Abstract
Inspection of over 250 hepatitis C virus (HCV) genome sequences shows that a threonine is strictly conserved at the P1 position in the NS3-NS4A (NS3-4A) autoproteolysis junction, while a cysteine is maintained as the P1 residue in all of the putative trans cleavage sites (NS4A-4B, NS4B-5A, and NS5A-5B). To understand why T631 is conserved at the NS3-4A junction of HCV, a series of in vitro transcription-translation studies were carried out using wild-type and mutant (T631C) NS3-4A constructs bearing native, truncated, and mutant NS4A segments. The autocleavage of the wild-type junction was found to be dependent on the presence of the central cofactor domain of NS4A (residues 21 to 34). In contrast, all NS3-4A T631C mutant proteins underwent self-cleavage even in the absence of the cofactor. Subgenomic replicons derived from the Con1 strain of HCV and bearing the T631C mutation showed reduced levels of colony formation in transfection studies. Similarly, replicons derived from a second genotype 1b virus, HCV-N, demonstrated a comparable reduction in replication efficiency in transient-transfection assays. These data suggest that the threonine is conserved at position 631 because it serves two functions: (i) to slow processing at the NS3-4A cleavage site, ensuring proper intercalation of the NS4A cofactor with NS3 prior to polyprotein scission, and (ii) to prevent subsequent product inhibition by the NS3 C terminus.
Hepatitis C virus (HCV), the etiologic agent of an often chronic and sometimes fatal form of hepatitis, constitutes a highly diverse genus within the Flaviviridae (32, 35). The viral NS3 protein, a 70-kDa bifunctional enzyme composed of a chymotrypsin-like serine protease domain (amino acids 1 to 180) and a helicase/NTPase (residues 181 to 631), is responsible for proteolytic cleavage of the polyprotein at the NS3-NS4A (NS3-4A), NS4A-4B, NS4B-5A, and NS5A-5B junctions (3, 14, 44). Although the NS3 protein has some basal proteolytic activity, catalysis is enhanced by, and for some substrates appears to require, the formation of a noncovalent complex with the 54-amino-acid NS4A (2, 5, 13, 21, 24, 27, 40, 42). The central cofactor residues (21-34) of NS4A have been shown to be sufficient to effect this enhancement and permit cleavage of the NS4A-dependent junctions (9, 28, 38, 40, 45). X-ray diffraction studies indicate that these NS4A residues intercalate into the N-terminal β-barrel of NS3, causing conformational changes which lead to improved alignment of the catalytic triad (20, 31, 48). It has been demonstrated that replacing the hydrophobic residues, especially Ile-25 and Ile-29, with alanine residues nearly abolished the proteolytic cleavage of the NS5A-5B protein (9). It has been speculated that, in addition to their cofactor role, the N-terminal 20 residues of NS4A form a hydrophobic transmembrane helix, which anchors the NS3-4A complex to the endoplasmic reticulum (ER) (16, 20, 36, 42), contributing to the formation of a replication complex (42).
The proteolytic cascade of HCV has been studied by using both vaccinia virus expression systems and in vitro transcription-translation (IVTT) methods (2, 4, 5). Pulse-chase studies suggest that cleavage between NS3 and NS4A occurs rapidly prior to the downstream cleavages at NS5A-5B, NS4B-5A, and NS4A-4B (2, 4, 12, 15). Dilution insensitivity of the NS3-4A cleavage reaction further implies that the process is unimolecular (i.e., occurs in cis). However, one detail that has remained unclear is whether intercalation of the central cofactor domain of NS4A precedes autoproteolysis or whether it occurs spontaneously following the scission event.
Cleavage by the NS3 protease is highly specific. Despite the tremendous variability of the HCV genome, a P1 cysteine residue is strictly conserved at all trans cleavage sites. In vivo polyprotein processing and in vitro peptide cleavage studies have shown that replacement of the P1 cysteine with other amino acids (including threonine) significantly reduces trans proteolysis of these junctions by 2 to 3 orders of magnitude (4, 46, 52). On the other hand, it is remarkable that threonine is equally invariant in the corresponding P1 position of the NS3-4A junction.
The conservation of threonine at P1 in the HCV NS3-4A junction was thought initially to be optimal for a modified S1 pocket predicted to be present in the NS3-4A precursor protein (52). However, more-recent structural analysis of a single-chain NS3-4A protein (49), which reflects the product complex of the autoproteolysis reaction, suggests that the active site probably does not undergo significant reorganization following scission of the NS3-4A scissile bond. These data make this explanation for the strict retention of threonine at position 631 less tenable. We present data that suggest an alternative hypothesis: that the P1 threonine in the NS3-4A junction retards autoproteolysis at the NS3-4A junction until the intercalation of the NS4A residues within the NS3 structure is complete. The P1 threonine may also help prevent product inhibition from the newly generated NS3 C terminus.
MATERIALS AND METHODS
Cells.
Huh7 cells, derived from a human hepatoma, were grown in Dulbecco's modified Eagle medium (DMEM; Cellgro, Herndon, Va.) with 10% fetal calf serum, 100 IU each of penicillin and streptomycin/ml, 4 mM l-glutamine, 1 mM sodium pyruvate, 0.13% sodium bicarbonate, and 1× nonessential amino acids. En5-3 cells (50) are stably transformed cells derived from Huh7 cells; they contain the sequence encoding secreted alkaline phosphatase (SEAP) placed under the transcriptional control of the long terminal repeat promoter of human immunodeficiency virus (HIV). They were maintained in culture as described previously (50).
Construction of plasmids and site-directed mutagenesis.
The plasmid BK 138-1, described previously (41) and encoding the entire NS3-4A region of the 1b/BK strain of HCV, was used as the PCR template. DNA fragments corresponding to amino acid sequences of NS31-631-NS4A1-54 and NS31-631-NS4A1-34 were amplified with Pfu DNA polymerase (Strategene, La Jolla, Calif.) and cloned between NheI and EcoRI restriction sites of the expression vector pET28b(+) (Novagen, Madison, Wis.) by standard methods (37) to generate plasmids pWW54 and pWW34 (Table 1). Mutations and truncations were introduced into the above constructs by site-directed mutagenesis as described elsewhere (47). Construction of p22 (NS31-631/S139A) and p24 (NS4A21-34-GSGS-NS33-181) has been described (Table 1) (43).
TABLE 1.
HCV NS3-4A constructs for in vitro translation studiesa
| Plasmid | Construct(s) | Protein |
|---|---|---|
| pWW54 | NS31-631-NS4A1-54 | T-54 |
| pWW34 | NS31-631-NS4A1-34 | T-34 |
| pWW20 | NS31-631-NS4A1-20 | T-20 |
| pWW34* | NS31-631-NS4A1-34/I25S, -I29S | T-34* |
| pWW54C | NS31-631/T631C-NS4A1-54 | C-54 |
| pWW34C | NS31-631/T631C-NS4A1-34 | C-34 |
| pWW20C | NS31-631/T631C-NS4A1-20 | C-20 |
| pWW34C* | NS31-631/T631C-NS4A1-34/I25S, -I29S | C-34* |
| pWW54C | NS31-631/S139A, T631C-NS4A1-54 | iC-54 |
| pWW34C | NS31-631/S139A, T631C-NS4A1-34 | iC-34 |
| pWW20C | NS31-631/S139A, T631C-NS4A1-20 | iC-20 |
| pWW34C* | NS31-631/S139A, T631C-NS4A1-34/I25S, -I29S | iC-34* |
| pWW75 | NS31-631/S139A-NS4A1-54/V4S, -L5Q, -V6S | MW-75 |
| pWW73 | NS31-631/S139A-NS4A1-34/V4S, -L5Q, -V6S | MW-73 |
| p22a | NS31-631/S139A | MW-70 |
| p24 | NS4A21-34-GSGS-NS33-181 | sc-NS33-181 |
See reference 43 for details of plasmid construction.
Subgenomic HCV replicons.
A dicistronic genotype 1b HCV replicon I377/NS3-3′, derived from the Con1 strain of HCV (GenBank accession number AJ242652) (30), was assembled and cloned from chemically synthesized DNA oligonucleotides (19) into a modified pUC18 vector. An XbaI site was introduced at the 3′ end of the replicon sequence to facilitate in vitro transcription of RNA from the DNA template. The cell culture adaptation mutation S1179I (7) was subsequently added to the I377/NS3-3′ cDNA to create a more-replication-efficient replicon genome. Point mutations were introduced to the NS3-4A cleavage site with a subcloned fragment of the domain (spanning nucleotides 3542 to 4630 of the replicon sequence) in pCR4Blunt-TOPO (Invitrogen) by the QuikChange method (Stratagene). Clones containing the desired mutations were confirmed with the CEQ DTCS-Quick Start kit for dye terminator cycle sequencing (Beckman Coulter, Inc., Fullerton, Calif.). A 120-bp fragment containing the desired mutation (nucleotides 4103 to 4223) was cloned into the full-length replicon cDNA.
Transient-RNA-replication assays were carried out with a second genotype 1b replicon, designated NTat-T631 for the purposes of this study, which was derived from the previously described replicon Ntat2ANeo(SI) (50). The downstream cistron of NTat-T631 contains an HCV-N sequence (6) (GenBank accession number AF139594) encoding NS3-NS5B, with several cell culture adaptation mutations within the NS3 and NS4A coding sequences. A hepatitis delta virus ribozyme sequence is placed downstream of the HCV sequence and enhances the replication competence of the RNA by generating an authentic 3′ HCV terminus (51; M. Yi and S. M. Lemon, unpublished data). The expression of the HIV Tat protein from the upstream cistron of NTat-T631 induces the secretion of SEAP from transfected En5-3 cells, with SEAP expression correlating closely with the abundance of the replicon RNA (50). The plasmid NTat-T631 was mutated at the bases encoding NS3 residue 631 by the QuikChange (Stratagene) method, as described above, and the small SphI-PmlI fragment of the mutant plasmids was religated into the NTat-T631 background to produce NTat-T631C, NTat-T631I, and NTat-T631V. The sequences of all DNA regions manipulated by PCR during this process were confirmed by DNA sequencing. A replication-defective variant of Ntat-T631, ΔGDD, containing a deletion within the NS5B polymerase protein (50), was included as a negative control in all transfection experiments.
In vitro transcription and translation of NS3 protease constructs.
Plasmids listed in Table 1 were linearized by HindIII digestion. Following phenol-chloroform extraction, the plasmids were ethanol precipitated and dissolved in water. One to 2 μg of the linearized plasmids was used to prime transcription of RNA using RiboMAX large-scale RNA production systems (Promega, Madison, Wis.). After 3.5 h at 30°C, 2 U of RNase-free DNase (Promega) was added to digest the plasmids. RNA was extracted with phenol-chloroform, precipitated with isopropanol, and dissolved in nuclease-free water (Promega). Samples of RNA were analyzed on nondenaturing agarose gels and quantified by UV spectrometry. One microgram of RNA was used to program a typical 50-μl translation reaction mixture containing rabbit reticulocyte lysate (Promega) and a [35S]methionine-[35S]cysteine mixture. Translated proteins were separated by electrophoresis on sodium dodecyl sulfate (SDS)-8% polyacrylamide gels (Novex, San Diego, Calif.) and analyzed by autoradiography and phosphorimaging. Digital images were processed with Photoshop, version 6.01 (Adobe Systems), and Canvas, version 7.01 (Deneba Systems).
Enhancement of autoproteolysis by NS4A cofactor peptide.
RNA was translated at 30°C for 60 min as described above and then incubated with RNase A (0.2 mg/ml) and cycloheximide (50 ng/ml) to remove RNA and stop translation, respectively. The NS4A peptide KK-NS4A21-34-KK (50 μM) was added to the lysate and incubated for an additional 60 min.
Dilution studies of the NS3-4A proteins.
Two strategies were used to correlate dilution of NS3-4A and the extent of autoprocessing. As a first strategy, in vitro translation reactions were carried out at 30°C for 15 to 20 min, followed by a 15-min incubation with RNase A and cycloheximide, as described above. Ten microliters of each reaction mixture was diluted with 0, 20, 90, and 490 μl of buffer A (25 mM HEPES [pH 7.2], 125 mM NaCl, 10% glycerol) or buffer B (buffer A plus 2 mM dithiothreitol). The samples were incubated at 30°C for another 60 min. All samples were brought to 500 μl in the appropriate buffers and precipitated with acetone overnight at −20°C. Pellets were collected by centrifugation, washed with acetone, resuspended in 50 μl of SDS-polyacrylamide gel electrophoresis (PAGE) loading buffer (Owl Separation Systems, Portsmouth, N.H.), and placed in a boiling water bath for 5 min. The samples were briefly centrifuged, and equal volumes of the supernatants were analyzed by SDS-PAGE and quantified by autoradiography and phosphorimaging. As an alternative approach, different quantities of RNA were used to prime the IVTT reaction as described above. After 30 or 60 min at 30°C, proteins were analyzed by standard methods. Quantitation of the detected proteins was performed by densitometry using the ImageQuant software, version 1.2 for the Macintosh (Molecular Dynamics).
trans cleavage of the NS3-4A junction by exogenous NS3 protease.
DNA constructs encoding proteolytically inactive proteases (bearing the S139A mutation) with cysteine replacing threonine 631 were used to program translation reactions as described above. Purified sc-NS4A21-32-GSGS-NS33-181 was added (final concentration, 250 nM) to 20-μl aliquots of lysates, and the samples were incubated for 60 min at 30°C. Proteolysis was monitored as described above.
In vitro transcription of replicon RNA.
Plasmid DNA was linearized with XbaI and treated with mung bean nuclease (New England Biolabs, Beverly, Mass.) to remove the remaining overhang strand (37). Purified DNA was used as the template for transcription with the MEGAscript system (Ambion, Austin, Tex.) and its protocol. Transcription reactions were terminated after 4 h by the addition of DNase I, and RNA products were precipitated with lithium chloride, according to the manufacturer's instructions (Ambion). The RNA concentrations were measured by spectrophotometry, and denaturing agarose gel electrophoresis was used to assess integrity.
Electroporation and selection of G418-resistant cell lines.
Transfection of replicon RNA and the subsequent selection of cells for G418 resistance were as previously described (30). Briefly, 5 μg of transcript RNA was mixed with a suspension of 5 × 106 Huh7 cells in 100 μl. Electroporation (25 μF, 350 V, infinite resistance, two pulses, and a 0.1-cm cuvette) was performed with a GenePulser system (Bio-Rad, Hercules, Calif.). Cells were immediately diluted into 10 ml of complete DMEM and seeded in a 10-cm-diameter cell culture dish. After 24 h, culture medium was replaced with complete DMEM containing 1 mg of G418 (GIBCO, Grand Island, N.Y.) per ml. Medium was changed twice a week, and colonies were observed after 3 to 4 weeks of selection. Cell colonies were isolated by lifting with filter paper soaked with 0.25% trypsin-0.1% EDTA (Cellgro). Replicon colonies isolated for amplification and further study were maintained under the same conditions as those for selection. Total RNA was extracted from recovered cell lines by the RNeasy method (Qiagen, Valencia, Calif.) in accordance with the manufacturer's instructions (11).
Colony-forming efficiency (CFE) assay.
Cell colonies, established as described above, were visualized by staining with crystal violet (29). Colony numbers from different experiments were adjusted relative to the number of colonies obtained from a transfection of 106 cells with 1 μg of input replicon RNA.
Transient-RNA-replication assay.
Transient-replication assays were carried out with En5-3 cells transfected with NTat-T631 RNA transcripts and transcripts derived from related mutants. Transfection of RNA transcripts was by electroporation, and RNA amplification was monitored by assay of cell culture supernatant fluids for SEAP activity, as described previously (50).
Analysis of replicating HCV RNA sequences.
The NS3 and NS4A coding regions of various I377/NS3-3′(SI) replicon RNAs were amplified with the Titan one-tube reverse transcription-PCR kit (Roche Diagnostics GmbH, Mannheim, Germany), starting with equal amounts of total RNA (final concentration, ≈4 ng/μl) from the cell lines. Products were directly analyzed by dye terminator cycle sequencing. Analysis of the wild-type and mutated sequences was carried out by pyrosequencing as described previously (23). PCR primers designed with the SNP primer design software, version 1.0.1 (Pyrosequencing AB, Uppsala, Sweden), to target the sequence surrounding the NS3-4A junction where mutations were introduced were as follows: forward PCR primer, 5′-AGGCTGGGAGCCGTTCAAA-3′; reverse PCR primer, 5′-biotin-AGTCCTAGCAGCTCTGGCCG-3′; sequencing primer, 5′-GTCGGCTGACCTGGAG-3′. The sequences to analyze were GTC GTC ACG AGC ACC TGG GTG CTG G (wild type) and GTC GTC TGC AGC ACC TGG GTG CTG G (T631C) (mutated nucleotides are underlined and in boldface).
Templates for pyrosequencing were produced as described previously (23). The amplification reactions were performed by using a temperature profile of 50°C for 30 min (reverse transcription); 94°C for 2 min; 50 cycles of 95°C for 15 s, 54°C for 30 s, and 72°C for 15 s (PCR); and 72°C for 5 min. To quantify the relative levels of the wild-type and mutant genotypes in the mixed population, results obtained from the pyrosequencing reactions were analyzed with the PSQ 96 MA software package, version 2.01 (Pyrosequencing).
RESULTS
A native NS4A cofactor region is required for autoproteolysis of the wild-type NS3-4A junction.
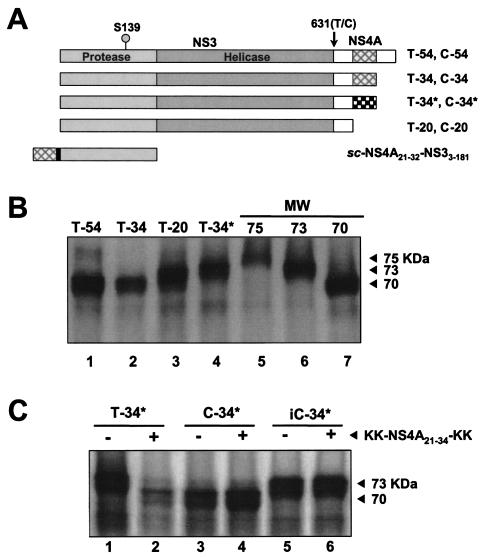
To study the impact of the NS4A cofactor domain on self-cleavage at the NS3-4A junction, four constructs (T-54, T-34, T-20, and T-34*; Table 1 and Fig. 1A) were translated and evaluated for their ability to efficiently undergo maturation. Following reactions in mixtures incubated for 60 min, the release of free NS3 was observed only with NS3-4A protein constructs harboring a functional cofactor peptide (i.e., T-54 and T-34, which contain the cofactor domain, NS4A21-34), while those bearing an altered, poorly binding cofactor peptide sequence (i.e., T-34*) or no cofactor peptide (i.e., T-20) showed no evidence of cleavage (Fig. 1B). Following the addition of exogenous cofactor peptide KK-NS4A21-34-KK and further incubation, processing was observed with T-34* (Fig. 1C, lanes 1 and 2) and T-20 (data not shown).
FIG. 1.
Self-cleavage of the wild-type NS3-4A junction is dependent on the presence of a functional NS4A central cofactor domain peptide (residues 21 to 34). (A) Schematic of expression constructs used for in vitro transcription and translation reactions. The protease and helicase domains of NS3 are labeled. Black box, cofactor domain of NS4A that intercalates into the structure of NS3. sc-NS4A21-32-GSGS-NS33-181 is a single-chain, proteolytically active variant of NS3 in which the NS4A cofactor domain is fused covalently to the N terminus of the NS3 sequence. The position of the S139A mutation, which renders the NS3 protease catalytically inactive, is shown, as is T631, which is at the C terminus of NS3 (P1 position of the NS3-4A junction). (B) RNA encoding constructs with the wild-type NS3-4A junction (i.e., T631), but bearing different NS4A sequences, was used to program translation in rabbit reticulocyte lysate at 30°C for 60 min, as described in Materials and Methods. Products were separated by SDS-8% polyacrylamide gels and analyzed by autoradiography. Translation products MW-75, -73, and -70 were used as molecular weight markers and migrate at 75, 73, and 70 kDa, respectively. (C) The construct T-34*, which contains a nonfunctional NS4A peptide domain (Table 1), undergoes cleavage only upon the addition of an exogenous cofactor peptide. RNA for T-34* was translated in the presence (+) and absence (−) of the peptide KK-NS4A21-34-KK. C-34* and iC-34* (which contains a catalytically inactive NS3 sequence) were used as positive and negative controls, respectively (see Results for details).
A native NS4A cofactor region is not required for autoproteolysis of a mutant (T631C) NS3-4A junction.
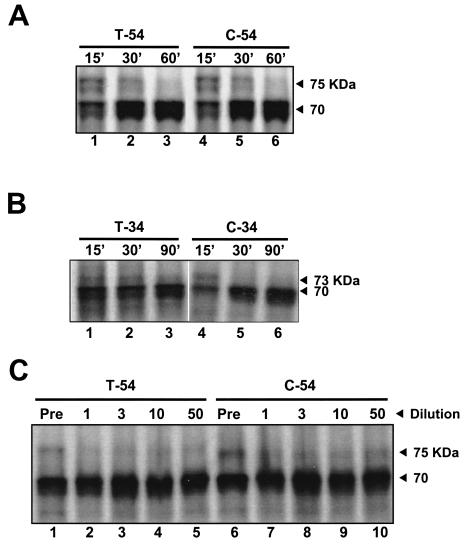
Since a cysteine residue is conserved at the P1 position of each NS3-4A trans cleavage site, it was of interest to determine the effects of replacing the equally well conserved NS3-4A junction P1 threonine residue with cysteine. To establish whether the NS4A cofactor domain would also be required for self cleavage of this mutant NS3-4A junction, a series of constructs identical to those described above but bearing a T631C substitution (Table 1 and Fig. 1A) were generated and evaluated for self cleavage. The constructs with the functional cofactor peptide (i.e., C-54 and C-34) were processed to completion in about 30 min (Fig. 2A and B, lanes 4 to 6), with the rate of proteolysis approximately equivalent to that for the T-54 and T-34 constructs (Fig. 2A and B, lanes 1 to 3). However, in contrast to what was found for constructs containing a P1 threonine residue, efficient processing was also observed for junctions in the construct containing the P1 cysteine without a cofactor sequence (i.e., C-20; Fig. 3A, lanes 1 to 3) or with the dysfunctional mutant NS4A cofactor sequence (i.e., C-34*; Fig. 3A, lanes 4 to 6). Nonetheless, a comparison of the extents of cleavage with and without functional cofactor sequence by phosphorimager analysis indicated that the NS4A domain facilitated the processing (i.e., at least 90% C-54 was processed after a 30-min translation, compared with 50% for C-20 and C-34* after 30 and 90 min, respectively).
FIG. 2.
Self-cleavage at the NS3-4A junction is insensitive to dilution. (A) One microgram of RNA encoding T-54 or C-54 was used to prime the in vitro translation reaction. Proteins were analyzed after 15, 30, and 60 min by electrophoresis in an SDS-8% polyacrylamide gel and autoradiography. The number above each lane refers to the reaction time (in minutes) for the sample. (B) Analysis of T-34 and C-34 constructs, processed in the same manner as for panel A. (C) Translation reactions programmed with T-54 and C-54 transcripts were carried out as described in Materials and Methods and were terminated after 15 min. The proteins were diluted 1-, 3-, 10- and 50-fold with buffer A and then incubated at 30°C for another 60 min before being precipitated with acetone and analyzed by electrophoresis. The numbers above the lanes indicate the dilution factors. Pre, starting translation product prior to dilution and the second incubation.
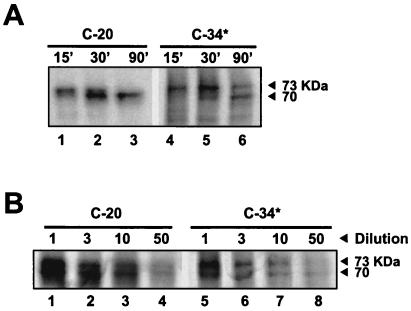
FIG. 3.
Self-cleavage of NS3-4A junctions with P1 cysteine is independent of the intercalation of a functional NS4A peptide. (A) One microgram of RNA encoding C-20 or C-34* was used to program the in vitro translation reaction. Proteins were analyzed as described for Fig. 2A. The number above each lane refers to the reaction time (in minutes) for the sample. (B) Plasmids (from 20 ng to 1 μg) encoding C-20 and C-34* proteins were used to program IVTT in a 50-μl reaction mixture. After 30 and 60 min of incubation, reactions were stopped and proteins were analyzed by SDS-PAGE and autoradiography. There was no significant change in the ratio of unprocessed to processed protein, as measured by densitometry.
Both NS4A-dependent and NS4A-independent cleavages are unimolecular.
Dilution studies of both wild-type and mutant constructs containing the full-length NS4A sequence (T-54 and C-54, respectively) showed the NS3-4A cleavage to be a dilution-insensitive intramolecular process (Fig. 2C), extending an earlier observation (2) to include the P1 cysteine variants. However, compared with a previously described construct encoding NS3′-4A-4B′ (2), both T-54 and C-54 exhibited faster self-processing, suggesting that an additional polyprotein sequence attached to the C terminus of NS4A may slow the folding and cleavage between NS3 and NS4A. As predicted, polypeptides with further C-terminal truncations of NS4A (i.e., T-34 and C-34) demonstrated an apparent increase in the proteolysis rate compared with results for the full-length T-54 and C-54 proteins (Fig. 2B).
To determine whether the cleavage at the mutant T631C junction (C-20 and C-34*) remained a unimolecular process, the translated proteins were diluted and further incubated to allow processing, as in the experiment described above. Dilution of the C-20 and C-34* proteins did not change either the ratio of unprocessed/processed protein (Fig. 3B) or the cleavage rates (data not shown), indicating that an intramolecular process was still dominant in the absence of a functional NS4A cofactor peptide fragment.
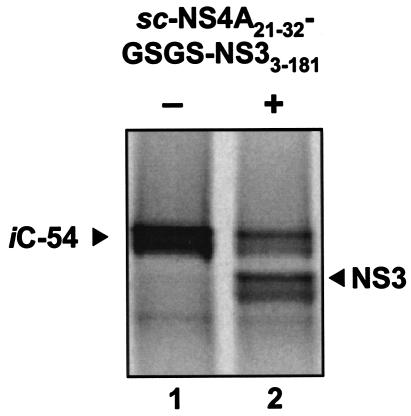
To unequivocally eliminate the possibility of trans cleavage, catalytically inactive counterparts of C-54, C-34, C-20, and C-34* were constructed and used as trans substrates for exogenous NS3 protease (iC-54, iC-34, iC-20, and iC-34*; Table 1). When these translated products were incubated with the active sc-NS4A21-32-GSGS-NS33-181 protease (Fig. 1A), only the iC-54 target was cleaved (Fig. 4). However, this trans reaction was less efficient than autoproteolysis (only 50% digestion after 1 h versus 90% self-digestion in 30 min) and required a high protease concentration (250 nM). These results suggest that only for the C-54 constructs might there have been any measurable contribution from trans cleavage under these reaction conditions.
FIG. 4.
Processing of the NS3-4A junction via a trans mechanism. Construct iC-54, with the S139A protease active-site mutation, was translated for 60 min. A 20-μl aliquot of the lysate was incubated with (+) or without (−) the purified sc-NS4A21-32-GSGS-NS33-181 protein (final concentration, 250 nM) for 60 min at 30°C. The products were separated by SDS-8% polyacrylamide gels and analyzed by autoradiography.
Effect of NS3-4A cleavage site mutations on replicon colony formation.
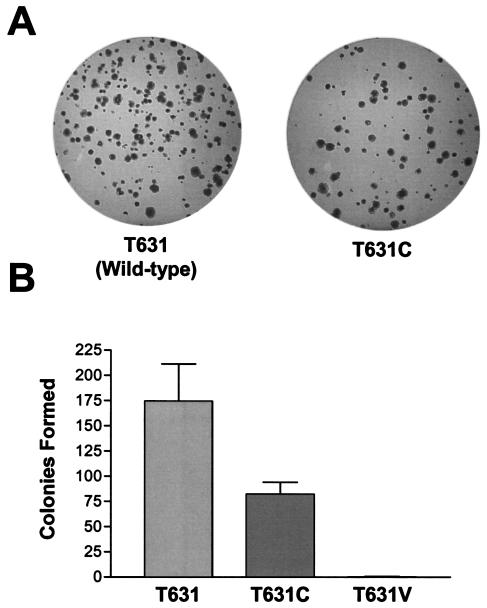
The results described above suggest that replacement of the NS3-4A junction P1 threonine with cysteine results in a substrate that undergoes autoproteolytic cleavage at least as efficiently as the highly conserved NS3-4A junction containing a P1 threonine. This suggests, indirectly, that the absolute conservation of the P1 threonine must result from an ability to enhance replication of the viral RNA or improve viral fitness by a mechanism other than facilitation of polyprotein processing at the NS3-4A junction. To assess this possibility, a cDNA clone of a subgenomic, genotype 1b replicon derived from the Con1 strain of HCV and bearing the T631C mutation was transcribed and transfected into Huh7 cells by electroporation, as previously described (30). This replicon contains a cell culture adaptation mutation within the NS5A sequence that promotes replication in Huh7 cells and leads to efficient colony formation under G418 selection pressure (see Materials and Methods). In three independent transfection experiments, the CFE was reduced for replicon sequences bearing the P1 cysteine mutation (49% ± 9% of replicon sequences that were identical except for encoding the wild-type T631 residue; Fig. 5). Despite the reduction in CFE, replicon RNAs encoding the T631C substitution were capable of establishing stable, G418-resistant cell lines. Single colonies from T631 or T631C transfections were amplified and further analyzed. Western blot analyses of lysates of the wild-type T631 and mutant T631C replicon cell lines demonstrated no apparent accumulation of unprocessed polyprotein (data not shown), suggesting that the change in the C terminus of NS3 did not significantly affect polyprotein processing. Sequencing results showed that the T631C mutation was retained in all isolates. However, other amino acid substitutions were present in the NS3 sequence of the T631C-encoding replicons at several different sites (Table 2). Significantly, a D441Y substitution was identified in three independently selected cell lines. Importantly, no similar NS3 mutations were identified in the colonies obtained following transfection of the wild-type T631 replicon RNA.
FIG. 5.
Effect of NS3-4A P1 cysteine mutation on Con1 replicon CFE. (A) The colony formation efficiency of an HCV replicon bearing the T631C mutation is reduced. Equal amounts of RNA transcripts of the wild-type or T631C mutant replicon were transfected into Huh7 cells by electroporation. After 24 h, cells were treated with G418 for selection, and, after 3 weeks, colonies were visualized by crystal violet staining. (B) Results of colony-forming assay comparing wild-type, T631C, and T631V replicons. Values are derived from three independent transfections, normalized to the number of colonies obtained from transfection of 106 cells with 1 μg of input replicon RNA. For the T631V replicon, the mean was less than 1 (0.3 ± 0.6).
TABLE 2.
Amino acid substitutions in the NS3 coding sequences of T631C replicon RNAs recovered from various stable G418-resistant cell lines
| RNA transfection | Amino acid substitution(s)c |
|---|---|
| T631C transfection (pooled)a | V318L, D441Y |
| Wild type + T631C cotransfection (pooled)b | D405Y, D441Y |
| T631C clone 1 | E533K |
| T631C clone 2 | E176G, D441Y, K589E |
Replicon colonies resulting from T631C replicon genome transfections were allowed to grow for 1 month posttransfection under G418 selection in a 10-cm dish before collection of the cells for RNA extraction.
Cells cotransfected with T631 and T631C replicon genomes were allowed to grow and were harvested as a pooled population, as described in footnote a.
The numbering system for the amino acids is based on the designation of the first true residue of NS3 as number 1. In the replicon I377/NS3-3′, an initiating start codon precedes residue 1 (i.e., M[−1]). Substitutions in boldface were found in most clones.
In parallel experiments, only one colony was recovered from three independent transfections with a Con1 replicon RNA encoding a T631V mutation (Fig. 5). This amino acid substitution is notably less permissive than T631C for NS3-4A processing in the context of a replicon sequence (Fig. 6A).
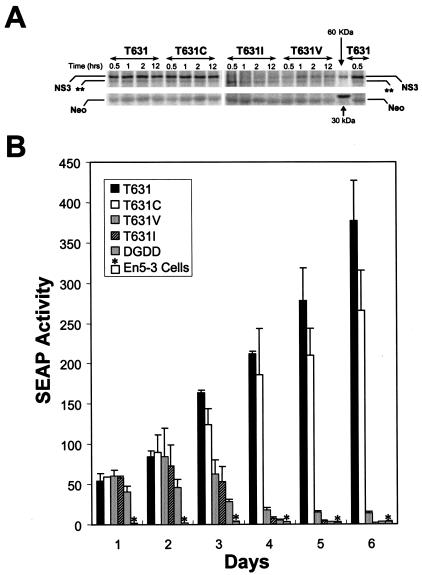
FIG. 6.
The T631C mutation degrades the replication capacity of a subgenomic replicon derived from the genotype 1b HCV-N virus that induces the expression of SEAP proportional to the intracellular abundance of replicon RNA. (A) In vitro translation reactions were programmed with replicon RNAs encoding the wild-type NS3-4A junction (NTat-T631) or cysteine, valine, or isoleucine replacements of the P1 threonine normally present at residue 631 and incubated for 0.5, 1, 2, or 12 h, followed by separation of the products by SDS-PAGE and autoradiographic visualization of the mature NS3 product of the NS3-4A cleavage. (Top) Translated mature NS3 protein. The band marked ** corresponds in apparent molecular mass to the predicted NS5B product. (Bottom) Neomycin phosphotransferase product (Neo) expressed from the upstream cistron of the replicon RNAs, which served as a translation control in these experiments. The positions of 30- and 60-kDa markers are shown. The T631 and T631C products underwent efficient processing at the NS3-4A junction, while the T631V and T631I products did not. (B) Transient-transfection assays demonstrate that the NTat-T631 replicon bearing the wild-type NS3-4A junction has greater replication capacity than NTat-T631C and much greater replication capacity than NTat-T641V and -T631I (whose replication capacity is indistinguishable from that of replication-defective NS5B deletion mutant ΔGDD). En5-3 cells were transfected with equal amounts of the indicated replicon RNAs by electroporation and then cultured in the absence of G418 selection for 6 days. Supernatant culture media were sampled at 24-h intervals and assayed for SEAP activity as a measure of the intracellular replicon RNA abundance (see Materials and Methods). Results shown represent the mean SEAP activities present in supernatant fluids at the times indicated. Error bars represent the ranges of values in replicate transfection experiments using RNA transcripts produced in separate reactions. T631, NTat-T631. *, SEAP expressed by nontransfected En5-3 cells.
Effect of the T631C mutation on replicon fitness.
As the T631C mutation was detrimental to CFE of the subgenomic Con1 replicons, this change might be expected to impact replicon fitness. To assess this, two different competition studies were performed. First, in an effort to examine directly intracellular competition between the mutant T631C and wild-type T631 replicons, mixed-transfection studies in which equimolar amounts of RNA transcripts were used to transform naive Huh7 cells were carried out. Once cell lines were established, replicon sequences were analyzed by capillary sequencing. Variations in peak heights inherent in capillary sequencing precluded quantitative assessment to identify the dominant species, but both the wild-type T631 and T631C sequences were present in pooled cell populations 1 month following transfection. The D441Y mutation was also observed in these populations, as was the wild-type D441 codon (Table 2).
As an alternative approach, equal numbers of clonally isolated, G418-resistant cells selected with the wild-type T631 and mutant T631C replicons were mixed and cocultured in the presence of G418. Aliquots of the cells were harvested at various time points and processed for total RNA extraction. The presence and relative proportion of the T631 and T631C codons in each sample were determined by pyrosequencing (23) (data not shown). Consistent with the results from the mixed transfections, the wild-type and mutant cells remained near their original ratios (data not shown), suggesting that the T631C mutation does not affect fitness appreciably, at least in the presence of potentially compensatory amino acid substitutions (second-site revertants) elsewhere within NS3.
Since the ability to determine the impact of the T631C substitution on the replication fitness of the subgenomic Con1 replicon used in the experiments described above is reduced by potentially confounding second-site mutations or the selection of Huh7 cell clones with clonally specific growth characteristics and/or permissiveness for viral RNA replication, additional studies were carried out with a subgenomic replicon derived from the genotype 1b HCV-N virus, which undergoes very efficient replication that can be easily monitored in Huh7 cells in the absence of G418 selection. The HCV-N replicon NTat-T631 is derived from a Tat-expressing replicon that induces the expression of SEAP in proportion to the abundance of intracellular replicon RNA in transfected En5-3 cells (Huh7 cells expressing SEAP under transcriptional control of the long terminal repeat promoter of HIV; see Materials and Methods). A cDNA copy of the NTat-T631 replicon encoding the conserved P1 threonine at the NS3-4A junction was mutated to encode T631C, T631I, or T631V substitutions at this residue. In vitro translation reactions were programmed with RNAs transcribed from these plasmid DNAs, and cleavage at the NS3-4A junction of the expressed polyproteins was assessed by demonstration of the mature NS3 protein by SDS-PAGE (Fig. 6A). These results confirmed equivalently efficient cleavage of the wild-type T631 and mutant T631C constructs at the NS3-4A junction. In contrast, processing was markedly decreased and delayed with the T631V construct and was virtually not detectable with the T631I mutant. These results are consistent with those described above.
To assess the impact of these substitutions on replication of the HCV-N replicon, these RNAs were transfected into En5-3 cells in the absence of G418 selection. Supernatant culture medium was collected at 24-h intervals to monitor the amplification of the transfected RNA. As shown in Fig. 6B, each of the transfected RNAs generated a high level of SEAP expression by 24 h following transfection. This represents the expression of Tat by translation of the transfected RNA, independent of RNA replication, and was observed also with an NTat-T631 variant (Fig. 6B; ΔGDD) encoding a replication-lethal deletion in the NS5B polymerase (51). Within 72 h, SEAP expression levels in cells transfected with replication-defective RNAs begin to fall, while they are significantly increased in cells transfected with efficiently replicating RNAs. As shown in Fig. 6B, cells transfected with the wild-type NTat-T631 expressed approximately one-third more SEAP than cells transfected with NTat-T631C. On the other hand, NTat-T631C induced SEAP expression approximately 10 times more efficiently than NTat-T631V, while NTat-T631I was indistinguishable from the replication-incompetent ΔGDD mutant. These results, which were reproduced in separate experiments with independent RNA transcripts, are remarkably similar to the differences in CFE observed with the Con1 replicons (Fig. 5) and confirm that the T631C substitution significantly reduces the replication fitness of HCV replicons, although it does not impede processing at the NS3-4A junction.
Using similar techniques, we also evaluated the effect of a D441Y substitution on the ability of NTat-T631 and NTat-T631C RNAs to replicate in En5-3 cells. Surprisingly, the inclusion of this substitution appeared to eliminate the replication capacity of these HCV-N derived RNAs, irrespective of whether the T631C mutation was present or not (data not shown). These preliminary results suggest that, if the D441Y substitution helps to compensate for the T631C mutation in the Con1 strain replicons studied in the experiments depicted in Fig. 5, it does so in a manner that is HCV strain specific and not tolerated by the HCV-N protein.
DISCUSSION
Positive-strand RNA viruses are noted for their high frequency of mutations. For HCV, this mutation rate (a frequency of misincorporated bases approximating 1 in 104 [11]), when considered in the context of its high level of replication (1012 genome copies per day in a typical infected individual [33]), suggests that the conservation of an amino acid residue within the polyprotein would require strong positive selection (10). The observation that the key residue of the recognition sequence in the autoproteolytic cis-active NS3-4A junction is different from that found in all three trans-active cleavage sites—in all 250 sequences reported to date—implies that this distinction is essential for viral fitness in vivo. The studies reported here were carried out in an effort to understand the nature of the selection pressures that compel this distinction without recourse to either an authentic viral culture system or a tractable animal model.
Insights from previous biochemical and biophysical studies (summarized in Tables 3 and 4) suggest that the critical P1 residue of the NS3 substrate can accommodate a variety of small amino acid residues but that cysteine is by far the optimal choice. Peptide cleavage studies, in a variety of contexts, consistently indicate that replacement of cysteine at the P1 residue of trans cleavage sites by threonine renders the substrate significantly more refractory to cleavage (46, 52). Threonine is among the preferred amino acids in the P1 position of the cis cleavage site of NS3-4A (4, 22, 26) and results in a substrate that is efficiently autoproteolytically processed. These results are similar to the studies presented above on the processing of the T-54 and C-54 in vitro translation products (Fig. 2A). However, a much more stringent requirement for the NS4A cofactor domain for proteolytic cleavage of the NS3-4A P1 threonine construct than for cleavage of the related P1 cysteine mutant was noted (Fig. 1C). Despite this, threonine is strictly conserved among all reported HCV sequences at the P1 position of the NS3-4A junction, while, with equal fidelity, a P1 cysteine is conserved at all three downstream trans-active cleavage targets. The only logical inference is that less-efficient cleavage at the NS3-4A junction is desired or that some other aspect of the conserved P1 threonine is so critical as to compel its selection. The studies reporting this finding, together with previous findings, suggest that both of these rationales may help explain the choice of threonine for the autoproteolytic site.
TABLE 3.
Summary of published results on P1 changes at HCV NS3-4A cis cleavage sites
| Authors (yr) | Model | Results
|
|
|---|---|---|---|
| Amino acid(s) | Effect on cleavagea | ||
| Leinbach et al. (1994) | In vitro-translated NS2-NS3-NS4 | Thr, Cys, Ser, Gly, Ala, Thr, Leu | Cleavage |
| Arg, Tyr | No cleavage | ||
| Kolykhalov et al. (1994) | Vaccinia virus T7 Pol + NS2-NS5B | Thr | 100 |
| Arg | No cleavage | ||
| Gly | ∼33 | ||
| Asn | ∼66 | ||
| Asp | <33 | ||
| Bartenschlager et al. (1995) | Vaccinia virus T7 Pol + NS3-NS5B | Thr | ++ |
| Ile | ± | ||
| Asn | ++ | ||
| Cys | ++ | ||
| Phe | + | ||
| Val | ++ | ||
| Ala | ++ | ||
TABLE 4.
Summary of published results on P1 changes at HCV NS3-4A trans cleavage sites
| Authors (yr) | Model | P1 residue | NS3 + NS4A
|
NS3 (no NS4A)
|
||||
|---|---|---|---|---|---|---|---|---|
| Km (μM) | kcat (min−1) | kcat/Km (M−1/s−1) | Km (μM) | kcat (min−1) | kcat/Km (M−1/s−1) | |||
| Urbani et al. (1997) | E. coli-produced NS3 + NS4A-4B peptide | Cys | 43 ± 4 | 1 ± 0.1 | 545 | 105 ± 20 | 0.3 ± 0.03 | 48 |
| hCysa | 54 ± 13 | 0.3 ± 0.01 | 96 | 312 ± 59 | 0.1 ± 0.01 | 5 | ||
| Abue | 97 ± 29 | 0.17 ± 0.01 | 30 | 288 ± 30 | 0.03 ± 0.01 | 2 | ||
| Thr | 235 ± 37 | 0.2 ± 0.01 | 15 | 244 ± 55 | 0.005 ± 0.001 | 0.3 | ||
| Val | 153 ± 54 | 0.009 ± 0.001 | 1 | NDb | ND | ND | ||
| Ser | NCd | NC | NC | NC | NC | NC | ||
| Zhang et al. (1997) | Baculovirus-produced sc-NS3(1-631)/4A(1-54) + NS5A-5B peptide | Cysc | 30 | 8.9 | 5,000 | |||
| hCysc | 70 | 1.0 | 240 | |||||
| Alac | 240 | 0.5 | 35 | |||||
| Thrc | 710 | 1.2 | 28 | |||||
| Metc | 78 | 0.1 | 21 | |||||
| Serc | 530 | 0.1 | 3 | |||||
hCys, l-homocysteine.
ND, not determined (kcat/Km < 0.05 M−1 s−1).
Substrate was modified at the P2 position (Cys to Ala) to minimize synthesis and oxidation problems.
NC, no cleavage.
Abu, α-aminobutyric acid.
Maturation of the nonstructural portion of the viral polyprotein and efficient assembly of the replicase complex on intracellular membranes are critical events in viral replication (1). Previous studies strongly suggest that the bifunctional NS3 protease-helicase is an integral part of the replicase and that it is most likely anchored to the ER by the N terminus of the cofactor NS4A (18). This is accomplished by the intercalation of residues 21 to 33 of NS4A into the N-terminal β-barrel of the protease domain (20, 31, 48). This intercalation simultaneously “perfects” the protease geometry and activates the protease, increasing its catalytic activity significantly (9, 28, 38, 40, 43, 45).
Results from the IVTT studies described in this report suggest that autoproteolysis requires the fully activated enzyme, i.e., intercalation of a functional NS4A cofactor region. Replacement of the threonine by the more proteolytically sensitive cysteine appears to allow autoproteolysis to occur even in the absence of a functional NS4A central cofactor domain (Fig. 1C), implying that in vivo proteolysis of such a mutant genome might occur without prior intercalation or activation of the protease. Without intercalation of the NS4A cofactor the NS3 protein would not be localized to the ER, but, more importantly, following scission of the NS3-NS4 junction the remainder of the polyprotein—the target of proteolytic activity—would no longer be compelled to associate with the NS3 protease. Because a catalytically refractory cleavage site is maintained at the protease C terminus, the full activation of the protease (i.e., intercalation of NS4A) is required; this ensures that NS3 remains associated with the ER and that its subsequent substrates remain associated with the protease, thereby rendering subsequent proteolytic reactions unimolecular (or at least constrained to a single, noncovalent molecular complex), which facilitates the complete processing of the polyprotein. The improved efficiency of such a “pseudo-cis” mechanism is incalculable, especially during the initial phase of infection, when polyprotein concentration is vanishingly low.
This argument is predicated on the notion that the IVTT studies reflect, to some extent, the differences in autoproteolytic rates one might observe in vivo. As indicated above, previous qualitative studies based on vaccinia virus expression of NS3-4A suggested that several amino acids, including cysteine, yield readily cleavable NS3-4A junctions (2, 4). This implies that polyprotein folding, rather than the P1 residue, may be the overriding force determining the efficiency of proteolysis. However, even though proximity effects may significantly enhance autoproteolysis, rendering many amino acids qualitatively acceptable in such a system, unimolecular reactions are nevertheless still affected by substrate specificity (8, 25). One must therefore assume that the replacement of threonine by the biochemically superior cysteine would further accelerate proteolysis, even in the context of true autoproteolysis.
Recent studies by Steinkuhler and colleagues (39) point to another possible deleterious aspect of the incorporation of the proteolytically more facile cysteine into the NS3-4A junction. These in vitro studies suggest that the P side fragments of the NS4A-4B, NS4B-5A, and NS5A-5B cleavages, which bear C-terminal cysteine residues, are significantly more inhibitory to the protease than is the threonine-bearing NS3 C-terminal peptide. This difference in affinity appears to be primarily due to the P1 residue, as evidenced by the difference in Km for peptide substrates in which the P1 cysteine residue has been replaced by threonine (46, 52) (Table 4). This suggests that a T631C NS3 protein, in addition to being catalytically enhanced, would also generate a C terminus that would be a more effective inhibitor of the protease, which in turn could compete with substrate binding for subsequent proteolytic events. The ability of the C terminus of NS3 to efficiently interact with and/or bind to the active site of the protease domain is suggested by the crystal structure of sc-NS4A21-32-GSGS-NS33-631 (49). Together, these observations provide two related and compelling rationales for why the catalytically superior cysteine residue is by no means preferred from an evolutionary standpoint. But can this be addressed experimentally?
The most germane system available for confirming the impact of mutations on viral fitness are the HCV replicons (7, 23, 30). While these dicistronic, subgenomic RNAs do not model all events in the viral life cycle, they do recapitulate the events involved in replication of the viral RNA, including the NS3-4A processing of the nonstructural segment of the polyprotein. Incorporation of the T631C mutation into a subgenomic replicon derived from the Con1 strain of HCV resulted in a modest, but statistically significant (P < 0.01), decrease in its ability to induce the selection of G418-resistant cell clones (Fig. 5). A quantitatively similar decrease in the replication capacity of a second HCV replicon derived from the HCV-N virus strain was observed. Although HCV-N is, like Con1, a genotype 1b virus, the NTat-T631 replicon has superior replication characteristics. Like that of its progenitor, NTat2ANeo(SI) (50), NTat-T631 replication can be quantitatively monitored by measurement of SEAP secreted from transfected cells and is sufficiently robust to be monitored directly after transfection in the absence of G418 selection. The decrease in CFE of the Con1 T631C mutant replicon (Fig. 5B) was comparable to the reduction of 27 to 45% in the expression of SEAP, compared with that for the wild-type NTat-T631 replicon, following transfection of the mutant NTat-T631C in replicate experiments (Fig. 6B). Other mutations, such as T631I and T631V, that strongly impeded cleavage of the NS3-4A junction (Fig. 6A) either completely eliminated or severely reduced SEAP expression and hence replication of the replicon RNA (Fig. 6B). In light of the similarities in the impact on replication fitness of the Con1 and HCV-N replicons, it is interesting that the amino acid sequences of the nonstructural proteins of these HCV strains differ at 6% of residues (17). The fact that the T631C mutation has a small, statistically significant, reproducible impact on replication fitness is in itself sufficient to suggest that over the course of repeated cycles of maturation and replication in vivo a dramatic survival advantage would be conferred (11) to genomes encoding threonine at position 631.
Taken together, the biochemical data, the impact on CFE of the Con1 replicon, and decreased replication of the NTat-T631C mutant, coupled with the absence of variation in position 631 in clinical isolates (34), make it appear that the catalytically optimal residue at the P1 position of the NS3-4A junction is not the best choice from the point of view of viral fitness. As with many evolutionary problems, definitive proof may never be found, but the supporting biochemical, biophysical, and ex vivo evidence clearly suggests that threonine is the residue of choice and that this is due to its ability to confer an optimal reduction in proteolytic susceptibility that will ensure NS3-4A assembly, polyprotein maturation, and replicase assembly.
Acknowledgments
We thank Nanhua Yao for his helpful discussions, and Nancy Butkiewicz for her technical advice. We also thank Xiao Tong for critical reading of the manuscript.
This work was supported in part by grants from the National Institute of Allergy and Infectious Diseases (U19-AI140035) and the Schering-Plough Research Institute (S.M.L.).
REFERENCES
- 1.Ali, N., K. D. Tardif, and A. Siddiqui. 2002. Cell-free replication of the hepatitis C virus subgenomic replicon. J. Virol. 76:12001-12007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartenschlager, R., L. Ahlborn-Laake, J. Mous, and H. Jacobsen. 1994. Kinetic and structural analysis of hepatitis C virus polyprotein processing. J. Virol. 68:5045-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartenschlager, R., L. Ahlborn-Laake, J. Mous, and H. Jacobsen. 1993. Nonstructural protein 3 of the hepatitis C virus encodes a serine-type proteinase required for cleavage at the NS3/4 and NS4/5 junctions. J. Virol. 67:3835-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartenschlager, R., L. Ahlborn-Laake, K. Yasargil, J. Mous, and H. Jacobsen. 1995. Substrate determinants for cleavage in cis and in trans by the hepatitis C virus NS3 proteinase. J. Virol. 69:198-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartenschlager, R., V. Lohmann, T. Wilkinson, and J. O. Koch. 1995. Complex formation between the NS3 serine-type proteinase of the hepatitis C virus and NS4A and its importance for polyprotein maturation. J. Virol. 69:7519-7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beard, M. R., G. Abell, M. Honda, A. Carroll, M. Gartland, B. Clarke, K. Suzuki, R. Lanford, D. V. Sangar, and S. M. Lemon. 1999. An infectious molecular clone of a Japanese genotype 1b hepatitis C virus. Hepatology 30:316-324. [DOI] [PubMed] [Google Scholar]
- 7.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 8.Bruice, T. C., and S. J. Benkovic. 2000. Chemical basis for enzyme catalysis. Biochemistry 39:6267-6274. [DOI] [PubMed] [Google Scholar]
- 9.Butkiewicz, N. J., M. Wendel, R. Zhang, R. Jubin, J. Pichardo, E. B. Smith, A. M. Hart, R. Ingram, J. Durkin, P. W. Mui, M. G. Murray, L. Ramanathan, and B. Dasmahapatra. 1996. Enhancement of hepatitis C virus NS3 proteinase activity by association with NS4A-specific synthetic peptides: identification of sequence and critical residues of NS4A for the cofactor activity. Virology 225:328-338. [DOI] [PubMed] [Google Scholar]
- 10.DeFilippis, V. R., and L. P. Villarreal. 2001. Virus evolution, p. 353-370. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 11.Domingo, E., and J. Holland. 1997. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 51:151-178. [DOI] [PubMed] [Google Scholar]
- 12.Failla, C., L. Tomei, and R. De Francesco. 1995. An amino-terminal domain of the hepatitis C virus NS3 protease is essential for interaction with NS4A. J. Virol. 69:1769-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Failla, C., L. Tomei, and R. De Francesco. 1994. Both NS3 and NS4A are required for proteolytic processing of hepatitis C virus nonstructural proteins. J. Virol. 68:3753-3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grakoui, A., D. W. McCourt, C. Wychowski, S. M. Feinstone, and C. M. Rice. 1993. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J. Virol. 67:2832-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hijikata, M., H. Mizushima, T. Akagi, S. Mori, N. Kakiuchi, N. Kato, T. Tanaka, K. Kimura, and K. Shimotohno. 1993. Two distinct proteinase activities required for the processing of a putative nonstructural precursor protein of hepatitis C virus. J. Virol. 67:4665-4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hijikata, M., H. Mizushima, Y. Tanji, Y. Komoda, Y. Hirowatari, T. Akagi, N. Kato, K. Kimura, and K. Shimotohno. 1993. Proteolytic processing and membrane association of putative nonstructural proteins of hepatitis C virus. Proc. Natl. Acad. Sci. USA 90:10773-10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ikeda, M., M. Yi, K. Li, and S. M. Lemon. 2002. Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells. J. Virol. 76:2997-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishido, S., T. Fujita, and H. Hotta. 1998. Complex formation of NS5B with NS3 and NS4A proteins of hepatitis C virus. Biochem. Biophys. Res. Commun. 244:35-40. [DOI] [PubMed] [Google Scholar]
- 19.Jayaraman, K., S. A. Fingar, J. Shah, and J. Fyles. 1991. Polymerase chain reaction-mediated gene synthesis: synthesis of a gene coding for isozyme c of horseradish peroxidase. Proc. Natl. Acad. Sci. USA 88:4084-4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, J. L., K. A. Morgenstern, C. Lin, T. Fox, M. D. Dwyer, J. A. Landro, S. P. Chambers, W. Markland, C. A. Lepre, E. T. O'Malley, S. L. Harbeson, C. M. Rice, M. A. Murcko, P. R. Caron, and J. A. Thomson. 1996. Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide. Cell 87:343-355. (Erratum, 89:159, 1997.) [DOI] [PubMed] [Google Scholar]
- 21.Koch, J. O., V. Lohmann, U. Herian, and R. Bartenschlager. 1996. In vitro studies on the activation of the hepatitis C virus NS3 proteinase by the NS4A cofactor. Virology 221:54-66. [DOI] [PubMed] [Google Scholar]
- 22.Kolykhalov, A. A., E. V. Agapov, and C. M. Rice. 1994. Specificity of the hepatitis C virus NS3 serine protease: effects of substitutions at the 3/4A, 4A/4B, 4B/5A, and 5A/5B cleavage sites on polyprotein processing. J. Virol. 68:7525-7533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lahser, F., J. Wright-Minogue, A. Skelton, and B. Malcolm. 2003. Quantitative estimation of viral fitness using pyrosequencing. BioTechniques 34:26-28. [DOI] [PubMed] [Google Scholar]
- 24.Landro, J. A., S. A. Raybuck, Y. P. C. Luong, E. T. O'Malley, S. L. Harbeson, K. A. Morgenstern, G. Rao, and D. J. Livingston. 1997. Mechanistic role of an NS4A peptide cofactor with the truncated NS3 protease of hepatitis C virus: elucidation of the NS4A stimulatory effect via kinetic analysis and inhibitor mapping. Biochemistry 36:9340-9348. [DOI] [PubMed] [Google Scholar]
- 25.Lau, E. Y., and T. C. Bruice. 1999. Consequences of breaking the Asp-His hydrogen bond of the catalytic triad: effects on the structure and dynamics of the serine esterase cutinase. Biophys. J. 77:85-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leinbach, S. S., R. A. Bhat, S. M. Xia, W. T. Hum, B. Stauffer, A. R. Davis, P. P. Hung, and S. Mizutani. 1994. Substrate specificity of the NS3 serine proteinase of hepatitis C virus as determined by mutagenesis at the NS3/NS4A junction. Virology 204:163-169. [DOI] [PubMed] [Google Scholar]
- 27.Lin, C., B. M. Pragai, A. Grakoui, J. Xu, and C. M. Rice. 1994. Hepatitis C virus NS3 serine proteinase: trans-cleavage requirements and processing kinetics. J. Virol. 68:8147-8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin, C., J. A. Thomson, and C. M. Rice. 1995. A central region in the hepatitis C virus NS4A protein allows formation of an active NS3-NS4A serine proteinase complex in vivo and in vitro. J. Virol. 69:4373-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lohmann, V., F. Korner, A. Dobierzewska, and R. Bartenschlager. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 75:1437-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 31.Love, R. A., H. E. Parge, J. A. Wickersham, Z. Hostomsky, N. Habuka, E. W. Moomaw, T. Adachi, and Z. Hostomska. 1996. The crystal structure of hepatitis C virus NS3 proteinase reveals a trypsin-like fold and a structural zinc binding site. Cell 87:331-342. [DOI] [PubMed] [Google Scholar]
- 32.Miller, R. H., and R. H. Purcell. 1990. Hepatitis C virus shares amino acid sequence similarity with pestiviruses and flaviruses as well as members of two plant virus supergroups. Proc. Natl. Acad. Sci. USA 87:2057-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neumann, A. U., N. P. Lam, H. Dahari, D. R. Gretch, T. E. Wiley, T. J. Layden, and A. S. Perelson. 1998. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science 282:103-107. [DOI] [PubMed] [Google Scholar]
- 34.Qiu, P., X. Y. Cai, L. Wang, J. R. Greene, and B. Malcolm. 25September2002, posting date. Hepatitis C virus whole genome position weight matrix and robust primer design. BMC Microbiol. 2:29. [Online.] http://www.biomedcentral.com/1471-2180/2/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reed, K. E., and C. M. Rice. 1998. Molecular characterization of hepatitis C virus. Curr. Stud. Hematol. Blood Transfus. 62:1-37. [DOI] [PubMed] [Google Scholar]
- 36.Rosa, C., S. Osborne, F. Garetto, S. Griva, A. Rivella, G. Calabresi, R. Guaschino, and F. Bonelli. 1995. Epitope mapping of the NS4 and NS5 gene products of hepatitis C virus and the use of a chimeric NS4-NS5 synthetic peptide for serodiagnosis. J. Virol. Methods 55:219-232. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Shimizu, Y., K. Yamaji, Y. Masuho, T. Yokota, H. Inoue, K. Sudo, S. Satoh, and K. Shimotohno. 1996. Identification of the sequence on NS4A required for enhanced cleavage of the NS5A/5B site by hepatitis C virus NS3 protease. J. Virol. 70:127-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steinkuhler, C., G. Biasiol, M. Brunetti, A. Urbani, U. Koch, R. Cortese, A. Pessi, and R. De Francesco. 1998. Product inhibition of the hepatitis C virus NS3 protease. Biochemistry 37:8899-8905. [DOI] [PubMed] [Google Scholar]
- 40.Steinkuhler, C., A. Urbani, L. Tomei, G. Biasiol, M. Sardana, E. Bianchi, A. Pessi, and R. De Francesco. 1996. Activity of purified hepatitis C virus protease NS3 on peptide substrates. J. Virol. 70:6694-6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takamizawa, A., C. Mori, I. Fuke, S. Manabe, S. Murakami, J. Fujita, E. Onishi, T. Andoh, I. Yoshida, and H. Okayama. 1991. Structure and organization of the hepatitis C virus genome isolated from human carriers. J. Virol. 65:1105-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanji, Y., M. Hijikata, S. Satoh, T. Kaneko, and K. Shimotohno. 1995. Hepatitis C virus-encoded nonstructural protein NS4A has versatile functions in viral protein processing. J. Virol. 69:1575-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taremi, S. S., B. Beyer, M. Maher, N. Yao, W. Prosise, P. C. Weber, and B. A. Malcolm. 1998. Construction, expression, and characterization of a novel fully activated recombinant single-chain hepatitis C virus protease. Protein Sci. 7:2143-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tomei, L., C. Failla, E. Santolini, R. De Francesco, and N. La Monica. 1993. NS3 is a serine protease required for processing of hepatitis C virus polyprotein. J. Virol. 67:4017-4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tomei, L., C. Failla, R. L. Vitale, E. Bianchi, and R. De Francesco. 1996. A central hydrophobic domain of the hepatitis C virus NS4A protein is necessary and sufficient for the activation of the NS3 protease. J. Gen. Virol. 77:1065-1070. [DOI] [PubMed] [Google Scholar]
- 46.Urbani, A., E. Bianchi, F. Narjes, A. Tramontano, R. De Francesco, C. Steinkuhler, and A. Pessi. 1997. Substrate specificity of the hepatitis C virus serine protease NS3. J. Biol. Chem. 272:9204-9209. [DOI] [PubMed] [Google Scholar]
- 47.Wang, W., and B. A. Malcolm. 1999. Two-stage PCR protocol allowing introduction of multiple mutations, deletions and insertions using QuikChange site-directed mutagenesis. BioTechniques 26:680-682. [DOI] [PubMed] [Google Scholar]
- 48.Yan, Y., Y. Li, S. Munshi, V. Sardana, J. L. Cole, M. Sardana, C. Steinkuehler, L. Tomei, R. De Francesco, L. C. Kuo, and Z. Chen. 1998. Complex of NS3 protease and NS4A peptide of BK strain hepatitis C virus: a 2.2 ÅA resolution structure in a hexagonal crystal form. Protein Sci. 7:837-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yao, N., P. Reichert, S. S. Taremi, W. W. Prosise, and P. C. Weber. 1999. Molecular views of viral polyprotein processing revealed by the crystal structure of the hepatitis C virus bifunctional protease-helicase. Struct. Fold Des. 7:1353-1363. [DOI] [PubMed] [Google Scholar]
- 50.Yi, M., F. Bodola, and S. M. Lemon. 2002. Subgenomic hepatitis C virus replicons inducing expression of a secreted enzymatic reporter protein. Virology 304:197-210. [DOI] [PubMed] [Google Scholar]
- 51.Yi, M., and S. M. Lemon. 2003. Structure-function analysis of the 3′ stem-loop of hepatitis C virus genomic RNA and its role in viral RNA replication. RNA 9:331-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang, R., J. Durkin, W. T. Windsor, C. McNemar, L. Ramanathan, and H. V. Le. 1997. Probing the substrate specificity of hepatitis C virus NS3 serine protease by using synthetic peptides. J. Virol. 71:6208-6213. [DOI] [PMC free article] [PubMed] [Google Scholar]