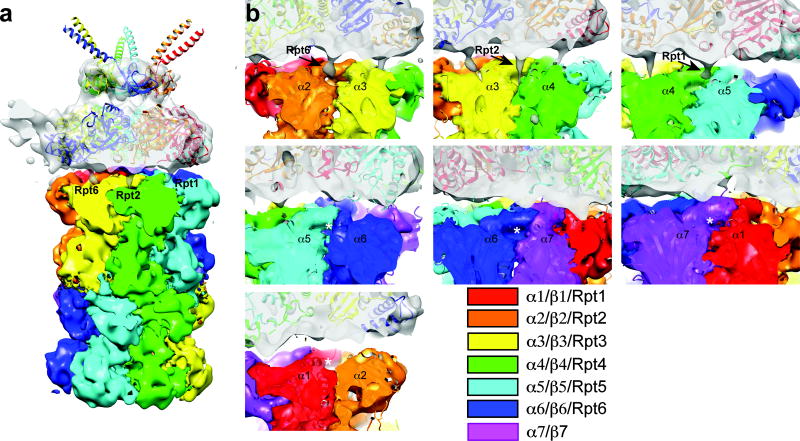
To investigate the base-CP interface more generally, we reconstituted this complex, using base from a wild-type (RPT6) strain of yeast, and subjected it to cryoEM (Supplementary Fig. 20). In the 3D reconstruction of base-CP (Fig. 5a), the rotational register between the Rpt ring and the CP, as well as the axial tilt of the Rpt ring from that of CP, are comparable to those in holoenzyme. Individual Rpt subunits were modeled into the structure as shown, based on previous characterization of the holoenzyme2 (Supplementary Fig. 20c,d).
Figure 5. 3D reconstruction of base-CP complex reveals an asymmetric interaction between Rpts and the α ring of the CP.
a, 3D reconstruction of singly-capped base-CP complex was determined by single particle cryoEM to a resolution of ~10Å (EMD-5617). CP subunits are rendered in different colors as indicated. A difference map was calculated between the original 3D reconstruction and one rotated 180° around the 2-fold CP symmetry axis. The positive difference density (grey) corresponds to base bound to CP. It shows prominent densities from C termini of Rpt6, Rpt2, and Rpt1, which are clustered on one side of the Rpt ring, bound to specific α pockets.
b, Each panel shows an α pocket. Thresholds of CP and base densities are set separately but are identical in all panels. C termini of Rpt6, Rpt2, and Rpt1 are seen to insert into α pockets.
A key feature of the base-CP complex is the existence of a strong contact between the Rpt6 tail and the α2/α3 pocket (Fig. 5), which is inherently specific for Rpt6 tail peptide. Since holoenzyme does not show a prominent Rpt6 tail contact2–4, the structural and functional data both suggest a transient role for the Rpt6 tail in assembly. Distinct contacts were also observed between Rpt1 and the α4/α5 pocket, as well as Rpt2 and the α3/α4 pocket (Fig. 5 and Supplementary Fig. 21), although the Rpt1 contact is relatively superficial. Based on the free tail peptide experiments of Fig. 3, the intrinsic specificity of Rpt2 and Rpt1 cannot explain the tail-pocket register of base-CP. This suggests Rpt6 as a significant determinant of tail-pocket register in base-CP.
The determination of tail-pocket register is inherently problematic because of the symmetry mismatch between the hexameric Rpt ring and heptameric α ring, which may underlie a tendency for ambiguous register at this interface3,4,22. Proper register may be achieved through global optimization of tail-pocket interactions, allowing for a subset of incorrect tail-pocket alignments to be rejected despite their being stronger than the correct alignment. However, our data suggest that tail-pocket register might be largely defined by dominant interactions of high specificity. The Rpt6 tail may perform such a function at an early stage of proteasome maturation, when the RP-CP interface is defined exclusively by Rpt tail-α pocket contacts. This role of Rpt6 is apparently not sustained in holoenzyme, perhaps because register is enforced by an alternative mechanism once the lid is incorporated into the complex. Subunit Rpn6 of the lid extends directly past Rpt6 to contact the CP2,23, and may substitute for Rpt6’s anchoring role.
Although the tail-pocket register of base-CP is consistent with that of holoenzyme, the dominant tail-pocket interactions are quite different. For holoenzyme2–4 these are thought to be Rpt3, Rpt2, and Rpt5. These tails alternate across the ring in the holoenzyme (Fig. 3c), while in base-CP the dominant tail contacts appear to be collected on one side of the ring (Fig. 5), in an arrangement resembling that of the archaeal PAN complex14.
An interesting feature of base-CP is that the neighboring17 Rpt6 and Rpt2 tails display strong pocket interactions. Because Rpt2’s tail has little inherent specificity, its insertion into the α3/α4 pocket may be facilitated by Rpt6. It is consistent with the symmetry mismatch between the Rpt and α rings that Rpt6 should preferentially promote tail insertion of its nearest neighbor, since more distant tails will fall out of phase with the CP pockets.
Negative regulation of tail-pocket interactions by chaperones may potentially help in temporal ordering of the assembly pathway, in suppressing out-of-register tail-pocket interactions, and in maintaining proteasome assembly intermediates in a highly dynamic state. Our data suggest that chaperone action may be coupled to the ATPase cycle of the Rpt ring, with nucleotide controlling the competition between chaperone and CP for base interaction. The mechanism may involve changes in positioning of the Rpt C-domain, which plays an integrative role in that it positions the Rpt C-terminal tail, while at the same time binding chaperone on its outer face and contacting nucleotide on its inner face.
The major forms of mature proteasome differ from the ATPγS-base-CP complex in that they are associated with a mixture of ATP and ADP24, whereas early intermediates in RP assembly are reported to have no detectable ATPase activity25, suggesting that an ADP-free species resembling the ATPγS-base-CP complex could potentially function as a transient assembly intermediate. The fate of this complex may be to undergo chaperone-dependent dissociation upon ATP hydrolysis, or alternatively the lid may join the complex prior to the first round of ATP hydrolysis, to impose new modes of CP binding and suppress complex dissociation.
METHODS SUMMARY
The GST-tagged CP used for cryoEM analysis was purified using a 3x FLAG tag appended to the Pre1 C terminus (β4). For the structure of CP complexed with peptide, ~0.5 mM peptide was incubated with 1.6 μM GST-tagged CP for 1h at 37°C directly before grid vitrification. Recombinant chaperones were purified from E. coli using a GST tag, which was removed with Prescission protease prior to biochemical assays. Single particle cryoEM studies were carried out as described13, with details given in Supplementary Information.
Supplementary Material
Acknowledgments
We thank M. Schmidt, T. Walz, C. Chen, and Finley laboratory members for suggestions, and C. Mann for antibodies. This work was supported in part by grants from NIH (5R01GM098672 and 1S10RR026814-01), UCSF Program for Breakthrough Biomedical Research (New Technology Award) to YC; the Johnson Cancer Research Center, the NCRR (5P20RR017708 and P20 RR016475) and NIH (8 P20 GM103420 and P20 GM103418) to JR; and grants from NIH to PC (R01GM045335) and DF (R37GM043601). SP was supported by the Charles A. King Trust Postdoctoral Research Fellowship Program of the Medical Foundation. Use of IMCA-CAT was supported by IMCA though a contract with the Hauptman-Woodward MRI. Use of the APS was supported by the U.S. Department of Energy (Contract No. DE-AC02-06CH11357).
Footnotes
Contributions
SP performed reconstitution of the base-CP complex and holoenzyme stability. XL performed all cryoEM experiments and analysis. HMK and RS generated yeast strains. HMK purified GST-fused CP and participated cryoEM experiments and analysis. RS performed purifications and MZ ultracentrifugation. KB and SL determined crystal structures, JR and GT performed structural analysis and modeling. MAH, HMK, and PC performed phenotypic and native gel analysis of Rpt6 mutations. JR wrote the supplement with contributions from all authors. The manuscript was drafted by DF and YC, and modified by all authors.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Kish-Trier E, Hill CP. Structural biology of the proteasome. Ann Rev Biophys. doi: 10.1146/annurev-biophys-083012-130417. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lander GC, et al. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482:186–191. doi: 10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lasker K, et al. Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc Natl Acad Sci USA. 2012;109:1380–1387. doi: 10.1073/pnas.1120559109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beck F, et al. Near-atomic resolution structural model of the yeast 26S proteasome. Proc Natl Acad Sci USA. 2012;109:14870–14875. doi: 10.1073/pnas.1213333109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roelofs J, et al. Chaperone-mediated pathway of proteasome regulatory particle assembly. Nature. 2009;459:861–865. doi: 10.1038/nature08063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park S, et al. Hexameric assembly of the proteasomal ATPases is templated through their C termini. Nature. 2009;459:866–870. doi: 10.1038/nature08065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Le Tallec B, Barrault MB, Guerois R, Carre T, Peyroche A. Hsm3/S5b participates in the assembly pathway of the 19S regulatory particle of the proteasome. Mol Cell. 2009;33:389–399. doi: 10.1016/j.molcel.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 8.Kaneko T, et al. Assembly pathway of the mammalian proteasome base subcomplex is mediated by multiple specific chaperones. Cell. 2009;137:914–925. doi: 10.1016/j.cell.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 9.Saeki Y, Toh EA, Kudo T, Kawamura H, Tanaka K. Multiple proteasome-interacting proteins assist the assembly of the yeast 19S regulatory particle. Cell. 2009;137:900–913. doi: 10.1016/j.cell.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 10.Funakoshi M, Tomko RJ, Jr, Kobayashi H, Hochstrasser M. Multiple assembly chaperones govern biogenesis of the proteasome regulatory particle base. Cell. 2009;137:887–899. doi: 10.1016/j.cell.2009.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forster A, Masters EI, Whitby FG, Robinson H, Hill CP. The 1.9A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions. Mol Cell. 2005;18:589–599. doi: 10.1016/j.molcel.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 12.Smith DM, et al. Docking of the proteasomal ATPases’ carboxyl termini in the 20S proteasome’s alpha ring opens the gate for substrate entry. Mol Cell. 2007;27:731–744. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rabl J, et al. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol Cell. 2008;30:360–368. doi: 10.1016/j.molcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith DM, et al. ATP binding to PAN or the 26S ATPases causes association with the 20S proteasome, gate opening, and translocation of unfolded proteins. Mol Cell. 2005;20:687–698. doi: 10.1016/j.molcel.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 15.Park S, Kim W, Tian G, Gygi SP, Finley D. Structural defects in the RP-CP interface induce a novel proteasome stress response. J Biol Chem. 2011;286:36652–36666. doi: 10.1074/jbc.M111.285924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kusmierczyk AR, Kunjappu MJ, Funakoshi M, Hochstrasser M. A multimeric assembly factor controls the formation of alternative 20S proteasomes. Nat Struct Mol Biol. 2008;15:237–244. doi: 10.1038/nsmb.1389. [DOI] [PubMed] [Google Scholar]
- 17.Tomko RJ, Jr, Funakoshi M, Schneider K, Wang J, Hochstrasser M. Heterohexameric ring arrangement of the eukaryotic proteasomal ATPases: implications for proteasome structure and assembly. Mol Cell. 2010;38:393–403. doi: 10.1016/j.molcel.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 19.Takagi K, et al. Structural basis for specific recognition of Rpt1, an ATPase subunit of 26S proteasome, by proteasome-dedicated chaperone Hsm3p. J Biol Chem. 2012;287:12172–12182. doi: 10.1074/jbc.M112.345876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barrault MB, et al. Dual functions of the Hsm3 protein in chaperoning and scaffolding regulatory particle subunits during the proteasome assembly. Proc Natl Acad Sci USA. 109:E1001–E1010. doi: 10.1073/pnas.1116538109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakamura Y, et al. Structural basis for the recognition between the regulatory particles Nas6 and Rpt3 of the yeast 26S proteasome. Biochem Biophys Res Commun. 2007;359:503–509. doi: 10.1016/j.bbrc.2007.05.138. [DOI] [PubMed] [Google Scholar]
- 22.Tian G, et al. An asymmetric interface between the regulatory particle and core particle of the proteasome. Nat Struct Mol Biol. 2011;18:1259–1267. doi: 10.1038/nsmb.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pathare GR, et al. The proteasomal subunit Rpn6 is a molecular clamp holding the core and regulatory subcomplexes together. Proc Nat Acad Sci USA. 2012;109:149–154. doi: 10.1073/pnas.1117648108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith DM, Fraga H, Reis C, Kafri G, Goldberg AL. ATP binds to proteasomal ATPases in pairs with distinct functional effects, implying an ordered reaction cycle. Cell. 2011;144:526–538. doi: 10.1016/j.cell.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson D, Hakala K, DeMartino GN. Subcomplexes of PA700, the 19S regulator of the 26S proteasome, reveal relative roles of AAA subunits in 26S proteasome assembly and activation and ATPase activity. J Biol Chem. 2009;284:24891–24903. doi: 10.1074/jbc.M109.023218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.