Abstract
Despite membrane-based separations offering superior alternative to packed bed chromatographic processes, there has been a substantial lacuna in their actual application to separation processes. One of the major reasons behind this is the lack of availability of appropriately modified or end-group modifiable membranes. In this paper, an affinity membrane was developed using a commercially available serine protease inhibitor, para-aminobenzamidine (pABA). The membrane modification was optimized for protein binding capacity by varying: i) the length of the spacer arm (SA; 5-atoms, 7-atoms, and 14-atoms) linking the ligand to membrane surface; ii) the affinity ligand (pABA) density on membrane surface (5–25 nmoles per cm2). Resulting membranes were tested for their ability to bind plasminogen activators (PAs) from mono- and multi- component systems in batch mode. The membrane containing pABA linked through 7-atoms SA but similar ligand density as in the case of 5- or 14- atoms long SA was found to bind up to 1.6-times higher amounts of PA per nmole of immobilized ligand from conditioned HeLa cell culture media. However, membranes with similar ligand densities but different lengths of SA, showed comparable binding capacities in monocomponent system. In addition, the length of SA did not affect the selectivity of the ligand for PA. A clear inverse linear correlation was observed between ligand density and binding capacity until the point of PA binding optima was reached (11±1.0 nmoles per cm2) in mono- and multi- component systems for 7- as well as 14- atoms SA. Up to 200-fold purification was achieved in a single step separation of PA from HeLa conditioned media using these affinity membranes. The issues of ligand leaching and reuse of the membranes were also investigated. An extensive regeneration procedure allowed the preservation of approximately 95% of the PA binding capacity of the membranes even after five cycles of use.
Keywords: Affinity membrane, Plasminogen Activator, para-Aminobenzamidine, Spacer arm, Ligand density
1. Introduction2
Membrane based chromatographic or batch separation processes have been discussed as promising alternatives for some time now [1] as they lead to more robust processes that are also very amenable to inclusion into a manufacturing process [2, 3]. While membranes provide a more easily accessible surface area for protein binding, they also help overcome the limitations of diffusion resistance, high pressure drop, need for high operating pressure/flow rate, and other usual problems related to traditional chromatographic process scale up [4–8]. This translates in many applications to simpler equipment and safer operations [8]. It has been determined that end-group modified membranes have capacities approaching those found in most synthetic chromatography beads [8]. However, the majority of downstream processes in pharmaceutical and other industrial sectors till date are based on traditional chromatographic media and one of the major reasons behind this has been a lack of availability of appropriately modified membranes.
It is important to understand that the membranes do not need to be developed to provide completely new separations [9, 10]; rather, the existing chemistries for modification of traditional chromatographic matrices need to be adapted to membrane modifications and further optimized with respect to length of spacer arm and ligand densities to develop efficient tools for separation processes. Affinity membranes in particular have been a very promising alternative to affinity bead based chromatographic media. Modification of membranes with ligands that have affinity and selectivity for the target protein provides a very effective tool for the separation of proteins that belong to a narrow size range [4, 11, 12]. In the process of making available new membranes for separation processes, several things need to be considered, such as the type of membrane, the length of spacer arm (SA), and the type of ligand. Membranes with hydrophilic surface properties, such as cellulose based membranes, have a very low propensity for nonspecific protein binding and are thus preferable for use in biological separations [8]. Furthermore, the length and nature of the SA also have a significant impact on support functionalization and ligand availability [6]. SA usually comprise of alkane chains and the optimal lengths are considered to be between 4 and 12 Carbon atoms [8]. A spacer molecule provides greater mobility, allowing the immobilized ligand to orient into the correct position for optimal binding of the target molecule [13]. As for ligands, while antibodies are very popular due to their promise of high selectivity, several reports have suggested small synthetic ligands to be superior alternatives due to their higher stability, lower costs, lower degree of leaching, and also lower probability of membrane fouling [6]. In the possible application of any affinity membrane to a separation scheme, one of the major considerations is the leaching of the ligand and this issue needs to be studied for any matrix in its evaluation for commercial applications [8]. The ligand density on membrane surface is also an important determinant of the membrane performance.
The study described in this manuscript was aimed at developing an affinity membrane for separation of plasminogen activators (PAs) from mammalian cell culture conditioned media. PAs activate plasminogen by cleaving a specific Arg-Val peptide bond located within the protease domain. The resulting plasmin dissolves clots (thrombus) [14]. PAs are thus administered as thrombolytic agents for the treatment of thrombovascular disorders such as myocardial infarction and stroke. However, their clinical use is marred by extremely high costs due to complicated downstream processes [2, 15]. The main source of PAs is conditioned media from mammalian cell cultures where they are secreted in very small quantities, thus making chromatographic processes particularly unsuitable for their purification. Also, different forms of PAs are usually secreted by a particular cell line and these forms can have different molecular sizes (40–70 kDa) which are difficult to separate from bovine serum albumin (BSA; 66 kDa) present in the conditioned media in high concentration. An affinity membrane based separation process could thus be a viable alternative for the isolation and purification of PAs from such media. Some recent studies have described attempts at improved PA isolation from crude extracts but these studies focus on developing better ligands [16, 17], while the study described in this manuscript experiments with the use of membranes instead of traditional chromatographic beads. An affinity membrane was thus developed using para-aminobenzamidine (pABA), a known inhibitor of plasminogen activators, as the affinity ligand [18]. Due to their hydrophilic surface properties and ease of use and chemical modification, Regenerated Cellulose (RC) membranes were used. The effects of length of SA, and ligand density on membrane surface were studied to determine the membrane parameters for optimal protein binding devoid of steric hindrance. The separation of PAs was performed from both mono- and multi-component systems. Results were analyzed with respect to the length of SA and ligand density on membrane surface. Leaching of ligand and the reusability of the membrane was determined. Despite the fact that batch operation mode is not considered economical in terms of industrial process development for affinity membranes [10], all the experiments in this study were performed in batch mode, as such studies are acceptable to determine the binding capacity and selectivity of the membrane [10], that being the main aim of this work.
2. Experimental
2.1. Materials
Regenerated cellulose (RC) membrane discs (pore size 0.45 µm, diameter 50 mm, thickness 160 – 200 µm) were purchased from Sartorius Stedium Biotech.
Para-aminobenzamidine dihydrochloride (pABA), urokinase type plasminogen activator (uPA), fibrinogen, thrombin, plasminogen, bovine serum albumin (BSA), D-Val-Leu-Lys-p-nitroanilide, plasminogen, bicinchoninic acid kit, were obtained from Sigma. Standard protein marker (Precision plus Protein™ standards unstained) was bought from Biorad Laboratories. All chemicals were of analytical grade purity and were used without further purification. Deionized water was used for the preparation of all solutions.
The conditioned media from HeLa cell cultures was provided by the group of Dr. Carlos Gonzalez (University of Puerto Rico - Rio Piedras).
2.2. Modification of RC membranes
RC membranes were modified to develop affinity membranes containing pABA as the affinity ligand for isolation of PAs. As a first step, SA of different lengths (5-atoms, 7-atoms and 14-atoms) were linked to different RC membranes using standard procedures [19, 20]. The free end of the SA on each of these three types of membranes was then linked to pABA. The methods for these modifications are described below.
2.2.1. Attachment of SA to the RC membranes
2.2.1.1. Functionalization with epichlorohydrin (5-atoms SA)
The RC membranes were immersed in a solution containing 19.0 mL of DMSO, 11.0 mL of epichlorohydrin, and 10.0 mL of 1.0N NaOH on a rocker (rpm 150) for 12 hours at room temperature. The activated functionalized RC membranes were washed with deionized water extensively prior to further modification.
2.2.1.2. Functionalization with glutaraldehyde (7-atoms SA)
The RC membranes were immersed in a mixture containing 30.0 mL of 0.1M phosphate buffer (pH 7.4). To obtain membranes with different spacer arm densities, varying quantities of 50% (v/v) aqueous glutaraldehyde solution (33.7, 15.0, 10.0, 5.0, or 1.0 mL) were added to membranes contained in different vessels. The membranes were gently rocked for 12 hours at room temperature. The membranes were then washed with deionized water to remove the excess glutaraldehyde.
2.2.1.3. Functionalization with 1,4-butanediol diglycidyl ether (14-atoms SA)
The reaction was performed as described in section 2.2.1.1, substituting epichlorohydrin with 1,4-butanediol diglycidyl ether. ). To obtain membranes with different spacer arm densities, varying quantities of 1,4-butanediol diglycidyl ether (25.7 mL, 2.6 or 0.3 mL) were added to membranes contained in different vessels.
2.2.2. Coupling of SA-linked membranes to affinity ligand (pABA)
2.2.2.1. Coupling of pABA to epoxy functionalized membranes
The epoxy (epichlorohydrin and 1,4-butanediol diglycidyl ether) activated RC membranes were added to a solution containing 608 mg of pABA hydrochloride in 50.0 mL of 0.1M phosphate buffer (pH 7.4). The mixture was allowed to rock at room temperature for 12 h. Functionalized membranes were extensively washed with acidic deionized water (pH 2.0) and deionized water to remove unbound pABA. The washing was continued until the absorbance of the spent wash at 280 nm was zero.
2.2.2.2. Coupling of pABA to glutaraldehyde activated membrane
RC membranes activated with glutaraldehyde were added to 50.0 mL of 0.1M phosphate buffer, pH 7.4 (containing 608 mg of pABA and 250 mg of sodium borohydride). The mixture was allowed to rock at room temperature for 12 h. Functionalized membranes were extensively washed with acidic deionized water (pH 2) and deionized water to remove unbound pABA. The washing was continued until the absorbance of the spent wash at 280 nm was zero.
2.3. Characterization of the modified RC membranes
2.3.1. FTIR characterization of the affinity ligand modified membranes
Small pieces were cut from the modified membranes (prepared as described in section 2.2) dried under vacuum, and analyzed through FT-IR spectroscopy. Pieces from three different areas of each membrane were cut and analyzed to ensure the reproducibility of the data. A Perkin Elmer Spectrum 100 FTIR spectrometer equipped with a Single reflection Diamond/ZnSe Standard Universal ATR Accessory was used. A total of 32 scans were acquired for each sample.
2.3.2. Quantification of ligand concentration on modified membrane surface
The concentration of pABA on membrane surface was determined by an adaptation of the glyoxal method developed by Jackson et al [21]. Square pieces were cut from three different areas of each of the membranes to be tested and treated with glyoxal reagent according to the published method. The resulting fluorescence on the membranes was read using a Shimadzu RF-1501 Spectrofluorometer equipped with a xenon lamp. The excitation wave length was 340 nm, and the emission wavelength was 500 nm. pABA (Sigma) was used as the standard for calculation of ligand densities.
2.3.3. Visualization of pABA bound to the RC membranes through confocal microscopy
The fluorescence labeling of the pABA linked membrane pieces as described in section 2.3.2, allowed their visualization through confocal microscopy. The microscope used was a Zeiss Axiovision Z1 with a Zeiss LSM510 META. The images were obtained using a 10x magnification EC Plan-Neofluar, 633nm excitation laser, and a band pass of 642nm-738nm emission filter.
2.4. Application of affinity ligand modified membranes to binding of PA from a monocomponent system
The membrane was equilibrated in binding buffer (Table 1) with two changes of buffer over a period of one hour. The uPA (Sigma) was dissolved in binding buffer to a concentration of 4.40 nM solutions and 10.0 mL of this solution was added to the membrane. The membrane was allowed to bind uPA for two hours. At the end of this period, spent uPA solution was withdrawn and the membrane washed with frequent changes of binding buffer until all the non-specifically adsorbed protein could be removed (as determined by absorbance at 280 nm). The elution of bound uPA was then performed with the glycine containing elution buffer (Table 1). Elution step was repeated with two successive rounds of fresh elution buffer to make sure that all the bound uPA was recovered. The entire process was performed on a rocking platform (Biorad) at 90 rpm and 4°C. All the eluate fractions were pooled, lyophilized, desalted, and then lyophilized again to be redissolved in 1X PBS to an appropriate concentration. The eluates, spent uPA solution, and wash were then analyzed for protein concentration and enzyme activity. Electrophoresis of the eluates was also performed (Section 2.7). RC membranes without any kind of modification were used as control.
Table 1.
Composition of different buffers
| Buffer | Composition |
|---|---|
| Binding Buffer | 0.05 M tris HCl containing 0.5 M NaCl, pH 7.4 |
| Elution Buffer | 0.05 M glycine, pH 3.0 |
| Regeneration Buffer | 0.05 M acetate buffer, 0.2% (v/v) Triton X-100, pH 4.0 |
2.5. Application of affinity ligand modified membranes to isolation of PA from a multicomponent system
The conditioned media from HeLa cultures was centrifuged at 1,500 × rpm for 15 minutes to pellet the cells. The cell pellet was discarded and the supernatant (10.0 mL) added to membranes that had been pre-equilibrated with the binding buffer. The membrane was allowed to bind PA from the media for two hours. At the end of this period, spent conditioned media was withdrawn and the membranes washed with frequent changes of binding buffer until all the non-specifically adsorbed protein could be removed. The elution of bound PA was then performed with glycine containing elution buffer (Table 1). Elution step was repeated with three successive rounds of fresh elution buffer to make sure that all bound PA was recovered. The entire process was performed on a rocking platform (Biorad) at 90 rpm at 4°C. All the eluate fractions were pooled, lyophilized, desalted, and then lyophilized again to be redissolved in 1X PBS to an appropriate concentration. The eluates, spent media and wash were then analyzed for protein concentration and enzyme activity. Electrophoresis of the eluates was also performed (Section 2.7). RC membranes without any kind of modification were used as control.
2.6. Regeneration of membrane
The used membranes were washed with three changes of deionized water over a period of 15 min. The water was then replaced with triton X-100 containing regeneration buffer (Table 1). The regeneration buffer was changed twice over a period of 45 min. The membrane was then washed again with deionized water with two changes over a period of 45 min. The entire process was performed on a rocking platform (Biorad) at 90 rpm at 4°C. The membranes were dried between folds of filter papers at room temperature and then stored at 4°C until further use.
2.7. Analytical methods for protein samples
The concentration of proteins was determined by the Bicinchoninic acid method as described by Smith et al [22]. The activity of plasminogen activator was measured by a chromogenic assay [23] using D-Val-Leu-Lys-paranitroanilide (V-7127, Sigma) as substrate, and also by fibrin plate assay [24] using pure uPA (Sigma) as the reference. PA activities were expressed as International Units (IU), where one IU activates an amount of plasminogen that will produce a change in A275 of 1.0 per mL per minute at pH 7.5 at 37 °C, when measuring HClO4 soluble products from α-casein (1 cm light path).
The analysis of purity of the eluates was performed by Sodium Dodecyl Sulphate -Polyacrylamide Gel Electrophoresis (SDS-PAGE) according to the method of Laemmli [25]. Protein bands on the gel were visualized by silver staining [26]. Enzymatically active protein bands were detected through fibrin zymography [27].
3. Results and Discussion
3.1. Modification of membranes with pABA
The affinity membranes were prepared by introducing SA of different lengths on RC membranes followed by end-group coupling to pABA. Depending upon the desired length of the SA (5-, 7-, or 14- atoms), the membranes were reacted with epichlorohydrin, glutaraldehyde, or 1,4-butanediol diglycidyl ether respectively, following known procedures [19, 20]. The atomic length of each SA was calculated considering the number of atoms between the carbon skeleton of cellulose and the aromatic ring of the affinity ligand. Coupling of pABA to SA was achieved through the reaction of the para-amino group of pABA with epoxy moiety for 5- and 14- atoms SA and aldehyde group for 7-atoms SA [19]. The synthetic scheme of the chemistry involved in membrane modification is shown in Figure 1.
Figure 1.
Preparation of the pABA affinity membranes with SA of different lengths.
Qualitative characterization of the membrane modification was done by FT-IR and confocal microscopy, while the quantification of the pABA attached to the membranes was done by modification of membrane according to the glyoxal method [21] followed by fluorescence spectroscopy.
3.1.1. FTIR Studies
3.1.1.1. pABA-5 atoms SA-RC membrane
The introduction of 5-atoms SA into the RC membrane could be seen in small changes in FT-IR spectrum of the functionalized membrane (Supplemental Figure 1b) as compared to the native cellulose membrane (Supplemental Figure 1a). The CH antisymmetric and symmetric stretching of epichlorohydrin appeared at 2924 cm−1 overlapping partially with the cellulose band and CH2 scissor vibration was seen as a shoulder of cellulose band at 1456 cm−1. The presence of epoxide was reflected in minor changes at 1232 cm−1 typical of epoxides and the band at 1135 and 1046 cm−1 attributed to C-O-C asymmetric stretching that appeared in an overlap with the strong band of cellulose. A small band at 1738 cm−1 was also seen indicating that part of epoxide was oxidized to aldehyde. The presence of pABA (Supplemental Figure 1c) was however not observed clearly due to the superimposition of aromatic amine and amidine bands with cellulose band in the 3000 and 1600 cm−1 area. However the disappearance of the aldehyde and epoxide bands was an evidence of the reaction. A comparison of these FTIR spectra was performed with that of the commercially available pABA Sepharose. Interestingly, the pABA signals could not be seen in pABA Sepharose either (Data not shown). Disappearance of the bands 2917 and 1456 cm−1 after pABA coupling could not be explained.
3.1.1.2. pABA-7 atoms SA-RC membrane
The functionalization of cellulose with glutaraldehyde (7-atoms SA) was clearly seen by the appearance of C=O stretching at 1728 cm−1 (Supplemental Figure 2b). The methylene chain was observed at 2956 and 1450 cm−1 as in the case of 5-atoms SA. Also in this case, the bands of pABA were not observed. However the disappearance of the aldehyde band suggested that the linking of pABA to the aldehyde was successful (Supplemental Figure 2c).
3.1.1.3. pABA-14 atoms SA-RC membrane
In case of 1,4-butanediol diglycidyl ether (14-atoms SA), functionalization was observed by a very small change at 1250 cm−1, typical of C-O stretching of epoxide (Supplemental Figure 3b). A more clear evidence of membrane modification was the appearance of the band at 1056 cm−1 attributable to C-O-C asymmetric stretching though partially overlapped by cellulose signal. Minor changes at 2956 cm−1 indicated the presence of CH2 while no shoulder at 1450 cm−1 was seen in this spectrum. Again in this case too, the pABA signals could not be observed (Supplemental Figure 3c).
Though the bands for pABA could not be seen in the FTIR spectra, the analysis of membranes by the glyoxal method (as described in the following sections 3.1.2 and 3.1.3) gave a clear proof of the successful linking of pABA to the membranes.
3.1.2. Quantitative estimation of pABA linked to the membrane surface
Ligand (pABA) density on the membrane surface was determined through an adaptation of the glyoxal method developed by Jackson et al. for quantification of aromatic amidines [13]. The exposed benzamidine groups on the membrane surface were allowed to react with glyoxal and benzaldehyde resulting in the formation of a fluorogenic compound (5-imidazolone) that can be estimated quantitatively by measuring its fluorescence. An unmodified RC membrane was used as the negative control. A change in color of the modified membranes from the natural white to yellow was seen after the reaction (Figure 2). The fluorescence intensities recorded for each of the membranes with different SA were translated into ligand (pABA) densities on the membrane surface using free pABA as standard. In the first batch of different SA linked membranes prepared by adding equal moles of different SA (3.5 M), the 14-atoms SA membranes showed highest ligand density (50.8 nmoles per cm2) followed closely by the 5-atoms SA (36.7 nmoles per cm2); while the 7-atoms SA linked membranes showed lowest degree of ligand immobilization (9.52 nmoles per cm2). The results were in agreement with the different intensities of yellow color seen on each membrane after the glyoxal method treatment, as the 14-atoms SA containing membrane showed the most intense yellow color among the three (Figure 2). To account for the possibility of a heterogeneous distribution of the ligand on membrane surface, square pieces were cut from membranes from three random locations on each membrane and ligand densities measured were averaged for a particular type of membrane. The ligand density was then optimized by a hit and trial approach (using different amounts of SA in the modification) to obtain similar ligand densities for the three different SA linked membranes (Table 2).
Figure 2.
Fluorescently labeled membranes for ligand density determination.
Table 2.
Ligand density: nmol of pABA per cm2 of membrane modified with SA of different lengths.
| Spacer arm |
Ligand density (nmol/ cm2 membrane) |
||||
|---|---|---|---|---|---|
| Before Use | After first use | After fifth use | |||
| HeLa | uPA | HeLa | uPA | ||
| 5-atoms | 11.70 ± 1.21 | 11.40 ± 0.50 | 11.00 ± 1.40 | N/A | N/A |
| 7-atoms | 12.01 ± 2.23 | 11.80 ± 0.03 | 11.49 ± 0.02 | N/A | N/A |
| 14-atoms | 11.40 ± 0.87 | 11.00 ± 0.48 | 11.30 ± 0.12 | 10.13 ± 0.23 | 11.00 ± 0.12 |
Leaching of the ligand has been a major consideration in application of affinity membranes to separation processes [8]. Hence, ligand densities were determined for all the three modified membranes before and after their use in the separation process. Results showed a variation in ligand density in the range of 2–5 % after a single use in each of three membranes (Table 2), which is within the range of experimental error of these measurements. The ligand density after five cycles of use was studied only for the 14-atoms SA containing membranes (Table 2) since these membranes showed the highest PA binding capacity. The extra cycles of separation however did not affect the ligand density significantly indicating that the 14-atoms SA is stably linked under the experimental conditions used.
3.1.3. Confocal Microscopic Analysis
The membranes modified by glyoxal method as described in section 3.1.2 were also observed through confocal microscopy (Figure 3). The distribution of ligand was found to be relatively heterogeneous on the membrane surface. The RC membrane linked to pABA through the 7-atoms SA showed big patches with very low fluorescence intensity indicating lower pABA binding in these areas. This observation was consistent with the fluorescence intensities measured on these membrane pieces through fluorescence spectroscopy, the 7-atoms SA membranes being the ones with lowest ligand densities in that case too among the membranes prepared in the first batch (section 3.1.2).
Figure 3.
Confocal microscopy images of the fluorescence labeled membranes.
3.2. PA binding capacity of the modified membranes
It has already been established in our laboratory that HeLa cells secrete PA into the media during their growth. The ability of the affinity modified membranes to bind PA and the role of the length of SA and ligand density in optimal protein binding was determined by allowing the membranes to incubate in pure uPA (Sigma) solution or alternatively in the conditioned media from HeLa cell cultures, in the batch mode for a period of 2h. At the end of this period the membranes were extensively washed to remove the non-specifically adsorbed proteins. The bound PA was then eluted using a glycine buffer (pH 3.0) and analyzed with respect to enzyme activity and protein concentration. In all reported experiments, the PA concentration in the starting solutions (both uPA solution and HeLa cells conditioned media) was found to be in excess of the binding capacity of the affinity membranes. The control membrane did not show any binding of PA.
Since the uPA from Sigma as well as the PAs secreted by HeLa cells, are a mixture of different molecular weight forms (most of which were enzymatically active, as confirmed by SDS-PAGE and zymography), it was not possible to calculate the ligand utilization efficiencies in terms of molar ratios. However the enzyme activity (IU) bound per mg ligand or alternatively enzyme activity (IU) bound per cm2 membrane surface area were determined as reported in Figures 4–5 and Table 3. All the reported values have been normalized with respect to the enzyme activity in the PA load solution in each case.
Figure 4.
A. Effect of membrane surface ligand density on the Plasminogen Activator binding efficiency of 7-atoms SA and 14-atoms SA consisting pABA linked RC membranes from unicomponent system (pure uPA (Sigma) solution; the amount of enzyme bound to the membrane was normalized with respect to the total enzyme in the load).
B. Effect of membrane surface ligand density on the Plasminogen Activator binding efficiency of 7-atoms SA and 14-atoms SA consisting pABA linked RC membranes from multicomponent system (HeLa cell conditioned media; the amount of enzyme bound to the membrane was normalized with respect to the total enzyme in the load).
Figure 5.
Effect of length of SA on Plasminogen Activator binding efficiency of pABA linked RC membranes (the amount of enzyme bound to the membrane was normalized with respect to the total enzyme in the load). The pABA density on each used membrane was 11±1.0 nmoles per cm2.
Table 3.
PA binding capacities of the pABA-SA-RC membranes
| Length of Spacer Arm |
PA bound (IU) per cm2 membrane (Normalized) |
PA bound (IU) per nmol ligand (Normalized) |
||
|---|---|---|---|---|
| uPA | HeLa | uPA | HeLa | |
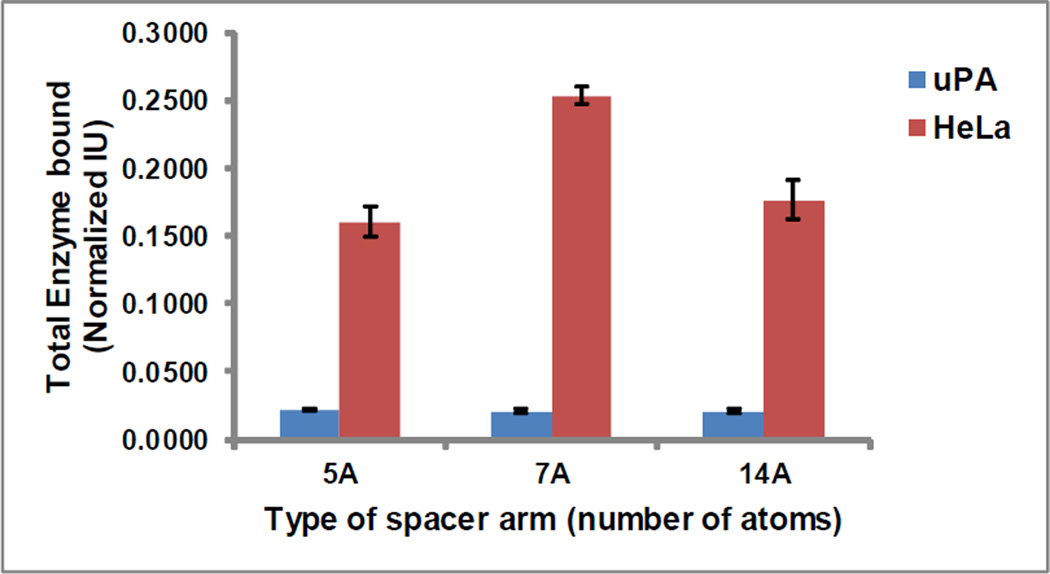
| 5-Atoms | 0.022 ± 0.001 | 0.160 ± 0.010 | 20.53 ± 0.41 | 50.01 ± 3.50 |
| 7-Atoms | 0.021 ± 0.001 | 0.253 ± 0.006 | 19.25 ± 0.77 | 84.55 ± 4.23 |
| 14-Atoms | 0.021 ±0.001 | 0.177 ± 0.014 | 21.99 ± 0.44 | 54.31 ± 4.34 |
3.2.1. Effect of ligand density
The modification of RC membranes was performed with serial dilutions of the different spacer arms (7-atoms SA and 14-atoms SA) followed by treatment with pABA to obtain affinity membranes with different ligand densities (5–25 nmoles per cm2).
The data obtained from PA binding experiments from pure uPA (Sigma) solution for these membranes are represented in Figure 4A. A very clear inverse linear relationship between ligand density on membrane surface and PA binding capacity of the membrane was seen for both SA. PA binding capacity in membranes with different SA but similar ligand densities appeared to be comparable. As can be seen clearly for both SA, a binding optima was obtained at a ligand density of 11.0 ± 1.0 nmoles per cm2. At still lower ligand densities, saturation in PA binding was apparent (the loading solution had an excess of PA in all experiments). These findings are an affirmation of the “crowding effect” as has been discussed in several published reports [8, 28– 30]. According to this theory, the binding efficiency, defined as the ratio of actual binding capacity to that calculated from ligand density, decreases when ligand coupling density is increased [8]. It is because the binding of a large ligate such as a protein molecule, requires space and orientational freedom which is not increased solely by increasing the ligand density in the microporous matrix [8, 19, 28, 29]. Results from similar experiments conducted for PA binding from multicomponent system (HeLa conditioned media) are summarized in Figure 4B. For each of the two SA (7-atoms and 14-atoms), membranes with three different ligand densities were used. Interestingly, in this case too, PA binding optima were obtained at ligand densities of 11.0 ± 1.0 nmoles per cm2, while a clear decrease in PA binding was seen at the tested lower and higher ligand densities in both cases.
3.2.2. Effect of length of SA
The length of SA has traditionally been considered a very critical determinant of the binding efficiency of any affinity matrix. As has been discussed by Urmenyi et al., SA shorter than an optimal length can hinder the ligand while longer spacer chains can bend or cross link therefore reducing the actual exposure of the ligand [13]. An optimal length of SA is one that provides less steric hindrance to conjugation and offers more active complexes [6]. This optimal length would depend upon the size of the ligand as well as the target species and hence needs to be determined for each system independently.
Generally, it is impossible to obtain identical ligand densities on membranes modified with different SA. However, using a modification process optimized through a hit and trial approach, we were able to obtain similar (not identical) ligand densities (11±1.0 nmoles per cm2) on the RC membranes linked to 5-atoms, 7-atoms, and 14-atoms SA (Table 2). These membranes were then used to study the effect of role of the length of SA in optimal protein binding from both mono-and multi-component systems (Table 3). The results obtained for PA binding from the multicomponent system (HeLa conditioned media) clearly showed a higher binding of PA per nmole of immobilized ligand in the presence of 7-atoms SA; while surprisingly, in case of the monocomponent system (pure uPA solution) no difference in binding capacities of the three different membranes was seen (Figure 5, Table 3). The binding behavior in case of HeLa conditioned media can easily be explained as the 7-atoms SA providing the optimal distance between the membrane and the affinity ligand to allow the PA binding to pABA with minimal steric hindrance. However why a similar effect was not seen in PA binding from monocomponent system remains a question to be answered and merits further investigation. No information in favor- or contrast- of our findings could be found in the literature.
Though both these effects (ligand density and length of SA) have been discussed individually in several papers [8] the authors could find only one study that was analyzed with respect to both the parameters. In the said study, Guo and Ruckenstein prepared macroporous cellulose membranes and activated them by introducing epoxy/ aldehyde groups with various spacer lengths (5-,7-, 10-, 12- atoms long) after which maltose was immobilized as an affinity ligand [19]. The membranes were then used for the affinity purification of Concavalin A (Con A). The results indicated that the adsorption capacity for Con A was the highest when 12-atoms SA was used as the activation agent even though the epoxy content generated in this case was the smallest. Their observation that the membrane with lowest epoxy content showed the highest Con A adsorption capacity, is partly in agreement with our findings, where too higher bidning capacities were observed at lower ligand densities. However, our study goes a step further by comparing the PA binding capacities of membranes with different spacer arms but similar ligand densities. We clearly established that binding capacity was maximized at an optimal ligand concentration (not necessarily the lowest or highest), and an optimal SA length (which is not necessarily the longest or shortest SA).
3.3. Selectivity of the affinity membranes for PA binding
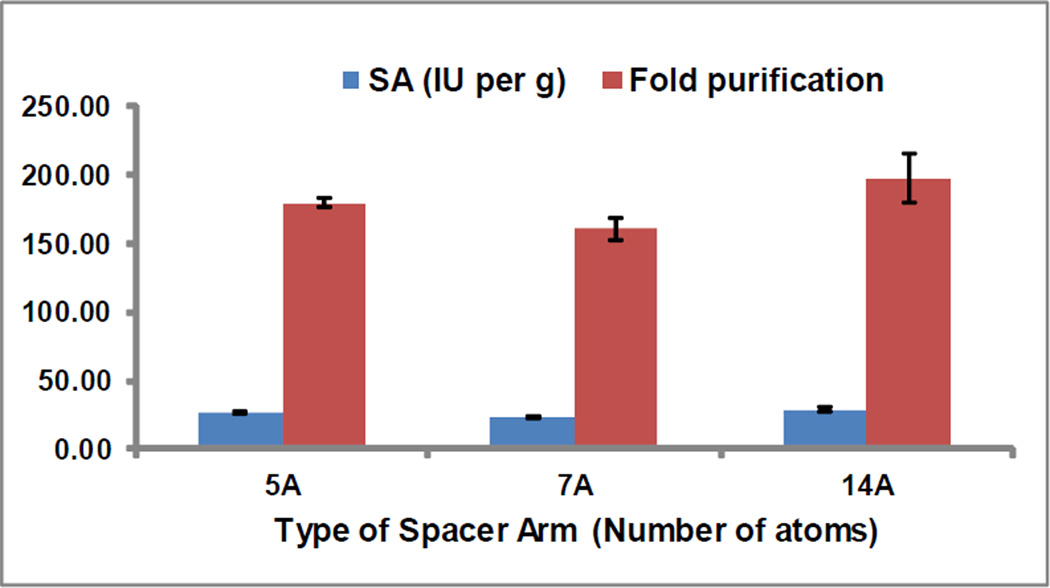
To determine the ability of the affinity modified membrane to bind PA selectively from a mixture of proteins, the data from the HeLa conditioned media studies, with membranes consisting of different types of SA but similar ligand densities, was analyzed further. This conditioned media consists of several different proteins, the major one being Bovine Serum Albumin (BSA, 66 kDa). Specific enzyme activities were determined for the eluates from all three types of membranes and fold-purification factors calculated as the ratio of specific enzyme activity in the eluates and the specific enzyme activity in the conditioned media from HeLa cells. The specific activities and fold purification factors were found to be similar for the three SA thus indicating that the the length of SA does not affect the selectivity of the ligand for the target protein (Figure 6). The amount of total protein adsorbed to the membranes in all three cases was found to vary in the range of 40 – 60 µg per cm2, directly proportional to the amount of PA bound. These protein adsorption capacities are lower than most of the published reports, the average adsorption capacities being in the range of 50 – 1,130 µg/cm2 membrane area [8, 10, 31– 33]. However, this is to be expected because most of these reports employed a filtration mode process. Instead, in the current study, batch method was employed since the main objective was to optimize the membrane modification in favor of binding capacity rather than develop a separation process. The effective capacity of a membrane is known to increase under filtration (dynamic binding capacity) conditions, while the static binding capacity (determined in batch mode) is generally much lower [34, 35].
Figure 6.
Effect of length of SA on selectivity of PA binding from HeLa cell conditioned media by pABA linked RC membranes. The pABA density on each used membrane was 11±1.0 nmoles per cm2.
Additionally, electrophoretic analysis of the eluates and HeLa conditioned media was performed to verify the purity of eluates from the different membranes. Multiple protein bands were seen in HeLa eluates from all the membranes on the SDS-PAGE visualized through silver staining (Figure 7A), which was to be expected in view of the presence normally of multiple forms of PAs in a wide molecular weight range in conditioned cell culture media. To identify the PA bands in these eluates, a western blot was performed (Figure 7B) which also revealed the presence of multiple bands in the HeLa conditioned media, though one band was predominant in all eluates (approximately 100 kDa). This band was clearly darker in both the SDS-PAGE and Western blot for the 7 atoms-SA membrane as compared to the other two SA confirming the results that the 7 atoms- SA containing membrane bound more PA. To obtain a better understanding of the eluates’ composition, fibrin zymography was performed (Figure 8). A big blob of BSA (66 kDa) was seen in the lane containing the crude conditioned media from HeLa cultures (as expected since BSA constitutes about 90% of the total protein content of the media). Multiple active bands were seen in this case, all of which belong to PA, as was confirmed by presence of multiple bands in the western blot. The most interesting observation, however was the brightness seen in the lane containing the eluate from 7-atoms SA containing membrane as compared to the other two eluates. This reconfirmed that the amount of PA was higher in this particular eluate. The degree of purification obtained in all cases (up to 200-fold) is much higher than the 3.3-fold purification reported in the only other published affinity membrane process for uPA separation by Hou and Zaniewski [32] who purified crude uPA from human urine using ion-exchange membranes. The purification in this study achieved is superior also to those reported in recent studies where higher affinity ligands were designed and used with traditional bead chromatographic matrices [16, 17].
Figure 7.
(A) SDS-PAGE, Lane 1: Molecular weight marker; HeLa conditioned media eluates from RC membrane linked to pABA through SA of: 5-atoms (Lane 2), 7-atoms (Lane 3), and 14-atoms (Lane 4); Lane 5: HeLa conditioned media. (B) Western blot, Lane 6: Molecular weight marker; HeLa conditioned media eluates from RC membrane linked to pABA through SA of: 5-atoms (Lane 7), 7-atoms (Lane 8), and 14-atoms (Lane 9); Lane 10: HeLa conditioned media.
Figure 8.
Fibrin Zymogram, Lane 1: HeLa conditioned media used as the load; Eluates from RC membrane linked to pABA through SA of: 5-atoms (Lane 2), 7-atoms (Lane 3), and 14-atoms (Lane 4).
3.4. Reusability of the membranes
The reusability of a separation device is extremely important for its favorable application in any laboratory- or industrial- scale process. Hence the affinity membranes developed in this work were regenerated and reused for separation from multi- as well as mono- component systems. The reuse experiments were performed only for the membrane with 14-atoms SA. A mild regeneration method was used initially wherein the membranes were washed with 0.1% (v/v) Triton X-100 containing regeneration buffer (Table 1). A progressive loss of PA binding was seen over five cycles of use for both uPA solution and HeLa conditioned media. In case of uPA solution, the membrane lost 20% of its binding capacity after the first use and up to 65% after the fifth use; while in case of HeLa conditioned media, the membrane lost 17% of its binding capacity after the first use and up to 90% after the fifth use (Data not shown). The regeneration procedure was then modified by increasing the Triton X-100 concentration to 0.2% (v/v), increasing the wash times, and adding a series of additional changes of buffer as well as more extensive washing with deionized water after each use. The results obtained from five repeated uses of the same set of membranes for PA binding from HeLa conditioned media and uPA solution, following the new regeneration procedure, are presented in Figure 9. As can be seen clearly, the change in regeneration procedure prevented the fouling of membranes to a significant extent. Relatively small decreases in binding capacity for PA were seen in case of both uPA and HeLa conditioned media (5–8%) during subsequent cycles of use.
Figure 9.
Reusability of the affinity (pABA-14-atoms SA-RC) membranes in isolation of PAs.
4. Conclusions
An affinity membrane was developed and used successfully for the isolation of PAs from conditioned cell culture media in a single step process. A detailed study of the PA binding capacity of the pABA-RC membranes consisting of different lengths of SA and different ligand densities revealed the PA binding capacity of the pABA-SA-RC membranes to be a function of the membrane surface ligand density as well as the length of spacer arm. The RC membrane linked to pABA through a SA of 7-atoms showed a higher binding of PA as compared to those linked through shorter or longer (5- or 14- atoms) SA with similar ligand densities, when used for PA separation from a multicomponent system (HeLa conditioned media). However the binding capacity of the membranes in case of monocomponent system appeared to be a function of only the ligand density on membrane surface, as it did not vary with change in length of spacer arm. An inverse linear correlation between the ligand density and protein binding was clearly established in all cases, with the binding optima occurring at a ligand density of 11±1.0 nmoles per cm2. It can thus be concluded, that the binding capacity of pABA-SA-RC membranes is highest in case of a SA length of 7-atoms, and at a ligand density of 11±1.0 nmoles per cm2. An extensive membrane regeneration process after each use allowed the preservation of the protein binding capacity of the membrane by preventing any fouling. The ligand was found to be linked stably to the membrane as evidenced by an absence of continuous leaching during the reuse/regeneration cycles. These membranes can be used for the development of stacked membrane devices for batch or continuous capture of PAs from reactors. Alternatively such membranes might be used as sensory devices in PA production. As has been discussed in several reviews and reports, in addition to improved module designs, there is a significant need for the development of activated, stable membranes that can be coupled by the end-user to provide the specific ligate capacity as per need [8, 11]. The study described in this manuscript is a step forward in this direction by providing a deeper understanding of issues that are critical in development of functional affinity membranes.
Supplementary Material
Highlights.
Chemically modified RC membrane
RC-pABA membrane binds PA
No membrane fouling
Length of spacer arm and ligand density important determinants of binding capacity
Acknowledgements
This project was supported by grants from the National Center for Research Resources (5P20RR016470-12) and the National Institute of General Medical Sciences (8 P20 GM103475-12) from the National Institutes of Health. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of the NCRR or NIH. Instrumentation facilities created through startup funds from the Institute of Functional Nanomaterials in Puerto Rico (NSF Grant No. 1002410), and startup funds (FIDI) from University of Puerto Rico at Cayey, were also used. The authors would also like to acknowledge the contribution by grants ISI0 RR-13705-01 and DBI-0923132 to establish and upgrade the Confocal Microscopy Facility at the University of Puerto Rico (CIF-UPR). The authors are grateful to Dr. Jose Lasalde and Lcdo. Bismark Madera for facilitating the use of Confocal Microscopy facility. The authors will also like to acknowledge the group of Dr. Carlos Gonzalez (University of Puerto Rico at Rio Piedras) for providing the conditioned media from HeLa cell cultures. Ms. Yiaslin Ruiz is the recipient of a research fellowship from Amgen-BIOMINDS program and Ms. Amaris Borges is the recipient of NIH-RISE fellowship at UPR-Cayey. Ms. Osiris Martinez is the recipient NIH-RISE fellowship at UPR-Humacao.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
PA: Plasminogen activator; RC: Regenerated cellulose; pABA: Para-aminobenzamidine; uPA: urokinase type plasminogen activator; DMSO: Dimethylsulfoxide; BSA: Bovine serum albumin; SA: Spacer arm.
REFERENCES
- 1.Zhang H, Wu C, Zhang Y, White C, Xue Y, Nie H, Zhu L. J. Mater. Sci. 2010;45:2296. [Google Scholar]
- 2.Bansal V, Roychoudhury PK. Protein Expres. Purif. 2006;45:1. doi: 10.1016/j.pep.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 3.Choi YW, Kim IS. Biotechnol. Bioprocess. Eng. 2008;13:25. [Google Scholar]
- 4.Ma Z, Ramakrishna S. J. Memb. Sci. 2008;319:23. [Google Scholar]
- 5.Ghosh R. J. Chromatogr. A. 2002;952:13. doi: 10.1016/s0021-9673(02)00057-2. [DOI] [PubMed] [Google Scholar]
- 6.Barroso T, Temtem M, Hussain A, Aguiar-Ricardo A, Roque ACA. J. Memb. Sci. 2010;348:224. [Google Scholar]
- 7.Castilho LR, Anspach FB, Deckwer W-D. J. Memb. Sci. 2002;207:253. [Google Scholar]
- 8.Klein E. J. Memb. Sci. 2000;179:1. [Google Scholar]
- 9.Liu J, Chen X, Shao Z, Zhou P. J. Appl. Polym. Sci. 2003;90:1108. [Google Scholar]
- 10.Suen SY, Liu YC, Chang CS. J. Chromatogr. B. 2003;797:305. doi: 10.1016/s1570-0232(03)00490-2. [DOI] [PubMed] [Google Scholar]
- 11.He LZ, Dong XY, Sun Y. Biotechnol. Prog. 1998;14:594. doi: 10.1021/bp980046k. [DOI] [PubMed] [Google Scholar]
- 12.He LZ, Sun Y. Biosyst. Bioprocess. Eng. 2002;25:155. doi: 10.1007/s00449-002-0288-7. [DOI] [PubMed] [Google Scholar]
- 13.Urmenyi AM, Poot AA, Wessling M, Mulder MHV. J. Memb. Sci. 2005;259:91. [Google Scholar]
- 14.Lopez-Sendon J, deLopez SE, Bobadilla JF, Rubio R, Bermejo J, Delcan JL. Rev. Esp. Cardiol. 1995;48:407. [PubMed] [Google Scholar]
- 15.Balaraman K, Prabakaran G. Indian J. Med. Res. 2007;126:459. [PubMed] [Google Scholar]
- 16.Wu F, Yu J, Li R. Biochem. Biophys. Res. Commun. 2007;355:673. doi: 10.1016/j.bbrc.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 17.Liu FF, Dong XY, Wang T, Sun Y. J. Chromatogr. A. 2007;1175:249. doi: 10.1016/j.chroma.2007.10.074. [DOI] [PubMed] [Google Scholar]
- 18.Billstrom A, Hartley-Asp B, Lecander I, Batra S, Astedt B. Intl. J. Cancer. 1995;61:542. doi: 10.1002/ijc.2910610419. [DOI] [PubMed] [Google Scholar]
- 19.Guo W, Ruckenstein E. J. Memb. Sci. 2001;182:227. [Google Scholar]
- 20.Hermanson GT. Bioconjugate techniques. 2nd ed. London: Academic Press; 2008. [Google Scholar]
- 21.Jackson DP, Kuhl WJ, Jr., Irvin JL. J. Biol. Chem. 1947;167:377. [PubMed] [Google Scholar]
- 22.Smith C. Nat. Methods. 2005;2:71. [Google Scholar]
- 23.Johnsen LB, Poulsen K, Kilian M, Petersen TE. Infect. Immun. 1999;67:1072. doi: 10.1128/iai.67.3.1072-1078.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haverkate F, Brakman P. In: Progress in Chemical Fibrinolysis and Thrombosis. Davidson JF, M SM, Desnoyers PC, editors. New York: Raven Press; 1975. p. 151. [Google Scholar]
- 25.Laemmli UK. Nature. 1970;227:680. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 26.Damerval C. Electrophoresis. 1994;15:1573. doi: 10.1002/elps.11501501226. [DOI] [PubMed] [Google Scholar]
- 27.Kim SH, Choi NS, Lee WY. Anal. Biochem. 1998;263:115. doi: 10.1006/abio.1998.2816. [DOI] [PubMed] [Google Scholar]
- 28.Chase HA. Chem. Eng. Sci. 1984;39:1099. [Google Scholar]
- 29.Kim M, Saito K, Furusaki S, Sato T, Sugo T, Ishigaki I. J. Chromatogr. A. 1991;585:45. doi: 10.1016/0021-9673(91)85055-k. [DOI] [PubMed] [Google Scholar]
- 30.Iwata H, Saito K, Furusaki S, Sugo T, Okamoto J. Biotechnol. Prog. 1991;7:412. doi: 10.1021/bp00011a005. [DOI] [PubMed] [Google Scholar]
- 31.Borcherding H, Hicke H-G, Jorcke D, Ulbricht M. Ann. NY Acad. Sci. 2003;984:470. doi: 10.1111/j.1749-6632.2003.tb06020.x. [DOI] [PubMed] [Google Scholar]
- 32.Hou KC, Zaniewski R. J. Chromatogr. A. 1990;525:297. doi: 10.1016/s0378-4347(00)83406-4. [DOI] [PubMed] [Google Scholar]
- 33.Jain P, Vyas MK, Geiger JH, Baker GL, Bruening ML. Biomacromolecules. 2010;11:1019. doi: 10.1021/bm9014792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boi C, Algeri C, Sarti GC. Biotechnol. Prog. 2008;24:1304. doi: 10.1002/btpr.42. [DOI] [PubMed] [Google Scholar]
- 35.Krause S, Kroner KH, Deckwer WD. Biotechnol. Tech. 1991;5:199. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.