SUMMARY
TET dioxygenases successively oxidize 5-methylcytosine (5mC) in mammalian genomes to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC). 5fC/5caC can be excised and repaired to regenerate unmodified cytosines by thymine-DNA glycosylase (TDG) and base excision repair (BER) pathway, but it is unclear to what extent and at which part of the genome this active demethylation process takes place. Here, we have generated genome-wide distribution maps of 5hmC/5fC/5caC using modification-specific antibodies in wild-type and Tdg-deficient mouse embryonic stem cells (ESCs). In wild-type mouse ESCs, 5fC/5caC accumulates to detectable levels at major satellite repeats but not at non-repetitive loci. In contrast, Tdg depletion in mouse ESCs causes marked accumulation of 5fC and 5caC at a large number of proximal and distal gene regulatory elements. Thus, these results reveal the first genome-wide view of iterative 5mC oxidation dynamics and indicate that TET/TDG-dependent active DNA demethylation process occurs extensively in the mammalian genome.
INTRODUCTION
Epigenetic modifications of DNA and histones play essential roles in regulating gene expression in development and diseases (Goldberg et al., 2007; Jaenisch and Bird, 2003; Sasaki and Matsui, 2008). The predominant epigenetic modification of DNA is methylation at the 5-position of cytosine (5mC), which is indispensable for normal mammalian embryogenesis and is implicated in a variety of human diseases (Baylin and Jones, 2011; Cedar and Bergman, 2012). DNA methylation pattern is established and maintained by DNA methyltransferases (DNMTs) and is relatively stable in somatic tissues (Bird, 2002; Jones, 2012). 5mC can be successively oxidized to 5hmC, 5fC and 5caC by Ten eleven translocation (TET/Tet) family of Fe(II) and 2-oxoglutarate-dependent DNA dioxygenases (He et al., 2011; Ito et al., 2010; Ito et al., 2011; Tahiliani et al., 2009) (Figure S1A). Different Tet enzymes (Tet1–3) exhibit distinct expression patterns in vivo and functional analyses of Tet-deficient mice indicate that they play important roles in diverse biological processes, including zygotic epigenetic reprogramming, germ cell development, pluripotent stem cell differentiation, and myelopoiesis (Cimmino et al., 2011; Dawlaty et al., 2013; Dawlaty et al., 2011; Gu et al., 2011; Koh et al., 2011; Marcucci et al., 2010; Wu and Zhang, 2011a; Yamaguchi et al., 2012).
The study of biological roles of Tet enzymes has been facilitated by the development of methods to specifically enrich or label 5hmC, a relatively abundant 5mC oxidation derivative detected in many tissues (Globisch et al., 2011; Kriaucionis and Heintz, 2009; Munzel et al., 2010). Immunostaining with antibodies specific for 5hmC have revealed that global erasure of paternal DNA methylation is first initiated by Tet3-mediated conversion of 5mC to 5hmC in the male pronucleus, followed by replication-dependent passive loss of 5hmC during preimplantation development (Gu et al., 2011; Inoue and Zhang, 2011; Iqbal et al., 2011; Wossidlo et al., 2011). Similar analysis also suggests a role of Tet1-mediated 5mC oxidation in epigenetic reprogramming during development of primordial germ cells (PGCs) and regulation of parental-origin specific imprinting (Dawlaty et al., 2013; Hackett et al., 2013; Seisenberger et al., 2012; Yamaguchi et al., 2013; Yamaguchi et al., 2012). Genome-wide 5hmC mapping studies of pluripotent stem cells and differentiated tissues using affinity enrichment-based methods or modified bisulphite sequencing (BS-seq) strategies indicate that 5hmC is enriched in highly transcribed gene bodies, as well as Polycomb repression complex bound promoters and distal cis-regulatory elements (Booth et al., 2012; Ficz et al., 2011; Mellen et al., 2012; Pastor et al., 2011; Song et al., 2011; Stroud et al., 2011; Szulwach et al., 2011a; Szulwach et al., 2011b; Williams et al., 2011; Wu et al., 2011a; Wu and Zhang, 2011b; Xu et al., 2011; Yu et al., 2012). Together, these studies not only confirm a functional role of Tet-mediated 5mC oxidation in regulating global DNA demethylation dynamics during specific embryonic stages (one-cell zygotes and developing PGCs), but also suggest that Tet-initiated DNA demethylation process may be more prevalent in the genome than previously anticipated.
In vitro biochemical studies show that DNA repair enzyme thymine-DNA glycosylase (TDG) can excise 5fC and 5caC to generate abasic sites (He et al., 2011; Maiti and Drohat, 2011; Nabel et al., 2012), which are repaired by base excision repair (BER) pathway. These observations suggest a mechanistic paradigm of active DNA demethylation in which Tet proteins first successively oxidize 5mC to 5hmC/5fC/5caC and TDG/BER pathways then excise 5fC/5caC and regenerate unmodified cytosines (Figure S1A). The demonstration that genetic inactivation of Tdg in mouse causes embryonic lethality (Cortazar et al., 2011; Cortellino et al., 2011), raises the possibility that TET/TDG-mediated active DNA demethylation process may be widespread in mammalian genomes and play an essential role in developmental gene regulation. However, it is currently unclear to what extent and at which part of the genome TDG-dependent 5fC/5caC excision followed by BER contributes to dynamic changes of DNA methylation patterns in vivo.
To directly address this question, we generated genome-wide maps of 5mC and its oxidation derivatives (5hmC/5fC/5caC) in wild-type and Tdg-deficient mouse ESCs. We reasoned that depletion of Tdg would block the DNA methylation/demethylation cycle, and causes accumulation of 5fC and 5caC, which can mark genomic loci actively undergoing TET/TDG-dependent 5mC oxidation dynamics. Our results reveal that TET/TDG-mediated cyclic changes of cytosine modification states occurs at a large cohort of gene regulatory regions and suggest that active DNA demethylation takes place more extensively than previously thought in mammalian cells.
RESULTS
Enrichment of 5fC and 5caC from genomic DNA by cytosine modification-specific antibodies
Genome-wide distribution of 5mC and 5hmC can be determined by affinity enrichment or bisulfite conversion-based methods (Song et al., 2012). However, reliable methods are yet to be developed to specifically enrich/label 5fC and 5caC for genome-wide mapping analysis. Antibody-based DNA immunoprecipitation followed by high throughput sequencing (DIP-Seq) represents a simple and reliable approach for profiling cytosine modifications (especially effective for detecting loci with clustered modified bases) if a highly specific antibody is available. A strategy for chemical labeling of 5fC with aldehyde-reactive probe (ARP) has previously been suggested (Pfaffeneder et al., 2011), but this approach may also label abasic sites, which are an intermediate product of endogenous DNA repair process and one of the most prevalent lesions in DNA (Nakamura et al., 1998; Raiber et al., 2012). Thus, proper controls or chemical blocking reactions need to be developed to allow ARP-based chemical labeling methods to distinguish 5fC from abasic sites (Raiber et al., 2012). More recently, modified BS-seq strategies have been developed to map 5hmC distribution at single-nucleotide resolution (Booth et al., 2012; Yu et al., 2012). However, current base-resolution mapping methods are not compatible for detecting 5fC/5caC and require substantially deeper sequencing depth to reliably detect low abundant 5hmC marks. Given that 5fC/5caC is present in the genome at much lower levels compared to 5hmC, it will be challenging to map 5fC/5caC at a genome-wide scale and at base-resolution. To better compare various approaches and identify effective methods for genome-wide mapping of 5fC/5caC, we first performed in-depth analysis comparing genome-wide 5hmC mapping results from antibody- or chemical labeling-based [e.g. GLIB (glucosylation, periodate oxidation and biotinylation)] methods with the base-resolution 5hmC map in mouse ESCs (Pastor et al., 2011; Yu et al., 2012). This analysis revealed that chemical labeling (GLIB) and 5hmC antibody-based methods respectively recovered 35.1% and 39.2% of all high-confidence 5hmC marks in the base-resolution map. Among 2.06 million 5hmC marks of the base-resolution map, 21.3% (0.44 million) of them are sparsely distributed (single 5hmC mark within 1kb). Interestingly, antibody-based method performed similarly as the chemical labeling method in terms of pulling down both clustered (48.1% for antibody and 43.1% for GLIB) and sparsely distributed 5hmC marks (6.4% for antibody versus 5.3% for GLIB) from in vivo genomic DNA (Figure S1B–C).
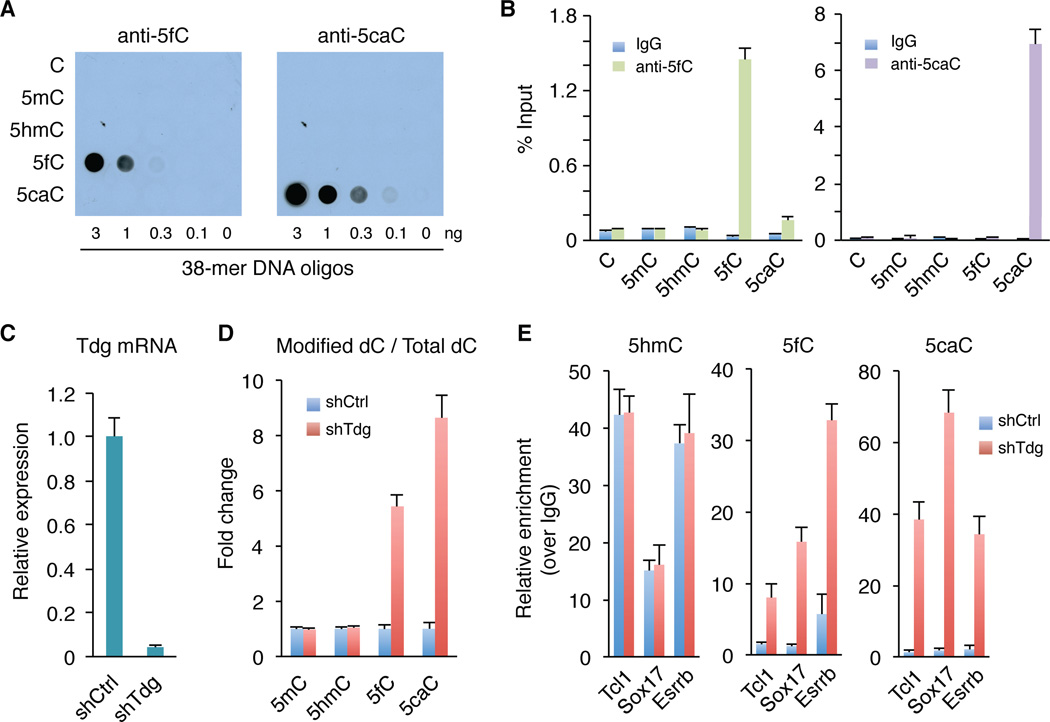
Given that the antibody-based method performs similarly as chemical labeling methods in 5hmC pull-down of genomic DNA, we focused our efforts on antibody-based DIP-Seq approach. The 5fC- and 5caC-specific antibodies we developed were previously used to examine global levels of 5fC/5caC by immunostaining (Inoue et al., 2011). After further confirmation of their specificity by dot blot analysis (Figure 1A), we tested their utility in DIP assays. This analysis indicated that these antibodies could pull-down 5fC- or 5caC-containing oligonucleotides specifically and efficiently, suggesting that they are suitable for DIP assays (Figure 1B).
Figure 1. Enrichment of 5fC and 5caC from genomic DNA by modification-specific antibodies.
(A) The 5fC and 5caC antibodies specifically recognize 5fC and 5caC-containing DNA oligos in dot-blot assays, respectively. Different amounts of 38-mer DNA oligonucleotides (oligos), where the cytosines in 9 CpGs are either C, 5mC, 5hmC, 5fC or 5caC, were spotted on membrane and probed with 5fC and 5caC antibodies, respectively.
(B) DIP-qPCR analysis demonstrates the specificity of the antibodies.
(C) RT–qPCR analysis of Tdg expression levels in control (shCtrl) and Tdg-deficient (shTdg) mouse ESCs.
(D) Mass spectrometric quantification of 5mC, 5hmC, 5fC, and 5caC in control and Tdg-deficient cells.
(E) DIP-qPCR analysis of 5hmC/5fC/5caC at three 5hmC enriched regions. Data are presented as mean ± SEM.
See also Figure S2.
Quantitative mass spectrometry analysis indicates that 5fC and 5caC levels are approximately 2% or 0.5% of the total level of 5hmC in wild-type mouse ESCs, respectively (Ito et al., 2011). Given that mouse ESCs possess high levels of Tet enzymatic activities, the relatively low abundance of 5fC/5caC suggests that 5fC and 5caC marks may be rapidly removed by TDG in vivo (He et al., 2011; Maiti and Drohat, 2011; Nabel et al., 2012). Thus, blocking TDG activity may result in accumulation of 5fC and 5caC, which allows the identification of genomic loci targeted by TDG activity. To test this possibility, we generated Tdg-deficient mouse ESCs by lentivirus-mediated knockdown (Figure 1C). Mass spectrometry analysis demonstrated that global levels of 5fC and 5caC increased by 5.6-fold and 8.4-fold, respectively, in response to Tdg knockdown (Figure 1D). In contrast, neither 5mC nor 5hmC showed significant change upon Tdg knockdown (Figure 1D). Consistent with previous results demonstrating that Tdg is not required for mouse ESC maintenance (Cortazar et al., 2011), neither the morphology nor the expression levels of pluripotent genes (Oct4, Sox2 and Nanog) or Tet genes were altered by Tdg knockdown (Figure S2).
We next tested 5fC and 5caC antibodies in immunoprecipitating genomic DNA fragments at three Tet1-bound and 5hmC-enriched regions (Tcl1, Sox17 and Esrrb) (Wu et al., 2011a; Wu et al., 2011b). Consistent with the fact that TDG does not excise 5hmC, 5hmC levels at these loci were not affected by Tdg knockdown (Figure 1E). In contrast, 5fC and 5caC levels at these loci were significantly increased in Tdg-deficient cells (Figure 1E). Given that marked elevation in 5fC- and 5caC-DIP signals are detected in Tdg knockdown ESCs, we conclude that 5fC and 5caC antibodies are highly specific and are potentially suitable for genome-wide 5fC/5caC-DIP analysis.
Preferential enrichment of 5fC/5caC at pericentric heterochromatin in mouse ESCs
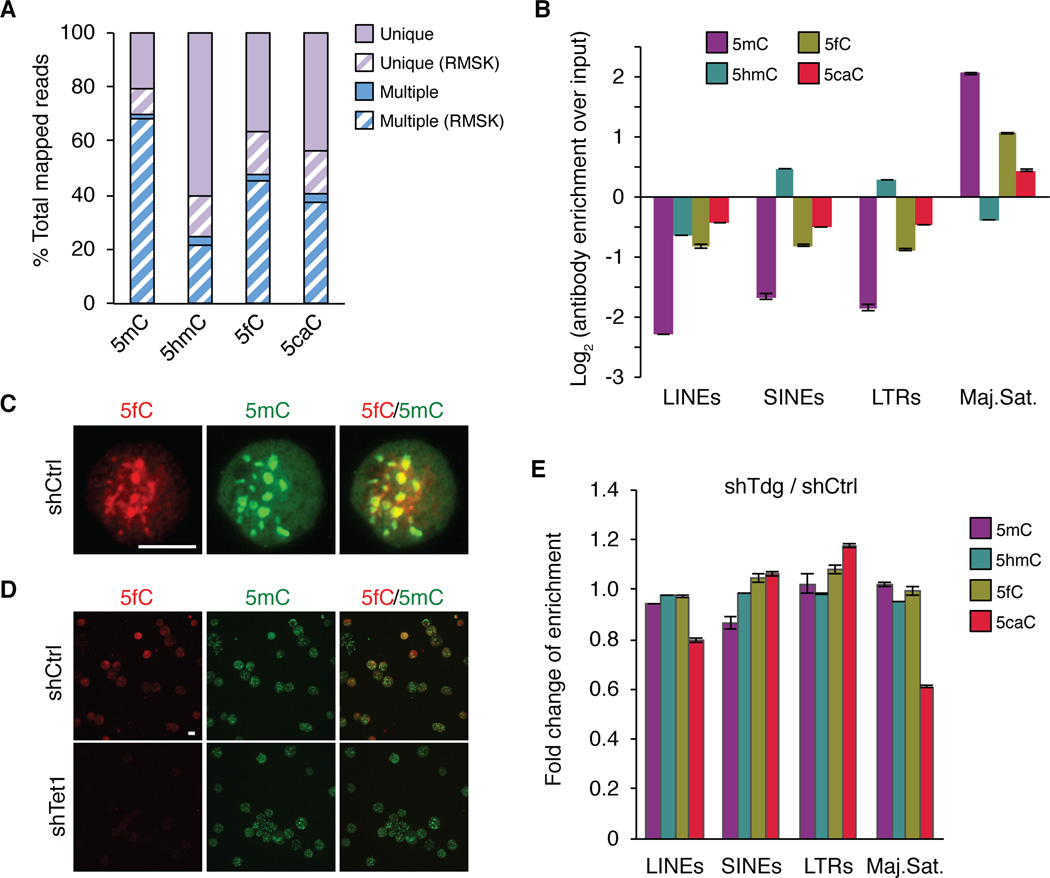
To map 5fC/5caC distribution, we performed 5fC and 5caC DIP-Seq experiments in replicates using genomic DNA of control and Tdg-deficient mouse ESCs (Figure S1E and Table S1). We also performed 5mC, 5hmC and mock IgG DIP-Seq experiments using the same genomic DNA (Figure S1E). Sequencing reads mapped to multiple genomic regions (multi-hit reads) generally represent repetitive sequences in the genome. Indeed, we found that 89–98% of the multiple mapped reads overlap with the UCSC RepeatMasker (RMSK) track (Dreszer et al., 2012), whereas only 20–31% of the uniquely mapped reads overlap with RMSK (Figure 2A). To evaluate the potential enrichment of 5fC/5caC at repetitive sequences, multi-hit reads were retained in the initial analysis. Interestingly, the percentage of multi-hit reads varies greatly among different cytosine modifications (Figures 2A). In control mouse ESCs, 48% 5fC reads and 41% 5caC reads are multi-hit reads, which is higher than that of 5hmC (25%), but lower than that of 5mC (70%). Thus, these results not only confirm that 5mC and 5hmC are relatively enriched and depleted from repetitive sequences respectively (Ficz et al., 2011; Williams et al., 2011; Yoder et al., 1997), but also suggest that 5fC and 5caC may accumulate to detectable levels at repetitive sequences in wild-type mouse ESCs.
Figure 2. 5fC and 5caC accumulate at major satellite repeats of the pericentric heterochromatin in wild-type mouse ESCs.
(A) Percentages of uniquely mapped and multi-hit reads in total mapped reads. Reads that overlap with the UCSC RepeatMasker (RMSK) track are highlighted by forward slash.
(B) Relative enrichment (log2 ratio of IP over input) for each cytosine modification at major classes of repetitive sequences in mouse ESCs. Values represent means of two biological replicates with ends of error bars corresponding to individual data points.
(C, D) Representative images of mouse ESC surface spreads co-stained with 5fC and 5mC antibodies. Same exposure time was used for comparing control (shCtrl) and Tet1 knockdown (shTet1) mouse ESCs in (D). Scale bar, 100 µm.
(E) Bar graph presentation of the fold change of the enrichment in each class of repetitive sequences upon Tdg knockdown.
See also Tables S1.
To further determine the types of repeats at which 5fC and 5caC are relatively enriched, we classified all sequencing reads on the basis of RMSK annotation, and calculated the number of reads in each repeat class. After correcting the relative percentage of various classes of repeats, 5mC, 5fC and, to a lesser extent, 5caC are found to be preferentially enriched at major satellite repeats, whereas 5hmC tends to accumulate at short interspersed nuclear elements (SINEs) and long-terminal repeats (LTRs) (Figures 2B). Furthermore, co-staining of mouse ESC surface spreads with 5fC and 5mC antibodies revealed significant overlap of 5fC signals with 5mC (Figure 2C), which is known to be enriched at pericentric heterochromatin (i.e., major satellite repeats). Immunostaining analysis further showed marked reduction of 5fC signals in the Tet1 knockdown cells (Figure 2D), validating the specificity of 5fC signals at pericentric regions. Tdg knockdown does not significantly alter the relative enrichment ratio of 5fC at major satellite repeats, but it significantly reduced that of 5caC at the major satellite repeats (Figure 2E), probably due to the increased level of 5caC at other genomic regions. Collectively, these results indicate that, 5fC and 5caC (to a less extent) tends to accumulate at major satellite repeats in wild-type mouse ESC, and TDG may not efficiently excise 5fC/5caC at pericentric heterochromatin.
Tdg-depletion results in accumulation of 5fC and 5caC in non-repetitive regions
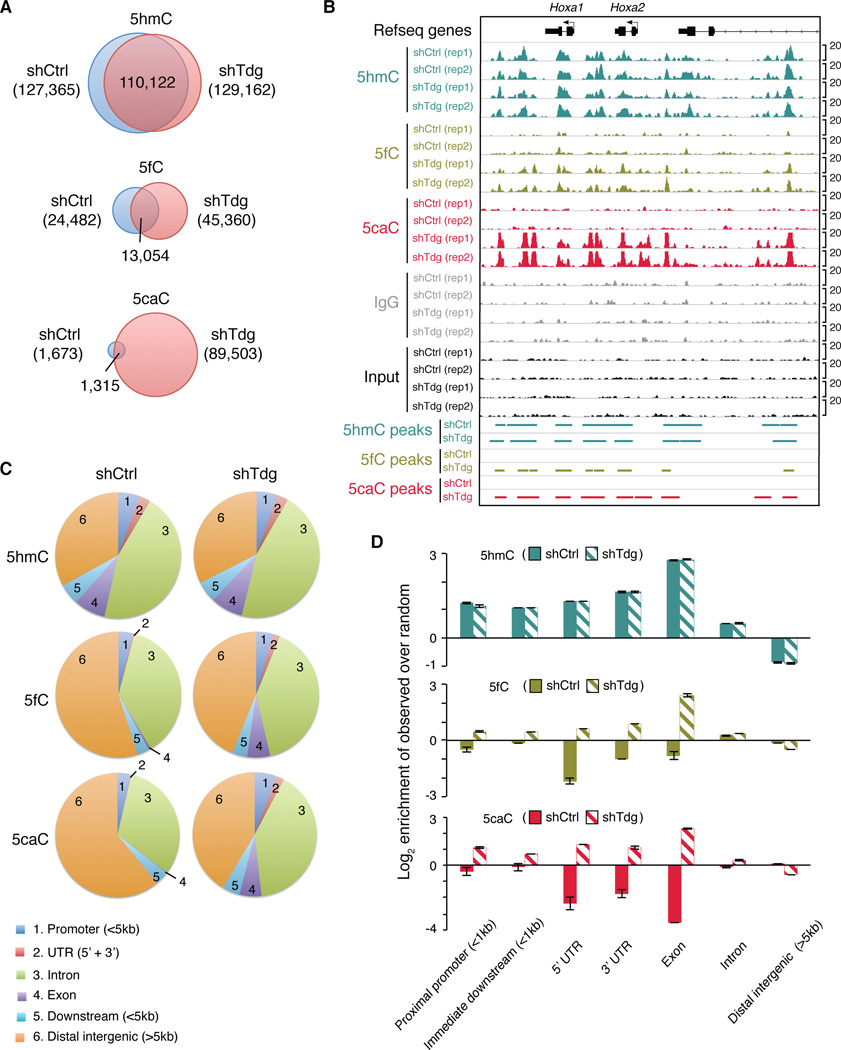
To further investigate Tdg-deficiency induced changes of 5fC/5caC signals at non-repetitive regions, we analyzed uniquely mapped reads. To identify genomic loci enriched for high-confidence 5fC/5caC signals, we first identified peak candidates using input genomic DNA as a negative control and then quantitatively filtered out peaks with relatively high levels of signals in IgG controls (see Extended Experimental Procedures). In wild-type mouse ESCs, we identified 1,673 regions enriched for 5caC (Figure 3A and Table S2). Upon Tdg knockdown, a marked increase in the number of 5caC peaks (n=89,503) was observed (Figure 3A and 3B; Table S3). Many newly appeared 5caC peaks co-localize with 5hmC peaks, which are largely unaffected by Tdg knockdown (Figure 3A and 3B). Tdg-depletion also leads to a less pronounced increase in the number of 5fC peaks (Figure 3A and 3B; Table S4 and S5). Notably, significantly more 5fC peaks (n=24,482) are detected in wild-type mouse ESCs relative to 5caC peaks (n=1,673) (Figure 3A), which is consistent with previous findings that 5fC is more abundant than 5caC in mouse ESCs (Ito et al., 2011; Pfaffeneder et al., 2011). Importantly, the observation that a large number of 5fC/5caC peaks are specifically detected in Tdg-knockdown cells underscores the sensitivity and specificity of the 5fC/5caC DIP-seq method.
Figure 3. Accumulation of 5fC and 5caC in non-repetitive regions in Tdg-deficient mouse ESCs.
(A) Venn diagrams showing the overlap of 5hmC, 5fC or 5caC peaks in control and Tdg-deficient mouse ESCs.
(B) Representative genomic loci (Hoxa1 and Hoxa2) showing 5hmC/5fC/5caC peaks in control and Tdg-deficient mouse ESCs.
(C) Pie chart presentation of the overall genomic distribution of 5hmC/5fC/5caC enriched regions.
(D) Enrichment (log2 ratios of observed over random) of 5hmC/5fC/5caC in Tdg-deficient cells relative to control at various genomic features. Values represent means of two biological replicates with ends of error bars corresponding to individual data points.
Next, we sought to determine where the newly appeared 5fC and 5caC peaks are preferentially located in the genome. Compared to random control regions, a large fraction of 5fC and 5caC peaks in Tdg-deficient mouse ESCs are located in distal intergenic regions (group 6 in Figures 3C), and the relative percentage of 5fC and 5caC peaks in genic regions (promoters: group 1; introns: group 3; exons: group 4 in Figure 3C) is increased compared to that in control cells (Figures 3D and S3A). Moreover, 5fC and 5caC signals are increased preferentially within exons (solid lines in Figure S3B) relative to introns (dash lines in Figure S3B) in response to Tdg knockdown. This finding is consistent with previous studies demonstrating the enrichment of 5hmC at exon/intron boundaries (Khare et al., 2012), suggesting that a potential role of TET/TDG-mediated generation and excision 5hmC/5fC/5caC in regulating transcriptional elongation and/or splicing.
5fC and 5caC exhibit common and unique distributions
Both Tet1 and Tet2 are highly expressed in mouse ESCs (Ito et al., 2010; Koh et al., 2011), and 5fC and 5caC levels are significantly reduced upon Tet1 depletion (Ito et al., 2011). To test whether Tet1 occupancy correlates with 5fC/5caC generation in vivo, we examined 5fC and 5caC signals at regions enriched for Tet1. In Tdg-deficient cells, Tet1 bound regions with medium-to-low CpG density are preferentially enriched for 5fC and 5caC signals (Figure S3C). In contrast, Tet1 bound regions with high CpG density tend to be depleted of 5fC/5caC in both control and Tdg-deficient mouse ESCs (Figure S3C), which is in agreement with the finding that CpG-rich regions are generally depleted of 5mC and 5hmC (Szulwach et al., 2011a; Wu et al., 2011a; Yu et al., 2012). Further analysis revealed that four cytosine modifications have both overlapping and unique distributions in the genome (Figure S3D). Notably, in Tdg-deficient mouse ESCs, 5fC peaks tend to co-localize with 5mC enriched regions, while 5caC peaks preferentially overlap with 5hmC-enriched regions (Figure S3E), suggesting that processivity of Tet proteins may be regulated by local sequence context and/or chromatin structure.
TDG-mediated 5fC/5caC excision occurs extensively at distal regulatory elements
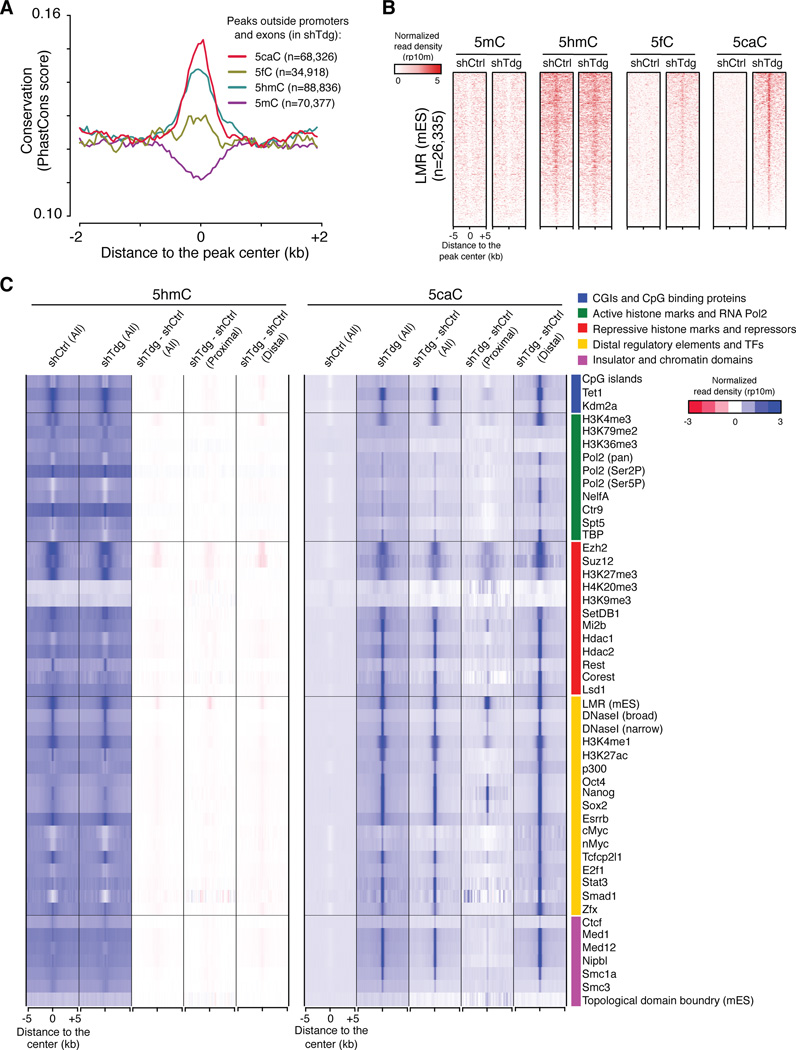
Further analysis of 5fC and 5caC peaks in Tdg-deficient mouse ESCs indicates that the majority of Tdg-depletion induced 5caC peaks (76.3% of all 5caC peaks, n=68,326) are located outside promoter or exonic regions. To investigate whether these distal regions are of functional relevance, we calculated the sequence conservation scores within these ectopic 5fC/5caC peaks. As expected, peaks overlapping with exons and proximal promoters show strong evolutionary conservation (Figure S4A). Interestingly, sequences overlapping with Tdg-depletion induced non-exonic and non-promoter 5caC peaks (to a less extent for ectopic 5fC peaks) are also relatively conserved compared to flanking regions (Figure 4A). Furthermore, 5caC peaks in Tdg-deficient mouse ESCs frequently overlap with low-methylated regions (LMRs) (Figure 4B), a unique group of genomic regions that display features of distal regulatory regions and are generally associated with intermediate (~30%, measured by BS-seq) DNA methylation level (Stadler et al., 2011). These results suggest that both 5mC oxidation and 5fC/5caC excision activity may be preferentially recruited to a large cohort of distal regulatory elements.
Figure 4. Tdg-depletion induced ectopic 5fC and 5caC accumulate at distal regulatory regions.
(A) Average conservation (phastCons) scores within regions flanking the center of 5mC/5hmC/5fC/5caC peaks (non-overlapping with exons or promoters) in Tdg-deficient mouse ESCs. The number of peaks that are located outside exons and proximal promoters (+/− 1kb flanking TSSs) for each cytosine modification is also shown.
(B) Heat maps of 5mC/5hmC/5fC/5caC levels (normalized read density) at centers of LMRs previously identified in mouse ESCs. The heat maps are ranked by the mean of 5caC signals in Tdg-deficient cells (top: highest, bottom: lowest).
(C) Heat maps of 5hmC and 5caC levels (normalized read density) in control and Tdg-deficient cells at centers of annotated genomic features or enriched regions for transcriptional regulators, histone modifications, pluripotency transcription factors (TFs) and distal regulatory regions (derived from published datasets in mouse ESCs). The difference in 5hmC and 5caC levels between control and Tdg-deficient cells (shTdg minus shCtrl) is also shown for all, proximal (overlapping with +/−1kb flanking TSSs) and distal features.
See also Figure S4.
To further characterize regions where TDG actively excises 5mC oxidation derivatives, we calculated the averaged 5hmC and 5caC signals within genomic features derived from published genome-wide mapping datasets for a number of DNA binding factors and major histone modifications. This analysis indicates that in Tdg-deficient mouse ESCs, 5caC accumulates at binding sites of pluripotency transcription factors (TFs) such as Oct4, Nanog, Sox2 and Esrrb (Figure 4C, yellow color group) (Chen et al., 2008; Marson et al., 2008). For instance, 5caC peaks in Tdg-deficient cells show a significant overlap (observed/expected=16.0, P<2.2×10−16, Fisher’s exact test) with Oct4 binding regions (+/−100bp flanking peak summits) compared to that expected by chance. Ectopic 5caC signals are also preferentially detected at peaks of factors (e.g. p300) and histone marks (e.g. H3K4me1 and H3K27ac) that are associated with distal enhancer elements (Figure 4C, yellow color group) (Creyghton et al., 2010; Shen et al., 2012). Notably, the presence of Tdg-depletion induced 5caC signals at these distal elements is not simply due to higher level of 5hmC, as Smad1 binding sites are not enriched for high levels of 5hmC, but are frequently associated with ectopic 5caC signals (Figure 4C). Furthermore, ectopic 5caC signals are also frequently detected at binding sites of the cohesion complex and mediator proteins, both of which are implicated in regulating interactions between promoter and enhancers (Figure 4C, pink color group) (Kagey et al., 2010). Many regions enriched for repressor complexes (e.g. LSD1, Hdac1/2) that are involved in decommissioning active ESC enhancers during differentiation also frequently overlap with ectopic 5caC peaks (Figure 4C, red color group) (Whyte et al., 2012). By contrast, regions corresponding to basal transcriptional machineries (e.g. RNA Pol2, TBP in green color group), insulators (CTCF in pink color group) and topological domain boundaries (in pink color group) are not enriched for ectopic 5caC peaks (Figure 4C) (Dixon et al., 2012; Kagey et al., 2010; Rahl et al., 2010). Notably, for most features analyzed in Figure 4C, distally located elements are preferentially associated with Tdg-depletion induced 5fC/5caC signals relative to proximally located ones (within +/−1kb regions flanking transcriptional start sites (TSSs)) (Figure 4C and S4B), suggesting that TET/TDG may be more active at or preferentially recruited to regions outside proximal promoters. Taken together, these results indicate that TET/TDG-mediated 5mC oxidation and 5fC/5caC excision actively take place at a large cohort of distal cis-regulatory elements.
TDG-mediated 5fC/5caC excision occurs preferentially at active enhancers in mouse ESCs
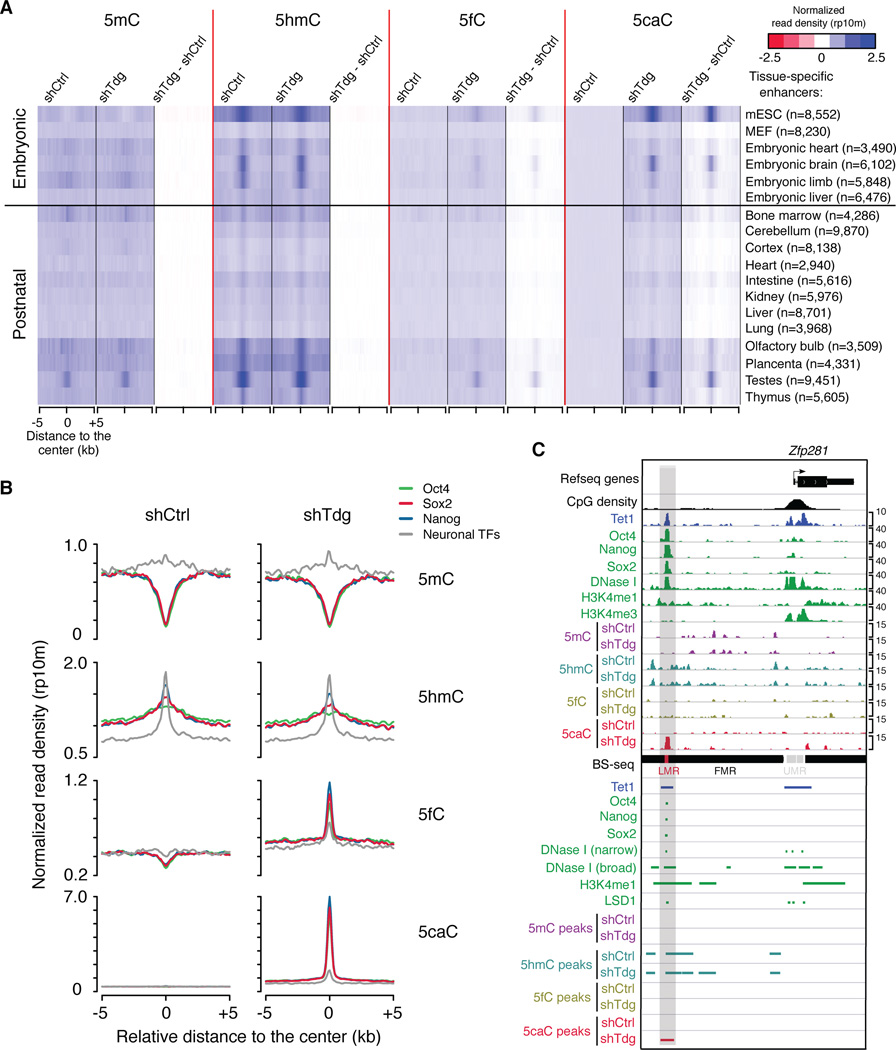
To further analyze distal cis-elements targeted by TDG activity in mouse ESCs, we calculated averaged 5hmC/5fC/5caC signals (in both control and Tdg-deficient mouse ESCs) at cell-type or tissue-specific enhancers identified by mouse ENCODE project (Shen et al., 2012). Interestingly, enhancers that are specifically active in mouse ESCs are associated with the highest level of ectopic 5caC signals in Tdg-deficient mouse ESCs (Figure 5A). These mouse ESC-specific enhancers are also preferentially bound by Tet1 and associated with DNase I hypersensitivity sites (Figure S5A), suggesting that cytosines within or surrounding active enhancer regions tend to undergo TET/TDG-mediated 5mC oxidation dynamics. Similarly, mouse ESC-specific LMRs are associated with higher levels of Tdg-depletion induced 5fC/5caC signals than neural progenitor (NP)-specific LMRs (Stadler et al., 2011) (Figure S5B). In addition, analysis comparing binding sites of pluripotency TFs and neuronal TFs indicates that in mouse ESCs (Kim et al., 2010; Marson et al., 2008), pluripotency TF bound regions are preferentially marked by ectopic 5fC/5caC signals in Tdg-deficient mouse ESCs (Figure 5B and S5C). As exemplified in Figure 5C and S5D, a cohort of distal regions bound by Oct4/Sox2/Nanog are associated with newly appeared 5caC peaks in Tdg-deficient mouse ESCs regardless of the presence of stable Tet1 occupancy. Together, these results suggest that TET/TDG-mediated 5mC oxidation dynamics in mouse ESCs may contribute to the regulation of active enhancer activity.
Figure 5. TET/TDG activities are preferentially recruited to active enhancers and distal pluripotency TF binding sites in mouse ESCs.
(A) Heat maps of 5mC/5hmC/5fC/5caC levels (normalized read density) in control and Tdg-deficient cells at centers of tissue-specific enhancers.
(B) Average 5mC/5hmC/5fC/5caC signals in control and Tdg-deficient mouse ESCs at the center of binding sites for pluripotency TFs (Oct4/Nanog/Sox2) and neuronal TFs.
(C) 5mC/5hmC/5fC/5caC levels in control and Tdg-deficient mouse ESCs at a representative locus (upstream of the Zfp281 gene) of binding sites of pluripotency TFs (Oct4/Nanog/Sox2). Other genomic features (e.g. DNase I hypersensitivity sites, H3K4me1 enriched regions, LMRs and enhancer-related epigenetic regulator LSD1) are also shown.
See also Figure S5.
TDG-mediated 5fC/5caC excision occurs preferentially at transcriptionally inactive gene promoters in mouse ESCs
Although only a small portion of ectopic 5fC/5caC peaks overlap with proximal promoters, we frequently observed 5caC accumulation at regions flanking gene promoters or 3’ gene bodies of transcribed genes in Tdg-deficient mouse ESCs. Previous studies suggest that distinct genic regions are associated with specific histone lysine methylation patterns (Barski et al., 2007; Bernstein et al., 2006; Mikkelsen et al., 2007; Whyte et al., 2012), which may in turn contribute to gene expression status (Figure S6A).
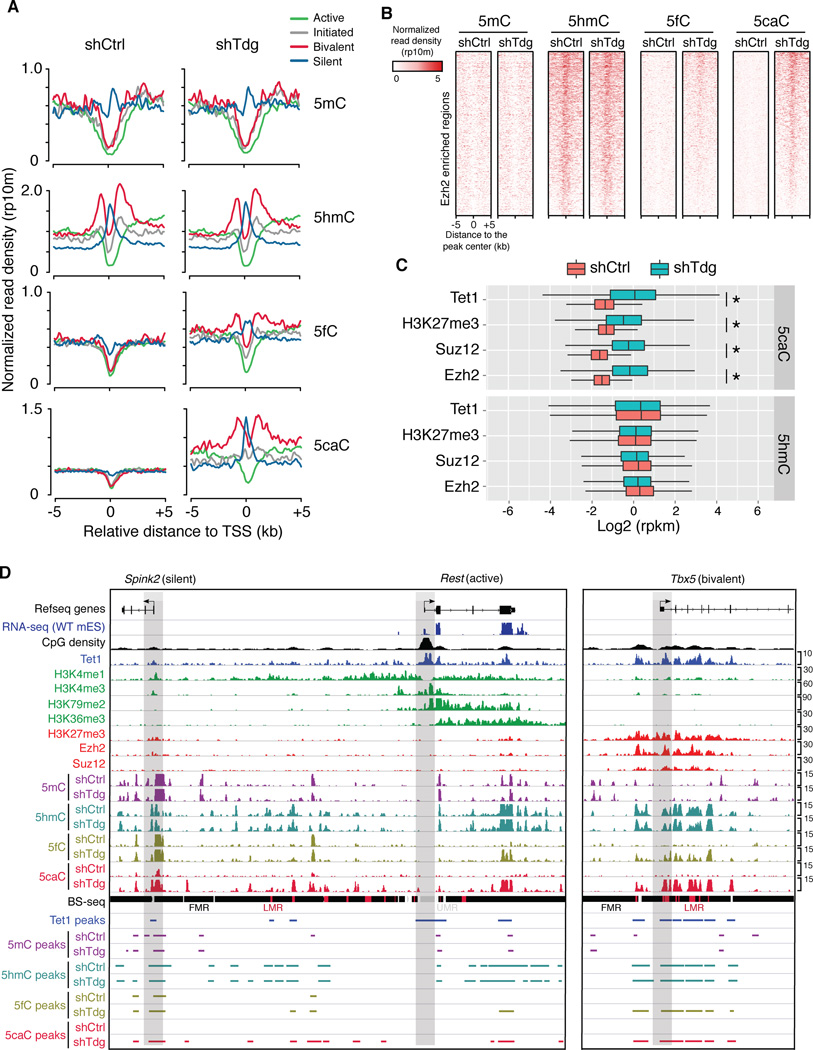
To explore the possibility that distinct chromatin states may influence the generation and excision of 5fC/5caC by TET/TDG, we compared average signal profiles of 5mC/5hmC/5fC/5caC at four groups of extended gene promoters (+/− 5kb relative to TSSs): 1) “Active”, characterized by the presence of high levels of H3K4me3 (active histone mark) at proximal promoters and H3K79me2/3 (indicative of elongation) at 5’ of gene bodies; 2) “Initiated”, only associated with promoter H3K4me3; 3) “Bivalent”, associated with both H3K27me3 (repressive histone mark) and medium-to-low levels of promoter H3K4me3; 4) “Silent”, lack of promoter H3K4me3. This analysis revealed that 5fC/5caC levels are relatively comparable between control and Tdg-deficient mouse ESCs at gene promoters that are associated with active transcription (green in Figure 6A, exemplified by Rest in Figure 6B and P2 promoter of Dnmt1 in supplementary Figure 6B) or transcription initiation (grey in Figure 6A). These observations suggest that TET/TDG-mediated 5mC oxidation dynamics is generally absent at these transcriptionally active/permissive promoters. By contrast, a substantial increase of 5fC/5cac levels was detected at genomic regions flanking bivalent promoters in the absence of Tdg (exemplified by Tbx5 in Figure 6B and HoxA cluster in supplementary Figure 6B). Considering that 5hmC is also enriched at bivalent domains (Figure 6A), these results suggest that bivalent domains are targeted by relatively high levels of TET/TDG activities in mouse ESCs. Silent promoters (blue in Figure 6A) were also associated with relatively high levels of ectopic 5fC/5caC signals(exemplified by Spink2 in Figure 6B and P1 promoter of Dnmt1 in supplementary Figure 6B). Because 5mC is also enriched at silent gene promoters (Figure 6A), it seems that silent gene promoters in mouse ESCs are simultaneously targeted by activities of DNMT/TET/TDG. Collectively, these results indicate that transcriptionally inactive (silent or bivalent/poised) gene promoters are preferentially regulated by TET/TDG activity and tend to undergo active DNA demethylation in mouse ESCs.
Figure 6. Tdg-depletion induced 5fC/5caC signals are enriched at bivalent and transcriptionally silent gene promoters in mouse ESCs.
(A) Average 5mC/5hmC/5fC/5caC signals in control and Tdg-deficient mouse ESCs at TSSs of four groups of gene promoters that are associated with distinct chromatin states (active: H3K4me3+/H3K79me2+; initiated: H3K4me3+ only; bivalent: H3K4me3+/H3K27me3+; silent: none).
(B) 5mC/5hmC/5fC/5caC levels in control and Tdg-deficient mouse ESCs at representative loci of gene promoters that are associated with different histone modification patterns. The gene promoters are highlighted by grey bars. Fully methylated regions (FMRs), low methylated regions (LMRs) and unmethylated regions (UMRs) were derived from previously published BS-seq datasets.
(C) Heat maps of 5mC/5hmC/5fC/5caC levels (normalized read density) in control and Tdg-deficient mouse ESCs at centers of Ezh2 binding sites.
(D) Box-plots of normalized 5hmC and 5caC levels (read per million reads and kilo bases, rpkm) in control and Tdg-deficient cells within genomic regions enriched for Tet1, Ezh2, Suz12 and H3K27me3. *, P<2.2e-16 (P-values were calculated by two-tailed t-test).
See also Figure S6.
H3K27me3 within bivalent domains are deposited by Polycomb repression complex 2 (PRC2) (Cao et al., 2002), and PRC2 binding to chromatin is antagonized by the presence of 5mC (Bartke et al., 2010; Wu et al., 2010). To directly examine the relationship between PRC2 binding and TET/TDG-mediated 5mC oxidation dynamics, we examined the ectopic 5fC/5caC levels within regions enriched for two core PRC2 subunits, Ezh2 and Suz12 (Ku et al., 2008). In Tdg-deficient cells, 5fC/5caC accumulates to a significant level within Ezh2 and Suz12 bound regions (Figure 6C–D and S6C), suggesting a potential role of TET/TDG proteins in regulating PRC2 activity or targeting.
TDG-mediated 5fC/5caC excision and gene expression
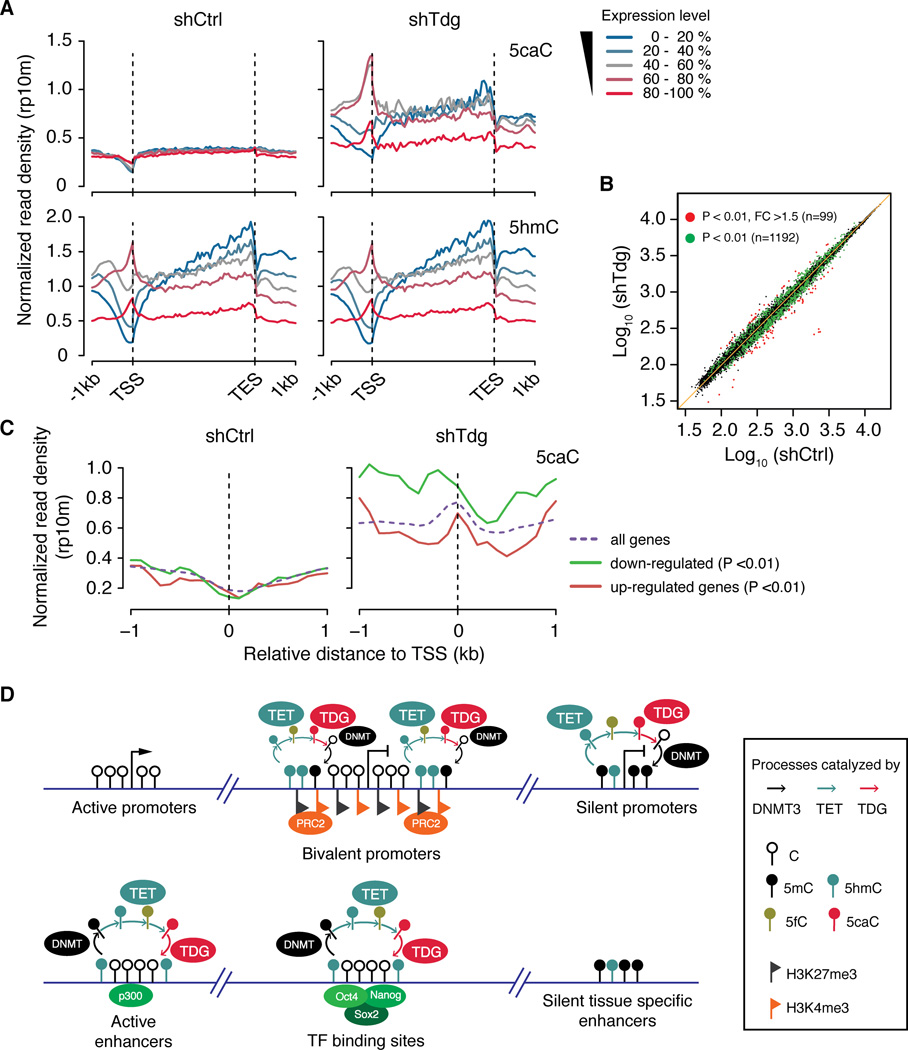
Next, we examined the relationship between 5fC and 5caC distribution and the global gene expression profile. Using published RNA-seq datasets of wild-type mouse ESCs (Ficz et al., 2011), ectopic 5fC/5caC signals are found to be depleted in the promoters of highly expressed genes, but were relatively enriched in the intragenic regions (especially 3’ end) of highly and moderately expressed genes (Figure 7A). In support of the notion that silent or repressed/poised promoters tend to be targeted by TET/TDG activity, promoters of gene with low-to-medium expression levels are enriched for ectopic 5fC/5caC signals (Figure 7A and S7A). Collectively, these results indicate that TET/TDG-dependent cytosine modification dynamics may play a complex role in transcriptional regulation, depending on their genomic location.
Figure 7. Complex relationship between gene expression and TET/TDG-mediated 5mC oxidation dynamics.
(A) Average signals of 5hmC and 5caC within genes expressed at different levels in control (left) and Tdg-deficient (right) mouse ESCs.
(B) Scatter plots comparing gene expression profiles of control and Tdg-deficient mouse ESCs. Green and red dots indicate differentially expressed genes at P < 0.01 and P < 0.01, FC >1.5, respectively.
(C) Average 5caC signals in control (left) and Tdg-deficient (right) mouse ESCs at the TSS of down-regulated and up-regulated genes (P < 0.01).
(D) Schematic diagram illustrating the relationship between transcriptional activity and DNMT/TET/TDG-mediated cytosine modification cycling at promoters and distal regulatory regions in mouse ESCs. Dynamic cyclic changes of cytosine modifications preferentially occur within bivalent and silent promoters, as well as active enhancers and pluripotency TF binding sites.
See also Figure S7.
To further study the potential role of cytosine modification cycling in regulating gene expression, we performed microarray analysis comparing gene expression in control and Tdg-deficient mouse ESCs. Consistent with the grossly normal phenotype of Tdg-deficient mouse ESCs (Figure S2), gene expression changes upon Tdg knockdown appear to be minor, with only 99 genes showing relatively marked expression change (P < 0.01 and fold change >1.5). More genes (n=1,192) exhibited small but significant change in expression (P < 0.01) in response to Tdg-depletion (Figure 7B). We then compared average signals for all cytosine modifications flanking TSSs of up-regulated (n=413) and down-regulated genes (n= 636). This analysis indicated that the 5fC/5caC signals at proximal promoters of down-regulated genes tend to increase more dramatically when compared to those of up-regulated genes in response to Tdg-depletion (Figure 7C and S7B), suggesting a transcriptional inhibitory role of 5fC/5caC at proximal promoters.
DISCUSSION
TET/TDG-mediated 5mC removal occurs extensively in the mammalian genome
In conjunction with DNMTs, the step-wise process of active DNA demethylation entailed by TET/TDG/BER in principle permits cyclic changes of modification state at all cytosine bases (predominantly in the context of CpG) in the genome. In contrast to the readily detectable 5hmC, 5fC and 5caC are present at much lower levels in mammalian cells. Several non-mutually exclusive mechanisms may be responsible for the observed scarcity of 5fC/5caC. First, oxidation of 5hmC to 5fC/5caC by TET proteins may be tightly regulated and is less efficient compared to conversion of 5mC to 5hmC. Second, 5hmC can be passively removed by replication-dependent dilution in proliferating cells or converted to other form of modifications (e.g. 5-hydroxymethyluracil or direct conversion of 5hmC to C) before 5hmC is further oxidized by Tet proteins (Chen et al., 2012; Guo et al., 2011; Inoue and Zhang, 2011). Third, TDG-mediated excision of 5fC/5caC is highly efficient, thus 5fC and 5caC are short-lived (Globisch et al., 2011; Ito et al., 2011). To better understand the relative contribution of TET/TDG-mediated active demethylation pathway (5mC oxidation and excision of 5fC/5caC) to DNA methylation dynamics, we applied antibody-based DIP-seq analysis to mouse ESCs and generated global distribution maps of 5hmC/5fC/5caC in the presence or absence of TDG activity. Comparative analysis of 5hmC/5fC/5caC distribution in control and Tdg-deficient mESCs has revealed that a large number of genomic loci are targeted by TET/TDG activities, suggesting that large-scale DNA methylation and demethylation dynamics may not be a unique feature for developing zygotes and PGCs, but rather a prevalent event that may takes place in the genome of diverse cell types. Because mouse ESCs are highly proliferative and 5hmC/5fC/5caC can also be regulated by the replication-dependent dilution mechanism, future studies of TET/TDG activity in terminally differentiated and post-mitotic cells will facilitate the study of functional roles of TET/TDG-dependent active DNA demethylation pathway in gene regulation.
Steady state accumulation of 5fC and 5caC at specific class of repetitive sequences
We observed that, much like 5mC, on a population average, 5fC and 5caC (to a lesser extent) are relatively enriched at repetitive sequences, particularly at major satellite repeats. The accumulation of 5fC and potentially 5caC in major satellite repeats can be explained by at least two mechanisms: (1) TET proteins tend to oxidize their substrates with a higher processivity in major satellite repeats due to the unique sequence context, local DNA methylation level or CpG density; (2) Tdg is less efficient in removing 5fC in major satellite repeats, which are located in pericentric heterochromatin, relative to other locations. In support of the second possibility, previous studies have shown that TDG is unable to associate with heterochromatized promoters (Cortazar et al., 2011).
TET/TDG-mediated 5mC oxidation dynamics at transcriptionally poised (bivalent) and silent gene promoters
Tet1 tend to be enriched at CpG-rich gene promoters through its CXXC domain (Tahiliani et al., 2009; Xu et al., 2011). However, both affinity enrichment-based and modified BS-seq (TAB-seq) analyses of 5hmC distribution indicate that 5hmC tends to be enriched at promoters with medium-to-low levels of CpG density, but depleted from CpG-rich promoters (Wu et al., 2011a; Yu et al., 2012). The discrepancy between Tet1 occupancy and the 5hmC level suggests that at CpG-rich promoters, either 5hmC is not efficiently generated by Tet1 due to lack of 5mC or 5hmC is rapidly oxidized to 5fC/5caC followed by TDG-mediated 5fC/5caC excision. The fact that, in Tdg-deficient cells, 5fC/5caC are not accumulated at CpG-rich, actively transcribed gene promoters, suggests that these CpG-rich, Tet1 bound promoters are generally not associated with active demethylation process. In contrast, a marked increase in 5fC/5caC level is detected at Tet1 bound bivalent domains flanking transcriptionally repressed/poised promoters (Figure 7D, upper panels). These bivalent promoters generally encode developmental regulators and lineage-specific transcription factors, thus TET/TDG-mediated active demethylation process may be required to maintain a transcriptionally poised state at these promoters. Interestingly, previous studies have suggested that bivalent promoters show a tendency of being DNA methylated in cancer cells (Baylin and Jones, 2011), so the dys-regulation of active demethylation process in tumors may contribute to the observed hypermethylation status at bivalent promoters.
TET/TDG-mediated 5mC oxidation dynamics at distal-regulatory regions
The ability to determine the genome-wide distribution of all 5mC oxidation derivatives offered a unique opportunity to assess the TET/TDG-mediated 5mC oxidation dynamics at various genomic features and regulatory elements. Unlike widespread distribution of 5mC and 5hmC, 5fC and 5caC in wild-type mouse ESCs are hardly detectable at non-repetitive regions. Upon depletion of Tdg, many ectopic 5fC and 5caC peaks appeared at distal, but not proximal regulatory elements. This observation agrees with recent findings from base-resolution mapping of 5mC and 5hmC in the mouse ESCs (Stadler et al., 2011; Yu et al., 2012) and suggests that TET/TDG-mediated active DNA demethylation occurs extensively at a large cohort of distal regulatory regions (Figure 7D, lower panels). Future studies are needed to elucidate the function of cyclic change of cytosine modifications at distal regulatory elements.
In summary, we have developed an affinity enrichment-based approach to determine genome-wide distribution of 5fC and 5caC and have generated 5fC and 5caC maps in both wild-type and Tdg-deficient mouse ESCs. Analysis of these datasets suggests that dynamic cytosine methylation/demethylation cycle occurs at an unexpectedly large number of genomic loci across the genome. Genome-wide mapping of all 5mC oxidation derivatives described in this study sets the stage to systematically study the function of DNA methylation and demethylation dynamics in development and diseases.
EXPERIMENTAL PROCEDURES
Cell culture and lentiviral knockdown of Tdg
Mouse E14Tg2A ESCs were cultured in feeder-free conditions. For Tdg knockdown, mouse ESCs were infected with lentiviruses expressing both the puromycin N-acetyl-tranferase and the short hairpin RNA (shRNA) targeting Tdg (5’-GCAAGGATCTGTCTAGTAA-3’). Infected cells were selected by puromycin for one week before being harvested for further experiments. Detailed procedures can be found in Extended Experimental Procedures.
5mC/5hmC/5fC/5caC DIP-Seq
The antisera for 5fC and 5caC were previously described (Inoue et al., 2011). For each DIP experiment, 10 µg of sonicated, adaptor ligated genomic DNA from control or Tdg knockdown mouse ESCs was used as input, and 5 µl of 5mC antibody (Eurogentec, BI-MECY-0500), 5 µL of 5hmC antibody (Active Motif, 39791), 1 µl of 5fC antiserum or 0.3 µl of 5caC antiserum was used to immunoprecipitate modified DNA as previously described (Wu et al., 2011a). Immunoprecipitated DNA was further amplified for high-throughput sequencing. Detailed DIP-Seq procedures as well as the following data analysis methods can be found in Extended Experimental Procedures.
Supplementary Material
HIGHLIGHTS.
Affinity enrichment-based approach for genome-wide mapping of 5fC and 5caC
The first genome-wide view of iterative 5mC oxidation dynamics in mouse ESCs
Tdg-depletion induced 5fC and 5caC accumulate at bivalent and silent promoters
TET/TDG activities are recruited to active enhancers and TF binding sites
ACKNOWLEDGEMENTS
We thank Drs. Jin He and Falong Lu for their helpful discussions. This project is supported by NIH grant U01DK089565 (to Y.Z.) and R01GM097253 (to K.Z.). H.W. is supported by a postdoctoral fellowship (Merck Fellow) from Jane Coffin Childs Funds for Medical Research. D.D. is a CIRM pre-doctoral fellow. Y.Z. is an investigator of the Howard Hughes Medical Institute. The DIP-seq and expression microarray datasets have been deposited in Gene Expression Omnibus (GEO) under the accession number GSE42250.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bartke T, Vermeulen M, Xhemalce B, Robson SC, Mann M, Kouzarides T. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell. 2010;143:470–484. doi: 10.1016/j.cell.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Booth MJ, Branco MR, Ficz G, Oxley D, Krueger F, Reik W, Balasubramanian S. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336:934–937. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. Programming of DNA methylation patterns. Annu Rev Biochem. 2012;81:97–117. doi: 10.1146/annurev-biochem-052610-091920. [DOI] [PubMed] [Google Scholar]
- Chen CC, Wang KY, Shen CK. The mammalian de novo DNA methyltransferases DNMT3A and DNMT3B are also DNA 5-hydroxymethylcytosine dehydroxymethylases. J Biol Chem. 2012;287:33116–33121. doi: 10.1074/jbc.C112.406975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Cimmino L, Abdel-Wahab O, Levine RL, Aifantis I. TET Family Proteins and Their Role in Stem Cell Differentiation and Transformation. Cell Stem Cell. 2011;9:193–204. doi: 10.1016/j.stem.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortazar D, Kunz C, Selfridge J, Lettieri T, Saito Y, MacDougall E, Wirz A, Schuermann D, Jacobs AL, Siegrist F, et al. Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature. 2011;470:419–423. doi: 10.1038/nature09672. [DOI] [PubMed] [Google Scholar]
- Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty MM, Breiling A, Le T, Raddatz G, Barrasa MI, Cheng AW, Gao Q, Powell BE, Li Z, Xu M, et al. Combined Deficiency of Tet1 and Tet2 Causes Epigenetic Abnormalities but Is Compatible with Postnatal Development. Dev Cell. 2013 doi: 10.1016/j.devcel.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty MM, Ganz K, Powell BE, Hu YC, Markoulaki S, Cheng AW, Gao Q, Kim J, Choi SW, Page DC, et al. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell. 2011;9:166–175. doi: 10.1016/j.stem.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreszer TR, Karolchik D, Zweig AS, Hinrichs AS, Raney BJ, Kuhn RM, Meyer LR, Wong M, Sloan CA, Rosenbloom KR, et al. The UCSC Genome Browser database: extensions and updates 2011. Nucleic Acids Res. 2012;40:D918–D923. doi: 10.1093/nar/gkr1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- Globisch D, Munzel M, Muller M, Michalakis S, Wagner M, Koch S, Bruckl T, Biel M, Carell T. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One. 2011;5:e15367. doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011 doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-Methylcytosine by TET1 Promotes Active DNA Demethylation in the Adult Brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett JA, Sengupta R, Zylicz JJ, Murakami K, Lee C, Down TA, Surani MA. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science. 2013;339:448–452. doi: 10.1126/science.1229277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Shen L, Dai Q, He C, Zhang Y. Generation and replication-dependent dilution of 5fC and 5caC during mouse preimplantation development. Cell Res. 2011;21:1670–1676. doi: 10.1038/cr.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011:334–194. doi: 10.1126/science.1212483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal K, Jin SG, Pfeifer GP, Szabo PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci U S A. 2011;108:3642–3647. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare T, Pai S, Koncevicius K, Pal M, Kriukiene E, Liutkeviciute Z, Irimia M, Jia P, Ptak C, Xia M, et al. 5-hmC in the brain is abundant in synaptic genes and shows differences at the exon-intron boundary. Nat Struct Mol Biol. 2012;19:1037–1043. doi: 10.1038/nsmb.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti A, Drohat AC. Thymine DNA Glycosylase Can Rapidly Excise 5-Formylcytosine and 5-Carboxylcytosine: POTENTIAL IMPLICATIONS FOR ACTIVE DEMETHYLATION OF CpG SITES. The Journal of biological chemistry. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucci G, Maharry K, Wu YZ, Radmacher MD, Mrozek K, Margeson D, Holland KB, Whitman SP, Becker H, Schwind S, et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2010;28:2348–2355. doi: 10.1200/JCO.2009.27.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munzel M, Globisch D, Bruckl T, Wagner M, Welzmiller V, Michalakis S, Muller M, Biel M, Carell T. Quantification of the sixth DNA base hydroxymethylcytosine in the brain. Angew Chem Int Ed Engl. 2010;49:5375–5377. doi: 10.1002/anie.201002033. [DOI] [PubMed] [Google Scholar]
- Nabel CS, Jia H, Ye Y, Shen L, Goldschmidt HL, Stivers JT, Zhang Y, Kohli RM. AID/APOBEC deaminases disfavor modified cytosines implicated in DNA demethylation. Nature chemical biology. 2012;8:751–758. doi: 10.1038/nchembio.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura J, Walker VE, Upton PB, Chiang SY, Kow YW, Swenberg JA. Highly sensitive apurinic/apyrimidinic site assay can detect spontaneous and chemically induced depurination under physiological conditions. Cancer Res. 1998;58:222–225. [PubMed] [Google Scholar]
- Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, McLoughlin EM, Brudno Y, Mahapatra S, Kapranov P, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffeneder T, Hackner B, Truss M, Munzel M, Muller M, Deiml CA, Hagemeier C, Carell T. The Discovery of 5-Formylcytosine in Embryonic Stem Cell DNA. Angew Chem Int Ed Engl. 2011;50:7008–7012. doi: 10.1002/anie.201103899. [DOI] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiber EA, Beraldi D, Ficz G, Burgess HE, Branco MR, Murat P, Oxley D, Booth MJ, Reik W, Balasubramanian S. Genome-wide distribution of 5-formylcytosine in embryonic stem cells is associated with transcription and depends on thymine DNA glycosylase. Genome Biol. 2012;13:R69. doi: 10.1186/gb-2012-13-8-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H, Matsui Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat Rev Genet. 2008;9:129–140. doi: 10.1038/nrg2295. [DOI] [PubMed] [Google Scholar]
- Seisenberger S, Andrews S, Krueger F, Arand J, Walter J, Santos F, Popp C, Thienpont B, Dean W, Reik W. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol Cell. 2012;48:849–862. doi: 10.1016/j.molcel.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CX, Yi C, He C. Mapping recently identified nucleotide variants in the genome and transcriptome. Nat Biotechnol. 2012;30:1107–1116. doi: 10.1038/nbt.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Scholer A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- Stroud H, Feng S, Morey Kinney S, Pradhan S, Jacobsen SE. 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol. 2011;12:R54. doi: 10.1186/gb-2011-12-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Li Y, Song CX, Han JW, Kim S, Namburi S, Hermetz K, Kim JJ, Rudd MK, et al. Integrating 5-hydroxymethylcytosine into the epigenomic landscape of human embryonic stem cells. PLoS Genet. 2011a;7:e1002154. doi: 10.1371/journal.pgen.1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Li Y, Song CX, Wu H, Dai Q, Irier H, Upadhyay AK, Gearing M, Levey AI, et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat Neurosci. 2011b;14:1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, Cowley SM, Young RA. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature. 2012;482:221–225. doi: 10.1038/nature10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wossidlo M, Nakamura T, Lepikhov K, Marques CJ, Zakhartchenko V, Boiani M, Arand J, Nakano T, Reik W, Walter J. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat Commun. 2011;2:241. doi: 10.1038/ncomms1240. [DOI] [PubMed] [Google Scholar]
- Wu H, Coskun V, Tao J, Xie W, Ge W, Yoshikawa K, Li E, Zhang Y, Sun YE. Dnmt3a–dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science. 2010;329:444–448. doi: 10.1126/science.1190485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, D’Alessio AC, Ito S, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 2011a;25:679–684. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, D’Alessio AC, Ito S, Xia K, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011b;473:389–393. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 2011a;25:2436–2452. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhang Y. Tet1 and 5-hydroxymethylation: A genome-wide view in mouse embryonic stem cells. Cell Cycle. 2011b;10 doi: 10.4161/cc.10.15.16930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, Barbera AJ, Zheng L, Zhang H, Huang S, et al. Genome-wide Regulation of 5hmC, 5mC, and Gene Expression by Tet1 Hydroxylase in Mouse Embryonic Stem Cells. Mol Cell. 2011;42:451–464. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Hong K, Liu R, Inoue A, Shen L, Zhang K, Zhang Y. Dynamics of 5-methylcytosine and 5-hydroxymethylcytosine during germ cell reprogramming. Cell research. 2013 doi: 10.1038/cr.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Hong K, Liu R, Shen L, Inoue A, Diep D, Zhang K, Zhang Y. Tet1 controls meiosis by regulating meiotic gene expression. Nature. 2012;492:443–447. doi: 10.1038/nature11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.