Abstract
African green monkeys (AGMs) infected by simian immunodeficiency virus (SIV) SIVagm are resistant to AIDS. SIVagm-infected AGMs exhibit levels of viremia similar to those described during pathogenic human immunodeficiency virus type 1 (HIV-1) and SIVmac infections in humans and macaques, respectively, but contain lower viral loads in their lymph nodes. We addressed the potential role of dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin (DC-SIGN; CD209) in viral dissemination. In previous studies, it has been shown that human DC-SIGN and macaque DC-SIGN allow transmission of HIV and SIVmac to T cells. Here, we looked at the ability of DC-SIGN derived from AGM lymph nodes to interact with SIVagm. We show that DC-SIGN-expressing cells are present mainly in the medulla and often within the cortex and/or paracortex of AGM lymph nodes. We describe the isolation and characterization of at least three isoforms of dc-sign mRNA in lymph nodes of AGMs. The predicted amino acid sequence from the predominant mRNA isoform, DC-SIGNagm1, is 92 and 99% identical to the corresponding human and rhesus macaque DC-SIGN amino acid sequences, respectively. DC-SIGNagm1 is characterized by the lack of the fourth motif in the repeat domain. This deletion was also detected in the dc-sign gene derived from thirteen animals belonging to five other African monkey species and from four macaques (Macaca fascicularis and M. mulatta). Despite three- to seven-amino-acid modifications compared to DC-SIGNmac, DC-SIGNagm1 allows transmission of SIVagm to T cells. Furthermore, AGM monocyte-derived dendritic cells (MDDC) expressed at least 100,000 DC-SIGN molecules and were able to transmit SIVagm to T cells. At a low multiplicity of infection (10−5 50% tissue culture infective doses/cell), viral transmission by AGM MDDC was mainly DC-SIGN dependent. The present study reveals that DC-SIGN from a natural host species of SIV has the ability to act as an efficient attachment and transmission factor for SIVagm and suggests the absence of a direct link between this ability and viral load levels in lymph nodes.
Dendritic cells (DC) residing in mucosal tissues are among the first cells targeted by human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV) SIVmac during pathogenic infections in humans and macaques, respectively (22, 51). DC can transmit these viruses to T cells in vitro (6, 17, 40). One mechanism by which DC enhance infection of T cells implies DC-SIGN (DC-specific ICAM-3-grabbing nonintegrin; CD209) (9, 13). DC-SIGN, a C-type (calcium-dependent) lectin, has been detected on monocyte-derived DC (MDDC). It also facilitates the formation of the immunological synapse between DC and T cells by binding to intercellular adhesion molecule 3 (ICAM-3) (14). In addition, HIV bound to DC-SIGN-positive cells can be transmitted to cells expressing CD4 and a coreceptor, resulting in efficient virus infection in trans (13, 38). DC-SIGN cannot replace CD4, CCR5, or CXCR4 in the process of HIV type 1 (HIV-1) entry but retains infectious particles for several days in culture (13) in a low-pH endosomal compartment (27). Similarly, DC-SIGN from macaques also acts as an attachment factor for SIVmac (2, 12, 25, 61). In addition, coexpression of human DC-SIGN with CD4 and an appropriate coreceptor can enhance infection of HIV and SIVmac in cis (28).
HIV-1 is thought to subvert the trafficking capacity of DC to gain access to the CD4+-T-cell compartment (19, 42, 53). It has been shown that SIVmac-infected DC are able to migrate from original sites of infection to draining lymph nodes (LNs) (22). Similar to “Trojan horses,” DC-SIGN expressing DC may serve as a shuttle for the transfer of HIV-1 from submucosa to secondary lymphoid organs (13, 52). During the formation of the immunological synapse with T cells, it is possible that infected DC then transmit the virus to permissive T cells within the secondary lymphoid organs (6, 18, 30, 40, 60).
To understand the role of DC-SIGN in viral dissemination to LNs and pathogenesis, we studied the function of DC-SIGN in a nonhuman primate model in which infection with a lentivirus is nonpathogenic. SIVagm infection in all four species of African green monkeys (AGM, Chlorocebus pygerythrus, C. aethiops, C. sabaeus, and C. tantalus) is associated with a lack of disease, although the associated viremia levels in plasma can reach those described for pathogenic HIV-1 and SIVmac infections (5, 10, 16, 21). In contrast to plasma viremia, the DNA and RNA viral loads in LNs of SIVagm-infected AGMs are generally significantly lower than those reported for HIV and SIVmac pathogenic infections (4, 5, 10). Here we examined whether potential structural and functional differences between DC-SIGNagm and human DC-SIGN and/or differences in the expression levels underlie the low viral load in AGM LNs.
MATERIALS AND METHODS
Animals and specimen collection.
The AGMs—C. sabaeus, C. tantalus, and C. pygerythrus—were kept in captivity at the Pasteur Institute in Senegal, at the Pasteur Institute in Bangui (Central African Republic), and at the Centre International de Recherches Médicales, in Franceville (Gabon), respectively. Patas monkeys (Erythrocebus patas) were held in captivity at the Pasteur Institute in Bangui. L'Hoest's monkeys (Cercopithecus lhoesti lhoesti), white-collared mangabeys (Cercocebus torquatus lunulatus), gray-cheeked mangabeys (Cercocebus albigena), and a naturally SIVcpz-infected chimpanzee (Pan troglodytes, Cam5) were housed in European and Cameroonian zoos (33, 34). Rhesus (Macaca mulatta) and cynomolgus (M. fascicularis) macaques were housed at the primate center of CEA, Fontenay aux Roses, France. All animals were held according to national guidelines and institutional policies. For the studies on genomic DNA, peripheral blood mononuclear cells (PBMC) of these nonhuman primates were Ficoll gradient purified, and total DNA was isolated. All of the studies on the characterization, the expression levels, and the function of DC-SIGN were performed with AGMs of the C. sabaeus species (animals 96028, 97008, 97026, 97029, 00017, 00021, 02001, 02007, 02008, 02013, and 02026). These animals were negative for SIV, except for 97026 and 98013, which were experimentally infected with SIVagm.sab92018 (10). Blood was collected from these animals, and excisional inguinal and axillar LN biopsies were performed. For AGMs 97026 and 98013, the axillar LNs were collected at days 7 and 120 postinfection, respectively.
Virus stocks.
We collected PBMC from the naturally SIVagm-infected animal D46 (32). SIVagm.sabD46 was isolated by coculture of the PBMC with SUPT1 cells, a human lymphoblastoid T-CD4+-cell line. For HIV-1, we used an HIV-1Lai isolate that was cultured on SUPT1 cells. Virus production was followed by the measurement of core antigens (SIV p27 and HIV p24; Coulter) and of reverse transcriptase activity in the supernatants. The infectious viral titer was determined by a limiting-dilution assay with SUPT1 cells as described previously (10). The infectious titer was expressed as the 50% tissue culture infectious dose (TCID50) determined by the Karber calculation method. The titers of HIV-1Lai and SIVagm.sabD46 stocks were 250 and 400 TCID50/ml, respectively. Concentrations of core antigens for HIV-1Lai and SIVagm.sabD46 stocks were 1,700 and 860 ng/ml, respectively.
Cells and cell lines.
SupT1 cells were cultured in RPMI 1640 with Glutamax (Invitrogen) supplemented with 10% fetal calf serum (FCS), penicillin-streptomycin (100 IU/ml each) and HEPES (10 mM; Invitrogen). HeLa cells were maintained in Dulbecco modified Eagle medium with Glutamax (glucose, 1,000 mg/liter; Invitrogen) supplemented with 10% FCS (Invitrogen), penicillin-streptomycin (100 IU/ml each), and HEPES (10 mM; Invitrogen). P4P cells are CD4+ CXCR4+ CCR5+ HeLa cells (59). These cells were maintained in the culture medium described above supplemented with puromycin (1 μg/ml; Sigma) and Geneticin G418 (0.5 mg/ml; Invitrogen).
To obtain MDDC, we first enriched monocytes from PBMC of human donors and of AGMs by a 1-h 30-min plastic adhesion step at 37°C, followed by extensive washing in prewarmed FCS-free culture medium in order to remove all nonadherent cells. The adherent cells were then cultured in RPMI 1640 supplemented with 5% FCS, granulocyte-macrophage colony-stimulating factor (1,000 IU/ml; Leucomax [Novartis]) and interleukin-4 (IL-4; 200 IU/ml; R&D Systems) for 4 to 5 days. The phenotype of the cells was checked by flow cytometry. The cells expressed the molecules typically found on immature MDDC, such as CD11c, major histocompatibility complex class II (DR), CD80, and CD86 (unpublished data). The purity of the cells was ≥95% as measured by anti-CD3 and anti-CD20 labeling.
Isolation and sequencing of dc-signagm mRNA.
Total mRNA of LN cells derived from AGM 96028 and 97026 was extracted and then reverse transcribed and amplified in one step by reverse transcription-PCR (RT-PCR; Superscript One-Step RT-PCR System; Invitrogen). The primers used for the RT-PCR step, designed by using a previously published human dc-sign cDNA sequence (9), were MDC1 (forward, 5′-CAGGAGTTCTGGACACTG-3′) and MDC2 (reverse, 5′-GATGGAGAGAAGGAACTGTAG-3′). To detect potential minor amplicons, we subjected the amplification products obtained by RT-PCR to a nested PCR. The primers used for the nested PCR were MDC3 (forward, 5′-CTGGGGGAGAGTGGGGTG-3′) and MDC4 (reverse, 5′-CTTAAAAGGGGGTGAAGTTC-3′). Annealing of primers occurs on the 5′ and 3′ ends of the coding region. Cycling conditions were as previously described (10). After the nested PCR, the amplicons were gel purified (gel extraction kit; Qiagen) and subcloned (TOPO TA Cloning kit; Invitrogen). The sequences were determined for both strands (BigDye terminator cycle sequencing ready reaction kit; Perkin-Elmer).
PCR on genomic DNA.
Total DNA was extracted from simian cells and submitted to PCR by using the primers DCS-EX4S (forward; 5′-CCCAGCTCCATAAGTCAGGAACAA-3′) and DCS-EX4AS (reverse; 5′-ACACAACGACCATCTCAGGCCCAAG-3′). Annealing of primers occurs on the 5′ part of exon III and the 5′ part of intron III of dc-sign when the organization of the human gene is considered (35). Cycling conditions were as previously described (10).
Assessment of DC-SIGNagm-mediated infection in trans.
The efficiency of DC-SIGNagm-mediated virus transfer was first assessed by using human cell lines. Nonpermissive HeLa cells were transduced transiently with an expression vector (pcDNA3.1/myc-his) containing either dc-signagm1 or dc-signhu cDNA (20) or with vector alone by using the Polyfect transfection reagent (Qiagen). After 48 h, the level of expression of DC-SIGN at the cell surface was determined by flow cytometry by using two mouse monoclonal antibodies (MAbs), 8A5 and 1B10, raised against DC-SIGNhu. 1B10 recognizes the carbohydrate recognition domain (CRD) and was described previously (20). Transduced-nonpermissive HeLa cells (5 × 105 cells) were exposed to low infectious doses of virus (multiplicity of infection [MOI] of 10−4 or 10−5 TCID50/cell). DC-SIGNhu- and DC-SIGNagm1-expressing cells were exposed for 2 h at 37°C to HIV-1Lai and to SIVagm.sabD46, respectively. The cells were then vigorously washed. After 24 h, cells were washed again and cultured with SupT1 cells (HeLa/SupT1 ratio = 0.5), permissive to both HIV-1Lai and SIVagm. Virus production was determined by measuring HIV p24 or SIV p27 core antigens in the supernatants. For inhibition of transmission, viral exposure was performed as described above after the cells were stained with neutralizing MAb 1B10 at 50 μg/ml (15 min, 4°C), 5 mM EGTA (Sigma), or mannan (Sigma) at 50 μg/ml.
To examine the capacity of primary AGM cells to transfer virus to T cells, 105 AGM MDDC were incubated with SIVagm.sabD46 (MOI of 10−3, 10−4, or 10−5 TCID50/cell) for 3 h at 37°C in a 5% CO2 incubator. As a positive control, we used human MDDC. After exposure to the virus, AGM and human MDDC were extensively washed with prewarmed Hanks balanced salt solution, resuspended in culture medium containing granulocyte-macrophage colony-stimulating factor and IL-4, and either immediately cocultured with permissive SupT1 cells or after 3 days washed again extensively and cocultured with SupT1 (MDDC/SupT1 ratio = 0.5). At 3- to 4-day intervals, supernatants were harvested and tested for core antigen by enzyme-linked immunosorbent assay to monitor virus replication. For inhibition assays, AGM MDDC were incubated with 65 μg of anti-DC-SIGNhu MAb (1B10) or control mouse immunoglobulin G2a (IgG2a)/ml for 20 min at 4°C prior to the addition of SIVagm.sabD46. AGM MDDC were also treated with mannan (50 μg/ml) prior to the addition of SIVagm.sabD46. To inhibit viral replication, 10 μM zidovudine (AZT; Sigma) was added to the MDDC 3 h before their exposure to virus and maintained until the start of the coculture with SupT1 cells. We have previously shown that 5 μM AZT blocks infection in human macrophages (29). In addition, it has been shown that 10 μM AZT inhibits HIV-1 replication in human Langerhans cells (24).
Quantification of DC-SIGN molecules.
MDDC were stained in phosphate-buffered saline supplemented with 2% FCS for 45 min at 4°C with an anti-DC-SIGNhu MAb, 1B10, at a final concentration of 65 μg/ml. The samples were washed and incubated with phycoerythrin-Cy5-conjugated goat anti-mouse Fab fragments (Dako; 1/20) for 15 min at 4°C and then washed and resuspended in buffer containing 1% paraformaldehyde. The samples were analyzed with an EPICS XL (Coulter) cell analyzer. For quantification, we used the Quantum Simply Cellular microbead kit (Sigma). The quantification was performed by converting the mean fluorescence intensity (MFI) into the antibody-binding capacity (ABC) per cell. For this, the kit contains a mixture of five microbead populations of uniform size, coated with goat anti-mouse antibodies that have different abilities to bind mouse antibodies (i.e., ranging from 0 to ∼215,000 molecules). All samples, including the microbeads, were processed separately and analyzed identically. The binding capacities of the stained microbeads were then regressed against the corresponding MFI of each microbead population. The MFI of the antigen analyzed on the cells was converted to ABC per cell by comparison with the regression curve generated. The MFIs of the samples were corrected by subtracting the MFI of the isotype control for each experiment, and the value was then converted to the ABC. This technology can accurately measure ABC values up to ca. 215,000. Values in excess of this amount fell outside of the standard values. Therefore, they should not be deemed as definitive.
Immunohistochemical analysis.
Sections (5 μm) of cryopreserved OCT-embedded tissues were analyzed by immunohistochemistry to determine whether DC-SIGN is expressed on cells in AGM LNs and to study the distribution of DC-SIGN+ cells within this tissue. Immunostaining was performed with the anti-DC-SIGN MAb clone 8A5 (2.4 μg/ml) or clone 1B10 (6.5 μg/ml), as well as with the anti-CD68 MAb (clone EBM11; Dako). Immunostaining with primary mouse antibody was revealed by using Dako EnVision horseradish peroxidase conjugated to a secondary antibody. Diaminobenzidine was used as a high sensitivity chromogenic substrate system. The slides were counterstained with Harris's hematoxylin for coloration of nuclei.
Nucleotide sequence accession numbers.
The sequences of the cloned DCSIGNagm1, DC-SIGNagm2, and DC-SIGNagm3 have been deposited in GenBank under accession numbers AY189945, AY189946, and AY189947, respectively. The nucleotide sequence of dc-signagm1 cDNA from AGM 97026 differs from that of AGM 96028 only by three synonymous mutations (data not shown). The latter sequence has been deposited in GenBank under the accession number AY189944.
RESULTS
Isolation and sequence of dc-signagm mRNA from AGM LNs.
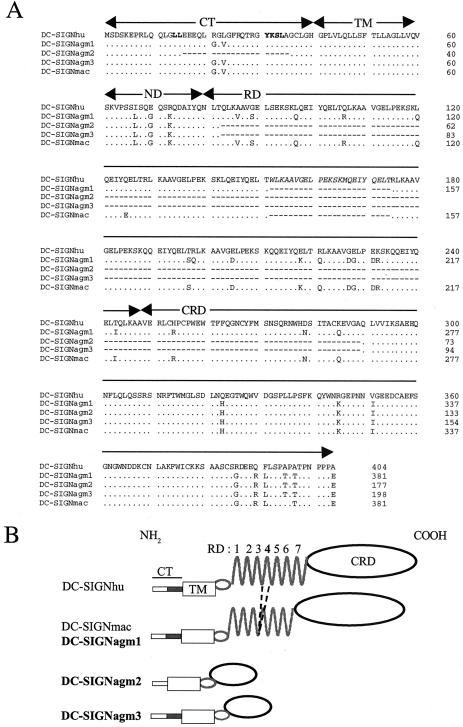
In order to assess the role of DC-SIGN in SIVagm infection, we first determined whether dc-sign mRNA is expressed in AGM LNs. Axillary and inguinal LNs were obtained from two AGMs (animals 96028 and 97026). Animal 96028 was negative for SIV, whereas 97026 had been experimentally infected with SIVagm.sab92018 (10). Total mRNA was extracted from LNs. Since a wide repertoire of transcripts of the human dc-sign has been reported (35), we designed a nested PCR to amplify potential minor transcripts of the dc-sign gene within AGM LNs. We detected a predominant 1,150-bp amplicon after a single round of PCR, as well as additional fragments with smaller sizes after the nested PCR (data not shown). Sequence analyses revealed three distinct cDNAs. Alignment of the predicted amino acid sequence of the predominant isoform, named DC-SIGNagm1, with DC-SIGNhu (9) and DC-SIGNmac (M. mulatta, allele 1) (2) revealed identities of 92.08 and 98.95%, respectively (Fig. 1A). The N-linked glycosylation sites (NxT/S) (38) and internalization motifs, such as LL and YxxL (50), as well as the transmembrane domain, are conserved in DC-SIGNagm1. The minor transcripts, called dc-signagm2 and dc-signagm3, both differ from dc-signagm1, dc-signhu, and dc-signmac by the absences of (i) exon Ic, encoding a small region in the cytoplasmic tail (CT); (ii) exon III, encoding the repeat domain (RD); and (iii) the 5′ part of exon IV encoding the first 40 amino-acids of the CRD (Fig. 1). dc-signagm2 and dc-signagm3 may have arisen from complex alternative splicing. Although transcripts encoding membrane-associated isoforms characterized by the deletion of exons III and IV have been described for humans, none showed a deletion of exon Ic (35). Furthermore, we have not detected mRNA deletions in exon II (TM) which could encode potentially soluble proteins. The latter have been detected in several human cells such as PBMC, DC derived from CD34+ peripheral blood hematopoietic progenitors, and resting CD14+ human monocytes (35). Finally, all DC-SIGNagm isoforms differ from DC-SIGNhu by the absence of 23 amino acids within the RD, corresponding to the fourth motif (Fig. 1).
FIG. 1.
(A) Alignment of predicted amino acid sequences of DC-SIGNagm1, DC-SIGNagm2, and DC-SIGNagm3 with reported sequences of DC-SIGNhu and DC-SIGNmac. The sequence of DC-SIGNhu (9) and the third reported sequence of DC-SIGN isolated from a macaque (M. mulatta) (61) wree used for the alignment. The three isoforms isolated from animal AGM 97026 are shown. The The dileucine (LL) and YxxL internalization motifs are in boldface. The fourth repeat motif is indicated in italics. Arrows above the sequences highlight the different domains of DC-SIGN: CT, cytoplasmic tail; TM, transmembrane; and ND, neck domain. Dots and dashes indicate, respectively, conserved amino acids and deletions. (B) Schematic representation of DC-SIGNagm1, DC-SIGNagm2, and DC-SIGNagm3. DC-SIGNhu and DC-SIGNmac were represented as proposed by Pöhlmann et al. (38). Note that DC-SIGNagm and DC-SIGNmac (2, 12, 44, 61) are characterized by the lack of the fourth motif repeat within the RD, in contrast to DC-SIGNhu. DC-SIGNagm isoforms 2 and 3 are characterized by the absence of exon III coding the RD and of the exon IV region encoding the 5′ part of the CRD. DC-SIGNagm2 also exhibits the absence of exon Ic encoding a small region in the CT.
Deletion within the region coding for the RD of the dc-signagm gene.
To determine whether the presence of 6.5 instead of 7.5 motifs is the result of differential splicing or a deletion in exon III (exon IV according to Soilleux et al. [48]) encoding the repeat region (35), we performed PCR on genomic DNA from AGM 96028 with primers annealing to the 5′ part of exon III and the 5′ part of intron IV. The expected size for the human fragment is 587 bp. The fragments amplified from AGM 96028 DNA were about 70 nucleotides shorter than those obtained from human DNA (Fig. 2), indicating that the lack of the fourth motif in the cDNA resulted from a deletion in the dc-sign gene rather than from alternative splicing.
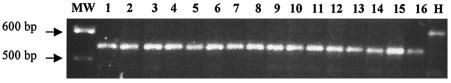
FIG. 2.
Amplification of the genomic region coding for the RD in simian and human dc-sign genes. PCR was performed with genomic DNA of 30 AGMs (14 C. sabaeus, 13 C. tantalus, 2 C. pygerythrus, and Vero cells), 4 patas monkeys (Erythrocebus patas), 2 l'Hoest monkeys (Cercopithecus lhoesti), 5 white-collared mangabeys (Cercocebus torquatus lunulatus), 1 gray-cheeked mangabey (Cercocebus albigena), 1 naturally SIVcpz-infected chimpanzee (Cam5), 2 rhesus macaques (M. mulatta), 2 cynomolgus macaques (M. fascicularis), and 1 human donor. Representative results obtained for one individual of each species and for nine AGMs are shown. The primers used were designed to amplify exon III encoding the RD, corresponding to a 587-bp fragment of the human dc-sign gene. Lanes: MW, molecular weight; 1, AGM, Sab96028; 2, AGM, C. sabaeus; 3, AGM, C. sabaeus; 4, AGM, C. sabaeus; 5, AGM, C. pygerythrus; 6, AGM, C. pygerythrus; 7, AGM, C. tantalus; 8, AGM, C. tantalus; 9, AGM, Vero cells; 10, Erythrocebus patas; 11, Cercopithecus lhoesti lhoesti; 12, Cercocebus torquatus lunulatus; 13, Cercocebus albigena; 14, Pan troglodytes troglodytes; 15, M. mulatta; 16, M. fascicularis; H, Homo sapiens sapiens.
Deletion in the dc-sign gene present in animals from 10 distinct Old World monkey species.
To study the frequency of occurrence of the deletion of the fourth motif in AGMs, we performed PCR on genomic DNAs from 28 other wild-born AGMs belonging to three distinct AGM species. The study included 22 SIVagm-infected animals. We also studied genomic DNA from the commonly used AGM cell line Vero, derived from a vervet. The same deletion was observed in DNAs from each AGM tested, as well as in DNA from Vero cells (Fig. 2).
Among African nonhuman primates studied to date, only dc-sign mRNA from one chimpanzee has been analyzed so far and it failed to show this deletion (12). In order to determine whether this deletion in exon III is present or not in other African nonhuman primates, we examined genomic DNA of 13 animals from five distinct species: patas monkey (Erythrocebus patas), l'Hoest monkey (Cercopithecus lhoesti), white-collared mangabey (Cercocebus torquatus lunulatus), gray-cheeked mangabey (Cercocebus albigena), and chimpanzee (Pan troglodytes troglodytes) (33). The latter corresponds to a naturally SIVcpz-infected chimpanzee (Cam5) (34). We observed again a smaller amplicon, indicating the same deletion in all of the animals studied (Fig. 2).
Since a deletion of this fourth motif was reported in DC-SIGN from macaques by some authors, whereas others did not observe it (2, 12, 44, 61), we analyzed genomic DNA from four macaques (two M. fascicularis and two M. mulatta macaques of Chinese origin). Again, only fragments of smaller sizes compared to the human fragment were detected in all four studied macaques. Our study therefore suggests that the allele with the 6.5 motifs in exon III is more frequent than the allele with the 7.5 motifs in Old World nonhuman primates, at least in those of African origin.
Reactivity of DC-SIGNagm1 with anti-DC-SIGNhu MAbs.
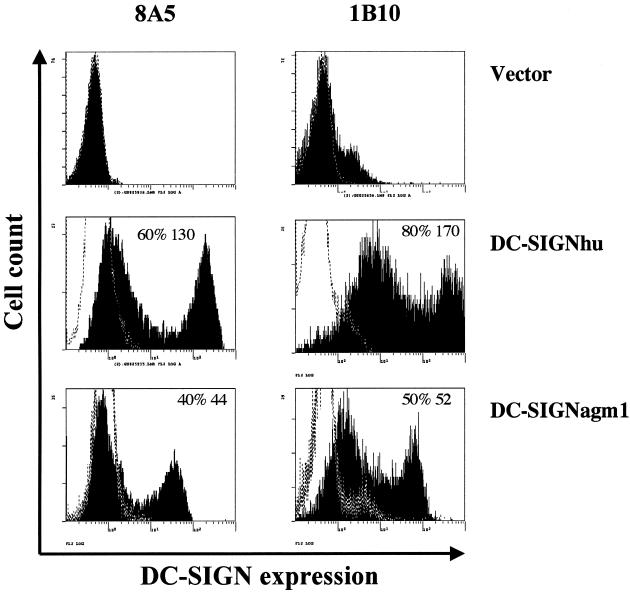
To study the function of DC-SIGN from AGM, we first analyzed DC-SIGNagm expression with MAb as a molecular tool. HeLa cells were transfected transiently with an expression vector (pcDNA3.1/myc-his) containing either dc-signagm1 or dc-signhu cDNA (20) or with vector alone. After 48 h, the level of expression of DC-SIGN at the cell surface was analyzed by flow cytometry using two mouse MAbs, 8A5 and 1B10 (20), raised against DC-SIGNhu. Cells transfected with either dc-signagm1 or dc-signhu cDNA stained positive, showing that both MAbs are cross-reactive against DC-SIGNagm from AGMs (Fig. 3). The staining was always higher in percentage and fluorescence intensity for DC-SIGNhu than for DC-SIGNagm due to either more efficient transfection and/or better affinity of the MAbs for DC-SIGNhu. Similar results were obtained with another MAb raised against DC-SIGNhu, AZND1 (13; data not shown).
FIG. 3.
Cross-reactivity of anti-DC-SIGNhu MAbs toward DC-SIGNagm1. HeLa and P4P cells, transiently expressing DC-SIGNhu or DC-SIGNagm1, were stained 48 h posttransfection with anti-DC-SIGNhu MAbs (either 8A5 or 1B10). On all histograms, the dotted curve represents staining with an isotype control antibody (IgG2a), whereas the black curve represents DC-SIGN staining. The percent positive cells and MFIs are indicated. The results obtained were similar with HeLa and P4P cells. One of four representative experiments with HeLa cells is shown. Both MAbs were cross-reactive with DC-SIGNagm1.
DC-SIGNagm1 ability of SIVagm transmission to T cells.
The presence of a shorter RD in DC-SIGN in both macaques and AGMs indicates that it cannot, by itself, be responsible for potential functional differences in viral transmission. However, DC-SIGNagm1 also differs by at least three amino acids from DC-SIGN of M. mulatta (Fig. 1A) and by seven amino acids from DC-SIGN of M. nemestrina (2, 12, 44, 61). Several differences were located within the CRD that is known to be involved in binding to HIV/SIVmac (15, 38). The CRD of DC-SIGNagm1 differs by at least one amino acid from the rhesus macaque CRD (Fig. 1A) and by three amino acids from the pigtailed macaque CRD (2). To evaluate whether any of these amino acid differences in DC-SIGNagm1 could affect its function as a viral attachment and transmission factor, we determined whether DC-SIGNagm1-expressing cells could transmit SIVagm to T cells
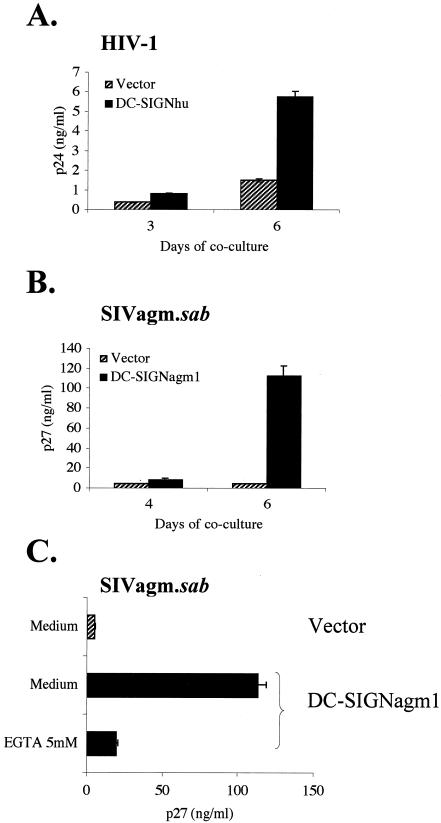
dc-signagm1 transiently transduced nonpermissive HeLa cells were exposed to low infectious doses of virus (MOIs of 10−4 and 10−5 TCID50/cell). DC-SIGNhu and DC-SIGNagm1 expressing cells were, respectively, exposed to HIV-1Lai and SIVagm.sabD46. After 24 h, cells were washed again and cultured with a human T-cell line (SupT1) permissive to both HIV-1 and SIVagm. As a positive control, we first confirmed the ability of DC-SIGNhu to mediate HIV-1 infection in trans (Fig. 4A). We then tested the properties of DC-SIGNagm. We consistently detected efficient SIVagm replication in SupT1 cells cultured with HeLa cells expressing DC-SIGNagm1 but not in SupT1 cells cultured with HeLa cells transfected with vector alone (Fig. 4B). Similar results were observed with another SIVagm.sab virus, SIVagm.sab92018 (data not shown). Moreover, transmission was largely inhibited by EGTA, a known inhibitor of C-type lectin-binding activity (Fig. 4C). The transmission was also inhibited by using mannan or an MAb raised against DC-SIGNhu, 1B10 (data not shown). These experiments demonstrate that DC-SIGNagm1 mediates transmission of SIVagm to T cells.
FIG. 4.
Ability of DC-SIGNagm to transmit SIVagm to T cells. Nonpermissive HeLa cells transiently expressing DC-SIGN were exposed to low infectious doses of virus (MOIs of 10−4 and 10−5) and cocultured for 24 h postexposure with SupT1 cells. Viral replication was determined by measuring the viral core antigen in supernatants every 2 or 3 days (HIV-1 p24 and SIV p27; Coulter). The results obtained with a virus MOI of 10−4 are shown. Experiments were performed in duplicate. The values measured at day 3 or 4 and day 6 of coculture are indicated. The results of one representative experiment of three is shown. We used HeLa cells transiently transduced with vector alone as a negative control. (A) Transmission of HIV-1Lai by DC-SIGNhu. (B) Cells transfected with vector containing dc-signagm1 cDNA or vector alone were exposed to SIVagm.sabD46. (C) The DC-SIGNagm1-expressing cells were exposed to SIVagm.sabD46 in the presence of 5 mM EGTA in order to test whether transmission of SIVagm from HeLa to SupT1 cells is dependent on Ca2+, a feature consistent with a C-type lectin activity. EGTA was added during the 2-h exposure of cells to the virus. P27 levels detected at day 6 of a 10-day culture are shown.
Coexpression of DC-SIGNagm1 along with CD4 and an appropriate coreceptor on HeLa cells enhances 3-fold SIVagm infection by permissive cells (data not shown). DC-SIGNagm1 therefore enhances infection in cis.
DC-SIGN expression on AGM MDDC.
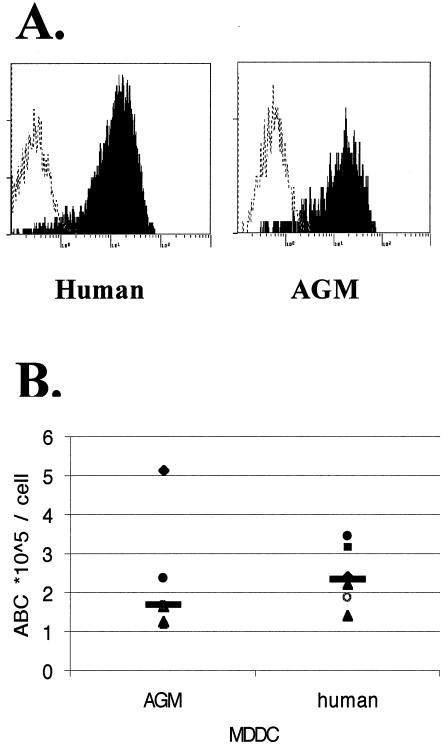
After we showed that the dc-sign mRNA in LNs codes for an efficient attachment and transmission molecule for SIVagm, we attempted to analyze whether primary AGM cells would be able to transmit SIVagm to T cells. We therefore determined whether AGM MDDC express DC-SIGN. Staining of AGM MDDC with MAb 1B10 revealed that DC-SIGN is expressed on their surface (Fig. 5A).
FIG. 5.
Quantitative measurement of DC-SIGN on MDDC. Immature MDDC were obtained from six uninfected AGM donors and six healthy human donors. We quantified the surface expression level of DC-SIGN by using a MAb raised against DC-SIGNhu (1B10) and a phycoerythrin-Cy5-coupled goat F(ab′)2 anti-mouse antibody in conjunction with the Quantum Simply Cellular microbead kit (Sigma). (A) DC-SIGN expression on AGM and human MDDC. On all histograms, the dotted curve represents staining with an isotype control antibody (IgG2a), whereas the solid curve represents DC-SIGN staining. (B) The values are expressed as the ABC per cell for each animal and each human donor. Each value is represented by a symbol, and horizontal bars show the median value.
It has been shown that the transmission efficiency depends on the expression levels of DC-SIGN and that 60,000 molecules per cell are necessary for efficient viral transmission (38). We therefore measured DC-SIGN expression on AGM MDDC by using a quantitative cell cytometry technology. The number of surface molecules on MDDC was determined for six SIVagm-seronegative AGM and six HIV-1-seronegative donors. All AGM and human MDDC studied expressed at least 100,000 molecules per cell. On AGM and human MDDC, respectively, we found a median of 168,000 (range, 118,000 to 517,000) and 244,000 (range, 141,000 to 347,000) DC-SIGN copies/cell (Fig. 5B). Statistical analysis did not reveal any significant differences between the levels of DC-SIGN expression on the analyzed AGM and human MDDC (Mann Whitney, P = 0.2623). The molecule numbers rather varied according to the donors, to similar degrees in humans and AGMs. Similar variations have also been previously reported in another study on human MDDC (1).
Capacity of AGM MDDC to transmit SIVagm to T cells and implication of DC-SIGN.
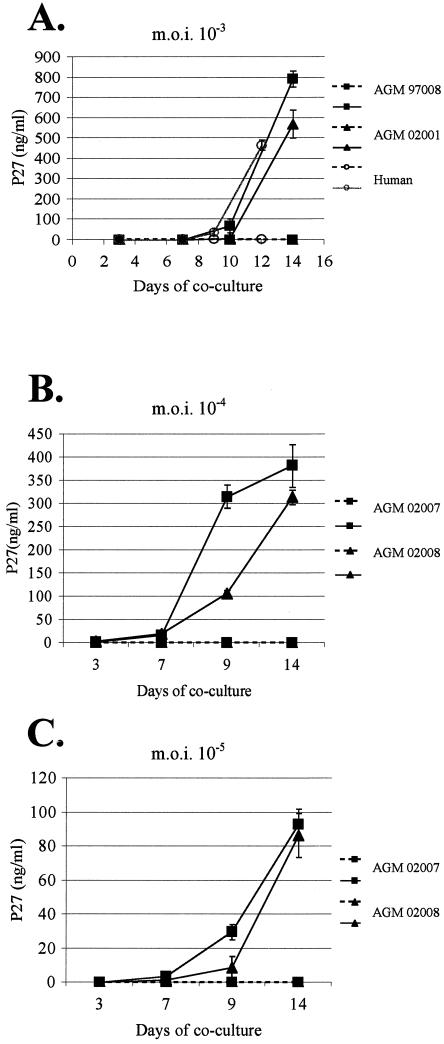
To determine whether AGM MDDC are able to transmit SIVagm to T cells, MDDC from two animals (97008 and 02001) on which the level of DC-SIGN expression had been assessed (516,000 and 166,000 copies/cell, respectively) were exposed to SIVagm (MOI of 10−3 TCID50/cell). It has been reported that DC-SIGN+ THP-1 human MDDC efficiently retain infectious HIV particles for 4 days (13). To increase the specificity of our assay, we increased the time between viral exposure and coculture with SupT1 cells by two more days compared to the assay with HeLa cells. MDDC were thus cultured with SupT1 cells only 3 days after viral exposure and after repeated washings. In parallel to AGM MDDC we treated human MDDC under the same experimental conditions. We detected a viral replication only in SIVagm-exposed human MDDC cocultured with SupT1 but not in SIVagm-exposed human MDDC alone (Fig. 6A). Thus, we show that human MDDC are able to transmit SIVagm. Furthermore, AGM MDDC are capable to transmit SIVagm to T cells (Fig. 6A). In AGM MDDC cultured alone, again we did not detect signs of SIVagm replication, as measured by p27 quantification in supernatants.
FIG. 6.
Assessment of SIVagm transmission by MDDC. Immature AGM MDDC were exposed to SIVagm.sabD46 (MOIs of 10−3, 10−4, and 10−5 TCID50/cell). Three days later, the cells were washed and cocultured with SupT1 cells. We exposed, in parallel, human MDDC to SIVagm.sabD46 (MOI of 10−3 TCID50/cell). The experiments were performed in duplicate. Virus production after the start of the coculture was monitored by SIV p27gag antigen assay. Plain and broken lines indicate levels of SIV p27gag antigen in the MDDC-SupT1 cocultures or in the cultures of MDDC alone, respectively. (A) SIVagm.sab transmission by AGM and human MDDC. The MDDC were exposed to a virus MOI of 10−3. (B) Transmission by AGM MDDC exposed to SIVagm at an MOI of 10−4. (C) Transmission by AGM MDDC exposed to SIVagm at an MOI of 10−5.
In order to further increase the specificity of the assay and to work with infectious doses even closer to physiological conditions in vivo, we also tested viral transmission by AGM MDDC with 10−4 and 10−5 MOI of SIVagm.sab. This assay was performed with MDDC derived from two animals (02007 and 02008) for which the number of DC-SIGN molecules expressed on their surface had also been determined (118,000 and 170,000 copies/cell, respectively). Due to the limitation of the amount of blood that can be taken from the animals, which are about 20 times smaller than humans, the number of generated DC is low. Consequently, it was impossible to perform all experiments with DC from a same animal. However, each assay was always repeated with at least two donors. AGM MDDC were again able to transmit SIVagm to T cells (Fig. 6B and C).
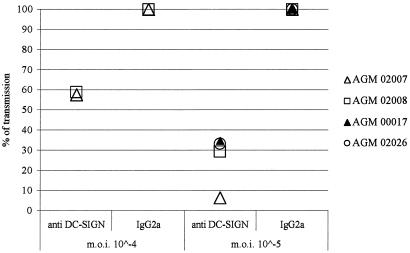
To address whether this SIVagm transmission was due to DC-SIGN, the MDDC derived from the two latter animals were also exposed to SIVagm.sab (MOIs of 10−4 and 10−5 TCID50/cell) in the presence of an anti-DC-SIGNhu MAb (1B10) or mannan. The latter is the most potent saccharide inhibitor of DC-SIGN (9). The presence of mannan reduced significantly SIVagm transmission at MOIs of 10−4 (60% inhibition) and 10−5 (85% inhibition) (data not shown). The presence of an anti-DC-SIGN MAb reduced the trans infection of T cells by MDDC of both animals. The inhibition was 40% and 60 to 95% at MOIs of 10−4 and 10−5, respectively (Fig. 7). At the lower MOI (10−5), the viral transmission was thus mainly DC-SIGN dependent. In order to confirm the observed results, we repeated the experiment at an MOI of 10−5 with MDDC from two other animals (00017 and 02026). Again, we observed significant inhibition (70%) of transmission (Fig. 7). Consequently, AGM MDDC sufficiently express high numbers of DC-SIGN molecules, retain SIVagm particles in an infectious state for at least 3 days after viral exposure, and promote SIVagm transmission to T cells through a mechanism that preferentially involves DC-SIGN when low doses of virus are used.
FIG. 7.
Inhibition of SIVagm transmission by AGM MDDC with an anti-DC-SIGNhu MAb. AGM MDDC were exposed to SIVagm.sabD46 in the presence of anti-DC-SIGNhu (1B10), a control IgG2a, or medium alone. Each experiment was performed in duplicate. In order to express percentage of inhibition, the p27gag antigen values of the IgG2a-treated cells, obtained for day 14, were normalized to 100%. The results obtained with MDDC from two AGM (animals 02007 and 02008) exposed to MOIs of 10−4 and 10−5 of SIVagm are shown. The results obtained with MDDC from two further animals (00017 and 02026) exposed to an SIVagm MOI of 10−5 are also presented here.
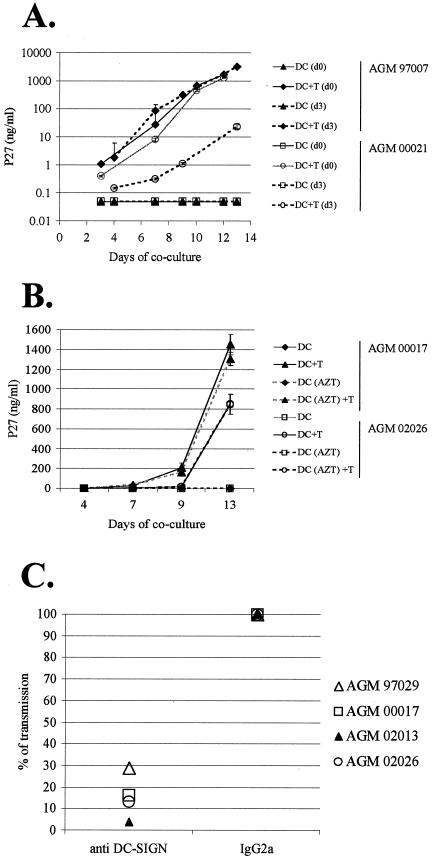
Nevertheless, at the lower MOI (10−5 TCID50/cell), the anti-DC-SIGN MAb failed to completely abolish viral transmission. One explanation might be the implication of others molecules with trans attachment activity (56). Another explanation could be that SIVagm productively infects AGM MDDC which could result in high virus replication in cocultures through infection of T cells independently of DC-SIGN, explaining the failure to completely inhibit viral transmission with anti-DC-SIGN MAb. We were not able to detect any significant production of SIV p27 antigen in the 14-day cultures of MDDC exposed to SIVagm.sab (Fig. 6), but we cannot exclude a very low level of virus replication undetectable by the enzyme-linked immunosorbent assay. To address this question, we performed two kind of approaches. First, we added the T cells immediately after exposure of MDDC to SIVagm to allow viral transmission before any potential viral de novo synthesis and compared p27 production to that determined when the T cells were added 3 days after virus exposure. The viral production in T cells cocultured immediately with MDDC was either higher (animal 00021) or equal to the levels observed in T cells cocultured 3 days after the SIVagm-exposure of MDDC (animal 97007) (Fig. 8A). The lower levels we detected if we cocultured MDDC with Supt1 after 3 days might be explained by a potential degradation of virus particles, as shown for HIV-1 particles (36). In any case, the 3-day time frame was associated with an increase of viral replication in DC-T cocultures. This suggested that, if low-level de novo synthesis of viral particles occurs within the 3-day period, it does not significantly add to viral transmission.
FIG. 8.
Viral transmission without lag time and in the absence of viral replication between exposure of MDDC and coculture with T cells. AGM MDDC were exposed to SIVagm.sabD46 (MOI of 10−5) and cocultured with SupT1 cells. Each experiment was performed in duplicate. Virus production was monitored by SIV p27gag antigen assay after the start of the coculture. (A) Viral replication in DC-T cocultures started immediately or 3 days after exposure of MDDC to SIVagm. Solid and broken lines indicate SIVagm replication in cocultures started immediately or with a 3-day lag after viral exposure of MDDC, respectively. (B) Assessment of AZT treatment of MDDC on viral transmission. DC-T cocultures were started 3 days after exposure of MDDC from four animals to SIVagm. An RT inhibitor (AZT) was added during the 3-day period to inhibit any potential SIVagm replication in MDDC. Broken and solid lines indicate MDDC treated with or without AZT, respectively. (C) Role of DC-SIGN in viral transmission by AZT-treated AGM MDDC. AGM MDDC from four animals treated with AZT before viral exposure and during the 3-day period before the start of the coculture were exposed to SIVagm in the presence of anti-DC-SIGNhu MAb (1B10), a control IgG2a, or medium alone. In order to express the percent inhibition, the p27gag antigen values of the IgG2a-treated cells, obtained for day 14, were normalized to 100%.
In order to confirm this, we blocked viral replication with an RT inhibitor. MDDC from two animals were treated with AZT during the 3 days before the start of the coculture. We observed that AZT treatment of MDDC did not reduce viral transmission compared to levels of viral replication in medium-treated MDDC (Fig. 8B). For both animals, levels of viral replication in DC-T cocultures were similar whether the DC were treated with AZT or not. We confirmed that virus transmission after 3 days is efficient even after treatment with AZT for MDDC from two further AGM donors (data not shown). Thus, virus transmission is efficient in the absence of early de novo synthesis. These experiments also indicate that infectious particles are maintained at least 3 days in AGM MDDC.
In order to confirm the implication of DC-SIGN in viral transmission, AZT-treated MDDC from four animals were exposed to SIVagm in the presence of anti-DC-SIGN MAb and cocultured with SupT1 cells 3 days after. In these conditions viral transmission was reduced between 70 and 95% (Fig. 8C) showing again that the viral transmission was promoted in a predominant DC-SIGN-dependent manner.
The presence of an RT inhibitor within the 3-day period thus failed to significantly reduce viral transmission, indicating that the failure to fully inhibit transmission by using an anti-DC-SIGN MAb was not due to an undetectable productive infection of MDDC. These findings support data showing that other molecules than DC-SIGN also contribute to viral transmission (55-57) albeit, as we show here, to a lower degree than DC-SIGN when low virus doses are used.
DC-SIGN-expressing cells in AGM LNs.
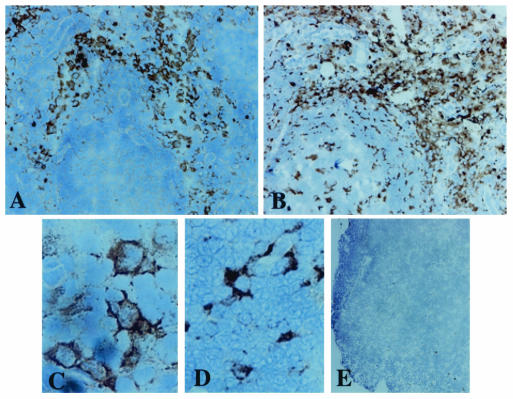
Our data demonstrate that AGM MDDC express high levels of DC-SIGN comparable to human MDDC and that DC-SIGN plays the main role in SIVagm transmission by AGM MDDC in the presence of lower doses of virus. The study also revealed that AGM LN cells abundantly express dc-sign mRNA, suggesting that DC-SIGN is expressed at significant levels in LN in vivo. To determine whether not only the mRNA but also the protein is expressed on the surface of cells within LNs, we performed immunohistochemistry analyses on LN sections derived from two SIVagm uninfected AGMs (animals 02001 and 00021) for which the level of DC-SIGN expression on MDDC had been evaluated (166,000 and 238,000 copies/cell, respectively) and from one chronically SIVagm-infected AGM (animal 98013). We labeled the sections with two distinct anti-DC-SIGN MAb (1B10 and 8A5) raised against to two distinct epitopes. The distribution of the DC-SIGN+ cells was similar whatever the MAb we used (data not shown). We detected DC-SIGN+ cells mainly within the medulla (Fig. 9A) and often within the cortex and/or paracortex (cortex/paracortex) in all three tested animals (Fig. 9A and data not shown). We did not detect any differences in the distribution of DC-SIGN+ cells between infected and uninfected animals (data not shown). DC-SIGN+ cells both in the medulla and in the cortex/paracortex exhibited an irregular morphology (Fig. 8C), and some DC-SIGN+ round cells were present within medullary sinuses (a finding consistent with sinusoidal macrophages) (data not shown). The distribution and the morphology of the DC-SIGN+ cells in AGM LNs were thus identical to that described in LNs of macaques (8, 23, 45, 62). In rhesus macaques, it has been previously described that in SIV-uninfected spleen and in SIV-infected LN, DC-SIGN+ cells with dendritic morphology were uniformly positive for the monocyte/macrophage lineage marker CD68 (8, 45). The latter remains abundant even after SIVmac infection (45). In AGM LNs, CD68 staining was also abundant in medulla and cortex/paracortex areas. The distribution of CD68+ and DC-SIGN+ cells differed in germinal centers (GC) (Fig. 9B). CD68+ cells were present in GC, whereas DC-SIGN+ cells remain always distributed outside the GC (Fig. 9A and B). Numerous CD68+ cells also exhibited an irregular morphology (Fig. 9D).
FIG. 9.
DC-SIGN+ cells in AGM LNs. Sections (5 μm) of cryopreserved OCT-embedded LN, harvested from two SIVagm-uninfected AGMs and from one experimentally SIVagm-infected AGM (120 days postinfection), were incubated with anti-DC-SIGNhu MAb (8A5 and 1B10), with anti-CD68hu MAb, or with control IgG2a. Staining was done by using horseradish peroxidase conjugated to a secondary antibody. Antigen staining appears in brown. Sections were counterstained with Harris's hematoxylin to stain nuclei, which appear in blue. Representative sections of axillary LNs from the experimentally SIVagm-infected AGM stained with the anti-DC-SIGN clone 8A5 are presented here. (A and C) Staining with anti-DC-SIGN (8A5); (B and D) staining with anti-CD68; (E) staining with the isotype control. Magnifications: ×100 (A, B, and E), ×400 (C and D).
DISCUSSION
This is the first study that has analyzed the function of DC-SIGN from a natural host species of SIV. Previous studies have only reported interactions between DC-SIGN from human and rhesus macaques expressed on THP-1 or 293T cells with HIV-luc pseudotyped with Env from SIVagm or SIVsm (38, 61). Importantly, interactions of SIV with human molecules or vice versa of HIV with simian molecules do not necessarily reflect interactions of the virus with the homologous host-specific molecule. It was shown, for instance, that SIVmac, although able to use human STRL33/Bonzo, does not efficiently use the macaque molecule for entry (39, 46). Similarly, HIV-1 uses AGM CCR5 only inefficiently due to the G163R difference between the human and AGM protein, whereas SIVagm is adapted to efficiently use the wild-type AGM CCR5 (26). It is thus important to analyze the interactions of receptors and viruses derived from the same species. Here we describe the efficient interaction between DC-SIGN derived from AGMs (C. sabaeus) and its host-specific SIV (SIVagm.sab). In order to be close to physiological conditions, we used a viral isolate and not a molecular clone. Furthermore, we performed infections with low viral doses (MOIs of 10−3 to 10−5). We showed on human cell lines that DC-SIGNagm can act as a viral attachment factor like its human and macaque counterparts despite structural differences among these molecules. Consequently, this suggests that the lower viral load in LNs during nonpathogenic SIVagm infection (10) compared to pathogenic HIV/SIVmac infections (4, 5) cannot be explained by structural differences in the AGM molecule. We cannot totally exclude variations either among SIVagm strains or among dc-signagm alleles that might be detected by analyzing more donors. However, even if such variations exist, they could not explain the consistently nonpathogenic outcome of SIVagm infection.
In view of the above results, additional factors must be involved in the low viral load in LNs during SIVagm infections. Efficiency of transmission to T cells in vitro depends on the levels of DC-SIGN and ICAM-1 expression on DC (38, 43). Alternative splicing of dc-sign mRNA might represent a mechanism that could regulate the expression of the dc-sign gene. Our study reveals a nonexhaustive repertoire of DC-SIGN in LNs of AGMs. We have not detected dc-sign mRNA encoding potentially soluble isoforms of DC-SIGNagm, whereas transcripts coding for these isoforms have been detected by using only a single RT-PCR step in human cells, such as PBMC, DC derived from CD34+ peripheral blood hematopoietic progenitors, and resting CD14+ monocytes (35). Indeed, the expression pattern of dc-sign transcripts may depend on the origin, as well as the maturation or activation state, of the cells (35). It remains unknown whether transcripts coding for potentially soluble isoforms also exist in human LN or in other nonhuman primates such as macaques. Although soluble isoforms do not seem to be able to mediate enhancement of infection (27), they may regulate expression of the membrane-associated forms of DC-SIGN (35). However, we demonstrate here that AGM MDDC show high surface-expressing levels of DC-SIGN protein. This contrasts with one study which reported very low levels of DC-SIGN on rhesus macaque MDDC (61). The expression levels of DC-SIGN may vary according to the primate species and/or the donor (25, 61). Numbers of DC-SIGN molecules on AGM MDDC did indeed vary according to the AGM donor, as shown for human MDDC (1). However, the AGM myeloid DC (MDDC) from all studied AGM donors expressed at least 100,000 DC-SIGN molecules and often more than 200,000 molecules per cell, a level sufficient to promote viral transmission to T cells in vitro.
In vivo, DC-SIGN+ cells could be easily detected within the medulla sinuses and the paracortex of AGM LNs. We cannot exclude the possibility that the detection of positive cells in the medulla sinuses could also be due to the related L-SIGN2 (3). However, the distribution of DC-SIGN+ cells in the LNs of AGMs was very similar to that described in macaques (8, 45, 62). Rhesus macaque antigen-presenting cells in the medulla and cortex/paracortex in organized lymphoid tissues (LNs and spleen) have been shown to express DC-SIGN, although the number of positive cells has not been quantified (8, 45, 62). The nature of the AGM DC-SIGN+ cells is unknown, but the cells exhibited a morphology characteristic of DC. It has been suggested in similar studies in humans and rhesus macaque tissues that the DC-SIGN+ cells likely correspond to interdigitating DC and sinusoidal macrophages (14, 45, 49, 62). The distribution of DC-SIGN+ cells in the medulla and paracortex was similar to that of cells expressing a marker of the monocyte/macrophage lineage (CD68). However, only CD68+ cells were detected in germinal center, showing that not all CD68+ cells express DC-SIGN. Furthermore, it has been shown that in macaque spleens and LNs all DC-SIGN+ cells exhibiting a dendritic morphology express CD68 (45). Altogether, our data show a similar morphology and distribution of DC-SIGN+ cells compared to these cells in humans and macaques. We also observed that mRNA coding for functional DC-SIGN is abundantly expressed in AGM LNs. This suggests that DC-SIGN expression on DC in AGM LN is not lower than in humans and macaques.
We cannot totally exclude a distinct regulation of DC-SIGN expression in response to SIVagm infection. The level of human DC-SIGN at the cell surface is indeed influenced by HIV-1 proteins. A dramatic increase of lymphocyte adhesion to HIV-1-infected DC in vitro due to an upregulation of DC-SIGN mediated by Nef has been reported (50). This phenomenon, however, is not consistently observed for all donors (31). It is not excluded that Nef encoded by SIVagm interacts differentially with host-specific cellular proteins compared to HIV/SIVmac Nef in human and macaque cells, which could result in lower levels of DC-SIGN expression and/or lower T-cell activation profiles in SIVagm-infected AGMs. Lower T-cell activation states are indeed observed in nonpathogenic SIV infections in their natural hosts (7, 11, 37, 47). However, there are no signs thus far of SIVagm replication within AGM MDDC with at least up to 10−3 TCID50/cell and after p27 values up to 2 weeks. This might also be dependent on the SIVagm strains used. Further studies are needed to determine the susceptibility of DC to SIV in nonpathogenic SIV infection models.
Finally, the expression level might depend on the cytokine environment in vivo after SIV infection. Several studies have reported that IL-4 induces DC-SIGN expression on human peripheral blood monocytes in vitro (35, 41). This IL-4-dependent expression is negatively regulated by alpha interferon (IFN-α), transforming growth factor β, and IFN-γ (41). Recently, it has been shown in another model of nonpathogenic SIVinfection, SIVsm infection in the sooty mangabeys, that the chronic phase of infection is characterized by higher levels of IL-4 and lower levels of IFN-γ production (47). Thus, there are as yet no data that could support any downregulation of DC-SIGN expression after SIV infection in the natural host.
We show here that MDDC from AGMs not only express high levels of DC-SIGN but also efficiently transmit SIVagm to T cells. This has been confirmed here for MDDC from 10 distinct AGM donors. Furthermore, as shown by inhibition assays with anti-DC-SIGN MAb, viral transmission by AGM MDDC is DC-SIGN dependent. This has been confirmed with MDDC from six distinct AGM donors. This result supports the findings of an earlier study of Chinese macaque species, M. fascicularis, in which viral transmission is also largely DC-SIGN dependent (25). However, it contrasts with a study of M. mulatta MDDC that indicated no discernible role of DC-SIGN in viral transmission (61). These controversial results might be explained by variations of DC-SIGN expression according to individuals, especially if only a few animals were studied, and/or according to monkey species. Interestingly, we observed that the viral transmission by AGM MDDC was significantly blocked by anti-DC-SIGN MAbs only when a low MOI (10−5) was used. It has been reported that the affinity of HIV-1 gp120 for DC-SIGNhu is higher than for CD4hu (9), and some data suggested that it is also higher than for the mannose receptor (56). Thus, the use of high viral infectious doses could probably allow attachment to other molecules as well. This might also explain why in previous studies anti-DC-SIGN MAb or mannan could not consistently block viral entry or Env/DC-SIGN interaction in MDDC. Indeed, in these previous studies, very high doses of HIV AD8 (MOI of 45) (57) or a high amount of gp120 (twofold more than the predetermined concentration for cellular saturation) (56) were used.
DC seem to be a “hiding place” for HIV-1 (58). However, the mechanism of viral transfer via DC-SIGN has yet to be fully elucidated. The function of DC-SIGN in the transmission of HIV-1 depends on its cellular context (54). DC-SIGN expressed by DC or the monocytic cell line THP-1 but not 293 and HOS cells internalizes and retains HIV-1Bal in a highly infectious state for more than 5 days (13, 54). Immature human MDDC promote HIV-1Bal transfer by a mechanism that is DC-SIGN dependent (40 to 100% according to the donors tested) at least up to 2 days after viral exposure (54). Recently, others suggest most endocytosed virus is already degraded after 24 h (55). We demonstrated here that sufficient amounts of infectious SIVagm particles remain intact in AGM MDDC up to 3 days, even when low doses of virus are used. This efficiency might be due to the fact that we used a virus isolate and MDDC to assess viral transfer to highly permissive T cells.
We analyzed here for the first time the virological functions of DC-SIGN from an animal species resistant to AIDS. The present study revealed similar activities for DC-SIGNagm regarding trans infection compared to previous reports on human and macaque DC-SIGN. We demonstrated that DC-SIGN+ cells are present in AGM LNs and that AGM MDDC express levels of DC-SIGN similar to those expressed by human MDDC. We showed that AGM MDDC are able to efficiently transmit SIVagm to T cells. Finally, we showed that SIVagm transmission is largely DC-SIGN dependent at low MOIs. Altogether, our data indicate that the virus-binding properties of DC-SIGN are not directly associated with viral load levels in LNs. In conclusion, additional factors, such as the frequency in LNs of major target cells (activated T CD4+ lymphocytes), are more likely determinating factors for viral replication levels in LNs.
Acknowledgments
We are indebted to Robert Bassin for critical reading of the manuscript. We thank Arnaud Moris for helpful discussion and Lisa An for contribution in editing of the manuscript. We thank Sandrine Dos Santos and M. C. Cumont for excellent technical help and Ahidjo Ayouba and veterinarians from the Cameroonian and European zoos for DNA samples.
This study was supported by grants from the French Agency for AIDS Research (ANRS). M.J.-Y.P. was a recipient of a scholarship from the Ministère de l'Education Nationale. L.M. and M.A.S. were recipients of ANRS fellowships.
REFERENCES
- 1.Baribaud, F., S. Pöhlmann, G. J. Leslie, F. Mortari, and R. W. Doms. 2002. Quantitative expression and virus transmission analysis of DC-SIGN on monocyte-derived dendritic cells. J. Virol. 76:9135-9142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baribaud, F., S. Pöhlmann, T. Sparwasser, M. T. Y. Kimata, Y. Choi, B. S. Haggarty, N. Ahmad, T. Macfarlan, T. G. Edwards, G. J. Leslie, J. Arnason, T. A. Reinhart, J. T. Kimata, D. R. Littman, J. A. Hoxie, and R. W. Doms. 2001. Functional and antigenic characterization of human, rhesus macaque, pigtailed macaque and murine DC-SIGN. J. Virol. 75:10281-10289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bashirova, A. A., L. Wu, J. Cheng, T. D. Martin, M. P. Martin, R. E. Benveniste, J. D. Lifson, V. N. KewalRamani, A. Hughes, and M. Carrington. 2003. Novel member of the CD209 (DC-SIGN) gene family in primates. J. Virol. 77:217-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beer, B., J. Scherer, J. Megede, S. Norley, M. Baier, and R. Kurth. 1996. Lack of dichotomy between virus load of peripheral blood and lymph nodes during long term simian immunodeficiency virus infection of African green monkeys. Virology 219:367-375. [DOI] [PubMed] [Google Scholar]
- 5.Broussard, S. R., S. I. Staprans, R. White, E. M. Whitehead, M. B. Feinberg, J. S. Allan, T. W. Chun, L. Stuyver, S. B. Mizell, L. A. Ehler, J. A. Mican, M. Baseler, A. L. Lloyd, M. A. Nowak, and A. S. Fauci. 2001. Simian immunodeficiency virus replicates to high levels in naturally infected African green monkeys without inducing immunologic or neurologic disease. J. Virol. 75:2262-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cameron, P. U., P. S. Freudenthal, J. M. Barker, S. Gezelter, K. Inaba, and R. M. Steinmann. 1992. Dendritic cells exposed to human immunodeficiency virus type-1 transmit a vigorous cytopathic infection to CD4+ cells. Science 257:383-387. [DOI] [PubMed] [Google Scholar]
- 7.Chakrabarti, L. A., S. R. Lewin, L. Zhang, A. Gettie, A. Luckay, L. N. Martin, E. Skulski, D. D. Ho, C. Cheng-Mayer, and P. A. Marx. 2000. Normal T-cell turnover in sooty mangabeys harboring active simian immunodeficiency virus infection. J. Virol. 74:1209-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi, Y. K., B. A. Fallert, M. A. Murphey-Corb, and T. A. Reinhart. 2003. Simian immunodeficiency virus dramatically alters expression of homeostatic chemokines and dendritic cell markers during infection in vivo. Blood 101:1684-1691. [DOI] [PubMed] [Google Scholar]
- 9.Curtis, B. M., S. Scharnowske, and A. J. Watson. 1992. Sequence and expression of a membrane-associated C-type lectin that exhibits CD4-independent binding of human immunodefficiency virus envelope glycoprotein gp120. Proc. Natl. Acad. Sci. USA 89:8356-8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diop, O. M., A. Gueye, M. Dias-Tavares, C. Kornfeld, A. Faye, P. Ave, M. Huerre, S. Corbet, F. Barré-Sinoussi, and M. C. Müller-Trutwin. 2000. High levels of viral replication during primary SIVagm infection are rapidly and strongly controlled in African green monkeys. J. Virol. 74:7538-7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Estaquier, J., T. Idziorek, F. de Bels, F. Barre-Sinoussi, B. Hurtrel, A. M. Aubertin, A. Venet, M. Mehtali, E. Muchmore, P. Michel, et al. 1994. Programmed cell death and AIDS: significance of T-cell apoptosis in pathogenic and nonpathogenic primate lentiviral infections. Proc. Natl. Acad. Sci. USA 91:9431-9435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geijtenbeek, T. B. H., G. Koopman, G. C. F. Van Duijnhoven, S. J. van Vliet, A. C. Van Schijndel, A. Engering, J. L. Heeney, and Y. Van Kooyk. 2001. Rhesus macaque and chimpanzee DC-SIGN act as HIV/SIV gp120 trans-receptor, similar to human DC-SIGN. Immunol. Lett. 79:101-107. [DOI] [PubMed] [Google Scholar]
- 13.Geijtenbeek, T. B. H., D. S. Kwon, R. Torensma, S. J. van Vliet, G. C. F. Van Duijnhoven, J. Middel, I. L. M. H. A. Cornelissen, D. R. Littman, C. G. Figdor, and Y. van Kooyk. 2000. DC-SIGN, a dendritic cell-specific HIV-1-Binding protein that enhances trans-infection of T cells. Cell 100:587-597. [DOI] [PubMed] [Google Scholar]
- 14.Geijtenbeek, T. B. H., R. Torensma, S. J. Van Vliet, G. C. F. Van Duijnhoven, G. J. Adema, and Y. Van Kooyk. 2000. Identification of DC-SIGN, a novel dendritic cell-specific ICAM-3 receptor that supports primary immune responses. Cell 100:575-585. [DOI] [PubMed] [Google Scholar]
- 15.Geijtenbeek, T. B. H., G. C. F. Van Duijnhoven, S. J. van Vliet, E. Krieger, G. Vriend, C. G. Figdor, and Y. Van Kooyk. 2002. Identification of different binding sites in the dendritic cell-specific receptor for ICAM-3 and HIV-1. J. Biol. Chem. 277:11314-11320. [DOI] [PubMed] [Google Scholar]
- 16.Goldstein, S., I. Ourmanov, C. R. Brown, B. E. Beer, W. R. Elkins, R. Plishka, A. Buckler-White, V. M. Hirsch, T. W. Chun, L. Stuyver, S. B. Mizell, L. A. Ehler, J. A. Mican, M. Baseler, A. L. Lloyd, M. A. Nowak, and A. S. Fauci. 2000. Wide range of viral load in healthy African green monkeys naturally infected with simian immunodeficiency virus. J. Virol. 74:11744-11753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Granelli-Piperno, A., E. Delgado, V. Finkel, W. Paxton, and R. M. Steinman. 1998. Immature dendritic cells selectively replicate macrophagetropic (M-tropic) human immunodefficiency virus type 1, while mature cells efficiency transmit both M- and T-tropic virus to T cells. J. Virol. 72:2733-2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Granelli-Piperno, A., V. Finkel, E. Delgado, and R. M. Steinman. 1999. Virus replication begins in dendritic cells during the transmission of HIV-1 from mature dendritic cells to T cells. Curr. Biol. 9:21-29. [DOI] [PubMed] [Google Scholar]
- 19.Grouard, G., and E. A. Clark. 1997. Role of dendritic and follicular dendritic cells in HIV infection and pathogenesis. Curr. Opin. Immunol. 9:562-567. [DOI] [PubMed] [Google Scholar]
- 20.Halary, F., A. Amara, H. Lortat-Jacob, M. Messerle, T. Delaunay, C. Houlès, F. Fieschi, F. Arenzana-Seisdedos, J.-F. Moreau, and J. Déchanet-Merville. 2002. Human cytomegalovirus binding to DC-SIGN is required for dendritic cell infection and target cell trans-infection. Immunity 17:653-664. [DOI] [PubMed] [Google Scholar]
- 21.Holzammer, S., E. Holznagel, A. Kaul, R. Kurth, and S. Norley. 2001. High virus loads in naturally and experimentally SIVagm-infected African green monkeys. Virology 283:324-331. [DOI] [PubMed] [Google Scholar]
- 22.Hu, J., M. B. Gardner, and C. J. Miller. 2000. Simian immunodeficiency virus rapidly penetrates the cervicovaginal mucosa after intravaginal inoculation and infects intraepithelial dendritic cells. J. Virol. 74:6087-6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jameson, B., F. Baribaud, S. Pöhlmann, D. Ghavimi, F. Mortari, R. W. Doms, and A. Iwasaki. 2002. Expression of DC-SIGN by dendritic cells of intestinal and genital mucosae in humans and rhesus macaques. J. Virol. 76:1866-1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawamura, T., F. O. Gulden, M. Sugaya, D. T. McNamara, D. L. Borris, M. M. Lederman, J. M. Orenstein, P. A. Zimmerman, and A. Blauvelt. 2003. R5 HIV productively infects Langerhans cells, and infection levels are regulated by compound CCR5 polymorphisms. Proc. Natl. Acad. Sci. USA 100:8401-8406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kimata, M. T. Y., M. Cella, J. E. Biggins, C. Rorex, R. White, S. Hicks, J. M. Wilson, P. G. Patel, J. S. Allan, M. Colonna, and J. T. Kimata. 2002. Capture and transfer of simian immunodeficiency virus by macaque dendritic cells is enhanced by DC-SIGN. J. Virol. 76:11827-11836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuhmann, S., N. Madani, O. M. Diop, E. J. Platt, J. Morvan, M. C. Müller-Trutwin, F. Barré-Sinoussi, and D. Kabat. 2001. Frequent substitution polymorphism in African green monkey CCR5 cluster at active sites for infections by SIVagm, implying ancient virus-host coevolution. J. Virol. 75:8449-8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwon, D. S., G. Gregorio, N. Bitton, W. A. Hendrickson, and D. R. Littman. 2002. DC-SIGN-mediated internalization of HIV is required for trans-enhancement of T-cell infection. Immunity 16:135-144. [DOI] [PubMed] [Google Scholar]
- 28.Lee, B., G. Leslie, E. Soilleux, U. O'Doherty, S. Baik, E. Levroney, K. Flummerfelt, W. Swiggard, N. Coleman, M. Malim, and R. W. Doms. 2001. Cis expression of DC-SIGN allows for more efficient entry of human and simian immunodeficiency viruses via CD4 and a coreceptor. J. Virol. 75:12028-12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marechal, V., M. C. Prevost, C. Petit, E. Perret, J. M. Heard, and O. Schwartz. 2001. Human immunodeficiency virus type 1 entry into macrophages mediated by macropinocytosis. J. Virol. 75:11166-11177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McDonald, D., L. Wu, S. M. Bohks, V. N. KewalRamani, D. Unutmaz, and T. J. Hope. 2003. Recruitment of HIV and its receptors to dendritic cell-T cell junctions. Science 300:1295-1297. [DOI] [PubMed] [Google Scholar]
- 31.Messmer, D., J.-M. Jacqué, C. Santisteban, C. Bristow, S.-H. Han, L. Villamide-Herrera, E. Mehlhop, P. A. Marx, R. M. Steinman, and M. Pope. 2002. Endogenously expressed nef uncouples cytokine and chemokine production from membrane phenotypic maturation in dendritic cells. J. Immunol. 169:4172-4182. [DOI] [PubMed] [Google Scholar]
- 32.Müller, M. C., N. K. Saksena, E. Nerrienet, C. Chappey, V. M. A. Hervé, J.-P. Durand, P. Legal-Campodonico, M.-C. Lang, J.-P. Digoutte, A. J. Georges, M.-C. Georges-Courbot, P. Sonigo, and F. Barré-Sinoussi. 1993. Simian immunodeficiency viruses from central and western Africa: evidence for a new species-specific lentivirus in tantalus monkeys. J. Virol. 67:1227-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Müller-Trutwin, M. C., S. Corbet, J. Hansen, M. C. Georges-Courbot, O. Diop, J. Rigoulet, F. Barré-Sinoussi, and A. Fomsgaard. 1999. Mutations in CCR5 coding sequences are not associated with SIV carrier status in African nonhuman primates. AIDS Res. Hum. Retrovir. 15:931-939. [DOI] [PubMed] [Google Scholar]
- 34.Müller-Trutwin, M. C., S. Corbet, S. Souquière, P. Roques, P. Versmisse, A. Ayouba, S. Delarue, E. Nerrienet, J. Lewis, P. Martin, F. Simon, F. Barré-Sinoussi, and P. Mauclère. 2000. SIVcpz from a naturally infected Cameroonian chimpanzee: biological and genetic comparison with HIV-1 N. J. Med. Primatol. 29:166-172. [DOI] [PubMed] [Google Scholar]
- 35.Mummidi, S., G. Catano, L. Lam, A. Hoefle, V. Telles, K. Begum, F. Jimenez, S. S. Ahuja, and S. K. Ahuja. 2001. Extensive repertoire of membrane-bound and soluble DC-SIGN1 and DC-SIGN2 isoforms: inter-individual variation in expression of DC-SIGN transcripts. J. Biol. Chem. 276:33196-33212. [DOI] [PubMed] [Google Scholar]
- 36.Nobile, C., A. Moris, F. Porrot, N. Sol-Foulon, and O. Schwartz. 2003. Inhibition of human immunodeficiency virus type 1 Env-mediated fusion by DC-SIGN. J. Virol. 77:5313-5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Onanga, R., C. Kornfeld, I. Pandrea, J. Estaquier, S. Souquière, P. Rouquet, V. Poaty Mavoungou, O. Bourry, S. M'Boup, F. Barré-Sinoussi, F. Simon, C. Apetrei, P. Roques, and M. C. Müller-Trutwin. 2002. High levels of viral replication contrast with only transient changes in CD4+ and CD8+ cell numbers during the early phase of experimental infection with simian immunodeficiency virus SIVmnd-1 in Mandrillus sphinx. J. Virol. 76:10256-10263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pöhlmann, S., F. Baribaud, B. Lee, G. J. Leslie, M. D. Sanchez, K. Hiebenthal-Millow, J. Münch, F. Kirchhoff, and R. W. Doms. 2001. DC-SIGN interactions with human immunodefficiency virus type 1 and 2 and simian immunodefficiency virus. J. Virol. 75:4664-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pöhlmann, S., B. Lee, S. Meister, M. Krumbiegel, G. Leslie, R. W. Doms, and F. Kirchhoff. 2000. Simian immunodeficiency virus utilizes human and sooty mangabey but not rhesus macaque STRL33 for efficient entry. J. Virol. 74:5075-5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pope, M., M. G. Betjes, N. Romani, H. Hirmand, P. U. Cameron, L. Hoffman, S. Gezelter, G. Schuler, and R. M. Steinman. 1994. Conjugates of dendritic cells and memory T lymphocytes from skin facilitate productive infection with HIV-1. Cell 78:389-398. [DOI] [PubMed] [Google Scholar]
- 41.Relloso, M., A. Puig-Kröger, O. Muniz Pello, J. L. Rodriguez-Fernandez, G. de la Rosa, N. Longo, J. Navarro, M. A. Munoz-Fernandez, P. Sanchez-Mateo, and A. L. Corbi. 2002. DC-SIGN (CD209) expression is IL-4 dependent and is negatively regulated by IFN, TGF-β, and anti-inflammatory agents. J. Immunol. 168:2634-2643. [DOI] [PubMed] [Google Scholar]
- 42.Rowland-Jones, S. L. 1999. HIV: the deadly passenger in dendritic cells. Curr. Biol. 9:R248-R250. [DOI] [PubMed] [Google Scholar]
- 43.Sanders, R. W., E. C. de Jong, C. E. Baldwin, J. H. N. Schuitemaker, M. L. Kapsenberg, and B. Berkhout. 2002. Differential transmission of human immunodeficiency virus type 1 by distinct subsets of effector dendritic cells. J. Virol. 76:7812-7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Santos, P. R., K. Petitprez, and C. Butor. 2002. Gene for Chinese rhesus macaque DC-SIGN predicts the existence of A but not B isoforms of the protein. AIDS Res. Hum. Retrovir. 18:977-981. [DOI] [PubMed] [Google Scholar]
- 45.Schwartz, A., X. Alvarez, and A. A. Lackner. 2002. Distribution and immunophenotype of DC-SIGN-expressing cells in SIV-infected and uninfected macaques. AIDS Res. Hum. Retrovir. 18:1021-1029. [DOI] [PubMed] [Google Scholar]
- 46.Sharron, M., S. Pöhlmann, K. Price, E. Lolis, M. Tsang, F. Kirchhoff, R. W. Doms, and B. Lee. 2000. Expression and coreceptor activity of STRL33/Bonzo on primary peripheral blood lymphocytes. Blood 96:41-49. [PubMed] [Google Scholar]
- 47.Silvestri, G., D. L. Sodora, R. A. Koup, M. Paiardini, S. P. O'Neil, H. M. McClure, S. I. Staprans, and M. B. Feinberg. 2003. Nonpathogenic SIV infection of sooty mangabeys is characterized by limited bystander immunopathology despite chronic high-level viremia. Immunity 18:441-452. [DOI] [PubMed] [Google Scholar]
- 48.Soilleux, E. J., R. Barten, and J. Trowsdale. 2000. DC-SIGN, a related gene, and CD23 form a cluster on 19p13. J. Immunol. 165:2937-2942. [DOI] [PubMed] [Google Scholar]
- 49.Soilleux, E. J., L. S. Morris, G. Leslie, J. Chemini, Q. Luo, E. Levroney, J. Trowsdale, L. J. Montaner, R. W. Doms, D. Weissman, N. Coleman, and B. Lee. 2002. Constitutive and induced expression of DC-SIGN on dendritic cell and macrophage subpopulations in situ and in vitro. J. Leukoc. Biol. 71:445-457. [PubMed] [Google Scholar]
- 50.Sol-Foulon, N., A. Moris, C. Nobile, C. Boccaccio, A. Engering, J.-P. Abastado, J.-M. Heard, Y. Van Kooyk, and O. Schwartz. 2002. HIV-1 Nef-induced upregulation of DC-SIGN in dendritic cells promotes lymphocyte clustering and viral spread. Immunity 16:145-155. [DOI] [PubMed] [Google Scholar]
- 51.Soto-Ramirez, L. E., B. Renjifo, M. F. McLane, R. Marlink, C. O'Hara, R. Sutthent, C. Wasi, P. Vithayasi, V. Vithayasi, C. Apichartpiyakul, P. Auewarakul, V. Pena Cruz, D. Chui, R. Osathamondh, K. Mayer, T. Lee, and M. Essex. 1996. HIV-1 Langherans cells tropism associated with heterosexual transmission of HIV. Science 271:1291-1293. [DOI] [PubMed] [Google Scholar]
- 52.Steinman, R. M. 2000. DC-SIGN: a guide to some mysteries of dendritic cells. Cell 100:491-494. [DOI] [PubMed] [Google Scholar]
- 53.Steinman, R. M., and K. Inaba. 1999. Myeloid dendritic cells. J. Leukoc. Biol. 66:205-208. [DOI] [PubMed] [Google Scholar]
- 54.Trumpfheller, C., C. Gyu Park, J. Finke, R. M. Steinman, and A. Granelli-Piperno. 2003. Cell type-dependent retention and transmission of HIV-1 by DC-SIGN. Int. Immunol. 15:289-298. [DOI] [PubMed] [Google Scholar]
- 55.Turville, S., J. Wilkinson, P. Cameron, J. Dable, and A. L. Cunningham. 2003. The role of dendritic cell C-type lectin receptors in HIV pathogenesis. J. Leukoc. Biol. 74:710-718. [DOI] [PubMed] [Google Scholar]
- 56.Turville, S. G., J. Arthos, K. Mac Donald, G. Lynch, H. Naif, G. Clark, D. Hart, and A. L. Cunningham. 2001. HIV gp120 receptors on human dendritic cells. Blood 98:2482-2488. [DOI] [PubMed] [Google Scholar]
- 57.Turville, S. G., P. U. Cameron, A. Handley, G. Lin, S. Pöhlmann, R. W. Doms, and A. L. Cunningham. 2002. Diversity of receptors binding HIV on dendritic cell subsets. Nat. Immunol. 3:975-983. [DOI] [PubMed] [Google Scholar]
- 58.Van Kooyk, Y., and T. B. Geijtenbeek. 2003. DC-SIGN: escape mechanism for pathogens. Nat. Rev. Immunol. 3:697-709. [DOI] [PubMed] [Google Scholar]
- 59.Verrier, F. C., P. Charneau, R. Altemeyer, S. Laurent, A. M. Borman, and M. Girard. 1997. Antibodies to several conformation-dependent epitopes of gp120/gp41 inhibit CCR5-dependent cell-to-cell fusion mediated by the native envelope glycoprotein of a primary macrophage-tropic HIV-1 isolate. Proc. Natl. Acad. Sci. USA 94:9326-9331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weissman, D., Y. Li, J.-M. Orenstein, and A. S. Fauci. 1995. Both a precursor and a mature population of dendritic cells can bind HIV: however, only the mature population that expressed CD80 can pass infection to unstimulated CD4+ T cells. J. Immunol. 155:4111-4117. [PubMed] [Google Scholar]
- 61.Wu, L., A. A. Bashirova, T. D. Martin, L. Villamide, E. Mehlhop, A. O. Chertov, D. Unutmaz, M. Pope, M. Carrington, and V. N. KewalRamani. 2002. Rhesus macaque dendritic cells efficiently transmit primate lentiviruses independently of DC-SIGN. Proc. Natl. Acad. Sci. USA 99:1568-1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zimmer, M. I., A. T. Larregina, C. M. Castillo, S. Capuano III, L. D. Falo, Jr., M. Murphey-Corb, T. A. Reinhart, and S. M. Barratt-Boyes. 2002. Disrupted homeostasis of Langerhans cells and interdigitating dendritic cells in monkeys with AIDS. Blood 99:2859-2868. [DOI] [PubMed] [Google Scholar]