Abstract
Aim of the study
Several epidemiological studies have attempted to demonstrate a relationship between increased serum level of insulin-like growth factor 1 (IGF-1) and an augmented risk of developing colorectal cancers (CRC).
The human IGF-1 gene is composed of 6 exons and demonstrated expression of 6 different splice variants (isoforms) of mRNA (IA, IB, IC, IIA, IIB and IIC).
The aim of the study was to evaluate the expression of different isoforms of IGF-1 mRNA in CRC and normal colon tissue.
Material and methods
13 paired tissue specimens (colorectal tumor and non-tumor tissues) were analyzed using both quantitative polymerase chain reaction (PCR) and immunocytochemistry methods (IHC). The expression of classes I and II and variants A, B, C of IGF-1 mRNA were measured.
Results
In CRC higher amounts of IGF-1 class II mRNA than class I mRNA were detected. Among A, B, C isoforms, A variant of IGF-1 mRNA prevailed. The amounts of IGF-1 class I and class II mRNAs and of IGF-1 variant B mRNA were lowered in CRC as compared to the control. In CRC significant correlations were detected between reciprocal expression of class I and class II as well as between I and II isoforms and A, B and C.
Conclusions
Expression of IGF-1 mRNA isoforms differs between normal and CRC tissues. Even if all isoforms of IGF-1 mRNA manifested correlations with each other in tissues of CRC, expression of all transcripts (except that of isoform A) was significantly decreased as compared to the control.
Keywords: colorectal carcinoma, IGF-1 isoforms, QPCR, immunocytochemistry
Introduction
Malignant tumors of the large intestine (colorectal cancers – CRC), almost 98% adenocarcinomas, present an extensive challenge to clinical medicine and basic sciences. In 2005, CRC constituted the second in men and the third in women most frequent cause of neoplastic deaths in industrialized European countries, including Poland [1, 2]. At the molecular level two pathogenetic pathways of CRC development have been characterized in details: the so-called APC/β-catenin pathway (early lesions) and the pathway with loss of TP53 function (terminal stages of carcinogenesis) [3–6]. In both pathways gene mutations may be inherited or acquired and probably affect DNA in stem cells of intestinal crypts [4, 5], and multiple molecular stages of CRC development in humans have been recognized. The incidence of this type of cancer continues to increase in Poland, while its curability shows no improvement [2, 3]. New cellular factors are being studied, which may promote development of this type of cancer. The recognized factors which stimulate normal and pathological cell growth in intestinal epithelium include insulin-like growth factor 1 (IGF-1) and IGF-2 [7]. They manifest structure and biological activity which are similar to those of insulin and play the role of pleiotropic hormones [8, 9]. Their activity depends on IGF-binding proteins 1-8 (IGF BPs), which extend IGFs half-life in blood, store the factors in selected tissue compartments and inhibit action of IGFs by making their receptors less accessible.
Insulin-like growth factor BPs may intensify mitogenesis and cell migration and participate in transport and interactions with other growth factors, including transforming growth factor β (TGF-β) [9–11]. Insulin-like growth factors affect the cell by their specific binding to three different membrane IGF receptors: of type I (IGF-1R), type II (IGF-2R) and insulin receptor (IR). Most activities of both IGFs are mediated by type I receptor. Insulin-like growth factor 1R itself is also engaged in cell transformation [12].
Both IGF-1 and IGF-2 proteins play roles as mediators in proliferation and growth of various cell types, in control of early stages of carcinogenesis, control of the cell cycle in phase G1, in persistence of neoplastic phenotype in cells, in metastasis and in apoptosis of neoplastic cells [13–15]. In culture, IGF-1 stimulates cell growth by increasing proliferation and by inhibiting tumor cell apoptosis [15].
The gene IGF-1 in the human genome is present in a single copy, located on the long arm of chromosome 12 (12q22-24.1) and encompassing a region of over 90 kbp. The gene structure includes 6 exons and 5 introns. In mammals transcription of the IGF-1 gene is controlled by two promoters (P1 and P2), situated before exons 1 and 2, respectively [16].
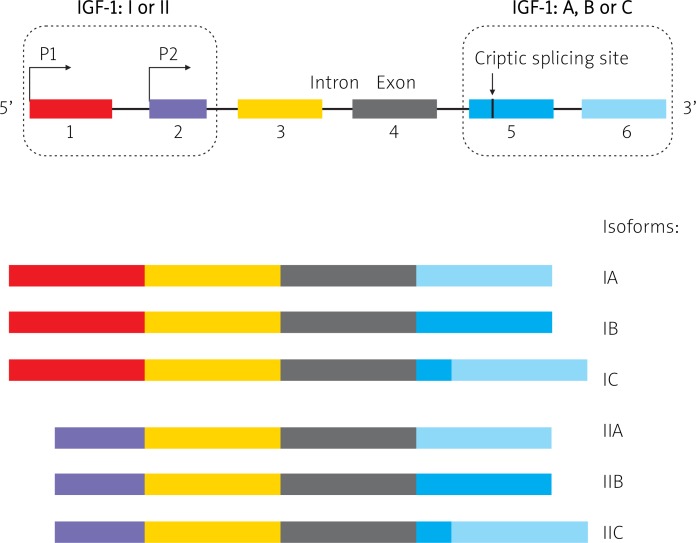
Differential exon 1 and 2 splicing of pre-mRNA IGF-1 [class I (1) and II (2)] and promoter usage result in expression of 6 different variants (isoforms) of mRNA: IA, IB, IC, IIA, IIB, and IIC [17, 18]. Expression and composition of nucleotides in exons 5 and 6 determine formation of isoform variants, including A (also termed Ea), B (Eb) and C (Ec) within IGF-1-I or IGF-1-II class [19] (Fig. 1). Isoforms of mRNA field as a result various isoforms of IGF-1 protein [19]. A mature molecule of IGF-1 protein is coded only by exons 3 and 4, while exons 1 and 2 encode leader peptides and exons 5 and 6 encode peptide E, present in the precursor molecule of the protein [20].
Fig. 1.
The structure of IGF-1 gene and the possible splicing of alternative isoforms. IGF-1 is composed of 6 exons. Exons 1 and 2 determine IGF-1 class I or II. Exons 3 and 4 are expressed in all known isoforms. Fluctuations in exons 5 and 6 determine class A, B or C (see details in the main text)
The physiological role of IGF-1 mRNA isoforms and of respective propeptides continues to be intensely studied, particularly in the muscular tissue. Insulin-like growth factor 1 isoforms are suggested to potentially exert distinct biological effects, employing various signaling pathways [22].
In several types of tumors, including CRC, an increased serum concentration of IGF-1 has been noted, suggesting a role of IGF-1 as a risk factor in development of such tumors [23, 24]. The studies conducted on large groups of patients with CRC and using sophisticated statistical methods have not always detected significant correlations of the suggested causal relationship [reviewed in 7].
Tissue expression of IGF-1 in carcinogenesis and, in particular, expression of IGF-1 splice variants (isoforms) has been described to date in only a few papers [21, 25]. No such studies on the involvement of IGF-1 and tissue expression of different isoforms have been focused on CRC.
Post-transcriptionally, IGF-1 isoforms are cleaved to give a mature 70-amino-acid core hormone (identical for all isoforms). Involvement of IGF-1 peptide cellular localization (cytoplasm/cell nucleus) in carcinogensis continues to be discussed. Although in most studies prevalence of cytoplasmic localization of IGF-1 was evident, Tan et al. (2002) have identified a nucleolar localization for an exon 5-encoded Eb isoform IGF-1 [26].
Due to the involvement, suggested in epidemiological studies, of IGF-1 in pathogenesis of CRC and the poorly recognized role of IGF-1 mRNA isoforms in this type of tumors, we attempted to demonstrate tissue expression of mature IGF-1 protein and to perform quantitative evaluation of expression levels manifested by various isoforms of IGF-I mRNA in large intestine cancers and in a fragment of normal (not neoplastically transformed) colon tissue.
Material and methods
Tissue material
The studies were performed on post-surgical tissue material originating from 13 patients (10 men, 3 women), 32 to 89 years of age, diagnosed and surgically treated (not treated previously with other forms of therapy) in the Chair of General, Gastroenterological and Endocrinological Surgery, Poznan University of Medical Sciences in Poznań, in the years 2010-2011.
The studies made use of 13 tissue material pairs, i.e. from fragments of colorectal cancers and of normal colorectal tissue in the same patients, verified by histopathological examination of hematoxylin and eosin-stained paraffin sections.
Histopathology of CRC confirmed presence of 12 adenocarcinomas (10 patients with G2, 2 patients with G3 grade) and one patient with adenocarcinoma in situ. The neoplastic lesions in 10 patients (77%) were located in the colon, in 2 patients in the rectum and in one patient in the caecum.
The Bioethical Commission of Poznan University of Medical Sciences granted permission for the studies.
Immunocytochemical studies
Tissue material was fixed in 4% buffered formalin and embedded in paraffin. Tissue sections of around 5 µm in thickness were deposited on SuperFrost/Plus microscope glasses. In order to qualify the material for molecular and immunocytochemical studies, routine staining of the sections with hematoxylin and eosin was also performed. For detection and cellular localization of IGF-1 we used anti-human mouse monoclonal antibodies (MAbs) specific for IGF-1 (R&D Systems; diluted 1 : 500) as well as the classical ABC (strept(avidin)-biotinylated peroxidase complex) according to Hsu et al. [27]. In short, stages of ABC immunocytochemical reactions involved deparaffinization and rehydration of preparations in, respectively, xylene and a row of alcohols of a decreasing concentration, blocking activity of endogenous peroxidase using incubation with 1% H2O2, incubation with a normal goat serum (1 : 20, DAKO) for 30 min, incubation with specific primary MAbs for approximately 18 hours at 4°C, incubation with secondary biotinylated antibodies (LSAB2, DAKO) (20 min), incubation with streptavidin-biotinylated peroxidase complex (LSAB2, DAKO) (20 min) and incubation with 3-3’-diaminobenzidine (DAB, DAKO). Following subsequent washes, the preparations were dehydrated in a row of alcohols of increasing concentrations, and equilibrated with xylene and closed under coverslips using Canada balsam. The preparations were counterstained using hematoxylin. Every experiment included negative controls in which specific antibodies were substituted by sera of a respective species in 0.05 M Tris-HCl, pH ∼7.6, supplemented with 0.1% bovine serum albumin (BSA) and 15 mM sodium azide.
Semiquantitative evaluation of insulin-like growth factor 1 expression
Insulin-like growth factor 1 expression was analyzed using the semi-quantitative 12-point IRS scale of Remmele and Stegner [28], in which the number of cells with positive reaction (A) and intensity of immunocytochemical reaction (B) were evaluated in 10 fields of an Olympus BH-2 light microscope, under a 40× objective. The final score represented the product of A × B. The final result for a patient was an average expression value for 10 visual fields.
Isolation of total RNA and synthesis of cDNA
The post-surgery tissue material was stored in RNA Stabilization Solution (RNAlater®, Applied Biosystems) at –80°C. The total RNA fraction was prepared using TRI Reagent® Solution (Applied Biosystems) and then purified on GeneMATRIX Universal RNA Purification Kit (EURx). Total RNA was determined by measuring optical density at 260 nm and purity was estimated by 260/280 nm absorption ratio, which was consistently > 1.8 (NanoDrop® ND-1000, ThermoScientific). RNA integrity was assessed by electrophoresis in 1% agarose gel with ethidium bromide. All RNA samples were stored in H2O at –80°C until used.
Reverse transcription was carried out on approximately 1.0 µg of total RNA following the manufacturer's protocol (First Strand cDNA Synthesis Kit, Fermentas). Template RNA, random hexamer primers (1 µl) and DEPC-treated water were mixed together to a total volume of 11 µl and pre-incubated at 65°C for 5 min in the Biometra thermocycler. Then the sample was chilled on ice, centrifuged to spin it down and moved to a fresh polymerase chain reaction (PCR) tube in order to prevent the permeability of the cup. 5X Reaction buffer (4 µl), RiboLock™ RNase Inhibitor 20 U/µl (1 µl), 10 mM dNTP Mix (2 µl) and M-MuLV Reverse Transcriptase 20 U/µl (2 µl) were added to the pre-incubated solution, mixed by pipeting and incubated at 42°C for 60 min (Biometra thermocycler). Finally, cDNA was stored in H2O at –20°C or immediately used for subsequent amplification reactions.
Absolute quantitation of mRNA copies
Construction of standard curves for copy number determination
cDNA of all IGF-1 isoforms (I, II, A, B, C, separately) (Fig. 1) was amplified using standard PCR using specific primers. Polymerase chain reaction products were analyzed in 2% agarose gel in order to confirm their specificity and then all PCR products were purified separately by GeneMATRIX PCR/DNA Clean-Up Purification Kit (EURx). The concentration of each DNA was estimated by measuring optical density at 260 nm (NanoDrop® ND-1000, ThermoScientific). The weight concentrations were converted to the corresponding DNA copy number the Avogadro constant:
A tenfold serial dilution series of corresponding DNA and specific primers, ranging from 1 × 107 to 10 copies per µl, was used to construct the standard curves. Threshold cycle (C t) values in each dilution were measured in duplicate and were plotted against the logarithm of their initial template copy numbers. Each standard curve was generated by a linear regression of the plotted points. From the slope of each curve, PCR amplification efficiency (E) was calculated according to the following equation:
E = 10-1/slope – 1
Real-time polymerase chain reaction
1 µl of a given cDNA or DNA was added to the reaction mixture composed of 12.5 µl 2× Maxima® SYBR Green/ROX qPCR Master Mix (Fermentas), 1 µl specific pair of primer (f.c. 0.3 µM) and 10.5 µl H2O. Primers for studies on expression of the vault-related gene and reference genes were taken from Koczorowska et al. [25] and purchased from the Laboratory of DNA Sequencing and Oligonucleotide Synthesis, Institute of Biochemistry and Biophysics, Polish Academy of Sciences, Warsaw.
The reactions were driven in twin.tec real-time PCR plates with PCR Film (Eppendorf) using Mastercycler ep-realplex 2 (Eppendorf). The PCR program was as followed: (1) initial denaturation, 95°C, 10 min, (2) denaturation, 95°C, 15 s, (3) annealing 60°C, 30 s, (4) extension 72°C, 30 s. The number of cycles was 40-50. Melting curves were made and 2% agarose gel electrophoresis was used to verify the amplification product specificity and size, respectively. All samples were amplified in duplicate or triplicate and in case that results varied more than 15% the reactions were repeated.
Absolute quantitation in real-time polymerase chain reaction
The absolute quantitation method was used to quantify mRNA copy numbers of all IGF1 isoforms (I, II, A, B, C, separately) (Fig. 1). Absolute quantitation determines the exact copy concentration of a target gene by relating the C t value to a standard curve. Prior to absolute quantitation, the C t values were normalized by comparison to the average of C t's obtained from five housekeeping genes (β-actin, B2M, GAPDH, HPRT1, MRLP19).
Evaluation of alterations in expression of IGF-1 mRNA isoforms involved a comparison of mRNA copy numbers for those isoforms per microgram RNA between each other in the tumor and in the control and reciprocal comparisons of the isoforms with each other in the tumor and in the control. Correlations were also tested between IGF-1 mRNA individual classes (I and II) and variants (A, B, C) within CRC and within normal tissues. Correlations were also examined between presence of immunocytochemically detectable IGF-1 protein and individual isoforms of IGF-1 mRNA.
Due to the prevalence of lesion localization in the colon (10/13 patients) and of grade 3 lesions (10/13 patients) in the examined group of patients no attempts were made to test correlations between isoform expression and the parameters.
Statistical analysis
First, the summary statistics were performed (arithmetic mean, standard deviation). Nonparametric statistical analysis of our data employed the Friedman test for multiple dependent samples and the Wilcoxon test for paired samples (tumor and control from the same patient). Spearman's rank correlation was applied to examine the association between variables. Differences and relationships were accepted to be statistically significant at the level of p less than 0.05. The statistical analysis was performed using Statistica PL v. 10 software.
Results
Cellular expression of insulin-like growth factor 1
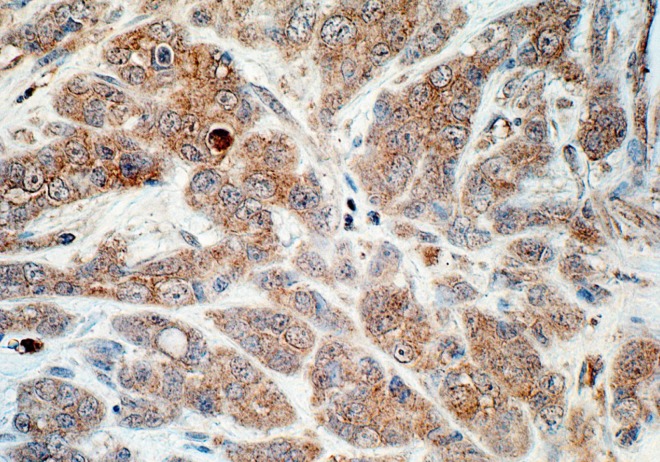
The immunocytochemical studies demonstrated cellular expression of IGF-1 in 9/13 (69%) patients. Extensive individual differences have been noted in intensity of the reaction, ranging from individual immunopositive cells (3-4 points on the semiquantitative scale) to an intense, diffuse reaction in the majority of neoplastic cells (12 points on the IRS scale) (data not shown). The cytoplasmic expression of the polypeptide prevailed even if the positive reaction was noted also in individual cell nuclei (Fig. 2). In tissue with no neoplastic lesions (control) reaction for the protein was absent or only individual IGF-1-positive cells were detected (data not shown).
Fig. 2.
Tissue expression of IGF-1 protein in patient with CRC (adenocarcinoma, G2). ABC method. Hemotoxylin counterstained. Objective magn. 40×
Expression of various insulin-like growth factor 1 mRNA isoforms
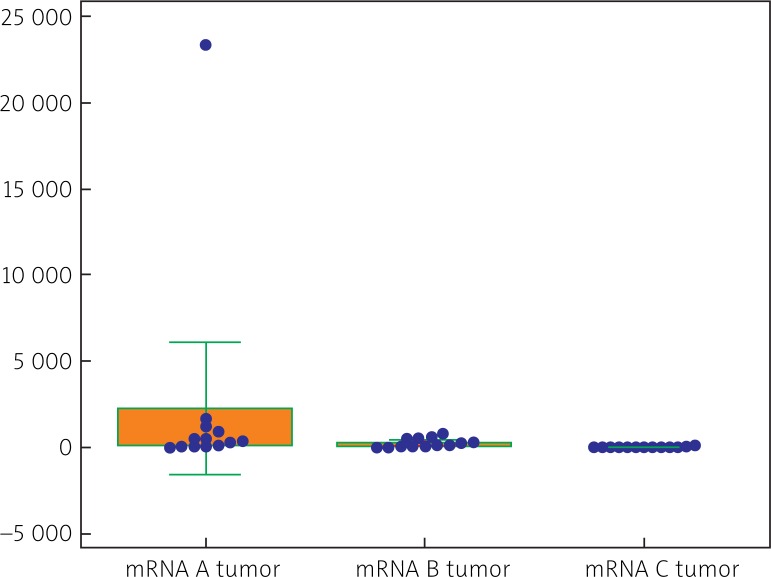
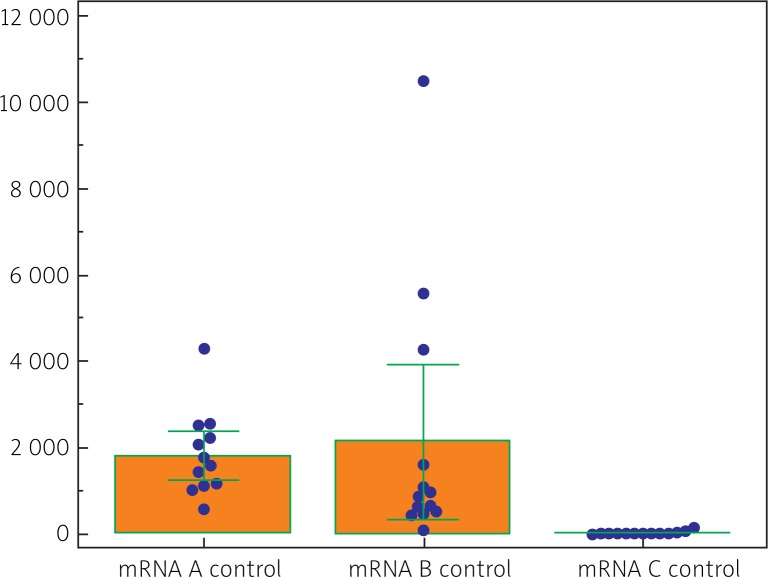
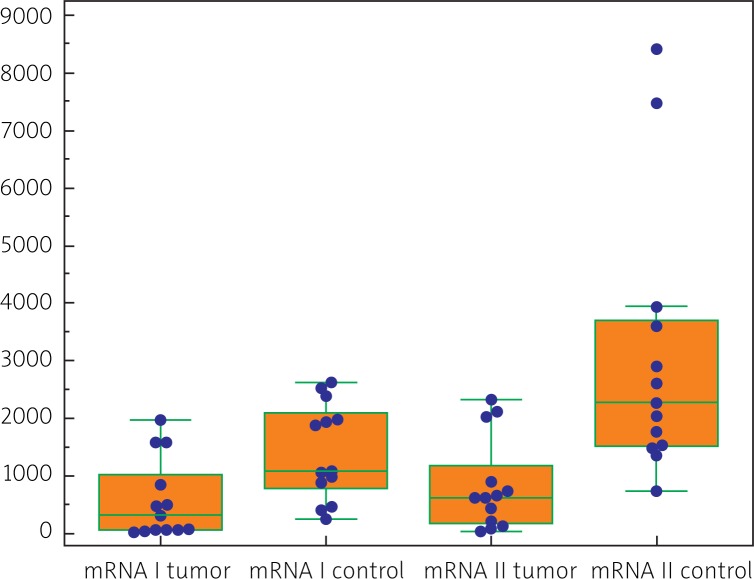
Both CRC tissues and control tissues manifested a quantitatively variable expression of all classes and variants (isoforms) of IGF-1 mRNA (i.e. I, II, A, B, C). In cases of CRC their quantitative analysis demonstrated prevalence of isoform II over isoform I (p < 0.02) (Table 1). Comparing amounts of isoforms A, B and C, isoform A proved to be present in the highest amounts, followed by isoforms B and C (Fig. 3). Also in the tumor-free tissue (control) quantitative prevalence of class II mRNA over class I mRNA was noted (p < 0.002) (Table 1). Out of isoform variants A, B, C, isoform B prevailed in the material over isoform A (p < 0.05) and over isoform C (p < 0.05) (Fig. 4).
Table 1.
Quantitative comparison between IGF-1 mRNA isoforms [number of copies/microgram of RNA in patients with CRC (tumor) and in unaltered tissue of the colon (control)]
| Parameter | Group | mean ± SD | p |
|---|---|---|---|
| Isoform A mRNA IGF-1 | tumor | 2252 ±6361 | 0.040 |
| control | 1816 ±963 | ||
| Isoform B mRNA IGF-1 | tumor | 266 ±256 | 0.003 |
| control | 2140 ±2982 | ||
| Isoform C mRNA IGF-1 | tumor | 22 ±45 | 0.146 |
| control | 40 ±36 | ||
| Isoform I mRNA IGF-1 | tumor | 582 ±695 | 0.054 |
| control | 1417 ±839 | ||
| Isoform II mRNA IGF-1 | tumor | 835 ±798 | 0.002 |
| control | 3084 ±2343 |
p – level of significance; bold numbers indicate significantly different values
Fig. 3.
Comparison of quantitative expression involving A, B and C variants (isoforms) of IGF-1 mRNA within neoplastic tissues of CRC (tumor) (data in numbers of mRNA copies/μg RNA). p < 0.05 between A and B, p = 0.05 between A and C; p = 0.05 between B and C
Fig. 4.
Comparison of quantitative expression involving A, B and C variants (isoforms) of IGF-1 mRNA in non-neoplastic tissues (control) (data in numbers of mRNA copies/μg RNA). p < 0.05 between A and B, A and C or B and C
In the case of IGF-1 class I and II mRNAs, significantly lower amounts were demonstrated in CRC, as compared to the morphologically unaltered tissue (Table 1, Fig. 5). In cases of the remaining isoform variants, isoform A proved to be significantly higher in CRC than in the control, isoform B was lower in CRC, and no significant difference was detected between the studied groups in the content of isoform C (Table 1).
Fig. 5.
Comparison of quantitative expression involving classes I and II of IGF-1 mRNA in CRC and control (data in numbers of mRNA copies/μg RNA). p = 0.054 between expression of class I in the tumor and in control; p = 0.002 between expression of class II in CRC and in control
Highly significant Spearman correlations were detected between classes I and II and between class I on one hand and isoforms A, B, C on the other as well as between class II and isoforms A, B, C in tissues of patients with CRC (Table 2). In the control material significant Spearman correlations were demonstrated only between class I and isoform B, between class II and isoforms A and C as well as between isoforms A and C No correlation could be shown between classes I and II (Table 2). No significant correlations could be disclosed between immunocytochemically detected protein expression on one hand and amounts of studied IGF-1 transcripts on the other (p > 0.05) (data not shown).
Table 2.
Spearman's correlation between reciprocal expression of all examined IGF-1 mRNA isoforms in colorectal cancers (tumor) and non-neoplastic tissue of the colon (control); bold numbers denote Spearman's rank correlation coefficients (r), at p ⩽ 0.05
| A | B | C | I | II | |||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
||||||
| Tumor | Control | Tumor | Control | Tumor | Control | Tumor | Control | ||
| A | – | 0.522 | 0.412 | 0.988 | 0.654 | 0.945 | 0.890 | 0.830 | 0.528 |
| B | – | – | 0.462 | 0.544 | 0.582 | 0.110 | 0.841 | 0.874 | |
| C | – | – | 0.957 | 0.681 | 0.777 | 0.462 | |||
| I | – | – | 0.824 | 0.231 | |||||
| II | – | – | |||||||
Discussion
In experimental models of CRC, autocrine release of all components of the IGF axis was demonstrated. Differences in proportion of produced IGFs depended on the type of process (proliferation vs. differentiation) and type of intestinal epithelial cells (intestinal crypts, enterocytes) [29].
The physiological role of differences between concentrations of circulating IGF-1 and the locally produced protein has not been recognized yet, particularly in tissues other than the liver. Increasingly, more numerous studies pertain to identification of various IGF-1 isoforms and evaluation of their biological activity [21, 22, 25, 29, 30].
Our immunocytochemical studies have confirmed local expression of IGF-1 in almost 70% patients with CRC. Studies on expression of various IGF-1 mRNA isoforms have allowed to be demonstrated IGF-1 gene transcripts in all CRC cases and in the control. Expression of all studied IGF-1 mRNA variants (isoforms) (i.e. of I, II, A, B, C) has been quantitated: both in CRC and in the control expression of IGF-1 mRNA class II isoforms prevailed (formed with involvement of P2 promoter) over expression of IGF-1 class I (formed using P1 promoter).
Considering the quantitative ratios of class I and class II isoforms between tumor and control tissue, mRNAs of the classes have manifested higher amounts in the control tissue material than in CRC. Among A, B, C variants of IGF-1 mRNA in CRC, mRNA isoform A has prevailed over the remaining isoforms and in control tissues isoform B has been the dominant isoform. Both in CRC and in control material the lowest amounts of IGF-1 mRNA representing isoform C have been detected.
The results related to prevalence of isoform A and the least pronounced expression of isoform C in adenocarcinoma coli are consistent with literature data although such results were demonstrated for another type of neoplastic tissues (uterine carcinomas). Also in the cases of uterine carcinoma the predominant splicing isoform was IGF-1 Ea (85-92%) and the expression of IGF-1 Ec isoform appeared to be at the lowest level [25]. In turn, studies on the role of the isoforms in cardiomyocytes demonstrated that locally acting mIGF-1 (comprises a Class 1 signal peptide and a C-terminal Ea extension peptide) protected cardiomyocytes from oxidative and hypertrophic stresses via SirT1 activity [30].
Our results allow to confirm a quantitatively variable expression of all IGF-1 mRNA isoforms both in the colorectal tissue involved by the neoplastic process and in the unchanged (normal) tissue of the organ.
Positive correlation between classes I and A, B, and C and between classes II and A, B, C indicates that in patients with CRC a parallel expression takes place of all the six IGF-1 mRNA isoforms. In the studied normal colon tissue material we have not been able to confirm all published relationships between IGF-1 mRNA variants.
In order to resolve whether the different isoforms of mRNA IGF-1 may play a role in CRC development, the studies should be continued using larger tissue material and/or examining expression of additional proteins in the IGF signaling pathway.
In conclusion: expression of IGF-1 mRNA isoforms differs between normal and CRC tissues. Although all IGF-1 isoforms of IGF-1 mRNA manifested correlation with each other in tissues of CRC, expression of all transcripts (except of isoform A) has been decreased, as compared to the control. Immunocytochemical studies have confirmed local cytoplasmic expression of mature IGF-1 protein in only 70% patients with colorectal cancer.
References
- 1.Boyle P, Ferlay J. Cancer incidence and mortality in Europe, 2004. Ann Oncol. 2005;16:481–8. doi: 10.1093/annonc/mdi098. [DOI] [PubMed] [Google Scholar]
- 2.Nowacki MP, Bujko K, Krzakowski M, Nowakowska D, Rutkowski A. Colon cancer. In: Diagnostic and therapeutic recommendations in malignant cancers treatment, Oncology in Clinical Practice. J Pol Clin Oncol Assoc. 2007;3(suppl. C):153–78. Krakowski M, Herman K, Jędrzejczak W, Kowalczyk JR, Podola-Dawidziak M, Reinfuss M. Via Medica, Gdańsk. [Google Scholar]
- 3.Crawford JM, Kumar V. Oral cavity and alimentary tract (in Polish) In: Kumar V, Cotran R, Robbins SL, Olszewski OT, editors. Robbins Patologia. Polish edition. Wrocław: Urban & Partner; 2008. pp. 621–77. [Google Scholar]
- 4.Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363:558–61. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 5.Chakravarthi S, Krishnan B, Madhavan M. Apoptosis and expression of p53 in colorectal neoplasms. Indian J Med Res. 1999;111:95–102. [Google Scholar]
- 6.Markowitz SD, Bertagnolli MM. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med. 2009;361:2449–60. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donovan EA, Kummar S. Role of insulin-like growth factor-1R system in colorectal carcinogenesis. Crit Rev Oncol Hematol. 2008;66:91–8. doi: 10.1016/j.critrevonc.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daughaday WH, Rotwein P. Insulin-like growth factors I and II, peptide, messenger ribonucleic acid and gene structures, serum and tissue concentrations. Endocr Rev. 1989;10:68–91. doi: 10.1210/edrv-10-1-68. [DOI] [PubMed] [Google Scholar]
- 9.Zarrilli R, Bruni CB, Riccio A. Multiple levels of control of insulin-like growth factor gene expression. Mol Cell Endocrinol. 1994;101:R1–14. doi: 10.1016/0303-7207(94)90253-4. [DOI] [PubMed] [Google Scholar]
- 10.Kostecka Y, Blahovec J. Insulin-like growth factor binding proteins and their functions (minireview) Endocr Regul. 1999;33:90–4. [PubMed] [Google Scholar]
- 11.Murphy LJ. Insulin-like growth factor-binding proteins: functional diversity or redundancy? J Mol Endocrinol. 1998;21:97–107. doi: 10.1677/jme.0.0210097. [DOI] [PubMed] [Google Scholar]
- 12.Baserga R. The insulin-like growth factor I receptor: a key to tumor growth? Cancer Res. 1995;55:249–55. [PubMed] [Google Scholar]
- 13.Alexia C, Fallot G, Lasfer M, Schweizer-Groyer G, Groyer A. An evaluation of the role of insulin-like growth factors (IGF) and of type-I IGF receptor signalling in hepatocarcinogenesis and in the resistance of hepatocellular cells against drug-induced apoptosis. Biochem Pharmacol. 2004;68:1003–15. doi: 10.1016/j.bcp.2004.05.029. [DOI] [PubMed] [Google Scholar]
- 14.Samani AA, Yakar S, LeRoith D, Brodt P. The role of the IGF system in cancer growth and metastasis: overview and recent insights. Endocr Rev. 2007;28:20–47. doi: 10.1210/er.2006-0001. [DOI] [PubMed] [Google Scholar]
- 15.Garrouste F, Remacle-Bonnet M, Fauriat C, Marvaldi J, Luis J, Pommier G. Prevention of cytokine-induced apoptosis by insulin-like growth factor-I is independent of cell adhesion molecules in HT29-D4 colon carcinoma cells-evidence for a NF-kappaB-dependent survival mechanism. Cell Death Differ. 2002;9:768–79. doi: 10.1038/sj.cdd.4401022. [DOI] [PubMed] [Google Scholar]
- 16.Adamo ML. Regulation of insulin-like growth factor I gene expression. Implications for normal and pathological growth. Diabetes Rev. 1995;3:2–27. [Google Scholar]
- 17.Rotwein P, Pollock KM, Didier DK, Krivi GG. Organization and sequence of the human insulin-like growth factor I gene. Alternative RNA processing produces two insulin-like growth factor I precursor peptides. J Biol Chem. 1986;261:4828–32. [PubMed] [Google Scholar]
- 18.Adamo ML, Neuenschwander S, LeRoith D, Roberts CT., Jr Structure, expression, and regulation of the IGF-I gene. Adv Exp Med Biol. 1993;343:1–11. doi: 10.1007/978-1-4615-2988-0_1. [DOI] [PubMed] [Google Scholar]
- 19.Temmerman L, Slonimsky E, Rosenthal N. Class 2 IGF-1 isoforms are dispensable for viability, growth and maintenance of IGF-1 serum levels. Growth Horm IGF Res. 2010;20:255–63. doi: 10.1016/j.ghir.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 20.LeRoith D, Roberts CT., Jr The insulin-like growth factor system and cancer. Cancer Lett. 2003;195:127–37. doi: 10.1016/s0304-3835(03)00159-9. [DOI] [PubMed] [Google Scholar]
- 21.Armakolas A, Philippou A, Panteleakou Z, Nezos A, Sourla A, Petraki C, Koutsilieris M. Preferential expression of IGF-IEc (MGF) transcript in cancerous tissues of human prostate: evidence for a novel and autonomous growth factor activity of MGF E peptide in human prostate cancer cells. Prostate. 2010;70:1233–42. doi: 10.1002/pros.21158. [DOI] [PubMed] [Google Scholar]
- 22.Matheny RW, Jr, Nindl BC, Adamo ML. Minireview: Mechano-growth factor: a putative product of IGF-I gene expression involved in tissue repair and regeneration. Endocrinology. 2010;151:865–75. doi: 10.1210/en.2009-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cohen P. Overview of the IGF-I system. Horm Res. 2006;65(Suppl 1):3–8. doi: 10.1159/000090640. [DOI] [PubMed] [Google Scholar]
- 24.Wu Y, Yakar S, Zhao L, Hennighausen L, LeRoith D. Circulating insulin-like growth factor-I levels regulate colon cancer growth and metastasis. Cancer Res. 2002;62:1030–5. [PubMed] [Google Scholar]
- 25.Koczorowska MM, Kwasniewska A, Gozdzicka-Jozefiak A. IGF1 mRNA isoform expression in the cervix of HPV-positive woman with pre-cancerous and cancer lesions. Exp Ther Med. 2011;2:149–56. doi: 10.3892/etm.2010.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan DS, Cook A, Chew SL. Nucleolar localization of an isoform of the IGF-I. BMC Cell Biol. 2002;3:17. doi: 10.1186/1471-2121-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsu SM, Raine L, Fanger H. Use of avidin-biotin-peroxidase complex (ABC) in immunoperoxidase techniques: a comparison between ABC and unlabeled antibody (PAP) procedures. J Histochem Cytochem. 1981;29:577–80. doi: 10.1177/29.4.6166661. [DOI] [PubMed] [Google Scholar]
- 28.Remmele W, Stegner HE. Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (ER-ICA) in breast cancer tissue. Pathologe. 1987;8:138–40. [PubMed] [Google Scholar]
- 29.Jehle PM, Fussgaenger RD, Blum WF, Angelus NK, Hoeflich A, Wolf E, Jungwirth RJ. Differential autocrine regulation of intestine epithelial cell proliferation and differentiation by insulin-like growth factor (IGF) system components. Horm Metab Res. 1999;31:97–102. doi: 10.1055/s-2007-978705. [DOI] [PubMed] [Google Scholar]
- 30.Vinciguerra M, Santini MP, Claycomb WC, Ladurner AG, Rosenthal N. Local IGF-1 isoform protects cardiomyocytes from hypertrophic and oxidative stresses via Sirt1 activity. Aging (Albany NY) 2009;2:43–62. doi: 10.18632/aging.100107. [DOI] [PMC free article] [PubMed] [Google Scholar]