Abstract
Thioredoxin reductase (TR) from Drosophila melanogaster(DmTR) is a member of the glutathione reductase (GR) family of pyridine nucleotide disulfide oxidoreductases and catalyzes the reduction of the redox-active disulfide bond of thioredoxin. DmTR is notable for having high catalytic activity without the presence of a selenocysteine (Sec) residue (which is essential for the mammalian thioredoxin reductases). We report here the X-ray crystal structure of DmTR at 2.4 Å resolution (Rwork = 19.8 %, Rfree = 24.7%) in which the enzyme was truncated to remove the C-terminal tripeptide sequence Cys-Cys-Ser. We also demonstrate that tetrapeptides equivalent to the oxidized C-terminal active sites of both mouse mitochondrial TR (mTR3) and DmTR, are substrates for the truncated forms of both enzymes. This truncated enzyme/peptide substrate system examines the kinetics of the ring opening step that occurs during the enzymatic cycle of TR. The ring opening step is 300-500 fold slower when Sec is replaced with Cys in mTR3 when using this system. Conversely, when Cys is replaced with Sec in DmTR, the rate of ring opening is only moderately increased (5-36 fold). Structures of these tetrapeptides were oriented in the active site of both enzymes using oxidized glutathione bound to GR as a template. DmTR has a more open subunit interface than the mouse enzyme and accommodates peptide Ser-Cys-Cys-Ser(ox) in a cis conformation that allows for protonation of the leaving group Cys by His464′, which helps to explain why this TR can function without the need for Sec. In contrast, mTR3 shows a narrower subunit interface. One possible result of this narrower interface is that the mammalian redox-active tetrapeptide Gly-Cys-Sec-Gly may adopt a trans conformation for a better fit. This places the Sec residue farther away from the protonating histidine residue, but the lower pKa of Sec in comparison to Cys eliminates the need for Sec to be protonated.
Thioredoxin reductase (TR)1 is a member of the glutathione reductase (GR) family of pyridine nucleotide-disulfide oxidoreductases. Members of this family accept reducing equivalents in the form of NADPH via an enzyme bound cofactor (FAD). These electrons are transferred to an enzyme disulfide (as a charge transfer complex) and then to the target substrate (1, 2). High Mr TRs, approximately 55 kDa per subunit, are unique in this family of enzymes as they contain a second reactive disulfide center at the C-terminus, which after becoming reduced, attacks the target substrate thioredoxin (Trx), a 12 kDa protein that plays a key role in cellular redox homeostasis.
The proteins of the GR family are homodimeric with a three domain modular architecture (3, 4). Each monomer contains a N-terminal FAD binding domain followed by a NAD(P) binding domain, each a variant of the Rossman βαβαβ fold. The two are oriented in a head to tail fashion to place the flavin and nicotinamide in proper orientation for electron transfer (3). The active site is located in an extended α-helical segment, connecting strand two to strand three in the parallel sheet of the FAD domain. The redox active site sequence Cys-Val-Asn-Val-Gly Cys (CVNVGC) is conserved within the family, with the N-terminal cysteine (Cys) responsible for thiol disulfide exchange and the C-terminal Cys forming the charge transfer complex with the flavin (5). The C-terminal domain is the dimerization domain for this class of proteins and is a five-stranded anti-parallel β-sheet with two α-helices on either side. Only the homodimer2 is physiologically relevant.
Oxidized diglutathione (GSSG), the substrate for GR, consists of two molecules of glutathione (GSH) that form an intermolecular disulfide bond. GSH I (one half of GSSG) is associated with chain A, while GSH II is associated with chain B. Two phases compose the catalytic cycle of GR: a reductive half-reaction and an oxidative half-reaction. During the reductive half reaction, the FAD-associated disulfide from chain A is reduced upon consumption of electrons from NADPH. The oxidative half-reaction is the reduction of GSSG that results in the re-oxidation of GR. The intermediate in the oxidative half-reaction is the mixed disulfide formed between the Cys of GSH I and Cys58 from chain A (6). This step is referred to as the interchange. Histidine467′ from chain B has been identified as the acid/base catalyst in GR (7) for protonation of the leaving group thiol of GSH and is conserved in high Mr TRs.
High Mr TRs can be mechanistically differentiated from GR by the presence of an extra thiol-disulfide exchange step, which is added to the enzyme before the target substrate can be reduced. This additional thiol-disulfide exchange step is analogous to the reduction of GSSG in GR and utilizes a 16 amino acid C-terminal extension characteristic of TR, which contains a vicinal Cys-Cys dyad that forms an intramolecular disulfide. Upon reduction of this dyad by the N-terminal redox center (on the opposite chain), the substrate (Trx) can then be reduced. Mammalian TRs are distinguished in this group since they contain the rare amino acid selenocysteine (Sec, U) as part of the C-terminal redox-active motif Gly-Cys-Sec-Gly (GCUG) (8, 9). Drosophilia melanogaster TR (DmTR), also a high Mr TR, lacks Sec as part of this motif and has the C-terminal sequence Ser-Cys-Cys-Ser (SCCS) (10, 11). Thus, high Mr TRs can be divided into enzymes that contain Sec and those that have a conventional Cys residue.
Replacement of the catalytic Sec residue in the mammalian enzyme with a Cys residue results in a large decrease in catalytic activity (12). It was therefore surprising to learn that DmTR (lacking Sec) has high activity towards its cognate Trx, thus demonstrating that the presence of a Sec residue is not required to catalyze the reduction of the catalytic disulfide bond of Trx (10, 11). Gromer et al.(10) proposed that it is the flanking serine (Ser) residues in the SCCS motif that aid in deprotonating the catalytic Cys residue, thus obviating the need for Sec in this enzyme. However, it was recently reported that the mitochondrial TR from C. elegans (which has a Gly-Cys-Cys-Gly motif) also has a high activity towards E. coli Trx (13). Since in this enzyme there are no flanking Ser residues to help explain its high activity then the question of how non-selenium containing TRs can function with high activity remains unanswered.
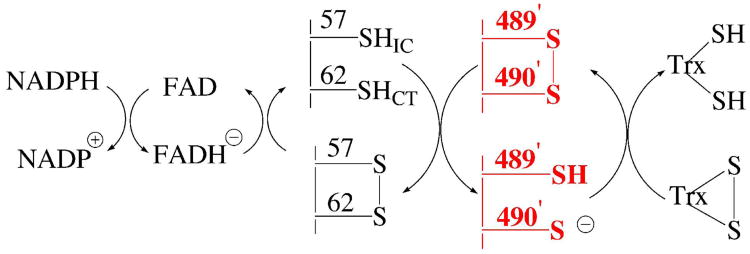
A general reaction mechanism for the reduction of Trx by DmTR is shown in Figure 1. Once the C-terminal Cys-Cys dyad becomes reduced, a thiolate nucleophile attacks the disulfide bond of Trx to initiate reduction of the substrate. In Figure 1, we show this nucleophile as residue 490′. Though there is no direct evidence in the field that Cys490′ is the attacking nucleophile, this residue occupies the same position as Sec in the mammalian enzyme. Because the presence of the Sec residue in mammalian TR is essential for its hydrogen peroxidase activity and is likely to be the nucleophilic residue in the peroxidase reaction, it is thought that Sec must be the attacking nucleophile in the reduction of Trx as well (12, 14). Thus by analogy to its mammalian counterpart, we show Cys490′ as the attacking nucleophile in Figure 1.
Figure 1.
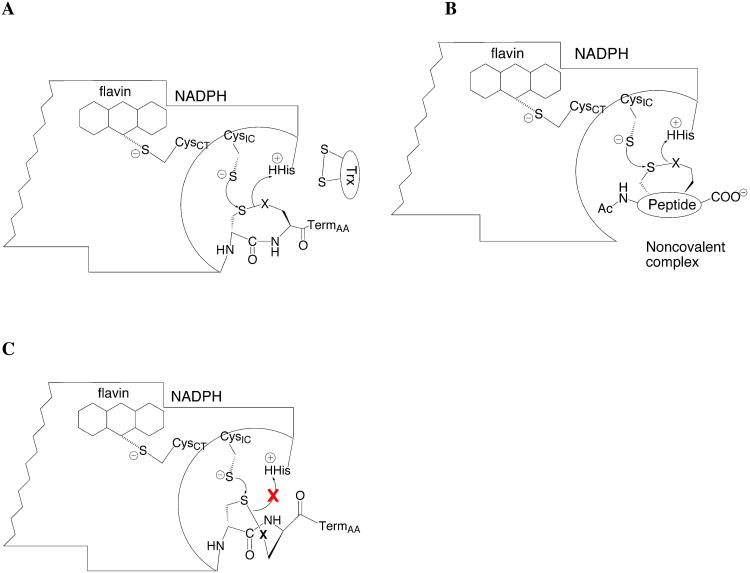
Proposed pathway for transfer of electrons to Trx by DmTR. Cys57 is the interchange residue (Cys57IC) and Cys62 is the charge transfer residue (Cys62CT). Once Cys57 becomes reduced by the flavin cofactor, it initiates attack on the 8-membered ring formed by adjacent Cys residues on the C-terminus on the opposing subunit. The reduction of this 8-membered ring is the additional thiol-disulfide exchange step not found in GR. We refer to this step in the reaction mechanism as the “ring opening” step and is highlighted in red in the diagram. Once this ring is reduced the attacking nucleophile (Cys490′) initiates attack on the disulfide bond of Trx. The “prime” designation indicates residues that are on the adjacent subunit.
In order to explain why mammalian TR requires Sec but DmTR can utilize Cys, we have solved the 2.4 Å X-ray crystal structure of DmTR. We demonstrate that tetrapeptides in the oxidized, monomeric-cyclic form, equivalent to the C-terminal active sites of both mammalianTR and DmTR, are substrates for the truncated forms of both enzymes missing the C-terminal redox center. We used the NMR structures of these tetrapeptides to orient the tetrapeptide motifs in the active site to propose an explanation for why DmTR can catalyze the reaction without a Sec residue.
Materials and Methods
Materials
Vent DNA Polymerase, Nde I, Kpn I, Eco RI, T4 DNA ligase, and chitin agarose beads were purchased from New England Biolabs Inc. (Ipswich, MA). The DEAE sephacel resin, DTNB, NADPH, NADP, N-methylmercaptoacetamide (NMA), and bovine pancreatic insulin was purchased from Sigma-Aldrich (St. Louis, MO). The Fast Flow 6 low substitution phenyl sepharose resin was purchased from Pharmacia-Amersham Biosciences (Uppsala, Sweden). All other chemicals were purchased from Fisher Scientific (Pittsburgh, PA) and were of reagent grade or better.
Cloning and Expression of TR from Drosophila
The Drosophila TR clone (accession number AF301144), was a generous gift from Stephan Gromer. For production of DmTR in E. coli we subcloned the coding region of DmTR (via PCR amplification) into plasmid pTYB1 of the Impact System from New England Biolabs, to generate an intein-fusion protein. Primers were purchased from Integrated DNA Technologies Inc. (Coralville, IA). The upstream primer was of sequence 5′ – AACAGACATATGGCGCCCGTGCAAGG – 3′. The downstream primer, 5′-ACAGCCGGTACCCTTGGCAAAGCAGCTGCAGCAGCTGGCCGG -3′, was designed for production of the full length TR containing the C-terminal tetrapeptide Ser-Cys-Cys-Ser (SCCS488-491). A second downstream primer, 5′-ACAGCCGGTACCCTTGGCAAAGCAGCTGGCCGGCGTGGGG-3′, was designed for production of a truncated TR, removing the C-terminal tripeptide CCS489-491.
The PCR reaction mixtures contained 100 pg template DNA, 50 pmol of each primer, 2 units of Vent DNA Polymerase, 2-3 mM magnesium chloride, in a volume of 100 μL Each PCR was performed on a GeneAmp PCR System 2400 from Perkin Elmer Life Sciences, Inc. (Boston, MA) using 25 cycles with the following parameters: 96 °C for 45 sec, 50 °C for 30 sec, and 72 °C for 180 sec. The product was analyzed by analytical agarose gel electrophoresis and then purified using the QIAquick Purification Kit from Qiagen (Valencia, CA). The PCR product and plasmid pTYB1 were each incubated with Kpn I and Nde I for 2 h at 37 °C, purified using the QIAquick kit, then ligated at 16 °C for 16 h using T4 DNA ligase at 37 °C. E. coli DH5α cells were made competent via the Inoue method (15) and then transformed with 1 μL of purified DNA. The culture was plated onto LB agar containing 200 μg/mL ampicillin and incubated at 37 °C. Individual colonies were used to inoculate 100 mL of LB containing 200 μg/mL ampicillin and allowed to shake at 37 °C overnight. Plasmids were purified using the QIAfilter Plasmid Prep Midi from Qiagen, screened by 1% analytical agarose gel electrophoresis, and verified by sequencing at the DNA Analysis Core Facility, Vermont Cancer Center, University of Vermont.
Cloning and Expression of Mouse Mitochondrial TR
The wild-type form of mouse mitochondrial TR (mTR3) (C-terminal sequence Gly-Cys-Sec-Gly), was produced by semisynthesis while a Sec to Cys mutant of mTR3 (C-terminal sequence Gly-Cys-Cys-Gly), was produced by mutagenesis as previously described (16). A second mutant of mTR3 with a C-terminal sequence of Ser-Cys-Cys-Ser, was also produced by mutagenesis (via PCR amplification). The downstream primer used for this mutant was of sequence 5′-ACAGCCGCTCTTCAGCAGGAACAGCAAGAAGTCACAGTAGGCTCC-3′.
Purification of TR
The detailed method for producing TR has been previously described and was utilized for both mouse and Drosophila TRs (16). Both TRs are expressed as a TR-intein-chitin binding domain fusion protein in E. coli ER2566 cells. The protein is affinity purified using chitin agarose beads with TR cleaved away from the intein using 70 mM NMA at pH 8.0. The TR purity was improved using phenyl sepharose chromatography with elution using an ammonium sulfate gradient (1.0 to 0 M), followed by DEAE sephacel chromatography using a linear gradient of 10 to 300 mM NaCl to elute the protein. Purity was judged as >95% by 12% SDS-PAGE and by the ratio of the absorbance at 280 nm to the absorbance at 460 nm. The concentration of homodimeric TR was determined using the flavin extinction coefficient of 22.6 mM−1cm−1 (17).
Enzymatic Characterization of TR
Each TR preparation was assayed for activity towards DTNB and Trx as described by Arnér et al. (17). All assays were performed on a Cary 50 UV/VIS spectrophotometer from Varian (Walnut Creek, CA) at 25 °C, pH 7.0, and were initiated by addition of enzyme. Activity was monitored over 2 min and the initial velocity was determined from a linear fit. Plots of Vo/ET in min−1 (mole of NADPH consumed per minute per mole of homodimeric TR) vs. substrate concentration were fit to the Michaelis-Menten equation using KaleidaGraph 4.02 from Synergy Software (Reading, PA).
The DTNB assay contained 0.2 mM NADPH and 10 mM EDTA in 100 mM potassium phosphate. For each concentration of DTNB, activity was background corrected by addition of buffer only. Activity was measured by the increase in absorbance at 412 nm and calculated using the extinction coefficient for thionitrobenzoic acid (13.6 mM−1cm−1). The concentration of TR in the assay was 2 nM for mTR3 and 5 nM for DmTR.
The Trx assay contained 0.15 mM NADPH, 1 mM EDTA, and 1 mg/mL insulin in 50 mM potassium phosphate buffer. Background activity was corrected at each concentration of Trx by measuring the ΔA340 upon addition of buffer only. Activity was measured by the decrease in absorbance at 340 nm for the consumption of NADPH and calculated using an extinction coefficient of 6200 M−1cm−1. The concentration of the enzyme in the assay was 25 nM for DmTR, 2 nM for semisynthetic mTR3, and 2 μM for each of the mTR3 Sec-Cys mutants.
Activity of truncated TR towards C-terminal Tetrapeptide Substrates
Tetrapeptides Ac-GCUG(ox), Ac-GCCG(ox), Ac-SCUS(ox), and Ac-SCCS(ox) were prepared by Fmoc solid-phase peptide synthesis and oxidized to the monomeric form using a newly described procedure for making disulfide bonds on-resin (18). The peptides were purified by preparative HPLC and verified as intramolecular disulfides by MALDI-TOF mass spectrometry using a Voyager-DE PRO from Applied Biosystems (Framingham, MA). The peptides were treated as substrates for the truncated DmTR and mTR3 using conditions similar to the DTNB assay and enzyme concentrations were adjusted to get a signal at 340 nm as is seen in the Trx assay. Activity was measured by the decrease in absorbance at 340 nm for the consumption of NADPH, corrected for background in the absence of enzyme and in the absence of substrate, and plotted as mole of NADPH consumed per minute per mole of homodimeric TR.
Crystallization of TR from Drosophila
Purified DmTR was dialyzed against 10 mM potassium phosphate pH 7.4 with 300 mM NaCl and 1 mM EDTA then concentrated to 26 mg/mL. Crystals were grown at 20 °C using the hanging drop diffusion method with a reservoir solution containing 200 mM succinate pH 5.5 and 22% PEG 6000. Hanging drops were 10 μL with 6 μL protein, 3 μL reservoir, and 1 μL of 10 mM NADP. Crystals (280 × 100 × 100 μm) were fully developed in 5 to 7 days. Cryoprotection was achieved by dipping the crystal in reservoir solution containing 4% increments of ethylene glycol for 4 min per step until a final concentration of 20% was achieved.
Data Collection, Structure Determination and Refinement
Data was collected at −169 °C using a Rigaku RU-H3R generator with a copper rotating anode on a Mar345 image plate detector. The data was indexed using DENZO and scaled and merged using SCALEPACK (19). The structure of rat TR1 (PDB 1H6V) was used for molecular replacement routines with CNS (v 1.1)(20). The cross-rotation search yielded a single peak 2.8σ above the mean and the translational search found a peak 2.1σ above the mean. Rigid-body refinement reduced the R-factor to 0.46 for data from 15 to 2.4 Å. Data were processed using space group R32. Model building was based upon simulated-annealing omit map interpretation with the graphics program O (21). Successive rounds of refinement were performed by simulated annealing, occupancy, and B-factor refinement using CNS and CCP4 (22). In the later stages of refinement, the position of the FAD as well as the NADP (minus the nicotinamide moiety) was determined by fitting omit maps. Final refinement was performed using REFMAC5 (23) in CCP4 version 5.0.2 with the entire model as a single TLS group.
C-terminal Peptide Structure and Docking
The oxidized tetrapeptides Ac-GCUG, Ac-GCCG, and Ac-SCCS were examined by NMR spectroscopy and found to exist as a population of four major conformers (Deker and Hondal, unpublished) as has been previously described for a similar peptide with a vicinal disulfide bridge (24). Each of the four conformers was placed at the reaction interface proximal to the conserved flavin-associated active site sequence CVNVGC of both DmTR and mTR3 (PDB 1ZKQ) (25). Because of the high similarity between the reaction mechanisms of GR and TR, we used the orientation of oxidized glutathione (GSSG) in the active site of GR (PDB 1GRA, (3)) to orient the structures of our oxidized peptides in the active site, tetrapeptide binding pocket, of TR. The disulfide bond of the oxidized peptides was placed in an orientation similar to that of the disulfide bond of GSSG and evaluated for the potential to form the interchange mixed-disulfide as well as for positioning of the leaving group in relation to the catalytic histidine of GR. Additional restrictions on the placement of the peptide were imposed by the covalent interchange mixed-disulfide structure of GR (PDB 1GRE(3)). Using the two GR structures, priority was given in the peptide modeling to the first step (interchange) of the GR GSSG-dependent oxidative half-reaction by super-positioning of the Cβ and sulfur (or selenium) atoms to the interchange cysteine of GSH I. Independent fits for each peptide were performed for both cysteines in peptide SCCS(ox), or the Cys and Sec in peptide GCUG(ox), as the interchange residue. Alignments were performed using the graphics program O (21).
Results
DTNB Reductase Activity
Truncated TRs missing the final three amino acids (abbreviated as DmTR-S and mTR3-G in the data tables), full-length DmTR and a semisynthetic version of mTR3 (containing the last three amino acids CUG) (16), were tested for their ability to reduce DTNB. A summary of this kinetic data is presented in Table 1. The results indicate that the truncated forms of both TRs retain quite high DTNB reductase activity (148% of full-length enzyme in the case of DmTR and 75% of the full-length enzyme in the case of mTR3).
Table 1.
DTNB Reductase Activity of Drosophila and mouse TRs.
| Enzyme | kcat (min−1) | Km (mM) |
|---|---|---|
| DmTR-SCCSa | 96 ± 2.9 | 0.17 ± 0.02 |
| DmTR-Sb | 142 ± 8.3 | 0.92 ± 0.15 |
| mTR3-GCUGc | 1250 ±70 | 0.47 ± 0.09 |
| mTR3-Gd | 935 ± 49 | 2.72 ± 0.43 |
The full-length Drosophila enzyme with C-terminal sequence SCCS.
This mutant is truncated (missing residues CCS from the C-terminus) so that Ser488 is the C-terminal amino acid.
The full-length mouse enzyme with C-terminal sequence GCUG.
This mutant is truncated (missing residues CUG from the C-terminus) so that Gly521 is the C-terminal amino acid.
Thioredoxin Reductase Activity
Wild-type DmTR, truncated DmTR, semisynthetic mTR3, truncated mTR3, and two mutants of mTR3 (containing C-terminal tetrapeptide sequences GCCG or SCCS) were also tested for activity towards a non-cognate Trx from E. coli, which has been used as a standard in the field for measuring TR activity (17). All of the kinetic data is summarized in Table 2 including the previously reported activity of our semisynthetic mTR3, which has very high activity towards E. coli Trx (16). Mutation of the catalytic Sec residue to Cys in the mammalian enzyme dramatically lowers the catalytic activity of the enzyme, which has been demonstrated previously by us (16) and others (12, 26). In light of the proposal by Gromer and coworkers (10), we made a second mutation to this enzyme in which the flanking Gly residues were replaced with Ser so that the C-terminal tetrapeptide sequence of mTR3 was SCCS, as in the enzyme from Drosophila. The results show that when the catalytic Sec residue of the mammalian TR is replaced with Cys, activity cannot be restored when the flanking Gly residues are mutated to Ser, in agreement with a recently published study (27).
Table 2.
Thioredoxin Reductase Activity of Drosophila and Mouse Enzymes.
| Enzyme | kcat (min−1) | Km (μM) |
|---|---|---|
| DmTR-SCCS | 346 ± 16 | 141 ± 12 |
| DmTR-S | No Activity | No Activity |
| mTR3-GCUGa | 2220 ± 78 | 67.6 ± 6 |
| mTR3-G | No Activity | No Activity |
| mTR3-GCCGb | 7.6 ± 0.7 | 123 ± 8 |
| mTR3-SCCSc | 5.0 ± 0.6 | 32 ± 8.8 |
Results for the semisynthetic enzyme have been previously been reported in (16).
The full-length mouse enzyme with C-terminal sequence GCCG.
The full-length mouse enzyme with C-terminal sequence SCCS.
It should be noted that DmTR has much higher activity towards its cognate Trx than E. coli Trx (11) so that when comparing cognate substrates, DmTR would have much more robust activity in comparing the two enzymes (DmTR still has 16% of the activity of the semisynthetic mouse enzyme, Table 2). The Km of DmTR for E. coli Trx is 141 μM, which is about a 20-fold increase over the cognate DmTRx-2 (10) and is not unexpected for a non-cognate substrate (13).
Truncated TR as a Disulfide Reductase
During the course of developing our semisynthetic technique for construction of mammalian TR, we discovered that oxidized peptides corresponding to the missing amino acids of the truncated enzyme could act as substrates for the truncated enzyme (16). This is reasonable because as part of the enzyme, the oxidized form of the C-terminal tetrapeptide must be reduced by the N-terminal redox-active dithiol during the enzymatic cycle. A disulfide bond formed between vicinal Cys-Cys (or Cys-Sec) residues results in the formation of an eight-membered ring and this assay examines the kinetics of the ring opening step that occurs during the enzymatic cycle of TR (see Figure 1). For example, the oxidized tripeptide Cys-Sec-Gly-OH(ox) is a substrate for mTR3 when added to the assay mixture containing NADPH and truncated TR (Supplemental Figure 1). When the selenium atom is replaced with a sulfur atom in the homologous peptide CCG(ox), there is very little activity (0.81 min−1). In contrast, peptide CUG(ox) is reduced at a rate of 260 min−1, indicating that the ring opening step is greatly affected by the replacement of a selenium atom with a sulfur atom in the mammalian enzyme.
We subsequently tested oxidized tetrapeptides as substrates for both truncated enzymes so that we could fully examine the effect of flanking residues on either side of the Cys-Cys or Cys-Sec dyad. This unique peptide complementation system allows us to investigate the electron transfer to the C-terminus from the conserved N-terminal redox-active center in a manner that is more analogous to GR and its cognate substrate GSSG. We tested tetrapeptides Ac-GCUG(ox), Ac-GCCG(ox), Ac-SCCS(ox), and Ac-SCUS(ox) (all as the oxidized, monomeric-cyclic peptides) as substrates for the truncated forms of DmTR and mTR3. A summary of the kinetic data for ring opening using the two different truncated TRs is given in Table 3A. The low affinity observed between all peptides and both enzymes, is probably the result of minimal binding contacts in this noncovalent TR/peptide system (as shown by the kinetic plots in Supplemental Figure 2).
Table 3.
Table 3A: Summary of the Relative Activities Toward Tetrapeptides for the Truncated Forms of Mouse and Drosophila TRsa.
| Enzyme | Peptide (turnover rate in min−1/mM) | |||
|---|---|---|---|---|
|
| ||||
| Ac-SCCS(ox) | Ac-SCUS(ox) | Ac-GCCG(ox) | Ac-GCUG(ox) | |
| mTR3-G | 0.21 | 107.3 | 0.06 | 18.5 |
| DmTR-Sb | 57.0 (271) | 322.6 (3) | 2.4 (40) | 87.0 (4.7) |
|
| ||||
| aThe rate of peptide turnover was determined at pH 7.0 in potassium phosphate buffer as described in the Methods. The relative activities for the individual peptides are the slopes from the titration curve in min−1/mM. | ||||
| bThe ratio of rates (fold difference) for DmTR-S and mTR3-G are given in parentheses in bold | ||||
| Table 3B: Ratio of Peptide Turnover Rates for the Truncated Forms of Mouse and Drosophila TRs. | |||||
|---|---|---|---|---|---|
|
| |||||
| Enzyme | SCUS/SCCS (Se/S) | GCUG/GCCG (Se/S) | GCUG/SCCS (Se/S) | SCCS/GCCG (S/S) | SCUS/GCUG (Se/Se) |
| mTR3-G | 511 | 308 | 88.1 | 3.5 | 5.8 |
| DmTR-S | 5.65 | 36.2 | 1.5 | 23.7 | 3.7 |
The ratio of peptide turnover rates for truncated mTR3 and DmTR are given in Table 3B. The data in Table 3B demonstrates a dramatic difference in the rate of peptide turnover in this step of the reaction mechanism. In the mammalian enzyme, we see that replacing the C-terminal Sec residue with a Cys residue (compare Ac-GCUG(ox) to Ac-GCCG(ox)) demonstrates that there is a 308 fold decline in activity, while replacing the C-terminal Cys residue of peptide Ac-SCCS(ox) with a Sec residue results in small increase in peptide turnover (5.65). If we compare the decline in the rate of ring opening to the ratio of kcat values for the reduction of Trx by mTR3-GCUG and mTR3-GCCG (292), we see that they are nearly equal. However, our semisynthetic wild-type enzyme is 91% full-length enzyme (as judged by selenium content), so the kcat value for the wild-type enzyme should be somewhat higher. If we use the kcat value of rat TR1 (2500 min−1) (12), then the ratio of kcat values increases to 329. A reasonable interpretation of this result is that much of the reduction in TR activity observed when Sec is replaced by Cys in the mammalian enzyme is due to the decline in the rate of the ring opening reaction.
Overall, these experiments indicate that replacement of Sec with Cys in the mammalian enzyme drastically decreases the rate of ring opening. Conversely, a Cys to Sec substitution in this reaction step for DmTR results in a slight increase in rate. The flanking serine residues of the SCCS tetepeptide motif in DmTR have a modest impact in increasing the rate of this step for both enzymes (3.5 fold for mTR3 and 23.7 fold for DmTR). This point will be further elaborated in the Discussion.
Crystal Structure of TR from Drosophila
A representative crystal used to collect the data is shown in Supplemental Figure 3. The crystal belongs to the space group R32, has a single monomer in the asymmetric unit, and the physiological dimer is generated by the two-fold symmetry axis. The final model has an Rfree of 24.7 % at a resolution of 2.4 Å (Table 4). Truncated DmTR has 488 residues of which residues 8-484 were observed definitively in the electron density and deposited in the final model (PDB 2NVK). Residues 485 to 488 showed discontinuous density and high B factors, which agrees with previous results where the electron density of the C-terminal tail containing the second redox active disulfide of the enzyme was either very weak or absent in the crystal structures of rat TR1 (28) and mTR3 (25). The overall fold is as expected for a GR family member with an N-terminal FAD-binding domain, a NAD(P)-binding central domain, and C-terminal dimerization domain (Figure 2). The position of the FAD and NADP were verified during refinement by multiple iterations of annealing while omitting the cofactors. The enzyme has low affinity for the oxidized form of the NADP cofactor, which is evident in the crystal structure by the absence of clear density for the nicotinamide moiety and high B factors for the remaining atoms of the cofactor.
Table 4.
Crystallographic statistics for Thioredoxin Reductase from Drosophila melanogaster.
| Cell parameters | |
|---|---|
| space group | R32 |
| a, b, c (Å) | 151.487 |
| 151.487 | |
| 134.259 | |
| Data collection statistics | |
| resolution range (Å) | 15 – 2.4 |
| unique reflections | 22,987 |
| completeness (%) | 98.7 (90.3)a |
| redundancy | 7.85 |
| Rmerge (%)b | 5.9 |
| I/σ | 36.8 (2.8) |
| Model statistics | |
| Rworkc | 19.8 |
| Rfreed | 24.7 |
| # Atoms, non-hydrogen | |
| Protein | 3684 |
| Ligand | 92 |
| Water | 121 |
| r.m.s. deviation, bonds (Å) | 0.011 |
| r.m.s. deviation, angles (°) | 1.359 |
| Ramachandran plot (%) | |
| most favored regions | 89.0 |
| additionally allowed | 9.6 |
| generously allowed | 0.7 |
| disallowed regions | 0.7 |
Number in parentheses denotes highest resolution shell.
Rmerge = ΣΣI(h)j − <I(h)>/ΣI(h), where I(h)j is the jth measurement of diffraction intensity of reflection h and <I(h)> is the average intensity of reflection h for all j measurements.
Rwork = Σ(|Fo | − |Fc|)/Σ|Fo |.
Rfree is calculated using a test set of 10% of the reflection excluded from refinement.
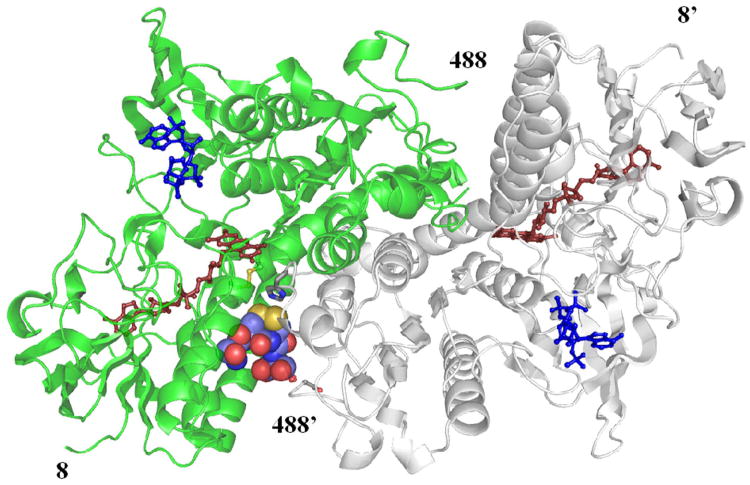
Figure 2.
The crystallographic structure of the TR from Drosophila. The homodimeric model of DmTR with chain A in green and chain B in gray. The SCCS(ox) tetrapeptide in the C+ is oriented with Cys489′ as the interchange and shown in space-filling model. The NADP without the nicotinamide moiety is indicated in blue, the FAD in dark red. The prime designation for residue numbers indicates the B chain of the TR homodimer and the resulting images were generated in PyMOL (50).
The DmTR model has a RMS residual of 0.84 Å upon least squares superposition of the Cα atoms to those of mTR3 (PDB 1ZKQ), 1.15 Å to rat TR1 (PDB 1H6V) (28), and 1.16 Å to human GR (PDB 1GRA). DmTR shares 55% sequence identity with mTR3 and the majority of sequence deviations are found on external loops, primarily in the NADP-binding domain. A distinct difference in the structures is observed at the tetrapeptide binding pocket where thiol-disulfide exchange takes place between the FAD-associated disulfide and the C-terminal disulfide motif of the adjacent monomer of TR. This represents the Rossman fold helical face of the FAD-domain where helix 1 is residues 19-29, helix 3 is residues 97-121, and helix 6 is residues 335-347. Helix 1 contains the conserved Lys26, previously suggested as a potential anchor for the carboxyl terminus (28), while helix 2 contains the conserved active site sequence CVNVGC.
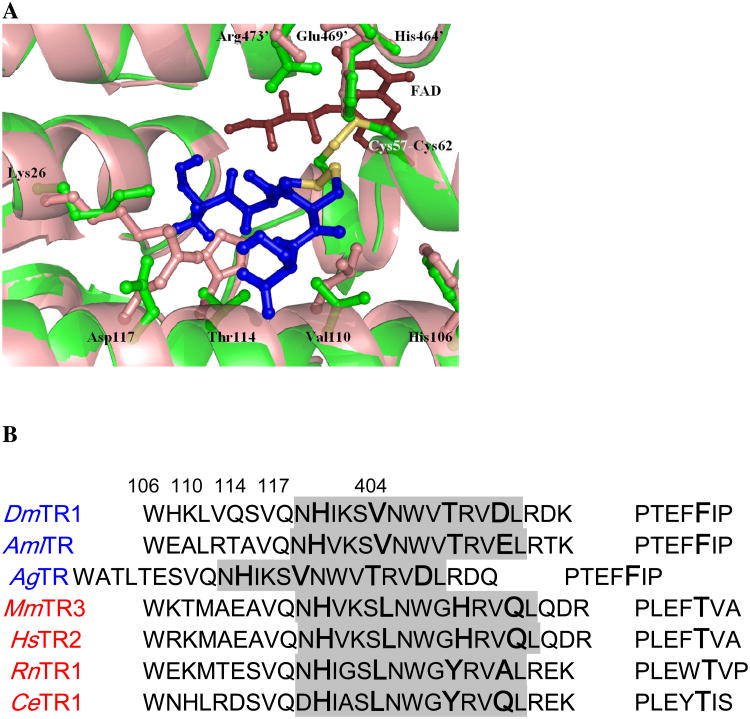
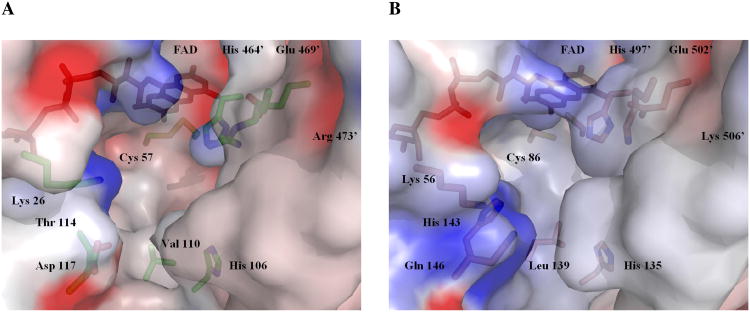
Helix 3 is of particular interest as the residues projecting toward the interface are each smaller compared to the mammalian enzymes containing Sec (Figure 3A). For DmTR the residues are Val110, Thr114, and Asp117 as is the case for A. gambie and A. mellifera, which have C-terminal sequences similar to that of DmTR (TCCS and SCCS, respectively). While valine occupies position 110 in DmTR, bulkier amino acids Leu or Ile are found at the equivalent position in mammalian TRs. Threonine114 of DmTR is replaced by either His or Tyr at this position in the mammalian enzyme (Figure 3B). The amino acid at position 117 is most variable when comparing sequences, Asp in DmTR, but is the bulkier Gln (numbered 146) in mTR3. Additionally in both mammalian TR structures (rat TR1 (28) and mTR3 (25)), these residues also project towards the interface.
Figure 3.
The tetrapeptide binding pocket where thiol-disulfide exchange occurs between the FAD-associated active site from chain A and the C-terminal disulfide of chain B for DmTR. Panel A is a close-up view of the SCCS(ox) tetrapeptide (blue) in the C+ conformation with Cys 489′ as the interchange residue positioned in the DmTR structure (green) with an overlay of the mTR3 (PDB 1ZKQ) colored in salmon. The residues for mTR3 are indicated in parentheses. Panel B is a multiple sequence alignment generated with ClustalW (51) with either the PDB ID or accession number in parentheses. Cys-containing TRs are lettered in blue and Sec-containing TRs are lettered in red. The sequences are TR1 from D. melanogaster (AF301144_1), TR from A. mellifera linguistica (AAP93583.1), TR from A. gambie (CAD30858.1), TR3 from M. musculus (PDB 1ZKQ), TR2 from H. sapiens (Q9NNW7), TR1 from R. norvegicus (PDB 1H6V), and TR1 from C. elegans (AF148217_1). The alignment shows helix 3 from Chain A with turns 3-6 in the shaded portion along with the loop from the dimerization domain of Chain B. The numbering is in accordance with the TR from Drosophila.
In addition to residue substitutions in helix 3, the widening of the tetrapeptide binding pocket is contributed to by the rotation of helix 3 away from the active site and towards the dimer interface. This is analogous to the rotation observed for the equivalent helix in trypanothione reductase when compared to GR for accommodation of the larger trypanothione substrate (29, 30). The movement is limited to turns 3-6 of the 7-turn helix from Asn105 to Leu118 (this is Asn134 to Leu147 in 1ZKQ) with a maximum shift of 1.68 Å at Val110. The source of this shift does not appear to be a function of residue substitutions at the packing between helix 2 and 3 but rather a packing interaction between His106 in helix 3 and Phe404′. This residue is within the helix between stands 3 and 4 of the dimerization domain of the second molecule and follows a conserved bulky hydrophobic residue, which packs within the dimerization domain. While His 106 is conserved in TR there is a Thr substituted for Phe404′ for the mammalian enzymes (Thr437′ in 1ZKQ). His106 was previously suggested to be a base catalyst for DmTR (28) however results from recent mutagenesis studies indicate a structural role instead (31).
Alignment of Tetrapeptides with GSSG
One of the key questions in the TR mechanism is how the C-terminal redox dyad is reduced by the N terminal redox center on the opposite subunit. The C-terminal redox center is either a Cys-Cys or Cys-Sec dyad as we introduced earlier. Our analysis begins with considering the known conformations of a Cys-Cys(ox) dyad (which are rare in protein structures) that have been reported previously. For example, the solution structure of Ac-Cys-Cys-NH2(ox) has been examined by NMR spectroscopy and found to exist as a mixture of four different conformers (24, 32). These conformers are designated as C+, C-, T-, and T′-. The designations “C” and “T” represent the geometry of the peptide bond as either cis or trans, whereas the designations “+” and “−” refer to the handedness of the disulfide bond (+ for right handed and - for left handed)3. It was observed that peptide Ac-Cys-Cys-NH2(ox) also had a second T- conformer that is defined by a larger separation of Cα atoms and this peptide was designated as T′-. Our NMR solution studies of peptides Ac-SCCS(ox), Ac-GCCG(ox), and Ac-GCUG(ox) also show that the same four major conformers described by Reitz and coworkers (24) are also the major conformers of these three peptides (Deker and Hondal, unpublished).
The observation that oxidized tetrapeptides can serve as substrate for the truncated forms of TR demonstrate that TR, like GR, may be considered a disulfide oxidoreductase where its own C-terminus is the cognate substrate. Using this relationship and the available structures for GR, we started by asking the question “Which of the four known conformers of Cys-Cys(ox) aligns with GSSG in the GR structure?”. We therefore aligned the four known conformations of peptide Ac-SCCS(ox) to the thiol-disulfide interchange step observed in GR (3). The formation of the mixed-disulfide between Cys57 and a Cys from the C-terminus of the second molecule of TR in the DmTR homodimer would represent the binding of GSSG in GR (PDB 1GRA) and the formation of the subsequent mixed-disulfide intermediate (PDB 1GRE). The comparison of these two GR structures shows the primary difference is the movement toward each other of the interchange Cys (Cys58 of GR) side chain and the Cys side chain from GSH I. Using GSH I of GSSG as a reference, each tetrapeptide was fit such that the position and angle of the interchange cysteines were equivalent.
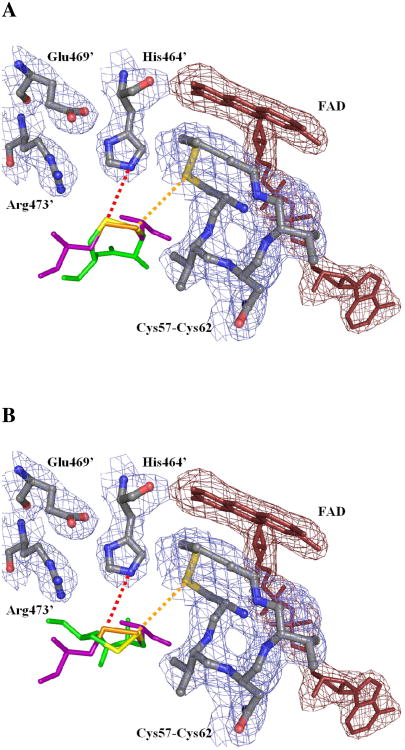
Altogether, we evaluated eight possible orientations for a given tetrapeptide in the active site of TR because each of the four conformations can be overlaid with the sulfur atoms of GSSG in two different orientations (by rotation of 180°), which would either place the N-terminal Cys in the interchange position or the C-terminal Cys in the interchange position. Upon alignment of the sulfur atoms of peptide SCCS(ox) with the sulfur atoms of GSSG, it is clear that peptides in the C+ conformation place the leaving group Cys (C-terminal Cys) in a favorable position for protonation from His464′ (His467′ in GR, His497′ in mTR3, see Figure 4A). This histidine has been shown to be essential for GR (7) and proposed as a conserved acid/base catalyst throughout the protein family (29, 33) including TR (34, 35). Maintaining the interchange position and angle for the trans (T- and T-′) peptides, however, results in a rotation of the leaving group Cys by approximately 90° (Figure 4B) as is also observed for the C− conformation. This moves it away from His464′ by increasing the distance from 3.5 Å to 5.1 Å and to an angle not reasonable for protonation. In order to align the C-terminal sulfhydryl of peptide SCCS(ox) in the T-conformation with the GSH II Cys so that it would be in a favorable position for protonation from His464′, the interchange cysteine (N-terminal Cys of SCCS(ox)) must be rotated away from the interchange residue (Cys57).
Figure 4.
Alignment of the tetrapeptide SCCS(ox) in the structure of TR from Drosophila. The tetrapeptide structures were determined by NMR spectroscopy and placed in the tetrapeptide binding pocket of TR in accordance to the position of GSH I (the interchange position) in the GR structure (PDB 1GRA). Shown is the FAD cofactor (dark red) and the helical loop of the conserved active site sequence CVNVGC, which are components of chain A. Also shown is Arg473′ from the B chain as well as His464′ and Glu469′, which are analogous to His467′ and Glu472′ in GR. The oxidized C-terminal disulfide of the B chain is reduced by the FAD-associated disulfide of the A chain during the enzymatic cycle. The residues from TR are in gray, oxidized glutathione is in purple, the tetrapeptide is in green, and omit electron density for the 2.4 Å structure is shown. Cys57 of DmTR is in position for interchange with GSH I and the Cys489 of the SCCS(ox) tetrapeptide, which is indicated by the orange dashed line. GSHII is in position for protonation from His464′ as is Cys490′ of the SCCS(ox) tetrapeptide when in the C+ conformation (Panel A), but not when in the T- conformation (Panel B) as indicated by the red dashed line. Only the cysteines for the SCCS(ox) peptide and GSSG are shown for simplicity.
Figures 5A and 5B model the tetrapeptide binding pocket where the oxidized tetrapeptide would be reduced by the N-terminal redox center in DmTR and mTR3, respectively. As can be seen in the Figure, the tetrapeptide binding pocket is significantly larger in the case of DmTR (Figure 5 A) compared to that of mTR3 (Figure 5B). This larger binding pocket in DmTR can accommodate the larger size of the C+ conformer of the SCCS(ox) tetrapeptide. The distance from the NH of the N-terminal Cys to the CO of the C-terminal Cys is larger in the C+ conformer than in the T- conformer. In addition, the larger binding pocket of DmTR can also more easily fit the bulkier side chains of Ser. The T- conformer is much more compact and forms a tight turn structure that is better accommodated in the binding pocket of mTR3. Thus, using this same analysis with the previously solved mouse TR structure (PDB 1ZKQ) in conjuction with our NMR structures of GCUG(ox) (Deker and Hondal, unpublished), we find that GCUG(ox) in the T- conformation with Cys in the interchange position appeared to be the most favorable conformation for interchange with Cys86 from chain A. With Sec oriented in the interchange position, an unfavorable interaction with the carbonyl from the adjacent Cys residue occurs with His497′, but we we cannot rule out the T- orientation with Sec in the interchange position. The fit of the GCUG(ox) tetrapeptide in the binding pocket of mTR3, tetrapeptide SCCS(ox) in the C+ conformation docked in the binding pocket of DmTR, and a stereo diagram of tetrapeptide GCUG(ox) in the C+ conformation positioned in the binding pocket of mTR3 is shown in the Supplemental Information.
Figure 5.
Electrostatic surface potential calculated using GRASP (52) for thioredoxin reductase showing a close-up of the tetrapeptide binding pocket where thiol-disulfide exchange occurs between the conserved N-terminal dithiol and the C-terminal tetrapeptide of the adjacent subunit containing the 8-membered ring that must be opened during redox cycling. (A) Interface for DmTR; (B) Interface for mTR3 (PDB 1ZKQ).
Discussion
An important question in elucidating the mechanism of high Mr TRs is how the Cys-Cys or Cys-Sec dyad becomes reduced by the N-terminal disulfide redox center. Using the conserved interchange step between TR and GR, the known conformations of an 8-membered ring formed by a vicinal disulfide, and the kinetics of ring opening, we present a model for how TR reduces this C-terminal redox center below.
Each of the various TRs catalyzes the same reaction, the reduction of the catalytic disulfide bond of their respective cognate Trx. A key difference among the various TRs is the sequence of the C-terminal tetrapeptide redox motif that is critical for this reaction. Kanzok and coworkers (11) were the first to demonstrate that the presence of a selenium atom is not required to catalyze this reaction. From a chemical perspective, why might a selenol be required to catalyze this reaction? A natural response would be the much lower pKa of the selenol of Sec (∼5.2) compared to that of a thiol in Cys (∼8.3) (36). However, there are many examples of Cys residues in proteins with greatly altered pKa values, such as Cys30 in DsbA which has a thiol pKa value of 3.42 (37). Thus proteins can enhance the chemical reactivity of a thiol using mechanisms such as the use of a general base, electrostatic interactions, and α-helical dipole moments without the need to place a Sec into the protein. A possible biological explanation for the presence of Sec in TR is due to the redox sensing function of the enzyme (38).
Regardless of the biological role of Sec in mammalian TRs, replacement of Sec with Cys in the mammalian enzyme results in a large loss of activity. However, the data in Table 1 demonstrates that the truncated form of the mammalian enzyme can still catalyze the reduction of reactive disulfides such as DTNB. Why is this aromatic disulfide a substrate for the truncated enzyme, while other simple disulfides such as cystine are not? A clue is given by comparing the rate of reduction of peptide CUG(ox) vs. peptide CCG(ox) by the truncated enzyme; The rates are 260 min−1 and 0.81 min−1, respectively. This 320 fold difference in the rate of ring opening can be explained by an impairment in the enzyme's ability to protonate the leaving group. Thus the large difference in the rate of ring opening of these two peptides can be understood in terms of leaving group pKas (ca. 1000 fold difference as discussed above). Based on this data we posit that DTNB is a substrate for the enzyme due to the very low pKa (4.75) of the leaving group thiol that is formed upon reduction of this disulfide (39), which is remarkably similar to the pKa of a selenol (5.2).
The issue of which sulfur atom is attacked in a thiol-disulfide exchange reaction was recently addressed very clearly by Arscott and coworkers (6) who discuss the example of which sulfur atom is attacked in a mixed disulfide of form R-S-S-TNB (TNB-S is 5-thio-2-nitrobenzoate). In such cases, TNB-S− will be the leaving group because of the strong electron withdrawing properties of the 2-nitrobenzoate group. Such mixed disulfides are also easier to break than a typical disulfide bond because they are highly polarized. The exact same situation exists in the peptide Ac-GCUG, which has a low pKa leaving group (Se) and the S-Se bond is also much weaker than a typical disulfide bond due to the polarization in the bond conferred by the Se atom. Thus our data using oxidized, monomeric-cyclic peptides as substrates supports the placement of Sec in the leaving group position in the ring opening reaction.
The use of oxidized tetrapeptides as substrates for the truncated enzyme allows us to isolate the kinetics of the ring opening step that occurs during the catalytic cycle of the enzyme. This point is illustrated by the generalized scheme of the TR reaction shown in Figures 6A and 6B. In the mammalian enzyme, replacement of Sec with Cys greatly impairs this ring opening step as demonstrated in Table 3B. For example, the ratio of turnover rates for peptides Ac-SCUS(ox) and Ac-SCCS(ox) is 511 fold, which is similar to the 308 fold difference between peptides Ac-GCUG(ox) and Ac-GCCG(ox). These data strongly support our hypothesis that the ability of the mammalian enzyme to protonate the leaving group in the ring opening step of the catalytic cycle is greatly impaired compared to DmTR. Our interpretation that proton transfer to the leaving group is partially rate limiting is in agreement with conclusions made about the GR mechanism, where proton transfer from His467′ to the thiol of GSH II has also been shown to be partially rate limiting (6, 40). Thus, we believe that this is strong evidence for the position of the tetrapeptide in the binding pocket and our proposed mechanism.
Figure 6.
(A) Diagrammatic model of one half of the TR dimer showing position of C-terminal tetrapeptide as it is undergoing ring opening. Once ring opening occurs, the reduced Cys is poised to attack the disulfide bond of Trx shown at right. If the C-terminal tetrapeptide is removed, then oxidized tetrapeptides can act as substrates for the truncated enzyme as shown in (B), and undergo a ring opening step analogous to the step in (A) when the tetrapeptide is covalently attached to the enzyme. If the C-terminal peptide is in a T- conformation as shown in (C), then proton transfer cannot take place from the catalytic His residue to the leaving group position. Whereas in (A) the peptide bond geometry is cis, allowing for proton transfer to the leaving group. In mammalian TRs that contain a selenium atom as the leaving group, protonation would be unnecessary. CysIC is the interchange Cys57 and CysCT is the charge-transfer Cys62 in DmTR, TermAA is the last amino acid of the tetrapeptide, and X is the leaving group residue (either Cys or Sec).
In DmTR, the presence of a Sec residue does not radically alter the rate of ring opening as it does for its mammalian counterpart. For example, if we compare the ratio of turnover rates for peptides Ac-SCUS(ox) and Ac-SCCS(ox) we see only a 5.65 fold difference. This ratio is increased to 36 fold when comparing peptides Ac-GCUG(ox) and Ac-GCCG(ox). This increase in peptide turnover is expected due to the lower bond dissociation energy for the S-Se bond if Se is in the leaving group position. A 24 fold increase is observed for DmTR when the effect of flanking residues of the tetrapeptide motif is examined by comparing the ratio of turnover for peptides Ac-SCCS(ox) and Ac-GCCG(ox). In the mammalian enzyme, the presence of flanking Ser residues on either side of the Cys-Cys or Cys-Se dyad only slightly (3 6 fold) improves the activity (Table 3B). These data indicate that the ability of the enzyme to protonate the leaving group is not impaired, as is the case for the mammalian enzyme. While the data in Table's 3A and 3B also indicate that the flanking serine residues of the tetrapeptide motif aid in deprotonating/protonating the catalytic Cys residues of the SCCS motif in DmTR (10), the effect is modest and we believe that the leaving group effect demonstrated here for the mammalian enzyme is striking. When this data is put in context with the structural data (as discussed in the next section), we can explain the loss of activity when Sec is replaced with Cys in the mammalian enzyme and why Cys-containing TRs (such as DmTR) can function with high activity.
An unusual feature of DmTR is the presence of the C-terminal catalytic disulfide bond between adjacent Cys residues. This vicinal disulfide bond has a very low frequency in the Brookhaven Protein Data Bank (PDB) as is discussed by Perczel and coworkers (41, 42). In the structures in which this type of disulfide is found, the intervening peptide bond torsion angle (ω) is strained, with values ranging from 159° to −133°. In two structures (human and bass hepcidin) it was noted that the intervenig peptide bond connecting the two half cystinyl -residues appears to be undergoing conformational exchange on the NMR time-scale, indicating that this portion of the molecule has high flexibility (43, 44). This suggests that, in some cases, where these unusual ring structures are found that they may be involved in conformational switching.
In this regard, DmTR has a more open, tetrapeptide binding pocket than the mammalian enzyme. The residues from helix 3 proximal to the FAD-associated active site are less bulky to more freely accommodate the hydroxyls of the adjacent serines (SCCS). The increase in the available space is likely to make the enzyme more amenable to the conformational switching of the SCCS peptide from T- (trans) to C+ (cis) necessary to protonate the leaving group Cys during interchange. In contrast, the mammalian TRs have evolved a more restricted binding pocket by incorporating bulkier residues, which project into the binding pocket from helix 3. The shape of the tetrapeptide binding pocket appears to define the final conformation of the oxidized tetrapeptide in the respective enzymes for the ring opening step of the enzymatic mechanism.
Our rationale for positioning the C-terminal tetrapeptide SCCS of DmTR in the active site as shown in Figure 4A is based upon several lines of reasoning. First, as described earlier, we used as a starting point the four known major conformers of dipeptide Ac-Cys-Cys-NH2 that were known to exist in solution. We verified that these same conformers were the major conformers in solution of peptides Ac-GCUG(ox) and Ac-SCCS(ox) using NMR spectroscopy (Deker and Hondal, unpublished). Secondly, as is clearly seen in Figure 4A, superposition of the sulfur atoms of GSSG in the GR structure with the tetrapeptide SCCS demonstrated that the best fit of the peptide in the active site was the C+ conformation. This seems like a logical way to proceed since in the other two TR structures (rat TR1 and mouse TR3) showed no electron density for the oxidized C-terminal tetrapeptide. Therefore the authors modeled the conformation of the C-terminal disulfide in the active site using undefined criteria in the case of rat TR1 (28), or in the case of mTR3, using the vicinal disulfide bond of methanol dehydrogenase (MDH) as a template (25). We would like to note that in the original structure of MDH, the peptide dihedral angle of the vicinal disulfide bond was assigned as a cis structure, but later was refined to a strained trans structure, with ω equal to −171° (45, 46). Lastly, since our data using oxidized tetrapeptides as substrates for the truncated mammalian enzyme indicated that leaving group ability was an important facet of the ring opening reaction, we placed the Sec residue of GCUG in the leaving group position, and by analogy we placed Cys490 (the C-terminal Cys of SCCS) in the leaving group position in DmTR. The resulting placement of the peptides is shown in Supplemental Figures 4 and 5. Our placement of the C-terminal Cys residue of the SCCS tetrapeptide of DmTR in the leaving group position, corresponding to Sec in the mammalian enzyme, is in agreement with the mechanism proposed for Sec-containing rat TR1 (see Figure 7 of reference (28)), but is in disagreement with the proposed placement of Sec for mTR3 as presented by Biterova and coworkers (25).
Our cumulative data has led us to propose a mechanism for reducing the Cys-Cys(ox) dyad of DmTR as depicted in Figure 6A. If the C-terminal tetrapeptide (lacking Sec) is in the C+ conformation, proton transfer can take place from His464′ to the leaving group thiol of the tetrapeptide. This would still be the case if Cys490 were the interchange residue as previously suggested (10, 47). If however, the C-terminal tetrapeptide is in the T- conformation, the transfer of the proton from the active site His to the leaving group position of the tetrapeptide is blocked (Figure 6C) and the rate of ring opening is greatly diminished, resulting in a much lower overall TR activity.
In mTR3 however, the low pKa of Sec in the T- conformation becomes essential due to its inability to be protonated by His497′ (PDB 1ZKQ). Upon reduction of the vicinal disulfide, the intervening peptide bond would be in the trans conformation. If the trans conformation is the conformation that is used for attack on the disulfide bond of Trx, then no conformational switching to the C+ conformation would be necessary to open the ring again. In the proposed mechanism of GR, His467′ would protonate the leaving group sulfur atom of GSH II, and then upon resolution of the interchange mixed disulfide, His467′ would also protonate the sulfur atom of GSH I. This same His residue is conserved in all high Mr TRs as well. Incorporation of Sec would eliminate the need for the leaving group protonation step and only require protonation of the adjacent Cys residue upon resolution of the interchange. Removing the cis/trans conformational switching and eliminating a protonation step may be a selective advantage for Sec incorporation in the mammalian TR.
Using this argument, the incorporation of selenium into the tetrapeptide may have been a way to stabilize the T- conformer. Selenium would stabilize the T- conformer of the eight-membered ring because a carbon-selenium bond is longer than a carbon-sulfur bond, and a sulfur-selenium bond is longer than a sulfur-sulfur bond. Thus the incorporation of selenium into the eight-membered ring makes the ring slightly larger, hence stabilizing the trans form of the ring.
If the general acid/base mechanism used by GR is also present in TR, then why is Sec needed in mammalian TRs? Much of the reason must undoubtedly be evolutionary pressure for a broad substrate range, including the ability to reduce peroxides (14) and ascorbate (48, 49). As argued above however, the incorporation of Se into the C-terminal 8-membered ring would have structural advantages as well that lead to further improvement in catalytic turnover beyond reduced pKa.
Finally, our proposal awaits further validation from a crystal structure of TR with the C-terminal redox center in the oxidized state, unequivocally answering the issues raised here of the geometry of the C-terminal tetrapeptide motif of TR.
Supplementary Material
Acknowledgments
We would like to thank the staff at the Center for X-Ray Crystallography at the University of Vermont and Vermont DOE-EPSCoR for use of facilities and generous support of this project. We acknowledge Ms. Katharine M. Harris for synthesis of peptides and Dr. P. Bruce Deker for determination of the peptide structures by NMR.
Footnotes
Abbreviations: CCG, the tripeptide Cys-Cys-Gly; CNS, Crystallography and NMR system; CUG, the tripeptide Cys-Sec-Gly; DEAE, diethylaminoethyl; DmTR, thioredoxin reductase from Drosophila melanogaster; DTNB, 5,5′-Dithio-bis(2-nitrobenzoic acid); EDTA, ethylenediamintetraacetic acid; FAD, flavin adenine dinucleotide; GCCG, the tetrapeptide Gly-Cys-Cys-Gly; GCUG, the tetrapeptide Gly-Cys-Sec-Gly; GR, glutathione reductase; GSH, reduced glutathione; GSH I, interchange glutathione; GSH II, first leaving group glutathione; GSSG, oxidized diglutathione; HPLC, high pressure liquid chromatography; LADH, dihydrolipoamide dehydrogenase; MALDI-TOF, matrix-assisted laser desorption ionization time of flight mass spectrometry; Mr, molecular weight; mTR3, mouse mitochondrial thioredoxin reductase; NADH, reduced nicotinamide adenine dinucleotide; NADP, oxidized nicotinamide adenine dinucleotide phosphate; NADPH, reduced nicotinamide adenine dinucleotide phosphate; NCS, non-crystallographic symmetry; NMA, N-methylmeraptoacetamide; NMR, nuclear magnetic resonance; PEG, polyethylene glycol; RMSD, root mean squared deviation; SCCS, the tetrapeptide Ser-Cys-Cys-Ser; Sec, selenocysteine; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; TR, thioredoxin reductase; Trx, thioredoxin; TryR, trypanothine reductase; U, the one-letter abbreviation for selenocysteine.
The subunits are referred to as chain A or chain B and residues from chain B are denoted with a “prime” designation.
In other literature, the term “helicity” is used to refer to the handedness of the disulfide bond. We refer the reader to references 24 and 32 for a description of the term “helicity”.
The coordinates (PDB 2NVK) will be released upon publication.
These studies were supported by National Institutes of Health grant GM070742 to RJH and by a Department of Energy EPSCoR grant DE-FG02-00ER45828 to RJH and SJE administered by Susan S. Wallace, Chair, Department of Microbiology and Molecular Genetics, University of Vermont.
Supporting Information Available: There are six figures available in the Supplementary Section. One figure shows a representative crystal used to generate the diffraction pattern for DmTR. There are two plots of kinetic data that describe the peptide complementation experiments. There are two figures that show the position of peptides SCCS(ox) and GCUG(ox) in the tetrapeptide binding pockets of DmTR and mTR3, respectively. Last, there is one stereodiagram showing peptide GCUG(ox) in the C+ conformation in the binding pocket of mTR3 and the resulting poor fit in this pocket. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Williams CH, Jr, Zanetti G, Arscott LD, McAllister JK. Lipoamide dehydrogenase, glutathione reductase, thioredoxin reductase, and thioredoxin. J Biol Chem. 1967;242:5226–31. [PubMed] [Google Scholar]
- 2.Zhong L, Arner ES, Holmgren A. Structure and mechanism of mammalian thioredoxin reductase: the active site is a redox-active selenolthiol/selenenylsulfide formed from the conserved cysteine-selenocysteine sequence. Proc Natl Acad Sci U S A. 2000;97:5854–9. doi: 10.1073/pnas.100114897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karplus PA, Schulz GE. Substrate binding and catalysis by glutathione reductase as derived from refined enzyme: substrate crystal structures at 2 A resolution. J Mol Biol. 1989;210:163–80. doi: 10.1016/0022-2836(89)90298-2. [DOI] [PubMed] [Google Scholar]
- 4.Schulz GE, Schirmer RH, Sachsenheimer W, Pai EF. The structure of the flavoenzyme glutathione reductase. Nature. 1978;273:120–4. doi: 10.1038/273120a0. [DOI] [PubMed] [Google Scholar]
- 5.Arscott LD, Thorpe C, Williams CH., Jr Glutathione reductase from yeast. Differential reactivity of the nascent thiols in two-electron reduced enzyme and properties of a monoalkylated derivative. Biochemistry. 1981;20:1513–20. doi: 10.1021/bi00509a016. [DOI] [PubMed] [Google Scholar]
- 6.Arscott LD, Veine DM, Williams CH., Jr Mixed disulfide with glutathione as an intermediate in the reaction catalyzed by glutathione reductase from yeast and as a major form of the enzyme in the cell. Biochemistry. 2000;39:4711–21. doi: 10.1021/bi9926431. [DOI] [PubMed] [Google Scholar]
- 7.Rietveld P, Arscott LD, Berry A, Scrutton NS, Deonarain MP, Perham RN, Williams CH., Jr Reductive and oxidative half-reactions of glutathione reductase from Escherichia coli. Biochemistry. 1994;33:13888–95. doi: 10.1021/bi00250a043. [DOI] [PubMed] [Google Scholar]
- 8.Gladyshev VN, Jeang KT, Stadtman TC. Selenocysteine, identified as the penultimate C-terminal residue in human T-cell thioredoxin reductase, corresponds to TGA in the human placental gene. Proc Natl Acad Sci U S A. 1996;93:6146–51. doi: 10.1073/pnas.93.12.6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bock A, Forchhammer K, Heider J, Leinfelder W, Sawers G, Veprek B, Zinoni F. Selenocysteine: the 21st amino acid. Mol Microbiol. 1991;5:515–20. doi: 10.1111/j.1365-2958.1991.tb00722.x. [DOI] [PubMed] [Google Scholar]
- 10.Gromer S, Johansson L, Bauer H, Arscott LD, Rauch S, Ballou DP, Williams CH, Jr, Schirmer RH, Arner ES. Active sites of thioredoxin reductases: why selenoproteins? Proc Natl Acad Sci U S A. 2003;100:12618–23. doi: 10.1073/pnas.2134510100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanzok SM, Fechner A, Bauer H, Ulschmid JK, Muller HM, Botella-Munoz J, Schneuwly S, Schirmer R, Becker K. Substitution of the thioredoxin system for glutathione reductase in Drosophila melanogaster. Science. 2001;291:643–6. doi: 10.1126/science.291.5504.643. [DOI] [PubMed] [Google Scholar]
- 12.Zhong L, Holmgren A. Essential role of selenium in the catalytic activities of mammalian thioredoxin reductase revealed by characterization of recombinant enzymes with selenocysteine mutations. J Biol Chem. 2000;275:18121–8. doi: 10.1074/jbc.M000690200. [DOI] [PubMed] [Google Scholar]
- 13.Lacey BM, Hondal RJ. Characterization of mitochondrial thioredoxin reductase from C. elegans. Biochem Biophys Res Commun. 2006;346:629–36. doi: 10.1016/j.bbrc.2006.05.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bjornstedt M, Hamberg M, Kumar S, Xue J, Holmgren A. Human thioredoxin reductase directly reduces lipid hydroperoxides by NADPH and selenocystine strongly stimulates the reaction via catalytically generated selenols. J Biol Chem. 1995;270:11761–4. doi: 10.1074/jbc.270.20.11761. [DOI] [PubMed] [Google Scholar]
- 15.Inoue H, Nojima H, Okayama H. High efficiency transformation of Escherichia coli with plasmids. Gene. 1990;96:23–8. doi: 10.1016/0378-1119(90)90336-p. [DOI] [PubMed] [Google Scholar]
- 16.Eckenroth B, Harris K, Turanov AA, Gladyshev VN, Raines RT, Hondal RJ. Semisynthesis and characterization of mammalian thioredoxin reductase. Biochemistry. 2006;45:5158–70. doi: 10.1021/bi0517887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arner ES, Zhong L, Holmgren A. Preparation and assay of mammalian thioredoxin and thioredoxin reductase. Methods Enzymol. 1999;300:226–39. doi: 10.1016/s0076-6879(99)00129-9. [DOI] [PubMed] [Google Scholar]
- 18.Harris KM, Flemer S, Hondal RJ. Studies on deprotection of cysteine and selenocysteine side chain protecting groups. J Pep Sci. 2006 doi: 10.1002/psc.795. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. In: Carter CW, Sweet RM, editors. Methods in Enzymology Part A. Academic Press; San Diego: 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 20.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallographica D Biological Crystallography. 1998;54:905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 21.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and location of errors in these models. Acta Crystallographica A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 22.Collaborative Computational Project, N. The CCP4 Suite: Programs for Protein Crystallography. Acta Crystallographica D Biological Crystallography D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 23.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr. 2001;57:122–33. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 24.Creighton CJ, Reynolds CH, Lee DH, Leo GC, Reitz AB. Conformational analysis of the eight-membered ring of the oxidized cysteinyl-cysteine unit implicated in nicotinic acetylcholine receptor ligand recognition. J Am Chem Soc. 2001;123:12664–9. doi: 10.1021/ja016505m. [DOI] [PubMed] [Google Scholar]
- 25.Biterova EI, Turanov AA, Gladyshev VN, Barycki JJ. Crystal structures of oxidized and reduced mitochondrial thioredoxin reductase provide molecular details of the reaction mechanism. Proc Natl Acad Sci U S A. 2005;102:15018–23. doi: 10.1073/pnas.0504218102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee SR, Bar-Noy S, Kwon J, Levine RL, Stadtman TC, Rhee SG. Mammalian thioredoxin reductase: oxidation of the C-terminal cysteine/selenocysteine active site forms a thioselenide, and replacement of selenium with sulfur markedly reduces catalytic activity. Proc Natl Acad Sci U S A. 2000;97:2521–6. doi: 10.1073/pnas.050579797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johansson L, Arscott LD, Ballou DP, Williams CH, Jr, Arner ES. Studies of an active site mutant of the selenoprotein thioredoxin reductase: The Ser-Cys-Cys-Ser motif of the insect orthologue is not sufficient to replace the Cys-Sec dyad in the mammalian enzyme. Free Radic Biol Med. 2006;41:649–56. doi: 10.1016/j.freeradbiomed.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 28.Sandalova T, Zhong L, Lindqvist Y, Holmgren A, Schneider G. Three-dimensional structure of a mammalian thioredoxin reductase: implications for mechanism and evolution of a selenocysteine-dependent enzyme. Proc Natl Acad Sci U S A. 2001;98:9533–8. doi: 10.1073/pnas.171178698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, Bond CS, Bailey S, Cunningham ML, Fairlamb AH, Hunter WN. The crystal structure of trypanothione reductase from the human pathogen Trypanosoma cruzi at 2.3 A resolution. Protein Sci. 1996;5:52–61. doi: 10.1002/pro.5560050107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bond CS, Zhang Y, Berriman M, Cunningham ML, Fairlamb AH, Hunter WN. Crystal structure of Trypanosoma cruzi trypanothione reductase in complex with trypanothione, and the structure-based discovery of new natural product inhibitors. Structure. 1999;7:81–9. doi: 10.1016/s0969-2126(99)80011-2. [DOI] [PubMed] [Google Scholar]
- 31.Jacob J, Schirmer RH, Gromer S. The conserved histidine 106 of large thioredoxin reductases is likely to have a structural role but not a base catalyst function. FEBS Lett. 2005;579:745–8. doi: 10.1016/j.febslet.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 32.Avizonis DZ, Farr-Jones S, Kosen PA, Basus VJ. Conformations and dynamics of the essential cysteinyl-cysteine ring derived from the acetylcholine receptor. J Am Chem Soc. 1996;118:13031–13039. [Google Scholar]
- 33.Benen J, van Berkel W, Dieteren N, Arscott D, Williams C, Jr, Veeger C, de Kok A. Lipoamide dehydrogenase from Azotobacter vinelandii: site-directed mutagenesis of the His450-Glu455 diad. Kinetics of wild-type and mutated enzymes. Eur J Biochem. 1992;207:487–97. doi: 10.1111/j.1432-1033.1992.tb17075.x. [DOI] [PubMed] [Google Scholar]
- 34.Gromer S, Wessjohann LA, Eubel J, Brandt W. Mutational studies confirm the catalytic triad in the human selenoenzyme thioredoxin reductase predicted by molecular modeling. Chembiochem. 2006;7:1649–52. doi: 10.1002/cbic.200600080. [DOI] [PubMed] [Google Scholar]
- 35.McMillan PJ, Arscott LD, Ballou DP, Becker K, Williams CH, Jr, Muller S. Identification of acid-base catalytic residues of high-Mr thioredoxin reductase from Plasmodium falciparum. J Biol Chem. 2006;281:32967–77. doi: 10.1074/jbc.M601141200. [DOI] [PubMed] [Google Scholar]
- 36.Huber RE, Criddle RS. Comparison of the chemical properties of selenocysteine and selenocystine with their sulfur analogs. Arch Biochem Biophys. 1967;122:164–73. doi: 10.1016/0003-9861(67)90136-1. [DOI] [PubMed] [Google Scholar]
- 37.Grauschopf U, Winther JR, Korber P, Zander T, Dallinger P, Bardwell JC. Why is DsbA such an oxidizing disulfide catalyst? Cell. 1995;83:947–55. doi: 10.1016/0092-8674(95)90210-4. [DOI] [PubMed] [Google Scholar]
- 38.Sun QA, Gladyshev VN. Redox regulation of cell signaling by thioredoxin reductases. Methods Enzymol. 2002;347:451–61. doi: 10.1016/s0076-6879(02)47045-0. [DOI] [PubMed] [Google Scholar]
- 39.Danehy JP, Elia VJ, Lavelle CJ. The Alkaline Decomposition of Organic Disulfides. IV. A Limitation on the Use of Ellman's Reagent, 2,2′Dinitro-5,5′-dithiodibenzoic Acid. J Org Chem. 1971;36:1003–1005. [Google Scholar]
- 40.Wong KK, Vanoni MA, Blanchard JS. Glutathione reductase: solvent equilibrium and kinetic isotope effects. Biochemistry. 1988;27:7091–6. doi: 10.1021/bi00418a063. [DOI] [PubMed] [Google Scholar]
- 41.Carugo O, Cemazar M, Zahariev S, Hudaky I, Gaspari Z, Perczel A, Pongor S. Vicinal disulfide turns. Protein Eng. 2003;16:637–9. doi: 10.1093/protein/gzg088. [DOI] [PubMed] [Google Scholar]
- 42.Hudaky I, Gaspari Z, Carugo O, Cemazar M, Pongor S, Perczel A. Vicinal disulfide bridge conformers by experimental methods and by ab initio and DFT molecular computations. Proteins. 2004;55:152–68. doi: 10.1002/prot.10581. [DOI] [PubMed] [Google Scholar]
- 43.Hunter HN, Fulton DB, Ganz T, Vogel HJ. The solution structure of human hepcidin, a peptide hormone with antimicrobial activity that is involved in iron uptake and hereditary hemochromatosis. J Biol Chem. 2002;277:37597–603. doi: 10.1074/jbc.M205305200. [DOI] [PubMed] [Google Scholar]
- 44.Lauth X, Babon JJ, Stannard JA, Singh S, Nizet V, Carlberg JM, Ostland VE, Pennington MW, Norton RS, Westerman ME. Bass hepcidin synthesis, solution structure, antimicrobial activities and synergism, and in vivo hepatic response to bacterial infections. J Biol Chem. 2005;280:9272–82. doi: 10.1074/jbc.M411154200. [DOI] [PubMed] [Google Scholar]
- 45.Blake CC, Ghosh M, Harlos K, Avezoux A, Anthony C. The active site of methanol dehydrogenase contains a disulphide bridge between adjacent cysteine residues. Nat Struct Biol. 1994;1:102–5. doi: 10.1038/nsb0294-102. [DOI] [PubMed] [Google Scholar]
- 46.Xia Z, Dai W, Zhang Y, White SA, Boyd GD, Mathews FS. Determination of the gene sequence and the three-dimensional structure at 2.4 angstroms resolution of methanol dehydrogenase from Methylophilus W3A1. J Mol Biol. 1996;259:480–501. doi: 10.1006/jmbi.1996.0334. [DOI] [PubMed] [Google Scholar]
- 47.Bauer H, Massey V, Arscott LD, Schirmer RH, Ballou DP, Williams CH., Jr The mechanism of high Mr thioredoxin reductase from Drosophila melanogaster. J Biol Chem. 2003;278:33020–8. doi: 10.1074/jbc.M303762200. [DOI] [PubMed] [Google Scholar]
- 48.May JM, Cobb CE, Mendiratta S, Hill KE, Burk RF. Reduction of the ascorbyl free radical to ascorbate by thioredoxin reductase. J Biol Chem. 1998;273:23039–45. doi: 10.1074/jbc.273.36.23039. [DOI] [PubMed] [Google Scholar]
- 49.May JM, Mendiratta S, Hill KE, Burk RF. Reduction of dehydroascorbate to ascorbate by the selenoenzyme thioredoxin reductase. J Biol Chem. 1997;272:22607–10. doi: 10.1074/jbc.272.36.22607. [DOI] [PubMed] [Google Scholar]
- 50.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos: 2002. [Google Scholar]
- 51.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Occena LGaS, D L. GRASP - A Prototype Interactive Graphic Sawing Program - (MU-IE Technical Report) MU-IE Technical Report 019501. 1995:1–17. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.