Abstract
The species Human enterovirus B (HEV-B) in the family Picornaviridae consists of coxsackievirus A9; coxsackieviruses B1 to B6; echoviruses 1 to 7, 9, 11 to 21, 24 to 27, and 29 to 33; and enteroviruses 69 and 73. We have determined complete genome sequences for the remaining 22 HEV-B serotypes whose sequences were not represented in public databases and analyzed these in conjunction with previously available complete sequences in GenBank. Members of HEV-B were monophyletic relative to all other human enterovirus species in all regions of the genome except in the 5′-nontranslated region (NTR), where they are known to cluster with members of HEV-A. Within HEV-B, phylogenies constructed from the structural (P1) and nonstructural regions of the genome (P2 and P3) are incongruent, suggesting that recombination had occurred. Similarity plots and bootscanning analysis across the complete genome identified multiple sites at which the phylogeny of a given strain's sequence shifted, indicating potential recombination points. These points are distributed in the 5′-NTR and throughout P2 and P3, but no sites with >80% bootstrap support were identified within the capsid. Individual sequence comparisons and phylogenetic analyses suggest that members of HEV-B have recombined with one another on multiple occasions, resulting in a complex mosaic of sequences derived from multiple parental viruses in the nonstructural regions of the genome. We conclude that RNA recombination is a common mechanism for enterovirus evolution and that recombination within the nonstructural regions of the genome (P2 and P3) has been observed only among members of the same species.
The human enteroviruses were originally classified into four categories on the basis of human disease and virulence/pathogenesis in intracranially inoculated suckling mice: (i) polioviruses (PV; agents of human poliomyelitis, PV are generally not pathogenic in mice), (ii) coxsackie A viruses (CAV; associated with human central nervous system disease and herpangina, CAV cause flaccid paralysis in mice), (iii) coxsackie B viruses (CBV; associated with human central nervous system and cardiac disease, CBV cause spastic paralysis in mice), and (iv) echoviruses (E; not pathogenic in mice and originally not associated with known human disease) (11, 43). It quickly became apparent, however, that this scheme was inadequate for the classification of human enteroviruses, because viruses pathogenic in mice were isolated that were serotypically identical to known echoviruses, and the echoviruses were soon shown to be associated with a wide range of human diseases (9, 11). Thereafter, new human enterovirus serotypes were simply named “enterovirus” and numbered sequentially, starting with EV68 (46, 80). A total of 64 serotypes are currently recognized (34), and additional serotypes have been proposed (53, 54, 57). In the present classification scheme, which takes into account both biological and molecular properties of the viruses, the human enteroviruses are divided among five species: (i) Poliovirus (PV1 to PV3), (ii) Human enterovirus A (HEV-A; CAV2 to CAV8, CAV10, CAV12, CAV14, CAV16, and EV71), (iii) HEV-B (CAV9, CBV1 to CBV6, E1 to E7, E9, E11 to E21, E24 to E27, E29 to E33, and EV69 and E73), (iv) HEV-C (CAV1, CAV11, CAV13, CAV15, CAV17 to CAV22, and CAV24), and (v) HEV-D (EV68 and EV70) (34). The close genetic relationship between the polioviruses and members of HEV-C suggests that they should be considered a single species (4, 26, 62).
The enterovirus genome is a single-stranded, polyadenylated, positive-sense RNA of ca. 7.4 kb, with a 22-amino-acid virus-encoded protein (3BVPg) covalently linked to the 5′ end. Flanked by 5′- and 3′-nontranslated regions (NTRs), the single long open reading frame encodes a polyprotein of ca. 2,200 amino acids that is processed during and after translation by viral proteases to yield the mature viral polypeptides. The P1 region encodes the capsid proteins 1A to 1D (VP4, VP2, VP3, and VP1, respectively). P2 encodes a protease, 2Apro, and two proteins involved in RNA replication and shutdown of host cell expression, 2B and 2C. The 3BVPg precursor (3AB), the major viral protease (3Cpro), and the RNA-dependent RNA polymerase (3Dpol) are encoded in the P3 region. Complete genome sequences were previously available for CAV9, for each of the six CBV serotypes, for echoviruses 1, 5, 6, 9, 11, 12, 18, and 30, and for EV73, but only partial sequences were available for the remaining 21 members of HEV-B. Phylogenetic analyses of available sequences have shown that the members of HEV-B are closely related to one another in multiple regions of the genome (26, 55, 56, 62), but the full extent and details of the relationship have not been described.
Picornavirus genetic recombination was originally identified in poliovirus, the prototype enterovirus (25, 36), and was also shown to occur in picornaviruses of other genera (63). Both replicative (template-switching) and nonreplicative (strand breakage and rejoining) mechanisms have been proposed, and both mechanisms are supported by in vitro experimental studies (20, 35). The most detailed studies of enterovirus recombination during natural infection and circulation have been with wild or vaccine-derived polioviruses (7, 12, 19, 28, 38). Analysis of available enterovirus complete genome sequences has suggested that recombination also plays a role in the evolution of the nonpolio enteroviruses (1, 58, 72). We present here the first analysis of the complete genome sequences of all members of HEV-B. Individual sequence comparisons and phylogenetic analyses suggest that members of HEV-B have recombined with one another on multiple occasions, resulting in a complex mosaic of sequences in the nonstructural regions of the genome.
MATERIALS AND METHODS
Viruses.
The prototype strains of echoviruses 2 to 4, 6 to 7, 13 to 17, 19 to 21, 24 to 27, and 29, 31 to 33 and of EV69 were obtained as National Institutes of Health research reference reagents from the National Institute of Allergy and Infectious Diseases (Bethesda, Md.) (Table 1) and propagated in cell culture by standard methods (52). These materials are now distributed by the American Type Culture Collection (Manassas, Va.).
TABLE 1.
Viruses analyzed
| Typea | Strain | Locationb | Yr | Accession no.c | Reference(s)d |
|---|---|---|---|---|---|
| CAV9 | Griggs | USA/MA | NKe | D00627 | 6, 45, 67 |
| CBV1 | Conn-5 | USA/CT | 1948 | M16560 | 14, 50 |
| CBV2 | Ohio-1 | USA/OH | 1947 | AF085363 | 14, 49 |
| CBV3 | Nancy | USA/CT | 1949 | M16560 | 14, 48 |
| CBV4 | JVB | USA/NY | 1951 | X05690 | 14, 74 |
| CBV5 | Faulkner | USA/KY | 1952 | AF114383 | 74 |
| CBV6 | Schmitt | PHL | 1953 | AF105342 | 23 |
| E1 | Farouk | EGY | 1951 | AF029859 | 42, 45, 47 |
| E2* | Cornelis | USA/CT | 1951 | AY302545 | 42, 45 |
| E3* | Morrisey | USA/CT | 1951 | AY302553 | 42, 45 |
| E4* | Pesacek | USA/CT | 1951 | AY302557 | 42 |
| E5 | Noyce | USA/ME | 1954 | AF083069 | 42, 45 |
| E6* | D'Amori | USA/RI | 1955 | AY302558 | 15, 42 |
| E7* | Wallace | USA/OH | 1953 | AY302559f | 64, 66, 78 |
| E9 | Hill | USA/OH | 1953 | X92886 | 64, 66, 78 |
| E11 | Gregory | USA/OH | 1953 | X80059 | 13, 64, 66, 78 |
| E12 | Travis | PHL | 1953 | X79047 | 10, 22 |
| E13* | Del Carmen | PHL | 1953 | AY302539 | 10, 24 |
| E14* | Tow | USA/RI | 1954 | AY302540 | 11, 44 |
| E15* | CH 96-51 | USA/WV | 1951 | AY302541 | 11, 59 |
| E16* | Harrington | USA/MA | 1951 | AY302542 | 11, 30 |
| E17* | CHHE-29 | MEX | NKg | AY302543 | 11, 78 |
| E18 | Metcalf | USA/OH | 1955 | AF317694 | 11, 65, 78 |
| E19* | Burke | USA/OH | 1955 | AY302544 | 11, 65 |
| E20* | JV-1 | USA/DC | 1956 | AY302546 | 70 |
| E21* | Farina | USA/MA | 1950 | AY302547 | 67 |
| E24* | DeCamp | USA/OH | 1956 | AY302548 | 79 |
| E25* | JV-4 | USA/DC | 1957 | AY302549 | 70 |
| E26* | Coronel | PHL | 1953 | AY302550 | 23 |
| E27* | Bacon | PHL | 1953 | AY302551 | 23 |
| E29* | JV-10 | USA/DC | 1958 | AY302552 | 60, 69 |
| E30 | Bastianni | USA/NY | 1958 | AF162711 | 60, 61 |
| E31* | Caldwell | USA/KS | 1955 | AY302554 | 60, 77 |
| E32* | PR-10 | PUR | 1961 | AY302555 | 3, 60 |
| E33* | Toluca-3 | MEX | 1959 | AY302556 | 68 |
| EV69* | Toluca-1 | MEX | 1959 | AY302560 | 51 |
| EV73 | Henderson | USA/CA | 1955 | AF241359 | 57 |
Strains sequenced in the present study are indicated by an asterisk.
Locations are indicated by three-letter country code and, for the United States (USA), by two-letter state codes. EGY, Egypt; MEX, Mexico; PHL, Philippines; PUR, Puerto Rico.
GenBank accession number for complete genome sequence.
Reference(s) for original isolation and characterization of the prototype strain.
NK, year not known. CAV9-Griggs (sometimes called Grigg) first appeared explicitly in the literature in 1955 (45).
Our E7-Wallace sequence differed from the published sequence AY036579 (8) at 11 sites: 1 in the 5′-NTR, 9 in the capsid region, and 1 in 2A. Four of the capsid differences were synonymous transitions. The remaining capsid differences and the difference in 2A resulted in amino acid changes, all of which were in regions that are highly variable among the viruses in HEV-B. The difference in the 5′-NTR also occurred at a site that is highly variable, suggesting that the discordant sequences may be due to differences in passage history of the viruses sequenced in the two laboratories.
NK, year not known. E17-CHHE-29 first appeared explicitly in the literature in 1957 (11).
Nucleotide sequencing.
Complete genomic sequences were determined for each of the 22 strains indicated in Table 1, including 21 serotypes not previously available and the E6 prototype strain, D'Amori. During the course of this work, a complete sequence also became available for E7-Wallace (8). Overlapping fragments representing each complete viral genome were amplified by reverse transcription-PCR with degenerate, inosine-containing primers designed to anneal to sites encoding amino acid motifs that are highly conserved among enteroviruses. Specific, nondegenerate primers were designed from preliminary sequences to close gaps between the original PCR products. The PCR products were purified for sequencing by using a High-Pure PCR product purification kit (Roche Molecular Biochemicals, Indianapolis, Ind.). Both strands were sequenced by automated methods with fluorescent dideoxy-chain terminators (Applied Biosystems, Foster City, Calif.). The complete genome sequences for CAV9, CBV1 to CBV6, E1, E5, E9, E11, E12, E18, and E30 were obtained from GenBank. The sequence of EV73 was previously determined in our laboratory (57).
Sequence analysis.
The pairwise sequence identities among the nucleotide and deduced amino acid sequences of all of the HEV-B serotypes were calculated by using the programs Gap and Distances (Wisconsin Sequence Analysis Package, version 10.2; Genetics Computer Group, Inc., Madison, Wis.). Nucleotide sequences were aligned by using the Pileup program (Wisconsin Package) and adjusted manually to conform to the optimized alignment of deduced amino acid sequences. The overall variability in the amino acid sequences was visualized by using the Wisconsin Package program, Plotsimilarity, with a window size of 10 residues advanced along the polyprotein alignment in one-residue steps. Phylogenetic relationships were inferred from the aligned nucleic acid sequences by the neighbor-joining method implemented in the programs DNADist and Neighbor (PHYLIP [Phylogeny Inference Package], version 3.57; University of Washington, Seattle) using the Kimura two-parameter substitution model (31) and a transition/transversion ratio of 10. Support for specific tree topologies was estimated by bootstrap analysis with 1,000 pseudoreplicate data sets. Branch lengths in consensus trees were calculated by the maximum-likelihood quartet-puzzling method, using the nucleotide substitution model of Tamura and Nei (76) as implemented in Tree-Puzzle 5.0 (75). Similarity plots depicting the relationships among the aligned nucleotide sequences were generated by using SimPlot, version 3.2 beta (39). Similarity was calculated in each window of 200 nucleotides by the Kimura two-parameter method (31) with a transition/transversion ratio of 10. The window was successively advanced along the genome alignment in 20-nucleotide increments. To assess potential recombinational relationships, aligned sequences were subsequently analyzed by using the bootscanning method implemented in SimPlot. Phylogenetic trees were generated for each 200-nucleotide window by the neighbor-joining method, with DNADist and Neighbor, and the bootstrap values for all possible sequence comparisons were plotted as a function of genome position.
Nucleotide sequence accession numbers.
The sequences reported here were deposited in the GenBank sequence database under accession numbers AY302539 to AY302560.
RESULTS
General genome features.
The 22 newly sequenced genomes vary in length from 7,394 nucleotides (E4 and E33) to 7,451 nucleotides (E14), which is within the length range among the previously sequenced HEV-B genomes (7,348 to 7,452 nucleotides). The genomes of all of the HEV-B viruses contain 46.4 to 48.7% G+C residues.
Noncoding region comparisons.
The 5′-NTR sequences are 738 to 750 nucleotides long and differ from one another by 5 to 23% (Table 2), with E12 and E32 the most distant from one another. Forty-seven percent of the 5′-NTR residues are invariant among all of the viruses, and almost 30% (101 of 356) of the variable sites are concentrated in the hypervariable region, the 80 to 110 residues immediately upstream of the initiation codon (data not shown). Structural elements that are important for the function of the internal ribosome entry site are well conserved among the HEV-B serotypes (data not shown). Previous studies have demonstrated the existence of two clusters of 5′-NTR sequences among the human enteroviruses (26). The viruses of HEV-C and HEV-D comprise 5′-NTR cluster I, whereas HEV-A and HEV-B make up cluster II. The HEV-B 5′-NTRs are 77 to 95% identical to one another and 79 to 88% identical to that of CAV16 (HEV-A) but only 68 to 73% identical to those of PV1 (HEV-C) and EV70 (HEV-D) (Table 2), a finding consistent with the previously described clusters. The 3′-NTRs of all of the viruses are similar in length, 102 to 109 nucleotides, and are 70 to 99% identical to one another but only 42 to 62% identical to those of representatives of other human enterovirus species (Table 2).
TABLE 2.
Nucleotide and amino acid sequence comparisons of HEV-B strains to one another and to representatives of HEV-A, HEV-C, and HEV-Da
| Regionb | % Identity (range)
|
|||
|---|---|---|---|---|
| HEV-B | HEV-A (CAV16) | HEV-C (PV1) | HEV-D (EV70) | |
| 5′-NTR | 76.9-95.0 | 79.2-88.0 | 68.2-73.0 | 68.0-72.9 |
| P1 (capsid) | 68.5-87.0 | 47.7-50.5 | 54.8-58.0 | 46.9-50.5 |
| VP4 | 69.6-100 | 53.6-60.9 | 59.4-71.0 | 46.4-58.0 |
| VP2 | 66.0-86.0 | 51.6-57.6 | 54.2-61.7 | 52.6-58.9 |
| VP3 | 65.8-87.0 | 44.4-51.1 | 53.8-61.8 | 42.7-50.4 |
| VP1 | 56.4-83.0 | 36.0-46.0 | 45.0-51.4 | 38.2-45.3 |
| P2 | 92.9-99.0 | 62.7-66.6 | 57.9-59.5 | 63.0-64.5 |
| 2A | 82.0-97.0 | 65.3-78.0 | 52.3-57.7 | 56.6-60.1 |
| 2B | 91.9-100 | 52.5-54.5 | 47.4-51.5 | 58.6-62.6 |
| 2C | 95.4-100 | 64.3-65.5 | 61.7-62.9 | 65.0-67.5 |
| P3 | 94.3-99.0 | 61.8-63.2 | 67.7-69.1 | 71.1-72.7 |
| 3AB | 88.3-100 | 53.7-56.5 | 56.0-59.6 | 66.7-70.3 |
| 3C | 92.9-100 | 55.7-56.8 | 59.6-61.2 | 62.3-63.9 |
| 3D | 94.6-99.0 | 66.0-67.7 | 73.5-74.8 | 75.5-76.8 |
| 3′-NTR | 70.3-99.0 | 41.7-51.8 | 45.1-62.3 | 42.7-51.2 |
Coding region comparisons.
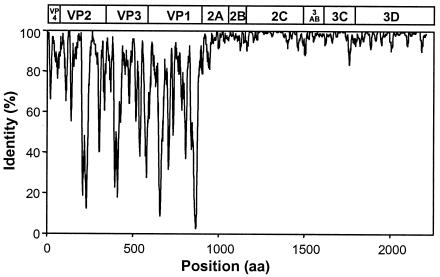
Similarity plots, pairwise sequence comparisons, multiple alignments, and phylogenetic reconstruction were used to examine the relationships among the sequences of the species B enteroviruses and to compare HEV-B sequences to those of other enterovirus species. The deduced HEV-B polyprotein sequences varied in length from 2,182 to 2,202 amino acids. The predicted proteolytic cleavage sites within the deduced polyproteins of the newly sequenced viruses are consistent with those predicted or experimentally determined for other enteroviruses (data not shown). The HEV-B deduced amino acid sequences differ from those of representatives of other enterovirus species by 42 to 53% in P1, 33 to 42% in P2, and 27 to 38% in P3 (Table 2). The HEV-B capsid sequences (P1) differ from one another by 13.0 to 31.5%, whereas the maximum difference for P2 and P3 sequences is only 7.1 and 5.7%, respectively. A similarity plot based on the alignment of all 37 complete amino acid sequences clearly demonstrates that the noncapsid proteins are much more highly conserved within HEV-B than are the capsid proteins (Fig. 1). The greatest sequence variation occurs among the individual capsid protein sequences, exemplified by the 17.0 to 43.6% difference among deduced HEV-B VP1 protein sequences (Table 2), as previously reported (55). The HEV-B viruses differ from one another by up to 34% in VP2 and VP3 and by up to 30% in the VP4 sequences. The lowest levels of identity within the capsid region are primarily localized at known points of variability, such as the surface determinants and in the amino and carboxyl termini of VP1. Unexpectedly, the deduced VP4 amino acid sequences of CBV1 and E19 are identical to one another, despite a nucleotide sequence identity of only 81.2%. The noncapsid proteins are fully colinear among all of the HEV-B viruses (P2 = 667 amino acids; P3 = 667 amino acids). The P2 and P3 regions are generally highly conserved within HEV-B, except for the deduced 2A proteins, which vary by up to 18.0% among viruses (Table 2). The other mature nonstructural proteins (2B, 2C, 3A, 3B, 3C, and 3D) vary by no more than 11.7% (3AB), and there are numerous examples of identical amino acid sequences for some nonstructural proteins among viruses of heterologous serotypes. The deduced 2C and 3D protein sequences are the most highly conserved, with no more than 5.5% variation in either protein (Table 2).
FIG. 1.
Similarity plot summarizing sequence identities among HEV-B polyproteins. Identities among the 37 aligned sequences were plotted in the center of a 10-residue window, and the window was progressively advanced in one-residue steps. The enterovirus genetic map is shown at the top.
Phylogenetic relationships.
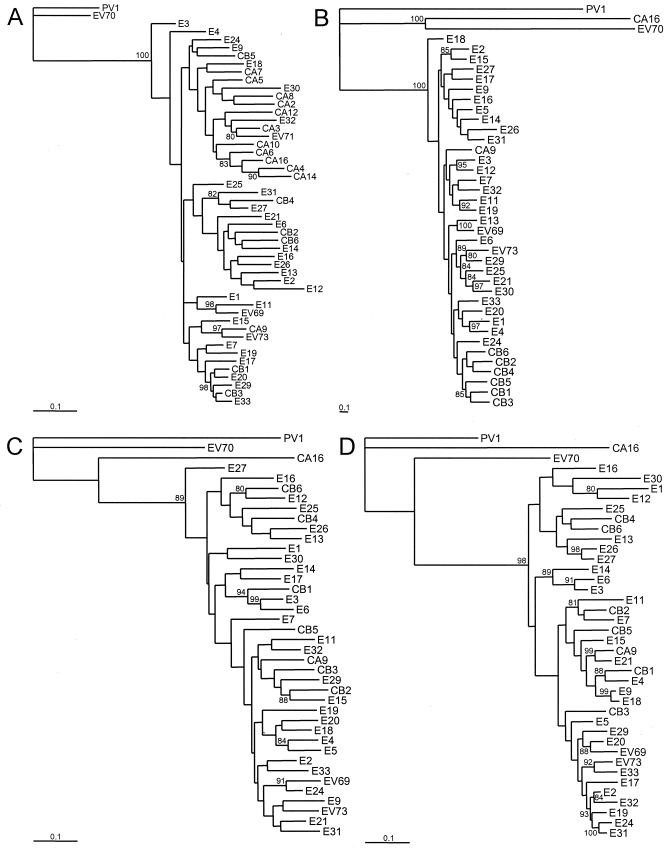
Neighbor-joining nucleotide sequence phylogenies were constructed separately for the 5′-NTR, the P1, P2, and P3 regions (Fig. 2), and for the regions encoding the mature capsid proteins (VP1 to -4), 2C, and 3D (Fig. 3). All HEV-B sequences are monophyletic relative to members of other human enterovirus species throughout the coding region (Fig. 2B to D). The 5′-NTR sequences, however, cluster with those of HEV-A viruses (Fig. 2A), as described above and in agreement with previously published phylogenies (26, 55, 56, 62). The six CBVs are monophyletic in the P1 region as a whole (Fig. 2B) and in the sequences that encode each of the individual external capsid proteins, VP1, VP2, and VP3 (Fig. 3B to D). Outside the capsid region, the CBVs are not monophyletic and, indeed, the overall tree topologies differ markedly from one region of the genome to another for most serotypes (Fig. 2C and D and 3E and F). For example, in the P1 region, five pairs of sequences cluster with >90% bootstrap support: E1-E4 (97%), E3-E12 (95%), E11-E19 (92%), E13-EV69 (100%), and E21-E30 (97%). In the P2 region, none of the five pairs remain clustered. Multiple additional examples are evident where viruses that are closely related in the P1 region (>90% bootstrap support) are not closely related in either the P2 or P3 regions, or both. For example, E3 is most closely related to E12 in the P1 region; however, it is closest to E6 in the P2 and P3 regions. Similarly, the corresponding closest related serotypes to E1 are E4, E30, and E12 in the P1, P2, and P3 regions, respectively. For E4 the closest related viruses in the three regions are E1, E5, and CBV1. There are no examples in this collection of prototype strains for which the closest pairwise serotypes maintain that relationship throughout all regions of the genome.
FIG. 2.
Phylogenetic trees based on HEV-B virus nucleotide sequences. Each of the major functional regions of the genome was analyzed independently. Bootstrap values (percentages of 1,000 pseudoreplicate data sets) supporting each cluster are shown at the nodes; for clarity, only values of > 80% are shown. CAV16, PV1, and EV70, representatives of HEV-A, HEV-C, and HEV-D, respectively, are included as outgroup taxa. All trees are plotted to the same scale, except for panel B (see scale bars). (A) 5′-NTR; (B) complete P1 region; (C) complete P2 region; (D) complete P3 region.
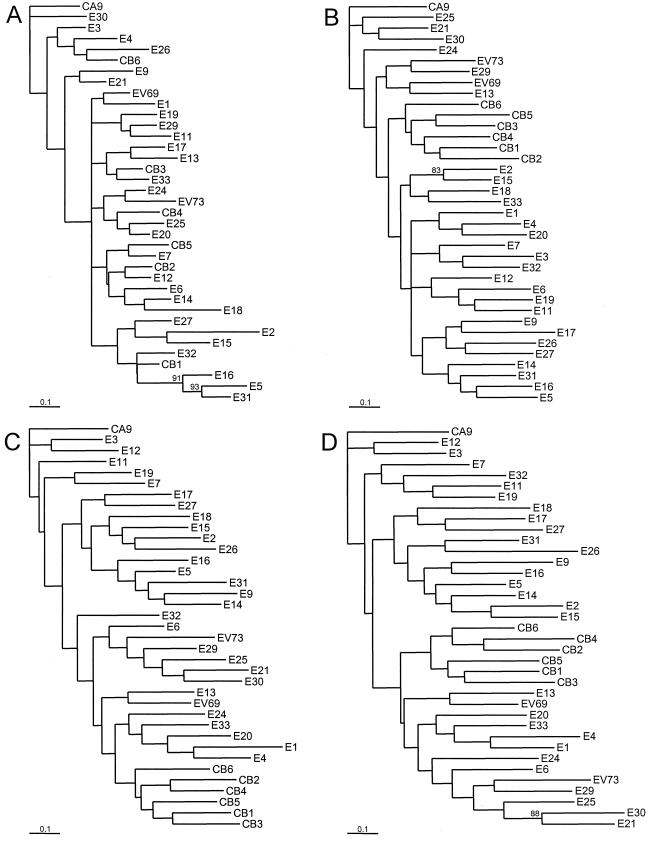
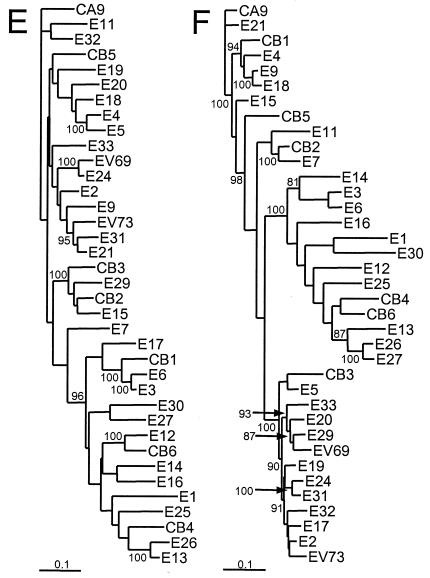
FIG. 3.
Phylogenetic trees based on HEV-B virus nucleotide sequences. The regions encoding each of the mature capsid proteins, the 2C protein, and the 3D protein were analyzed independently. Bootstrap values (percentages of 1,000 pseudoreplicate data sets) supporting each cluster are shown at the nodes; for clarity, only values of >80% are shown. All trees are plotted to the same scale (see scale bars). (A) 1A (VP4); (B) 1B (VP2); (C) 1C (VP3); (D) 1D (VP1); (E) 2C; (F) 3D.
Evidence for recombination.
The radically incongruent tree topologies between the structural and nonstructural regions and even between different proteins across the nonstructural region of the genome suggested that recombination might have played a significant role in the evolution of the prototype strains examined. To address the issue of recombination in more detail, we analyzed the aligned HEV-B complete genome sequences by examining the similarity among sequences in a sliding window of 200 residues by using the program SimPlot (39). The sequence of each strain was used as the query sequence and compared to those of all other serotypes, resulting in 37 separate similarity plots. The P1 regions are highly dissimilar, a finding in agreement with the overall similarity plot shown in Fig. 1, whereas parts of P2 and P3 are well conserved among some of the strains (Fig. 4 and data not shown).
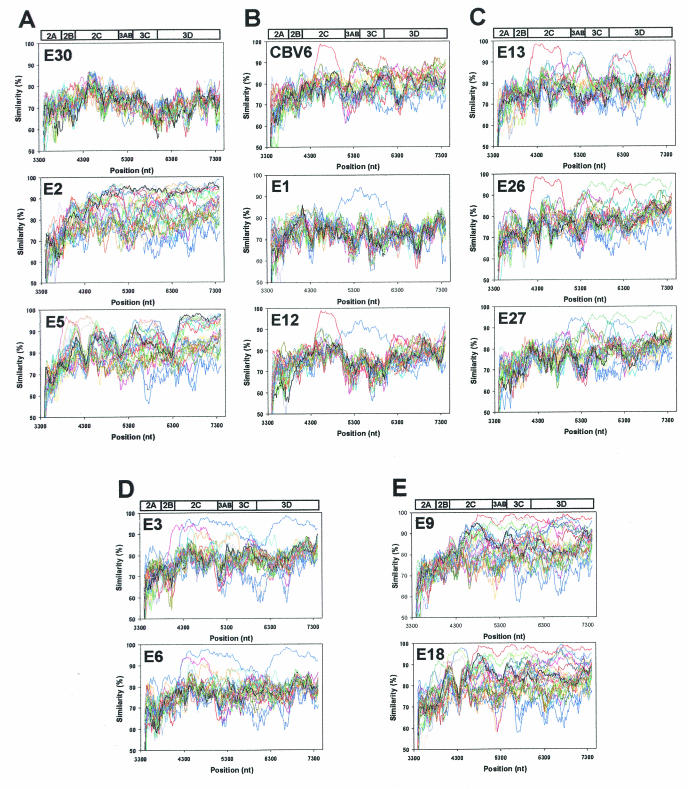
FIG. 4.
Representative similarity plots of HEV-B virus P2 and P3 nucleotide sequences calculated by SimPlot 3.2 beta (39). Each point represents the similarity between the query sequence and a given heterologous sequence, within a sliding window of 200 nucleotides centered on the position plotted, with a step of 20 residues between points. Positions containing gaps were excluded from the analysis. The enterovirus genetic map is shown at the top of each panel. For each plot, the identity of the query sequence is indicated in the upper left corner. Within panels B to E, homologous peaks are depicted in the same color. (A) E30-Bastianni, E2-Cornelis, and E5-Noyce; (B) CBV6-Schmitt, E1-Farouk, and E12-Travis; (C) E13-Del Carmen, E26-Coronel, and E27-Bacon; (D) E3-Morissey and E6-D'Amori; and (E) E9-Hill and E18-Metcalf.
There are four main patterns of similarity in P2 and P3, as depicted in the representative plots in Fig. 4: (i) relatively low similarity (ca. 60 to 80%) to all other prototype strains; (ii) a high degree of similarity (80 to 95%) to many other strains, throughout most of P2 and P3; (iii) high similarity to many other strains, but in discrete regions of P2 and P3; and (iv) low similarity to most prototype strains, but with a high degree of similarity to one or a few other strains in discrete regions of P2 and P3. As an example of the first pattern, the E30-Bastianni sequence is <80% similar to all other sequences, except for a small portion of 2C surrounding the cis-acting replication element (cre) (Fig. 4A). In contrast, E2-Cornelis is an example of the second pattern and is highly similar to at least 11 other strains, with the region of high similarity (85 to 95%) extending from 2B to the end of 3D (Fig. 4A). As an example of the third pattern, E5-Noyce is closely related in 3D to the same 11 strains as E2-Cornelis, but it is highly similar to different strains in 2B-2C and in 3AB-3C (Fig. 4A).
In contrast, several of the prototype strains show a high degree of similarity to only one or a few other strains (the fourth pattern). Each of these relationships was confirmed by bootscanning, in which bootstrap values are calculated for phylogenetic trees constructed in a 200-nucleotide window that is progressively moved across the genome alignment (data not shown). The pattern for E1-Farouk resembles that of E30-Bastianni, except that E1-Farouk is ca. 90% similar to E12-Travis over a stretch of ca. 1,200 nucleotides extending from the end of 2C to the beginning of 3D, with 95 to 100% bootstrap support in the bootscan analysis (Fig. 4B and data not shown). Similarly, CBV6-Schmitt is >95% similar to E12-Travis in 2C, with nearly 100% bootstrap support, but it is not highly related to E1-Farouk (Fig. 4B and data not shown). CBV6-Schmitt is also related to E26-Coronel at the 3B-3C junction and to E27-Bacon at the 3C-3D junction, but in both cases, the region of similarity is relatively small (Fig. 4B and C).
E13-Del Carmen, E26-Coronel, and E27-Bacon exhibit complex relationships with one another (Fig. 4C). E26-Coronel and E27-Bacon are highly similar to one another throughout 3C and 3D. E13-Del Carmen is closely related to E26-Coronel in 2C and to E27-Bacon in 3AB. It is related to both E26-Coronel and E27-Bacon in the 5′ end of 3D, to approximately equal degrees. Similarly, E3-Morrisey is related to E6-D'Amori over most of P2 and P3, with strong bootstrap support; however, the regions of similarity are not contiguous (Fig. 4D). E3-Morrisey is also related to CBV1-Conn-5 in the 3′ end of 2B and in 2C, and to E14-Tow in 3C, whereas E6-D'Amori is related to CBV1-Conn-5 in 2C but not in 2B.
The relationship between E9-Hill and E18-Metcalf has been described previously using 10 strains of six serotypes in the SimPlot analysis (1), but inclusion of all 37 HEV-B serotypes in the analysis reveals that the relationship is more complex than previously appreciated (Fig. 4E). Although the two viruses are 90 to 98% similar to one another in a span of nearly 3,000 nucleotides from the middle of 2C to the 3′ end of 3D, each is also related to other prototype strains to an almost equal degree in several subregions through this part of the genome (Fig. 4E). E18-Metcalf is also related to E24-DeCamp, E5-Noyce, and E4-Pesacek in 2B and 2C, upstream of its region of similarity with E9-Hill. The bootscanning analysis mirrors these findings, revealing five relatively narrow peaks of strong support for the E9-E18 relationship (85 to 100% bootstrap support) interspersed with regions of very low bootstrap support (<30%) (data not shown).
Reconstruction of hypothetical ancestral relationships.
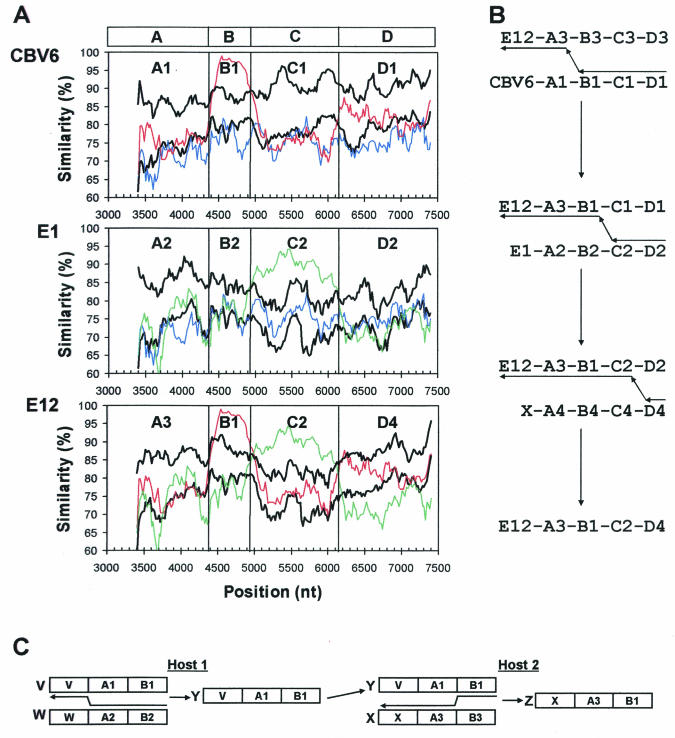
In some cases, it may be possible to partially reconstruct the recombinational history of certain clinical isolates by using the simplots to infer the minimum set of recombination events that are required to produce a given set of observed isolates. As an example of a relatively simple case, we have analyzed the relationships among CBV6-Schmitt, E1-Farouk, and E12-Travis (Fig. 5). To facilitate the analysis, we consider four ancestral strains of four different serotypes: CBV6, E1, E12, and X, where X is any HEV-B serotype. The ancestral strains are not necessarily identical to the observed prototype strains of the same serotypes. The ancestral genomes may be considered to be composed of five distinct regions: capsid, A, B, C, and D, based on the relationships between the observed (prototype) CBV6, E1, and E12 strains, as shown in Fig. 4B and summarized in Fig. 5A. Within each of these genome regions, distinct sequences are indicated by a different serotype designation (for the capsid) or by alleles 1 to 4 (e.g., A1, A2, A3, and A4), and similar sequences are given the same allele designation (e.g., B1 in both CBV6 and E12). If we consider a simple scenario (only a small number of steps needed to produce all of the observed isolates), we will assume that the parental (donor) CBV6 and E1 strains are identical in genome segment structure to the observed (prototype) strains (Fig. 5B). Recombination of CBV6-A1-B1-C1-D1 with E12-A3-B3-C3-D3 between regions A and B produces E12-A3-B1-C1-D1, thereby associating the E12 capsid with the B1 allele. Subsequent recombination of E12-A3-B1-C1-D1 with E1-A2-B2-C2-D2 between regions B and C produces E12-A3-B1-C2-D2 and associates allele C2 with the E12 capsid. Since the E12-Travis D region is distinct from that of either CBV6-Schmitt or E1-Farouk, recombination with an unobserved isolate that carries allele D4 is required to produce the observed E12-Travis isolate with the allele structure E12-A3-B1-C2-D4.
FIG. 5.
Putative recombination pathways for CBV6-Schmitt, E1-Farouk, and E12-Travis, deduced from similarity plots and bootscanning analysis and a simple, schematic representation of recombination involving three hypothetical parental strains. (A) Similarity plots, as in Fig. 4, comparing the relationships among CBV6-Schmitt, E1-Farouk, and E12-Travis with their relationships to the other prototype strains. The curves depicting comparisons between CBV6, E1, and E12 are color-coded as follows: blue, CBV6-E1; red, CBV6-E12; and green, E1-E12. For each query sequence, the average similarity to strains other than CBV6-Schmitt, E1-Farouk, and E12-Travis is plotted in the lower black curve; the upper black curve indicates three standard deviations above the mean. The boundaries of regions A, B, C, and D, at the top, indicate sites where the relationships change. Within each of these regions, distinct alleles are labeled A1, A2, A3, etc. (B) Minimum recombination pathway to produce the observed virus isolates. Capsid identities are indicated by the serotype designations, CBV6, E1, E12, and X, where X is an unobserved strain of any HEV-B serotype, as described in the text. Regions and alleles are indicated as in panel A. Arrows pointing from right to left indicate minus strand synthesis and template switching to produce chimeric RNAs. (C) Coinfection of host 1 with strains V and W (recombination results in strain Y) and coinfection of host 2 with strains X and Y (recombination results in strain Z). Arrows between the parental strains indicate hypothetical template-switch points during minus strand synthesis.
DISCUSSION
Both intra- and intertypic recombination have been shown to occur among poliovirus vaccine strains (5, 18, 28, 37) and between polioviruses and other, unidentified, donor strains (21, 38). There is also evidence for intertypic recombination between certain nonpolio enteroviruses (1, 58, 72). Enteroviruses evolve rapidly (29) and, even though most nucleotide substitutions are synonymous, amino acid changes do occur. Recombination may eliminate multiple accumulated deleterious mutations in a single step, restoring fitness and permitting continued circulation. Our HEV-B phylogenetic trees and simplots reveal evidence of mosaicism in a number of HEV-B strains; that is, their pairwise relationships change, depending on genome position, suggesting that recombination has shuffled sequences between strains of different serotype (Fig. 2 to 4). Furthermore, the complexity of the pattern of relationships suggests that intertypic recombination has occurred repeatedly within HEV-B.
The relatively high sequence diversity among the capsid-coding sequences of HEV-B serotypes suggests that nucleotide substitution is the dominant evolutionary mechanism in this region of the genome. That is, evidence of recombination, if it occurs at all, is quickly obscured by rapid accumulation of nucleotide substitutions. Our results are consistent with those of previous studies that have suggested that interserotypic recombination within the capsid is relatively rare (32, 33). In several cases, our bootscanning analysis identified peaks in the capsid region that suggested possible recombination sites, but all of these peaks were narrow, and the bootstrap values were <80% (data not shown), making it difficult to distinguish recombination from localized convergent evolution or conservation due to common function, such as the use of the same cellular receptor. It is also possible that these peaks represent primordial relationships that have been partially obscured by subsequent point substitutions. Presumably, some regions must evolve more slowly due to functional requirements so that serotypes that are more closely related evolutionarily might retain some vestige of a common ancestral sequence. Peaks of sequence similarity were generally consistent with the overall P1 phylogeny, in that the peaks tended to identify virus pairs that were clustered with high bootstrap support (e.g., E1-E4, E2-E15, E3-E12, and E11-E19), as shown in Fig. 2B. Alternatively, they could be the result of convergent evolution to maintain the required capsid β-barrel structure or be sites of receptor interaction. Intratypic recombination has been shown to occur between wild and vaccine poliovirus strains near the 3′ end of the capsid region, near the end of VP1 (38). Intertypic recombination between poliovirus vaccine strains has also been observed in this same region (2). The surprising identity between the CBV1 and E19 VP4 amino acid sequences may be evidence for intertypic recombination near the 5′ end of the capsid region.
A detailed analysis of natural enterovirus recombination is difficult because one can never isolate all intermediates between any two temporally and geographically distinct strains. In practice, only a very small fraction of all circulating enteroviruses are actually isolated (probably no more than 0.01% of the total even under ideal surveillance) because (i) generally fewer than 1% of all enterovirus infections result in symptoms that might bring the infected individual to medical attention; (ii) among those who seek medical care, virus isolation is usually attempted only from patients with severe symptoms; (iii) even specimens from severe cases may not always yield virus; and (iv) all virus isolates that are obtained in different laboratories are not usually available for analysis at a single site. Despite these limitations, we have shown that it may occasionally be possible to analyze similarity plots, such as those shown in Fig. 4, to infer recombinational relationships based on available isolates and sequences (Fig. 5).
Although it is not prominent in Fig. 4, our SimPlot and bootscan analysis showed that the CBV6, E12, E13, E26, and E27 prototypes shared sequences in P2 and P3 in complex combinations (Fig. 4B and C), suggesting that these five strains are closely related to one another through multiple recombination events. In addition, E12-Travis and E1-Farouk share a large segment of P3 (Fig. 4B and 6A), suggesting that they too are closely related by recombination. CBV6-Schmitt, E12-Travis, E13-Del Carmen, E26-Coronel, and E27-Bacon were all isolated from specimens obtained from healthy children in 1953 on or near Clark Air Force Base on the island of Luzon in the Philippines (22-24). Their presence in the same community at the same time would have provided ample opportunity for coinfection and recombination. E1-Farouk was isolated in Egypt in 1951, but E1 also circulated in the population studied in the Philippines in August to November 1953 (22, 24). In one reported case, both E1 and E13 (as well as poliovirus type 3) were isolated from a single rectal swab specimen from one patient, clearly demonstrating coinfection by E1 and E13 (24). One might speculate that an E1 strain carrying a P3 sequence related to that of E1-Farouk was introduced into the Philippines shortly before the fall of 1953 and that this strain recombined with a progenitor of E12-Travis as they cocirculated in the community. Of course, given that recombination appears to occur at a very high frequency, the P3 sequence could have passed through another serotype(s) prior to introduction into the E12-Travis lineage.
It was recently shown that the E9 and E18 prototype strains (Hill and Metcalf, respectively) share a high degree of sequence similarity in their P3 nonstructural genes, suggesting a recombinational relationship, whereas E9-Hill and another E9 strain (Barty) clearly carry distinct P3 sequences (1). The authors of that study postulated that progenitors of E9-Barty and E18-Metcalf recombined to produce E9-Hill, but we consider that scenario to be unlikely. E9-Hill and E18-Metcalf were both isolated in the same community (Cincinnati, Ohio), in 1953 and 1955, respectively (64, 65), whereas E9-Barty was isolated in Milwaukee, Wis., in 1957 (17). There are no comprehensive surveillance data available for this period, but it is likely that E18 strains (i.e., a Metcalf progenitor) were present in Cincinnati during the E9 outbreak of 1953, providing an opportunity for genetic exchange between the two serotypes and thereby producing a virus (Metcalf) with an E18 capsid and a P3 region derived from an E9 strain (Hill). Given that enterovirus VP1 capsid sequences fix mutations at a rate of about 1% per year (29), the high degree of divergence between Hill and Barty (21% difference in VP1 sequence [81]) would argue that Barty and Hill were derived from epidemiologically distinct sources. Therefore, E9-Barty is either not directly descended from E9-Hill or, if it is descended from Hill, the Hill/Metcalf nonstructural genes have been exchanged out by recombination some time between the isolation of E9-Hill in 1953 and the isolation of E9-Barty in 1957.
Both replicative (template-switching) and nonreplicative (strand breakage and rejoining) mechanisms have been proposed for enterovirus recombination (20, 27, 35). Template switching is thought to occur during negative-strand synthesis, resulting in a chimeric negative-strand that becomes the template for the synthesis of chimeric plus strands. In classical genetic recombination between DNA genomes, individual recombination events (crossovers) are largely discrete, independent events. That is, there is no systematic cooperativity between crossovers at multiple sites, and the probability of multiple crossovers is generally the product of the probabilities of the individual events. An enterovirus replication complex may contain many template RNA molecules, providing opportunities for interaction between genetically distinct RNAs if the cell is coinfected with two different viruses (16, 40). It has been proposed that the accumulation of RNA during replication may lead to an increase in recombination frequency due to an increase in the local concentration of donor and acceptor RNA molecules (27). In the present study several prototype strains of different serotypes exhibit genomic structures that are consistent with multiple recombination events. For example, E3-Morrisey and E6-D'Amori are clearly related to one another in two regions that are separated by distinct sequences (Fig. 4D). A similar pattern was observed for the relationships among E13-Del Carmen, E26-Coronel, and E27-Bacon (Fig. 4C). Multiple template switches between two heterologous parental molecules would result in the observed pattern. Alternatively, template switching during copying of the newly synthesized, chimeric positive strand (or in subsequent rounds of minus strand synthesis) could result in replacement of some or all of the sequence that was removed during the original template switch. It is also possible that these patterns represent multiple, sequential recombination events that occur in multiple hosts, with both parental serotypes continuously present in the community, providing numerous occasions for interaction.
That two prototype strains are related by recombination does not mean these two serotypes were simultaneously present in the same host or that all isolates of these two serotypes are related to one another outside of the capsid region. For example, consider three hypothetical viruses of serotypes V, W, and X, with noncapsid regions A1-B1, A2-B2, and A3-B3, respectively (Fig. 5C). Viruses V and W simultaneously infect host 1 and recombine to produce virus Y. Viruses X and Y then coinfect a new host (host 2) and recombine to produce virus Z (Fig. 5C). Part of the noncapsid sequence of virus Z (serotype X) is derived from virus V, even though viruses of serotypes V and X were never simultaneously present in the same host. Viruses V and Y are both of serotype V, and yet their noncapsid regions are completely different. Continued transmission of viruses derived from virus Y will result in many additional recombination events and the accumulation of nucleotide substitutions, further obscuring the linkage between the “V” capsid and the “A1-B1” noncapsid sequences that were present in virus V. Furthermore, the number of discrete “regions” in the noncapsid portion of the genome is not restricted to the two depicted in Fig. 5C (or the four shown in Fig. 5A and B). Rather, it may theoretically equal the number of P2-P3-3′-NTR nucleotides, since recombination might occur at any site. Likewise, recombination may also delink a given capsid sequence (serotype) from its original 5′-NTR, explaining why this region is generally unsuitable as a target for molecular methods of serotype identification (41, 73).
Any recombination mechanism requires that the two parental RNA molecules are simultaneously present in the same cell. At least four different cellular receptors are used for cell entry by members of HEV-B: coxsackievirus-adenovirus receptor, decay accelerating factor, integrin αvβ3, and integrin α2β1 (71). Therefore, in order for recombination to occur, the two parental viruses must either use the same receptor to enter the cell or two different receptors must be present on the same cell. Although four HEV-B receptors have been identified, the receptors used by many serotypes remain unknown. The availability of complete genome sequences for all members of HEV-B will facilitate a correlation between capsid sequences and receptor utilization. If a pattern of receptor usage and interserotypic recombination exists, then the genomic patterns described here may suggest specific hypotheses for receptor utilization in serotypes for which this information is currently lacking.
In general, an enterovirus might be viewed as a capsid sequence in search of noncapsid sequences of the highest fitness to provide a selective replicative advantage. Alternatively, an enterovirus may be viewed as a replicon in search of a capsid in order to enter a host cell and thereby gain access to the cytoplasmic milieu needed for replication. Only the capsid, however, is inherited as a single unit, suggesting that the capsid is the primary determinant of enterovirus identity. Regardless of perspective, it is not possible to identify serotype-specific sequences in the P2 or P3 regions. Rather, the P2-P3 sequences of a given isolate represent only a snapshot of that particular isolate or of a closely related lineage, within a narrow temporal and geographic window. This view of the role of recombination in enterovirus evolution would predict that the specific genomic combinations and sequences in the P2-P3 regions of the prototype strains are not likely to be present in currently circulating strains of the same serotype. Conversely, sequences related to those of a given prototype strain may be found in different serotypes within the same species among currently circulating enteroviruses. Analysis of capsid and noncapsid sequences from additional clinical isolates may allow estimation of the rate of enterovirus recombination during natural circulation in the human population and may shed additional light of the role of recombination in the evolution of viruses in HEV-B.
REFERENCES
- 1.Andersson, P., K. Edman, and A. M. Lindberg. 2002. Molecular analysis of the echovirus 18 prototype: evidence of interserotypic recombination with echovirus 9. Virus Res. 85:71-83. [DOI] [PubMed] [Google Scholar]
- 2.Blomqvist, S., A.-L. Bruu, M. Stenvik, and T. Hovi. 2003. Characterization of a recombinant type 3/type 2 poliovirus isolated from a healthy vaccinee and containing a chimeric capsid protein VP1. J. Gen. Virol. 84:573-580. [DOI] [PubMed] [Google Scholar]
- 3.Branche, W. C., V. M. Young, F. M. Houston, and L. W. Koontz. 1965. Characterization of prototype virus ECHO-32. Proc. Soc. Exp. Biol. Med. 118:186-190. [DOI] [PubMed] [Google Scholar]
- 4.Brown, B. A., K. Maher, M. S. Oberste, and M. A. Pallansch. 2003. Complete genomic sequencing shows that polioviruses and members of human enterovirus species C are closely related in the non-capsid coding region. J. Virol. 77:8973-8984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cammack, N., A. Phillips, G. Dunn, V. Patel, and P. D. Minor. 1988. Intertypic genomic rearrangements of poliovirus strains in vaccinees. Virology 167:507-514. [PubMed] [Google Scholar]
- 6.Chang, K. H., P. Auvinen, T. Hyypiä, and G. Stanway. 1989. The nucleotide sequence of coxsackievirus A9: implications for receptor binding and enterovirus classification. J. Gen. Virol. 70:3269-3280. [DOI] [PubMed] [Google Scholar]
- 7.Cherkasova, E. A., E. A. Korotkova, M. L. Yakovenko, O. E. Ivanova, T. P. Eremeeva, K. M. Chumakov, and V. I. Agol. 2002. Long-term circulation of vaccine-derived poliovirus that causes paralytic disease. J. Virol. 76:6791-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chua, B. H., P. C. McMinn, L. S. K., and K. B. Chua. 2001. Comparison of the complete nucleotide sequences of echovirus 7 strain UMMC and the prototype (Wallace) strain demonstrates significant genetic drift over time. J. Gen. Virol. 82:2629-2639. [DOI] [PubMed] [Google Scholar]
- 9.Committee on Enteroviruses. 1962. Classification of human enteroviruses. Virology 16:501-504. [Google Scholar]
- 10.Committee on the ECHO viruses. 1955. Enteric cytopathogenic human orphan (ECHO) viruses. Science 122:1187-1188. [PubMed] [Google Scholar]
- 11.Committee on the Enteroviruses. 1957. The enteroviruses. Am. J. Public Health 47:1556-1566. [PMC free article] [PubMed] [Google Scholar]
- 12.Cuervo, N. S., S. Guillot, N. Romanenkova, M. Combiescu, A. Aubert-Combiescu, M. Seghier, V. Caro, R. Crainic, and F. Delpeyroux. 2001. Genomic features of intertypic recombinant Sabin poliovirus strains excreted by primary vaccinees. J. Virol. 75:5740-5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dahllund, L., L. Nissinen, T. Pulli, V. P. Hyttinen, G. Stanway, and T. Hyypia. 1995. The genome of echovirus 11. Virus Res. 35:215-222. [DOI] [PubMed] [Google Scholar]
- 14.Dalldorf, G. 1953. The coxsackie virus group. Ann. N. Y. Acad. Sci. 56:583-586. [DOI] [PubMed] [Google Scholar]
- 15.Davis, D. C., and J. L. Melnick. 1956. Association of echo virus type 6 with aseptic meningitis. Proc. Soc. Exp. Biol. Med. 92:839-843. [DOI] [PubMed] [Google Scholar]
- 16.Egger, D., and K. Bienz. 2002. Recombination of poliovirus RNA proceeds in mixed replication complexes originating from distinct replication start sites. J. Virol. 76:10960-10971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eggers, H. J., and A. B. Sabin. 1959. Factors determining pathogenicity of variants of echo 9 virus for newborn mice. Proc. Soc. Exp. Biol. Med. 110:951-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furione, M., S. Guillot, D. Otelea, J. Balanant, A. Candrea, and R. Crainic. 1993. Polioviruses with natural recombinant genomes isolated from vaccine-associated paralytic poliomyelitis. Virology 196:199-208. [DOI] [PubMed] [Google Scholar]
- 19.Georgescu, M.-M., F. Delpeyroux, and R. Crainic. 1995. Tripartite genome organization of a natural type 2 vaccine/nonvaccine recombinant poliovirus. J. Gen. Virol. 76:2343-2348. [DOI] [PubMed] [Google Scholar]
- 20.Gmyl, A. P., E. V. Belousov, S. V. Maslova, E. V. Khitrina, A. B. Chetverin, and V. I. Agol. 1999. Nonreplicative RNA recombination in poliovirus. J. Virol. 73:8958-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guillot, S., V. Caro, N. Cuervo, E. Korotkova, M. Combiescu, A. Persu, A. Aubert-Combiescu, F. Delpeyroux, and R. Crainic. 2000. Natural genetic exchanges between vaccine and wild poliovirus strains in humans. J. Virol. 74:8434-8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hammon, W. M., E. H. Ludwig, R. A. Pavia, L. W. McCloskey, and G. E. Sather. 1957. Problems raised by certain ECHO viruses in the attempted detection of poliomyelitis virus infection. Ann. N. Y. Acad. Sci. 67:304-310. [DOI] [PubMed] [Google Scholar]
- 23.Hammon, W. M., D. S. Yohn, and R. A. Pavia. 1960. Isolation and characterization of prototype viruses ECHO-26, ECHO-27, and coxsackie B-6. Proc. Soc. Exp. Biol. Med. 103:164-168. [DOI] [PubMed] [Google Scholar]
- 24.Hammon, W. M., D. S. Yohn, R. A. Pavia, and G. Sather. 1959. Echo virus 13. I. Isolation and characteristics. Proc. Soc. Exp. Biol. Med. 100:425-429. [DOI] [PubMed] [Google Scholar]
- 25.Hirst, G. 1962. Genetic recombination with newcastle disease virus, polioviruses, and influenza. Cold Spring Harbor Symp. Quant. Biol. 27:303-309. [DOI] [PubMed] [Google Scholar]
- 26.Hyypiä, T., T. Hovi, N. J. Knowles, and G. Stanway. 1997. Classification of enteroviruses based on molecular and biological properties. J. Gen. Virol. 78:1-11. [DOI] [PubMed] [Google Scholar]
- 27.Jarvis, T. C., and K. Kirkegaard. 1992. Poliovirus RNA recombination: mechanistic studies in the absence of selection. EMBO J. 11:3135-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kew, O. M., V. Morris-Glasgow, M. Landaverde, C. Burns, J. Shaw, Z. Garib, J. André, E. Blackburn, C. J. Freeman, J. Jorba, R. Sutter, G. Tambini, L. Venczel, C. Pedreira, F. Laender, H. Shimizu, T. Yoneyama, T. Miyamura, H. G. van der Avoort, M. S. Oberste, D. R. Kilpatrick, S. Cochi, M. Pallansch, and C. de Quadros. 2002. Outbreak of poliomyelitis in Hispanola associated with circulating type 1 vaccine-derived poliovirus. Science 296:356-359. [DOI] [PubMed] [Google Scholar]
- 29.Kew, O. M., M. N. Mulders, G. Y. Lipskaya, E. E. da Silva, and M. A. Pallansch. 1995. Molecular epidemiology of polioviruses. Semin. Virol. 6:401-414. [Google Scholar]
- 30.Kibrick, S., L. Meléndez, and J. F. Enders. 1957. Clinical associations of enteric viruses with particular reference to agents exhibiting properties of the ECHO group. Ann. N. Y. Acad. Sci. 67:311-325. [DOI] [PubMed] [Google Scholar]
- 31.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucelotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 32.King, A. M. Q. 1988. Genetic recombination in positive strand RNA viruses, p. 149-165. In E. Domingo, J. J. Holland, and P. Alquist (ed.), RNA genetics, vol. 2. CRC Press, Inc., Boca Raton, Fla.
- 33.King, A. M. Q. 1988. Preferred sites of recombination in poliovirus RNA: an analysis of 40 intertypic cross-over sequences. Nucleic Acids Res. 16:11705-11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.King, A. M. Q., F. Brown, P. Christian, T. Hovi, T. Hyypiä, N. J. Knowles, S. M. Lemon, P. D. Minor, A. C. Palmenberg, T. Skern, and G. Stanway. 2000. Picornaviridae, p. 657-678. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, Inc., San Diego, Calif.
- 35.Kirkegaard, K., and D. Baltimore. 1986. The mechamism of RNA recombination in poliovirus. Cell 47:433-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ledinko, N. 1963. Genetic recombination with polioviryus type 1. Studies of crosses between a normal horse serum-resistant mutant and several guanidine-resistant mutants of the same strain. Virology 20:107-119. [DOI] [PubMed] [Google Scholar]
- 37.Lipskaya, G. Y., A. R. Muzychenko, O. K. Kutitova, S. V. Maslova, M. Equestre, S. G. Drozdov, R. Perez Bercoff, and V. I. Agol. 1991. Frequent isolation of intertypic poliovirus recombinants with serotype 2 specificity from vaccine-associated polio cases. J. Med. Virol. 35:290-296. [DOI] [PubMed] [Google Scholar]
- 38.Liu, H.-M., D.-P. Zheng, L.-B. Zhang, M. S. Oberste, M. A. Pallansch, and O. M. Kew. 2000. Molecular evolution of a type 1 wild-vaccine poliovirus recombinant during widespread circulation in China. J. Virol. 74:11153-11161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lole, K. S., R. C. Bollinger, R. S. Paranjape, D. Gadkari, S. S. Kulkarni, N. G. Novak, R. Ingersoll, H. W. Sheppard, and S. C. Ray. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lyle, J. M., E. Bullit, K. Bienz, and K. Kirkegaard. 2002. Visualization and functional analysis of RNA-dependent RNA polymerase lattices. Science 296:2218-2222. [DOI] [PubMed] [Google Scholar]
- 41.Manayani, D. J., R. V. Shaji, G. J. Fletcher, T. Cherian, N. Murali, N. Sathish, T. Solomon, C. Gnanamutha, and G. Sridraran. 2002. Comparison of molecular and conventional methods for typing of enteroviral isolates. J. Clin. Microbiol. 40:1069-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Melnick, J. L. 1954. Application of tissue culture methods to epidemiological studies of poliomyelitis. Am. J. Public Health 44:571-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Melnick, J. L. 1996. Enteroviruses: polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses, p. 655-712. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 44.Melnick, J. L. 1957. Problems associated with viral identification and classification in 1956. Ann. N. Y. Acad. Sci. 67:363-382. [DOI] [PubMed] [Google Scholar]
- 45.Melnick, J. L. 1955. Tisssue culture techniques and their application to original isolation, growth, and assay of poliomyelitis and orphan viruses. Ann. N. Y. Acad. Sci. 61:754-773. [DOI] [PubMed] [Google Scholar]
- 46.Melnick, J. L., V. I. Agol, H. L. Bachrach, F. Brown, P. D. Cooper, W. Fiers, S. Gard, J. H. Gear, Y. Ghendon, L. Kasza, M. LaPlaca, B. Mandel, S. McGregor, S. B. Mohanty, G. Plummer, R. R. Rueckert, F. L. Schaffer, I. Tagaya, D. A. Tyrrell, M. Voroshilova, and H. A. Wenner. 1974. Picornaviridae. Intervirology 4:303-316. [DOI] [PubMed] [Google Scholar]
- 47.Melnick, J. L., and K. Ågren. 1952. Poliomyelitis and coxsackie viruses isolated from normal infants in Egypt. Proc. Soc. Exp. Biol. Med. 81:621-624. [DOI] [PubMed] [Google Scholar]
- 48.Melnick, J. L., and N. Ledinko. 1950. Immunological reactions of the coxsackie viruses. I. The neutralization test: technique and application. J. Exp. Med. 92:463-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Melnick, J. L., N. Ledinko, A. S. Kaplan, and L. M. Kraft. 1950. Ohio strains of a virus pathogenic for infant mice (coxsackie group): simultaneous occurrence with poliomyelitis virus in patients with “summer grippe.” J. Exp. Med. 91:185-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Melnick, J. L., E. W. Shaw, and E. C. Curnen. 1949. A virus isolated from patients diagnosed as non-paralytic poliomyelitis or aseptic meningitis. Proc. Soc. Exp. Biol. Med. 71:344-349. [DOI] [PubMed] [Google Scholar]
- 51.Melnick, J. L., I. Tagaya, and H. von Magnus. 1974. Enteroviruses 69, 70, and 71. Intervirology 4:369-370. [DOI] [PubMed] [Google Scholar]
- 52.Melnick, J. L., H. A. Wenner, and C. A. Phillips. 1979. Enteroviruses, p. 471-534. In E. H. Lennette and N. J. Schmidt (ed.), Diagnostic procedures for viral, rickettsial, and chlamydial infections, 5th ed. American Public Health Association, Washington, D.C.
- 53.Norder, H., L. Bjerregaard, L. Magnius, B. Lina, M. Aymard, and J.-J. Chomel. 2003. Sequencing of “untypable” enteroviruses reveals two new types, EV-77 and EV-78, within human enterovirus type B and substitutions in the BC loop of the VP1 protein for known types. J. Gen. Virol. 84:827-836. [DOI] [PubMed] [Google Scholar]
- 54.Oberste, M. S., K. Maher, M. R. Flemister, G. Marchetti, D. R. Kilpatrick, and M. A. Pallansch. 2000. Comparison of classic and molecular approaches for the identification of “untypable” enteroviruses. J. Clin. Microbiol. 38:1170-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oberste, M. S., K. Maher, D. R. Kilpatrick, and M. A. Pallansch. 1999. Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification. J. Virol. 73:1941-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oberste, M. S., K. Maher, and M. A. Pallansch. 1998. Molecular phylogeny of all human enterovirus serotypes based on comparison of sequences at the 5′ end of the region encoding VP2. Virus Res. 58:35-43. [DOI] [PubMed] [Google Scholar]
- 57.Oberste, M. S., D. Schnurr, K. Maher, S. al-Busaidy, and M. A. Pallansch. 2001. Molecular identification of new picornaviruses and characterization of a proposed enterovirus 73 serotype. J. Gen. Virol. 82:409-416. [DOI] [PubMed] [Google Scholar]
- 58.Oprisan, G., M. Combiescu, S. Guillot, V. Caro, A. Combiescu, F. Delpeyroux, and R. Crainic. 2002. Natural genetic recombination between co-circulating heterotypic enteroviruses. J. Gen. Virol. 83:2193-2200. [DOI] [PubMed] [Google Scholar]
- 59.Ormsbee, R. A., and J. L. Melnick. 1957. Biologic and serologic characteristics of ECHO viruses from west Virginia. J. Immunol. 79:384-392. [PubMed] [Google Scholar]
- 60.Panel for Picornaviruses. 1963. Picornaviruses: classification of nine new types. Science 141:153-154. [DOI] [PubMed] [Google Scholar]
- 61.Plager, H., and W. Decher. 1963. A newly-recognized enterovirus isolated from cases of aseptic meningitis. Am. J. Hyg. 77:26-28. [DOI] [PubMed] [Google Scholar]
- 62.Pöyry, T., L. Kinnunen, T. Hyypia, B. Brown, C. Horsnell, T. Hovi, and G. Stanway. 1996. Genetic and phylogenetic clustering of enteroviruses. J. Gen. Virol. 77:1699-1717. [DOI] [PubMed] [Google Scholar]
- 63.Pringle, C. R. 1965. Evidence of genetic recombination in foot-and-mouth disease virus. Virology 25:48-54. [DOI] [PubMed] [Google Scholar]
- 64.Ramos-Alvarez, M., and A. B. Sabin. 1954. Characteristics of poliomyelitis and other enteric viruses recovered in tissue culture from healthy American children. Proc. Soc. Exp. Biol. Med. 87:655-661. [DOI] [PubMed] [Google Scholar]
- 65.Ramos-Alvarez, M., and A. B. Sabin. 1958. Enteropathogenic viruses and bacteria: role in summer diarrheal diseases of infancy and early childhood. JAMA 167:147-156. [DOI] [PubMed] [Google Scholar]
- 66.Ramos-Alvarez, M., and A. B. Sabin. 1956. Intestinal viral flora of healthy children demonstrable by monkey kidney tissue culture. Am. J. Public Health 46:295-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Robbins, F. C., J. F. Enders, T. H. Weller, and G. L. Florentino. 1951. Studies on the cultivation of poliomyelitis viruses in tissue culture. V. The direct isolation and serologic identification of virus strains in tissue culture from patients with nonparalytic and paralytic poliomyelitis. Am. J. Hyg. 54:286-293. [DOI] [PubMed] [Google Scholar]
- 68.Rosen, L., and J. Kern. 1965. Toluca-3, a newly recognized enterovirus. Proc. Soc. Exp. Biol. Med. 118:389-391. [DOI] [PubMed] [Google Scholar]
- 69.Rosen, L., J. Kern, and J. A. Bell. 1964. Observations on a group of viruses (JV-5, JV-6, and JV-10) comprising a newly recognized enterovirus serotype. Am. J. Hyg. 79:7-15. [DOI] [PubMed] [Google Scholar]
- 70.Rosen, L., J. Kern, and J. A. Bell. 1964. Observations on an outbreak of infection with a newly recognized enterovirus (JV-4). Am. J. Hyg. 79:1-6. [DOI] [PubMed] [Google Scholar]
- 71.Rossmann, M. G., Y. He, and R. J. Kuhn. 2002. Picornavirus-receptor interactions. Trends Microbiol. 10:324-331. [DOI] [PubMed] [Google Scholar]
- 72.Santti, J., T. Hyypia, L. Kinnunen, and M. Salminen. 1999. Evidence of recombination among enteroviruses. J. Virol. 73:8741-8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Siafakas, N., P. Markoulatos, and G. Stanway. 2002. Variability in molecular typing of coxsackie A viruses by RFLP analysis and sequencing. J. Clin. Lab. Anal. 16:59-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sickles, G. M., P. Feorino, and H. Plager. 1955. Isolation and type determination of coxsackie virus, group B, in tissue culture. Proc. Soc. Exp. Biol. Med. 88:22-24. [DOI] [PubMed] [Google Scholar]
- 75.Strimmer, K., and A. von Haeseler. 1996. Quartet puzzling: a quartet maximum-likelihood method for reconstructing tree topologies. Mol. Biol. Evol. 13:964-969. [Google Scholar]
- 76.Tamura, K., and M. Nei. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10:512-526. [DOI] [PubMed] [Google Scholar]
- 77.Wenner, H. A., M. E. Soergel, P. S. Kamitsuka, P. Perine, T. D. Y. Chin, and T. Y. Lou. 1963. The Caldwell group of enteric viruses. Am. J. Hyg. 78:247-259. [PubMed] [Google Scholar]
- 78.Wigand, R., and A. B. Sabin. 1962. Antigenic purity and plaque properties of the prototype strains of ECHO virus types 7 to 11 and 17 and 18. Arch. Gesamte Virusforsch. 11:708-717. [DOI] [PubMed] [Google Scholar]
- 79.Wigand, R., and A. B. Sabin. 1961. Properties of ECHO types 22, 23, and 24 viruses. Arch. Ges. Virusforsch. 11:224-247. [DOI] [PubMed] [Google Scholar]
- 80.Wildy, P. (ed.). 1971. Classification and nomenclature of viruses, vol. 5. S. Karger, Basel, Switzerland.
- 81.Zimmermann, H., H. J. Eggers, and B. Nelsen-Salz. 1996. Molecular cloning and sequence determination of the complete genome of the virulent echovirus 9 strain Barty. Virus Genes 12:149-154. [DOI] [PubMed] [Google Scholar]