Abstract
By using particle-associated reverse transcriptase (RT) activity as an assay for Pol incorporation into human immunodeficiency virus type 1 (HIV-1) Gag virus-like particles (VLPs), it has been found that truncated, protease-negative, Gag-Pol missing cis Gag sequences is still incorporated into Gag VLPs, albeit at significantly reduced levels (10 to 20% of the level of wild-type Gag-Pol). In this work, we have directly measured the incorporation of truncated Gag-Pol species into Gag VLPs and have found that truncated Gag-Pol that is missing all sequences upstream of RT is still incorporated into Gag VLPs at levels approximating 70% of that achieved by wild-type Gag-Pol. Neither protease nor integrase regions in Pol are required for its incorporation, implying an interaction between Gag and RT sequences in the Pol protein. While the incorporation of Gag-Pol into Gag VLPs is reduced 12-fold by the replacement of the nucleocapsid within Gag with a leucine zipper motif, this mutation does not affect Pol incorporation. However, the deletion of p6 in Gag reduces Pol incorporation into Gag VLPs four- to fivefold. Pol shows the same ability as Gag-Pol to selectively package tRNALys into Gag VLPs, and primer tRNA3Lys is found annealed to the viral genomic RNA. These data suggest that after the initial separation of Gag from Pol during cleavage of Gag-Pol by viral protease, the Pol species still retains the capacity to bind to both Gag and tRNA3Lys, which may be required for Pol and tRNA3Lys to be retained in the assembling virion until budding is completed.
During human immunodeficiency virus type 1 (HIV-1) assembly, the major tRNALys isoacceptors in mammalian cells, tRNA3Lys and tRNA1,2Lys (44), are selectively packaged into the virion (28), where tRNA3Lys serves as the primer for the reverse transcriptase (RT)-catalyzed synthesis of minus-strand cDNA (37). The annealing of tRNA3Lys to the primer binding site of the viral RNA genome is proportional to the amount of tRNA3Lys packaged in the viral population (14). tRNALys packaging into HIV-1 depends upon the presence of the precursor protein Gag-Pol (36) and may depend directly upon sequences in the RT thumb domain. Thus, C-terminal deletions of Gag-Pol which include sequences coding for integrase and the RNase H and connection domain of RT do not affect tRNALys packaging, but larger deletions that also include the thumb domain inhibit packaging (31). While deletion of the RT thumb domain could induce conformational changes affecting tRNALys binding elsewhere in the molecule, cross-linking studies between purified HIV-1 RT and tRNA3Lys indicate a direct interaction between the thumb domain and tRNA3Lys (12).
Extracellular virus-like particles (VLPs) can be produced by HIV-1 Gag alone (17, 21, 29, 50), and putative regions of interactions between Gag molecules have been identified within the C-terminal half of Gag and include the C-terminal half of capsid (CA) (3, 15, 34, 38), p2 (1, 32, 40), nucleocapsid (NC) (6, 7, 10, 49), and p6 (16). It is generally assumed that in order to obtain the interactions required for assembly, the Gag molecules must first be concentrated at a cellular site. Membrane has been proposed to play a role in the concentration and alignment of Gag molecules, and in HIV-1-transfected COS cells, almost all steady-state Gag is membrane bound (22, 24, 52). In many retroviruses, the Gag precursor protein attaches to the lipid membrane via a myristic acid moiety added posttranslationally to the amino terminus of the matrix (4, 18, 23, 45, 48) and through ionic interactions between the phospholipids in the membrane and a sequence of basic amino acid residues within the matrix (46, 47, 53, 56). However, a minimal, 16-kDa Gag construct lacking matrix has been shown to be capable of forming VLPs in vivo and is composed of an N-terminal myristylation signal, the C-terminal half of CA, p2, a leucine zipper domain of yeast GCN4 replacing NC, and a PPPPY motif replacing p6 (1).
The incorporation of Gag-Pol into extracellular particles requires the participation of Gag, since the expression of Gag-Pol alone does not readily generate such particles (22). Gag appears to be responsible for bringing Gag-Pol to the membrane, since unmyristylated Gag or Gag-Pol molecules can also be rescued into assembly complexes by myristylated Gag (39, 41, 50). The direct interaction of Gag with Gag-Pol has been demonstrated by immunoprecipitation of this complex from lysates of HIV-1-transfected cells with anti-integrase (22). While RNA appears to be important in facilitating Gag-to-Gag interactions (2, 7, 19), RNA is not required for the direct interaction of Gag-Pol with multimeric Gag (30).
The amino acid sequences within Gag and Gag-Pol that facilitate their interaction have been less studied than those involved in Gag/Gag multimerization. It is believed that Gag sequences within Gag-Pol play an important role in the interaction of Gag-Pol with Gag. In particular, deletions of the major homology region and the adjacent C-terminal half of the capsid region in Gag-Pol were shown to inhibit Gag-Pol incorporation into Gag particles (25, 51). More recent work has indicated that the Gag sequences in Gag-Pol may not be essential for Pol incorporation into Gag VLPs in either murine leukemia virus or in HIV-1 (5, 8). In HIV-1 (8), the incorporation of both protease (PR)-positive and PR-negative Gag-Pol constructs into Gag VLPs was examined. By using PR-positive constructs, Pol incorporation was determined by measuring the amount of processed viral proteins in the particle. However, in cells overexpressing viral proteins, there is premature processing of viral proteins in the cytoplasm, and their presence in VLPs is therefore not necessarily a direct measure of incorporated Gag-Pol. By using PR-negative virions, the presence of RT activity in the viral particle was used to indicate Pol incorporation, even though no processed, mature RT was present in the Gag VLPs. The authors concluded that after removal of Gag, or Gag plus PR, from Gag-Pol, the truncated Pol species were still incorporated. Incorporation was, however, only 10 to 20% of that of wild-type Gag-Pol. On the other hand, when murine leukemia virus Gag and Pol were coexpressed from separate plasmids in cells, RT activity associated with Gag VLPs was about 25% lower when Pol was incorporated than when Gag-Pol was incorporated (5). The incorporation of Pol into retroviral Gag particles has a natural precedent. In human foamy virus, Pol is made from a separate spliced mRNA than that used for producing Gag (54), i.e., no Gag-Pol species is found. Pol is incorporated into human foamy virus, and while tRNA1,2Lys appears to be the primer for reverse transcription in this virion (33), no studies have been done yet upon whether it is selectively packaged into the virus.
The interaction of Gag with Gag-Pol is clearly an important part of the retroviral life cycle since Gag-Pol carries essential enzymes involved in viral replication, as well as the tRNA3Lys required for priming reverse transcription. The nature of this interaction is therefore of interest, both from a theoretical and a therapeutic viewpoint. However, because of difficulties in purifying Gag-Pol, the interaction between Gag and Gag-Pol is most easily examined in vivo by studying the effects of mutations in Gag-Pol upon its interaction with Gag and its packaging into Gag VLPs. In the report herein, we have directly measured the incorporation of truncated forms of Gag-Pol and observe that even after removal of Gag and PR from Gag-Pol, the truncated species are incorporated at levels equal to 70% of that of wild-type and, furthermore, still retained the biological function of selectively packaging tRNALys.
Specific incorporation of Pol into HIV-1 is independent of Gag within Gag-Pol.
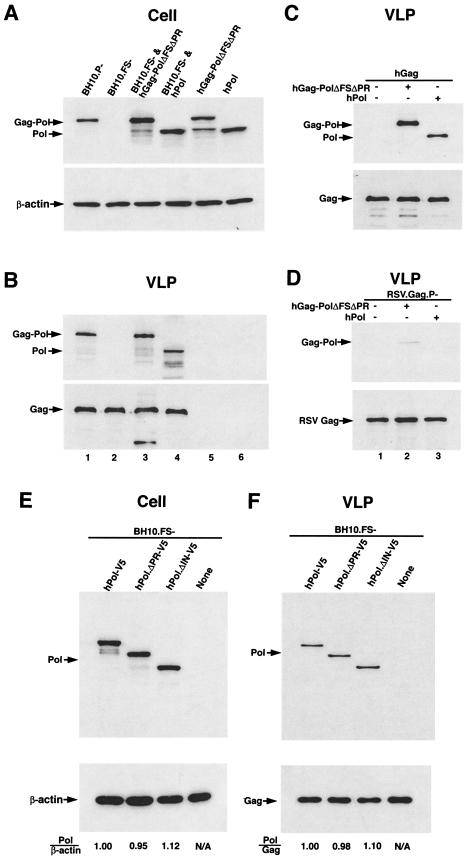
In this report, HIV-1 proteins were produced in transfected HEK-293T cells in the absence of an active viral PR. The production of Gag alone is sufficient to produce extracellular VLPs. Lysates of cells transfected with various plasmids (see Table 1) or VLPs produced by these cells were analyzed by Western blots (Fig. 1A and B) probed with anti-RT (upper panels), anti-β-actin (Fig. 1A, lower panel), or anti-CA (Fig. 1B, lower panel). Lanes 1 and 2 show cell or VLP proteins produced from cells transfected, respectively, with BH10.P−, a plasmid which codes for all wild-type HIV-1 proteins except PR, which is inactive, or with the BH10.FS− plasmid, which codes for all HIV-1 proteins except Gag-Pol. Lanes 3 and 4 show cell or VLP proteins produced from cells cotransfected with BH10.FS− and either hGag-PolΔFSΔPR−, which codes for Gag-Pol (Fig. 1A and B, lanes 3) or hPol, which codes for Pol (Fig. 1A and B, lanes 4). In Gag VLPs, Pol is packaged approximately 70% as efficiently as Gag-Pol. Cells transfected with a single plasmid coding for either Gag-Pol (Fig. 1A and B, lanes 5) or Pol (lanes 6) alone did not produce either Gag or extracellular VLPs, although they are effectively produced in the cell as shown by the Western blots of cell lysate.
TABLE 1.
Plasmids used in this study
| Plasmida | Codon usage | Protein expressed |
|---|---|---|
| BH10.P− | virus | Gag and Gag-Pol |
| BH10.FS− | virus | Gag-Pol |
| hGag-PolΔFSΔPR | humanized | Gag-Pol |
| hPol | humanized | Pol |
| hGag | humanized | Gag |
| ZWt | virus | Mutant Gag |
| ZWt-p6 | virus | Mutant Gag |
| RSV.Gag.P− | virus | RSV Gag |
No plasmid expresses an active viral PR.
FIG.1.
Incorporation of Pol and Gag-Pol into Gag VLPs. 293T cells were transfected or cotransfected with different plasmids coding for wild-type Gag and wild-type or mutant Gag-Pol proteins, using Lipofectamine. The plasmids used are listed along the top of each panel. BH10.P− is a simian virus 40-based vector that contains full-length wild-type HIV-1 proviral DNA containing an inactive viral PR (D25G) and was a gift from E. Cohen, University of Montreal. BH10.FS− contains mutations at the frameshift site, i.e., nucleic acid sequence 2082-TTTTTT-2087 replaces 2082-CTTCCT-2087, which prevents frameshifting during the translation of the Gag protein and generates particles that contain only Gag, not Gag-Pol (35). hGag-PolΔFSΔPR was constructed by deleting five thymidines in the frameshift site and codes for Gag-Pol. The humanized proteins have identical amino acid sequences to their viral counterparts, but the mRNAs coding for them have had their codons optimized for mammalian cell codon usage, which results in more efficient translation and protein production and also makes nuclear export of these mRNAs Rev-independent through modification of the multiple inhibitory sequences (26, 43). Both hPol and hGag-PolΔFSΔPR contain an inactive PR due to an R42G mutation in the active site. hPol-V5, hPol.ΔPR-V5, and hPol.ΔIN-V5 code for Pol, Pol missing PR, and Pol missing integrase, respectively; and all proteins contain a C-terminal V5 tag. Plasmid pSV.Myr1.3 h was a gift from R. Craven and J. Wills (University of Pennsylvania, Philadelphia) and was constructed as previously described (9). We have renamed it RSV.Gag.P−. It encodes a truncated Myr1 protein that carries RSV Gag sequences and only the first seven amino acids of RSV PR, followed by one foreign amino acid, and produces RSV extracellular Gag VLPs containing Gag but no Gag-Pol. HEK-293T cells were cultured as previously described (20), transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, Calif.) according to the manufacturer's instructions. At 48 h posttransfection, Gag VLPs were purified from the media as previously described (14). Cells or Gag VLPs were lysed in radioimmunoprecipitation assay buffer (10 mM Tris [pH 7.4], 100 mM NaCl, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 1% NP-40, 2 mg of aprotinin/ml, 2 mg of leupeptin/ml, 1 mg of pepstatin A/ml, 100 mg of phenylmethylsulfonyl fluoride/ml). Proteins in the cell or viral lysates were resolved by SDS-PAGE (10% acrylamide), followed by blotting onto nitrocellulose membranes (Amersham Pharmacia). Detection of protein by Western blotting utilized monoclonal antibodies that are specifically reactive with HIV-1 CA (Zepto Metrocs Inc.), RT (National Institutes of Health, AIDS Research and Reference Reagent Program) and β-actin (Sigma). Rabbit antiserum to RSV was a kind gift from R. Craven (University of Pennsylvania). Detection of proteins was performed by enhanced chemiluminescence (NEN Life Sciences Products) by using as secondary antibodies anti-mouse (for CA, RT, and β-actin) and anti-rabbit (for RSV), both obtained from Amersham Life Sciences. (A and B) Lysates of cells transfected with various plasmids or VLPs produced by these cells were analyzed by Western blots probed with anti-RT (upper panels), anti-β-actin (A, lower panel), or anti-CA (B, lower panel). (C) Western blots of Gag VLPs, probed with anti-RT (upper panel) or anti-CA (lower panel). (D) Western blots of lysates of RSV Gag VLPs probed with either RSV anti-CA (lower panel) or HIV anti-RT (upper panel). (E and F) Lysates of cells transfected with various plasmids or Gag VLPs produced by these cells were analyzed by Western blots probed with anti-V5 and either anti β-actin (E) or anti-CA (F). Bands in Western blots were quantitated with the UN-SCAN-IT gel automated digitizing system.
The lower efficiency of incorporation of Pol into Gag VLPs is found even when viral Gag is replaced with Gag synthesized from humanized Gag plasmid (hGag). Cells were transfected with the hGag plasmid coding for Gag alone or cotransfected with the hGag plasmid and a humanized plasmid coding for either Gag-Pol (Fig. 1C, lane 2) or Pol (lane 3). The Western blots shown, probed with anti-RT (upper panel) or anti-CA (lower panel), indicate that Pol protein missing the Gag sequences is still incorporated into Gag VLPs, albeit with only about 70% efficiency compared to levels for Gag-Pol.
To test for the specificity of interaction between Gag and Pol, similar transfection experiments were performed in which we replaced the plasmids coding for HIV-1 Gag with one coding for Gag from Rous sarcoma virus (RSV). This plasmid (RSV.Gag.P−) codes for RSV Gag but not Gag-Pol (9). Figure 1D shows Western blots of lysates of RSV Gag VLPs probed with either anti-CA (for RSV) or anti-RT (for HIV-1). While the lysates of the VLPs show a strong signal for RSV capsid, little or no Gag-Pol or Pol is incorporated into the particles, indicating that this incorporation is specific for HIV-1 Gag and not due to random incorporation.
The sequences in Pol required for incorporation into Gag VLPs were determined by cotransfecting HEK-293T cells with BH10.FS− as the source of Gag, with a plasmid coding for either Pol (hPol-V5), Pol protein lacking PR (hPol.ΔPR-V5), or Pol protein lacking integrase (hPol.ΔIN-V5). Lysates of cells transfected with various plasmids (Fig. 1E) or Gag VLPs produced by these cells (Fig. 1F) were analyzed by Western blots probed with anti-V5 and either anti-β-actin (Fig. 1E) or anti-CA (Fig. 1F). The Pol/Gag ratios indicate that neither PR nor integrase sequences are required for incorporation of Pol sequences into Gag VLPs. Since we have previously reported that the incorporation of both Gag-Pol and tRNALys into HIV-1 does not require the C-terminal Gag-Pol sequences coding for integrase or the RNase H and connection subdomains of RT (31), it seems likely that the Pol sequences that bind to Gag lie somewhere within the fingers, palm, or thumb subdomain of RT.
The role of NC and p6 in Gag interactions with Gag-Pol and Pol.
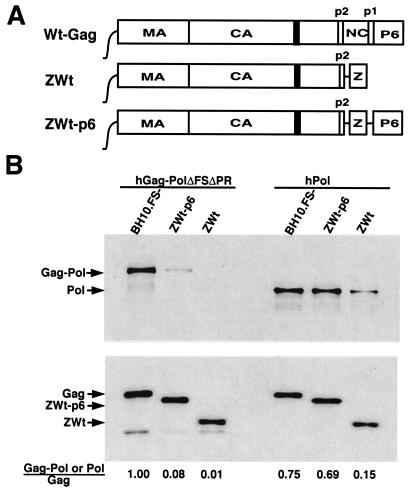
The ability of Gag-Pol or Pol to be incorporated into wild-type or mutant forms of Gag VLPs was monitored. HEK-293T cells were cotransfected with either hGag-PolΔFSΔP− (coding for Gag-Pol, but not Gag) or hPol and with a second plasmid coding for wild-type Gag (BH10.FS−) or mutant Gag (ZWt or ZWt-p6). The Gag expressed from each plasmid is shown in the results in Fig. 2. BH10.FS− codes for wild-type HIV-1 proteins (with an inactive viral PR), including Gag; ZWt-p6 codes for a mutant Gag in which the NC sequence has been replaced with a yeast leucine zipper domain to allow for protein-to-protein interactions; ZWt codes for a mutant Gag similar to ZWt-p6, except for the further deletion of the p6 region. These two mutant Gag species remain efficient in forming VLPs (1), even with the deletion of p6, which can play an important role in viral release. However, several studies have shown that a role for p6 in viral budding can be overridden in high-efficiency transfection systems and in virions containing an inactive PR (for a review, see reference 13). Western blots of lysates of viruses produced from cells cotransfected with these different plasmids were probed with either anti-RT or anti-CA, and the results are shown in the upper and lower parts of Fig. 2B, respectively. These results indicate that there is a 12-fold reduction in the incorporation of Gag-Pol into VLPs composed of mutant Gag in which NC has been replaced by a yeast leucine zipper domain, while the incorporation of Pol into these Gag VLPs is not significantly altered. This result indicates that NC plays a more prominent role in the incorporation of Gag-Pol into Gag VLPs than in the incorporation of Pol. On the other hand, Pol incorporation into Gag VLPs is reduced four- to fivefold in the absence of p6 in Gag (Fig. 2B).
FIG. 2.
Sequences in Gag binding Gag-Pol or Pol. 293T cells were cotransfected with a plasmid coding for wild-type or mutant Gag and with a plasmid coding for either hGag-Pol (hGag-PolΔFSΔPR) or hPol (hPol). ZWt and ZWt-p6 were a gift of Heinrich Göttlinger (Dana- Farber Cancer Institute) and were constructed as previously described (1). The plasmids used are listed along the top of each panel. At 48 h posttransfection, purified Gag VLPs were lysed in radioimmunoprecipitation assay buffer, and Gag and Gag-Pol proteins were detected with Western blots probed with antibodies to HIV-1 RT or CA. (A) Cartoon of wild-type and mutant Gag expressed. (B) Lysates of viruses analyzed by Western blots probed with anti-RT (upper panel) or anti-CA (lower panel). Bands were quantitated with the UN-SCAN-IT gel automated digitizing system.
Pol can facilitate the selective packaging of tRNALys into Gag VLPs and its annealing onto the viral RNA genome.
The incorporation of tRNALys isoacceptors into HIV-1 requires Gag-Pol (36). C-terminal deletions which include sequences coding for integrase and the RNase H and connection domains of RT do not inhibit tRNALys packaging, but further deletions into the thumb domain of RT inhibit tRNALys selective packaging (31). We have investigated whether Pol sequences alone can facilitate the selective incorporation of tRNALys isoacceptors into the virus. Cells were transfected with BH10.P− alone or BH10.FS− alone or were cotransfected with BH10.FS− and either hGag-PolΔFSΔPR or hPol. Western blot analysis similar to that shown in Fig. 1 was used to determine the Gag-Pol or Pol/Gag ratios in the VLPs produced from these transfected cells, and these ratios are listed in Table 2. As previously demonstrated by the results shown in Fig. 1, Pol incorporation into Gag VLPs is somewhat lower (70 to 80%) than Gag-Pol incorporation.
TABLE 2.
The effect of Gag-Pol or Pol incorporation upon selective packaging and annealing of tRNA3Lys in HIV-1a
| Plasmid | Ratio of Gag-Pol or Pol to Gagb | tRNA3Lys packagingc | tRNA3Lys annealingd |
|---|---|---|---|
| BH10.P− | 1.00 | 1.00 | 1.00 |
| BH10.FS− | 0.00 ± 0.00 | 0.11 ± 0.02 | 0.04 ± 0.01 |
| BH10.FS− and hGag-PolΔFSΔPR | 1.05 ± 0.07 | 0.96 ± 0.06 | 0.94 ± 0.04 |
| BH10.FS− and hPol | 0.75 ± 0.04 | 0.65 ± 0.05 | 0.30 ± 0.04 |
All values are normalized to the values found for BH10.P−. The results are the means ± standard deviations of experiments performed three or more times. The values for packaging and annealing are based on the ratios of tRNA3Lys to genomic RNA.
Values are based on the data shown in Fig. 1.
Values are based on the data shown in Fig. 3A.
Values are based on the data shown in Fig. 3C.
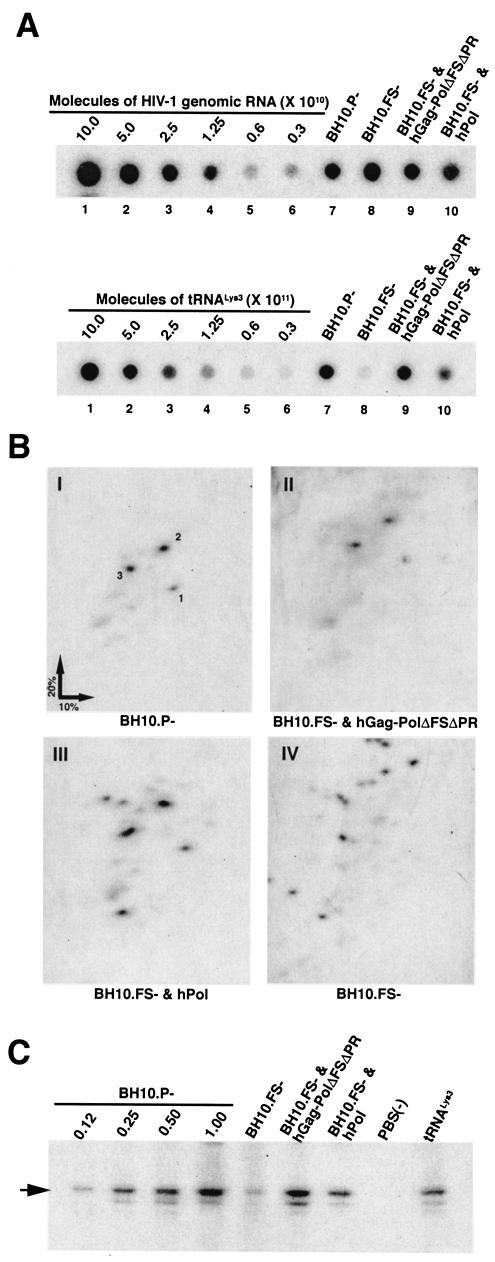
Total RNA was then isolated from virions, and analyzed by dot blot hybridization with probes specific for tRNA3Lys or viral genomic RNA, as previously described (14). These results are shown in Fig. 3A, which also includes standard curves of different concentrations of genomic RNA or tRNA3Lys to show that the dot blot signals of samples fall within the linear range of the curves. The ratios of tRNA3Lys/genomic RNA were then determined, and these ratios, normalized to BH10P−, are also listed in Table 2. It can be seen that the lower amount of tRNA3Lys packaged reflects the lower amount of Pol incorporated into VLPs, indicating that Pol sequences can efficiently replace Gag-Pol in facilitating tRNA3Lys packaging into VLPs.
FIG. 3.
Incorporation of tRNA3Lys into Gag VLPs and its annealing to viral RNA. HEK-293T cells were transfected or cotransfected with different plasmids coding for wild-type Gag and wild-type or mutant Gag-Pol proteins with Lipofectamine. The plasmids used are listed along the top of each panel. At 48 h posttransfection, Gag VLPs were purified, and total viral RNA was extracted using guanidiumisothiocynate, as previously described (28). (A) Incorporation of tRNA3Lys into virions. Dot blots of viral RNA were hybridized with DNA probes complementary to either viral genomic RNA (upper panel) or tRNA3Lys (lower panel), as previously described (14). Hybridization signals were analyzed by phosphorimaging, and the ratio of tRNA3Lys/genomic RNA was determined for each sample (see Table 1). The standard curves shown in the left part of each blot contain known amounts of in vitro transcribed HIV-1 genomic RNA (5′ end fragment) or in vitro transcribed tRNA3Lys and were hybridized with the DNA probes complementary to either tRNA3Lys or genomic RNA to show the linearity of the signal. (B) 2D PAGE analysis of low-molecular-weight viral RNA. Total viral RNA was extracted from virions, 3′-end-labeled with [32P]pCp and electrophoresed in 11% polyacrylamide in the first dimension and 20% polyacrylamide in the second dimension, as previously described (28). Only low-molecular-weight RNA moves into the gel and is detected by autoradiography. Spot 3, tRNA3Lys; spots 1 and 2, tRNA1,2Lys. The text below each panel lists plasmids used to express each viral type analyzed. (C) tRNA3Lys annealing to viral RNA. Total viral RNA was used as the source of primer tRNA3Lys/genomic RNA template in an in vitro reverse transcription reaction, carried out in the presence of [α-32P]dGTP, dCTP, dTTP, and ddATP, as previously described (27). This process results in a six-base extension product since the first six bases incorporated are CTGCTA. Products were resolved by one-dimensional PAGE, with different samples containing equal amounts of genomic RNA. The standard curve shown in the left part of the blot in panel C contains a dilution series of total BH10 viral RNA, which is used as the source of primer or template to show the linearity of the signal.
The ability of Pol sequences to selectively package tRNALys isoacceptors into Gag VLPs was further analyzed by using two-dimensional polyacrylamide gel electrophoresis (2D-PAGE), as previously described (28). Total viral RNA was 3′-end-labeled with [32P]pCp and analyzed by 2D-PAGE. Only low-molecular-weight RNA can enter the gel, and Fig. 3B shows the 2D-PAGE patterns of this RNA. Panel I shows the pattern of RNA in virions produced from cells transfected with the BH10.P− plasmid. The labeled spots have previously been identified (28); spot 3 is tRNA3Lys, the primer tRNA for RT, and spots 1 and 2 represent tRNA1,2Lys. A similar pattern of low-molecular-weight RNA is seen in VLPs produced from cells cotransfected with BH10.FS−, and either hGag-PolΔFSΔPR (Fig. 3B, panel II), or hPol (panel III). In virions produced from cells transfected only with BH10.FS−, no selective packaging of tRNALys isoacceptors is seen (panel IV).
The ability of Pol to selectively package primer tRNA3Lys is also reflected in the ability of the total viral RNA to support reverse transcription. We have shown that the annealing of tRNA3Lys to viral genomic RNA is proportional to the viral concentration of tRNA3Lys (14). To measure the amount of tRNA3Lys annealed to the primer binding site in vivo, total viral RNA is used as the source of primer or template in an in vitro reverse transcription reaction with exogenous HIV-1 RT, dCTP, dTTP, α-32P-dGTP, and ddATP. This assay measures the amount of extendable tRNA3Lys placed on the viral genome (it is not known if all annealed tRNA3Lys is extendable). Since the sequence of the first six dNTPs incorporated is CTGCTA, annealed primer tRNA3Lys will be extended by only six bases in the presence of ddATP, and the extended tRNA3Lys can be resolved and detected by one-dimensional PAGE. The electrophoretic results are shown in Fig. 3C and listed in Table 2. The first four lanes in Fig. 3C use increasing amounts of genomic RNA in the viral RNA to show that the signal undergoes a linear increase with increasing RNA. The amount of viral genomic RNA labeled 1.00 in lane 4 is the amount of genomic RNA used in the remaining five lanes. The ratios of extension of primer tRNA3Lys/genomic RNA, relative to that found for BH10P−, are listed in Table 2. tRNA3Lys placement in Gag VLPs containing Pol is significantly reduced compared to VLPs containing Gag-Pol. In part this result is probably due to the reduced incorporation of tRNA3Lys, since we have shown that the amount of tRNA3Lys annealed to viral genomic RNA is proportional to the amount of tRNA3Lys packaged (14). However, the reduction in tRNA3Lys annealing is greater than expected, which may reflect factors other than the concentration of tRNA3Lys in the virion, including the presence of Pol rather than Gag-Pol in the virion.
In this report, we have shown that Pol can be efficiently incorporated into Gag VLPs independently of upstream Gag sequences in Gag-Pol. Although this process has been studied in a cellular system in which larger amounts of viral proteins are being synthesized compared to normally infected T cells, it is unlikely that Pol incorporation is the result of random packaging. Thus, the results of the experiment shown in Fig. 1D demonstrate that Rous sarcoma Gag VLPs do not package HIV Pol species. Also, the results of the experiments shown in Fig. 2 demonstrate a specific requirement for particular Gag sequences. Replacement of NC in Gag with a leucine zipper motif (plasmid Zwtp6) prevents Gag-Pol, but not Pol, incorporation into Gag VLPs, while an additional deletion of p6 in Gag (plasmid Zwt) reduces Pol incorporation into Gag VLPs four- to fivefold.
It is possible, however, that with lower viral protein concentrations in the cell, the interaction of Gag-Pol with Gag p6 plays a minor role in the incorporation of Gag-Pol into virions, with homologous interactions between Gag and Gag (Gag-Pol) being sufficient for this process. The interaction between Gag-Pol and Gag p6 may be more important at a later stage of assembly. Thus, previous reports have presented data that suggest that the HIV-1 Gag p6 domain functions to retain the Pol domain proteins within the assembling virus after the initial activation of the viral PR (11, 55). Current evidence suggests that prior to viral budding, the viral PR-induced cleavage of Gag-Pol at the cell membrane is initiated in trans within a Gag-Pol dimer to produce an N-terminal Gag fragment terminating in p2 and a C-terminal fragment containing NC-transframe-PR-RT-integrase (42). Thus, this first PR cleavage will separate that part of Gag sequences within Gag-Pol which contain major sites of binding for Gag, i.e., the major homology region and C-terminal portion of capsid. The binding of Pol sequences to p6 sequences in Gag could therefore be important for retaining Pol sequences and tRNALys until viral budding is complete. This interpretation is further supported by the observation reported above that Pol can facilitate the selective packaging of tRNALys isoacceptors with an efficiency reflecting the incorporation of Pol into the Gag particles (approximately 70% of that found using intact Gag-Pol) (Table 2).
Acknowledgments
This work was supported by grants from the Canadian Institutes for Health Research and the Canadian Foundation for AIDS Research.
We thank Heinrich Göttlinger and Rebecca Craven for the gift of plasmids used in this work.
REFERENCES
- 1.Accola, M. A., B. Strack, and H. G. Göttlinger. 2000. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J. Virol. 74:5395-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berkowitz, R., J. Fisher, and S. P. Goff. 1996. RNA packaging, p. 177-218. In H. G. Krausslich (ed.), Morphogenesis and maturation of retroviruses, vol. 214. Springer-Verlag, New York, N.Y.
- 3.Berthet-Colominas, C., S. Monaco, A. Novelli, G. Sibai, F. Mallet, and S. Cusack. 1999. Head-to-tail dimers and interdomain flexibility revealed by the crystal structure of HIV-1 capsid protein (p24) complexed with a monoclonal antibody Fab. EMBO J. 18:1124-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryant, M., and L. Ratner. 1990. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc. Natl. Acad. Sci. USA 87:523-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchschacher, G. L., Jr., L. Yu, F. Murai, T. Friedmann, and A. Miyanohara. 1999. Association of murine leukemia virus pol with virions, independent of Gag-Pol expression. J. Virol. 73:9632-9637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burniston, M. T., A. Cimarelli, J. Colgan, S. P. Curtis, and J. Luban. 1999. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J. Virol. 73:8527-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell, S., and V. M. Vogt. 1995. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J. Virol. 69:6487-6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiu, H., S. Yao, and C. Wang. 2002. Coding sequences upstream of the HIV-1 reverse transcriptase domain in Gag-Pol are not essential for incorporation of the Pr160gag-pol into virus particles. J. Virol. 76:3221-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Craven, R. C., R. P. Bennett, and J. W. Wills. 1991. Role of the avian retroviral protease in the activation of reverse transcriptase during virion assembly. J. Virol. 65:6205-6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Craven, R. C., and L. J. Parent. 1996. Dynamic interactions of the Gag polyprotein. Curr. Top. Microbiol. Immuol. 214:65-94. [DOI] [PubMed] [Google Scholar]
- 11.Dettenhofer, M., and X. F. Yu. 1999. Proline residues in human immunodeficiency virus type 1 p6Gag exert a cell type-dependent effect on viral replication and virion incorporation of Pol proteins. J. Virol. 73:4696-4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dufour, E., J. Reinbolt, M. Castroviejo, B. Ehresmann, S. Litvak, L. Tarrago-Litvak, and M. L. Andreola. 1999. Cross-linking localization of a HIV-1 reverse transcriptase peptide involved in the binding of primer tRNALys3. J. Mol. Biol. 285:1339-1346. [DOI] [PubMed] [Google Scholar]
- 13.Freed, E. O. 2002. Viral late domains. J. Virol. 76:4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gabor, J., S. Cen, H. Javanbakht, M. Niu, and L. Kleiman. 2002. Effect of altering the tRNA3Lys concentration in human immunodeficiency virus type 1 upon its annealing to viral RNA, GagPol incorporation, and viral infectivity. J. Virol. 76:9096-9102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gamble, T. r., S. Yoo, F. Vajdos, U. K. Von Schwedler, J. McCutcheon, and W. I. Sundquist. 1997. Structure of the carboxy-terminal dimerization domain of HIV-1 capsid protein. Science 278:849-853. [DOI] [PubMed] [Google Scholar]
- 16.Garnier, L., L. Ratner, B. Rovinski, S. X. Cao, and J. W. Wills. 1998. Particle size determinants in the human immunodeficiency virus type 1 Gag protein. J. Virol. 72:4667-4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gheysen, D., E. Jacobs, F. de Foresta, C. Thiriart, M. Francotte, D. Thines, and M. De Wilde. 1989. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell 59:103-112. [DOI] [PubMed] [Google Scholar]
- 18.Göttlinger, H. G., J. G. Sodroski, and W. A. Haseltine. 1989. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 86:5781-5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gross, I., H. Hohenberg, C. Huckhagel, and H. G. Krausslich. 1998. N-terminal extension of human immunodeficiency virus capsid protein converts the in vitro assembly phenotype from tubular to spherical particles. J. Virol. 72:4798-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo, F., S. Cen, M. Niu, H. Javanbakht, and L. Kleiman. 2003. Specific inhibition of the synthesis of human Lysyl-tRNA synthetase results in decreases in tRNALys incorporation, tRNA3Lys annealing to viral RNA, and viral infectivity in HIV-1. J. Virol. 77:9817-9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haffar, O., J. Garrigues, B. Travis, P. Moran, J. Zarling, and S. L. Hu. 1990. Human immunodeficiency virus-like, nonreplicating, gag-env particles assemble in a recombinant vaccinia virus expression system. J. Virol. 64:2653-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Halwani, R., A. Khorchid, and L. Kleiman. 2003. Rapid localization of Gag/GagPol complexes to detergent-resistant membrane during the assembly of HIV-1. J. Virol. 77:3973-3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henderson, L. E., H. C. Krutzsch, and S. Oroszlan. 1983. Myristyl amino-terminal acylation of murine retrovirus proteins: an unusual post-translational proteins modification. Proc. Natl. Acad. Sci. USA 80:339-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hermida-Matsumoto, L., and M. D. Resh. 2000. Localization of human immunodeficiency virus type 1 Gag and Env at the plasma membrane by confocal imaging. J. Virol. 74:8670-8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang, M., and M. A. Martin. 1997. Incorporation of Pr160gag-pol into virus particles requires the presence of both the major homology region and adjacent C-terminal capsid sequences within the Gag-Pol polyprotein. J. Virol. 71:4472-4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang, Y., W.-P. Kong, and G. J. Nabel. 2001. Human immunodeficiency virus type 1-specific immunity after genetic immunization is enhanced by modification of Gag and Pol expression. J. Virol. 75:4947-4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang, Y., J. Mak, Q. Cao, Z. Li, M. A. Wainberg, and L. Kleiman. 1994. Incorporation of excess wild type and mutant tRNA34ys into human immunodeficiency virus type 1. J. Virol. 68:7676-7683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang, M., J. Mak, A. Ladha, E. Cohen, M. Klein, B. Rovinski, and L. Kleiman. 1993. Identification of tRNAs incorporated into wild-type and mutant human immunodeficiency virus type 1. J. Virol. 67:3246-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karacostas, V., K. Nagashima, M. A. Gonda, and B. Moss. 1989. Human immunodeficiency virus-like particles produced by a vaccinia virus expression vector. Proc. Natl. Acad. Sci. USA 86:8964-8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khorchid, A., R. Halwani, M. A. Wainberg, and L. Kleiman. 2002. Role of RNA in facilitating Gag/Gag-Pol interaction. J. Virol. 76:4131-4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khorchid, A., H. Javanbakht, M. A. Parniak, M. A. Wainberg, and L. Kleiman. 2000. Sequences within Pr160gag-pol affecting the selective packaging of tRNALys into HIV-1. J. Mol. Biol. 299:17-26. [DOI] [PubMed] [Google Scholar]
- 32.Kräusslich, H.-G., M. Fäcke, A.-M. Heuser, J. Konvalinka, and H. Zentgraf. 1995. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J. Virol. 69:3407-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leis, J., A. Aiyar, and D. Cobrinik. 1993. Regulation of initiation of reverse transcription of retroviruses, p. 33-47. In A. M. Skalka and S. P. Goff (ed.), Reverse transcriptase, vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N. Y.
- 34.Li, S., C. P. Hill, W. I. Sundquist, and J. T. Finch. 2000. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature 407:409-413. [DOI] [PubMed] [Google Scholar]
- 35.Liang, C., L. Rong, N. Morin, E. Cherry, Y. Huang, L. Kleiman, and M. A. Wainberg. 1997. The roles of human immunodeficiency virus type 1 Pol protein and the primer binding site in the placement of tRNA(3Lys) onto viral genomic RNA. J. Virol. 71:9075-9086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mak, J., M. Jiang, M. A. Wainberg, M.-L. Hammarskjold, D. Rekosh, and L. Kleiman. 1994. Role of Pr160gag-pol in mediating the selective incorporation of tRNALys into human immunodeficiency virus type 1 particles. J. Virol. 68:2065-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mak, J., and L. Kleiman. 1997. Primer tRNAs for reverse transcription. J. Virol. 71:8087-8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Momany, C., L. Kovari, A. Prongay, W. Keller, R. Gitti, B. Lee, A. Gorbalenya, L. Tong, J. McClure, L. Ehrlich, M. Summers, C. Carter, and M. Rossmann. 1996. Crystal structure of dimeric HIV-1 capsid protein. Nat. Struct. Biol. 3:763-770. [DOI] [PubMed] [Google Scholar]
- 39.Morikawa, Y., S. Hinata, H. Tomoda, T. Goto, M. Nakai, C. Aizawa, H. Tanaka, and S. Omura. 1996. Complete inhibition of human immunodeficiency virus Gag myristoylation is necessary for inhibition of particle budding. J. Biol. Chem. 271:2868-2873. [DOI] [PubMed] [Google Scholar]
- 40.Morikawa, Y., D. J. Hockley, M. V. Nermut, and I. M. Jones. 2000. Roles of matrix, p2, and N-terminal myristoylation in human immunodeficiency virus type 1 Gag assembly. J. Virol. 74:16-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park, J., and C. D. Morrow. 1992. The nonmyristylated Pr160gag-pol polyprotein of human immunodeficiency virus type 1 interacts with Pr55gag and is incorporated into virus-like particles. J. Virol. 66:6304-6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pettit, S. C., S. Gulnik, L. Everitt, and A. H. Kaplan. 2003. The dimer interfaces of protease and extra-protease domains influence the activation of protease and the specificity of GagPol cleavage. J. Virol. 77:366-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qiu, J. T., R. Song, M. Dettenhofer, C. Tian, T. August, B. K. Felber, G. N. Pavlakis, and X. F. Yu. 1999. Evaluation of novel human immunodeficiency virus type 1 Gag DNA vaccines for protein expression in mammalian cells and induction of immune responses. J. Virol. 73:9145-9152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raba, M., K. Limburg, M. Burghagen, J. Katz, M. Simsek, J. Heckman, U. Rajbhandary, and H. Gross. 1979. Nucleotide sequence of three isoaccepting lysine tRNAs from rabbit liver and SV40-transformed mouse fibroblasts. Eur. J. Biochem. 97:305-318. [DOI] [PubMed] [Google Scholar]
- 45.Rein, A., M. R. McClure, N. R. Rice, R. B. Luftig, and A. M. Schultz. 1986. Myristylation site in Pr65gag is essential for virus particle formation by Moloney murine leukemia virus. Proc. Natl. Acad. Sci. USA 83:7246-7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Resh, M. D. 1994. Myristylation and palmitylation of Src family members: the fats of the matter. Cell 76:411-413. [DOI] [PubMed] [Google Scholar]
- 47.Rhee, S. S., and E. Hunter. 1987. Structural role of the matrix protein of type D retroviruses in Gag polyprotein stability and capsid assembly. J. Virol. 64:4383-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rhee, S. S., and E. Hunter. 1987. Myristylation is required for intracellular transport but not for assembly of D-type retrovirus capsids. J. Virol. 61:1045-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sandefur, S., V. Varthakavi, and P. Spearman. 1998. The I domain is required for efficient plasma membrane binding of human immunodeficiency virus type 1 Pr55gag. J. Virol. 72:2723-2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith, A. J., M. I. Cho, M. L. Hammarskjöld, and D. Rekosh. 1990. Human immunodeficiency virus type 1 Pr55gag and Pr160gag-pol expressed from a simian virus 40 late replacement vector are efficiently processed and assembled into virus-like particles. J. Virol. 64:2743-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Srinivasakumar, N., M.-L. Hammarskjöld, and D. Rekosh. 1995. Characterization of deletion mutations in the capsid region of human immunodeficiency virus type 1 that affect particle formation and Gag-Pol precursor incorporation. J. Virol. 69:6106-6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tritel, M., and M. D. Resh. 2000. Kinetic analysis of human immunodeficiency virus type 1 assembly reveals the presence of sequential intermediates. J. Virol. 74:5845-5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wills, J. W., R. C. Craven, R. A. Weldon, Jr., T. D. Nelle, and C. R. Erdie. 1991. Supression of retroviral MA deletion by the amino-terminal membrane-binding domain of p60src. J. Virol. 65:3804-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu, S. F., D. N. Baldwin, S. R. Gwynn, S. Yendapalli, and M. L. Linial. 1996. Human foamy virus replication: a pathway distinct from that of retroviruses and hepadnaviruses. Science 271:1579-1582. [DOI] [PubMed] [Google Scholar]
- 55.Yu, X. F., L. Dawson, C. J. Tian, C. Flexner, and M. Dettenhofer. 1998. Mutations of the human immunodeficiency virus type 1 p6Gag domain result in reduced retention of Pol proteins during virus assembly. J. Virol. 72:3412-3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou, W., L. J. Parent, J. W. Wills, and M. D. Resh. 1994. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J. Virol. 68:2556-2569. [DOI] [PMC free article] [PubMed] [Google Scholar]